Abstract
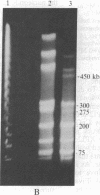
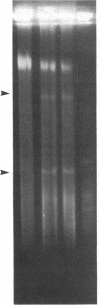
The circular genome of Staphylococcus aureus was cut into two fragments by a simple enzymatic method that cleaves a 10-base-pair site. The recognition sequence, A-T-C-G-mA decreases T-C-G-mA-T, was created by the combined use of the methylase M.Cla I (A-T-C-G-mA-T) and the restriction endonuclease Dpn I (G-mA decreases T-C). This technique is insensitive to CpG methylation and in human DNA is predicted to produce fragments that, on average, are greater than five million base pairs. The ability to create such long pieces of DNA should facilitate mapping of large, complex chromosomes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Botstein D., White R. L., Skolnick M., Davis R. W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980 May;32(3):314–331. [PMC free article] [PubMed] [Google Scholar]
- Brooks J. E., Blumenthal R. M., Gingeras T. R. The isolation and characterization of the Escherichia coli DNA adenine methylase (dam) gene. Nucleic Acids Res. 1983 Feb 11;11(3):837–851. doi: 10.1093/nar/11.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. H., Sigman D. S. Chemical conversion of a DNA-binding protein into a site-specific nuclease. Science. 1987 Sep 4;237(4819):1197–1201. doi: 10.1126/science.2820056. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Green P., Helms C., Cartinhour S., Weiffenbach B., Stephens K., Keith T. P., Bowden D. W., Smith D. R., Lander E. S. A genetic linkage map of the human genome. Cell. 1987 Oct 23;51(2):319–337. doi: 10.1016/0092-8674(87)90158-9. [DOI] [PubMed] [Google Scholar]
- Gardiner K., Laas W., Patterson D. Fractionation of large mammalian DNA restriction fragments using vertical pulsed-field gradient gel electrophoresis. Somat Cell Mol Genet. 1986 Mar;12(2):185–195. doi: 10.1007/BF01560665. [DOI] [PubMed] [Google Scholar]
- Hartley D. A., Davies K. E., Drayna D., White R. L., Williamson R. A cytological map of the human X chromosome--evidence for non-random recombination. Nucleic Acids Res. 1984 Jul 11;12(13):5277–5285. doi: 10.1093/nar/12.13.5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T. T., Huang T. C., Teng M. H. 5-Methylcytosine replacing cytosine in the deoxyribonucleic acid of a bacteriophage for Xanthomonas oryzae. J Mol Biol. 1968 Jul 14;34(2):373–375. doi: 10.1016/0022-2836(68)90263-5. [DOI] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. A deoxyribonuclease of Diplococcus pneumoniae specific for methylated DNA. J Biol Chem. 1975 Jun 10;250(11):4060–4066. [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977 Jul;114(1):153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- McClelland M., Hanish J., Nelson M., Patel Y. KGB: a single buffer for all restriction endonucleases. Nucleic Acids Res. 1988 Jan 11;16(1):364–364. doi: 10.1093/nar/16.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Ivarie R. Asymmetrical distribution of CpG in an 'average' mammalian gene. Nucleic Acids Res. 1982 Dec 11;10(23):7865–7877. doi: 10.1093/nar/10.23.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Jones R., Patel Y., Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 1987 Aug 11;15(15):5985–6005. doi: 10.1093/nar/15.15.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Kessler L. G., Bittner M. Site-specific cleavage of DNA at 8- and 10-base-pair sequences. Proc Natl Acad Sci U S A. 1984 Feb;81(4):983–987. doi: 10.1073/pnas.81.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Nelson M. Enhancement of the apparent cleavage specificities of restriction endonucleases: applications to megabase mapping of chromosomes. Gene Amplif Anal. 1987;5:257–282. [PubMed] [Google Scholar]
- McClelland M. Purification and characterization of two new modification methylases: MClaI from Caryophanon latum L and MTaqI from Thermus aquaticus YTI. Nucleic Acids Res. 1981 Dec 21;9(24):6795–6804. doi: 10.1093/nar/9.24.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. Recognition sequences of type II restriction systems are constrained by the G + C content of host genomes. Nucleic Acids Res. 1988 Mar 25;16(5):2283–2294. doi: 10.1093/nar/16.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. Site-specific cleavage of DNA at 8-, 9-, and 10-bp sequences. Methods Enzymol. 1987;155:22–32. doi: 10.1016/0076-6879(87)55006-6. [DOI] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Nelson M., McClelland M. Purification and assay of type II DNA methylases. Methods Enzymol. 1987;155:32–41. doi: 10.1016/0076-6879(87)55007-8. [DOI] [PubMed] [Google Scholar]
- Orbach M. J., Vollrath D., Davis R. W., Yanofsky C. An electrophoretic karyotype of Neurospora crassa. Mol Cell Biol. 1988 Apr;8(4):1469–1473. doi: 10.1128/mcb.8.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction enzymes and their isoschizomers. Nucleic Acids Res. 1988;16 (Suppl):r271–r313. doi: 10.1093/nar/16.suppl.r271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Turmel C., Lalande M. Resolution of Schizosaccharomyces pombe chromosomes by field inversion gel electrophoresis. Nucleic Acids Res. 1988 May 25;16(10):4727–4727. doi: 10.1093/nar/16.10.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Campa A. G., Springhorn S. S., Kale P., Lacks S. A. Proteins encoded by the DpnI restriction gene cassette. Hyperproduction and characterization of the DpnI endonuclease. J Biol Chem. 1988 Oct 15;263(29):14696–14702. [PubMed] [Google Scholar]