Abstract
Age-related hearing loss (AHL) in common inbred mouse strains is a genetically complex quantitative trait. We found a synonymous single-nucleotide polymorphism in exon 7 of Cdh23 that shows significant association with AHL and the deafness modifier mdfw (modifer of deafwaddler). The hypomorphic Cdh23753A allele causes in-frame skipping of exon 7. Altered adhesion or reduced stability of CDH23 may confer susceptibility to AHL. Homozygosity at Cdh23753A or in combination with heterogeneous secondary factors is a primary determinant of AHL in mice.
Inbred mouse strains vary greatly in their susceptibility to age-related (AHL) and noise-induced hearing loss (NIHL; refs. 1–3). BALB/cByJ, BUB/BnJ and C57BL/6J strains develop early- or late-onset sensorineural hearing impairment and are highly susceptible to acoustic overstimulation2,4; in comparison, CBA/CaJ and MOLF/Ei have normal hearing throughout life and are fairly resistant to noise trauma. Quantitative and qualitative linkage analyses linked predisposition to AHL and NIHL to the ahl locus on chromosome 10 (ref. 4,5). The ahl interval coincides with the map location of the deafness modifier mdfw (modifier of deafwaddler). The recessive mdfw allele accelerates hearing loss in heterozygous Atp2b2 (plasma membrane Ca2+ ATPase 2)-deficient BALB/cByJ-Atp2b2dfw-2J/+ and C57BL/6J-Atp2b2dfw-2J/+ mice; on wild-type backgrounds, mdfw has little effect6. Genetic complementation tests have shown allelism between ahl and mdfw7.
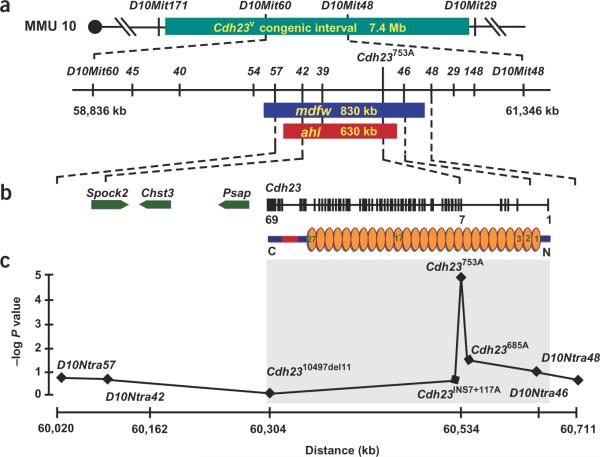
We localized mdfw to a 830-kb and ahl to a 630-kb interstitial genomic region between markers D10Ntra57 and D10Ntra46 (Fig. 1a). Four genes localize to this interval: Spock2, Chst3, Psap and Cdh23 (encoding cadherin 23; Fig. 1b). Mutations in Cdh23 cause deafness in humans and in mouse models8. We screened for nucleotide differences by sequencing all exons of these genes and flanking intronic sequences (≤20 bp) in CBA/CaJ and C57BL/6J mice. We found two sequence changes, both in Cdh23: a deletion of 11 bp in the 3′ untranslated region of exon 69 (Cdh2310497del11) and a G→A transition at nucleotide 753 in exon 7 (Cdh23753G→A). To investigate association with AHL, we sequenced exon 7 and exon 69 in an additional 54 inbred strains. We found Cdh2310497del11 in strains R/J and C57BL/6J only. The 753G→A polymorphism showed nearly perfect correlation with AHL (P = 2 × 10−5 by χ2 test; Fig. 1c and Supplementary Table 1 online). Of 31 strains classified with AHL, 27 carry the 753A allele, and of 25 AHL-negative strains, 22 segregate the 753G variant. All strains for which we genetically linked AHL to the ahl interval and those for which we showed allelism with mdfw carry the Cdh23753A allele. Cby-Atp2b2dfw-2J, which segregates mdfw, also has the Cdh23753A variant. The few strains that did not correlate may show incomplete penetrance, may develop hearing loss later in life (BDP/J, SEC/Re1J, SHR/GnJ), may segregate a mutation in Ednrb (I/LnJ) or may have acquired susceptibility allele(s) other than ahl (MRL/MpJ, C3H/HeSnJ, YBR/Ei). At seven marker loci across the ahl interval, laboratory strains share the same haplotype, which is derived from an ancient Mus musculus domesticus chromosome (Supplementary Table 2 online). Given the origin of these strains from a few founder mice, the association of the Cdh23753A allele with one common haplotype argues in favor of an ancestral mutation.
Figure 1.
Positional cloning of ahl and mdfw. (a) Physical map of ahl and mdfw. The Cdh23v congenic interval (7.4 Mb) is defined by D10Mit171 and D10Mit29. Positions of newly developed SNP markers (D10Ntra45, D10Ntra40, D10Ntra54, D10Ntra57, D10Ntra42, D10Ntra39, D10Ntra46, D10Ntra48, D10Ntra29 and D10Ntra148; stems omitted in figure) in relation to mdfw and ahl intervals are shown. D10Ntra54 and D10Ntra48 are recombinant (D10Ntra54 - 0.26 ± 0.18 cM - mdfw - 0.13 ± 0.13 cM - D10Ntra48) with mdfw. The highest lod score for ahl (108.4) was in the region between D10Ntra57 and D10Ntra46. Primer sequences are available on request. (b) Four genes localize to the critical interval: sparc/osteonectin 2 (Spock2), carbohydrate sulfotransferase 3 (Chst3), prosaposin (Psap) and cadherin 23 (Cdh23). Transcription orientation is indicated. Genomic structure, from telomere to centromere, of Cdh23 (black vertical lines) and the domain structure of cadherin 23, including transmembrane domain (red) and ectodomains (orange, 1–27), are shown. (c) Profile of probability scores. Negative logarithm of P is plotted against marker location on the physical map (available at http://genome.cse.ucsc.edu; February 2002 assembly).
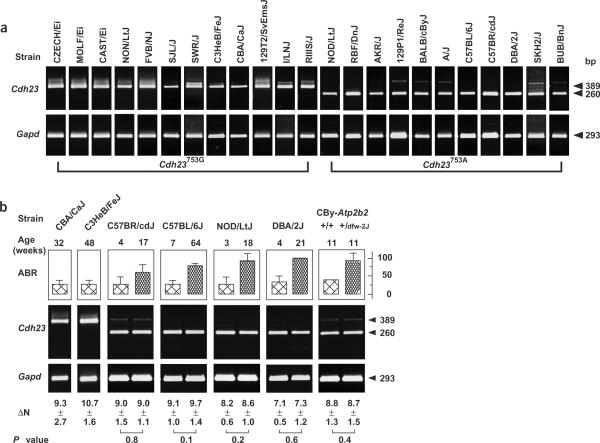
The synonymous G→A substitution occurs at the last position of exon 7. To test whether the substitution alters splicing, we carried out PCR analysis on reverse-transcribed cochlea mRNA. In 11 strains tested, the Cdh23753A allele perfectly correlated with in-frame skipping of exon 7 (Fig. 2a). Strains with the Cdh23753G allele preferentially produced wild-type transcripts. We next asked whether increasing levels of alternatively spliced transcripts parallel the onset and progression of AHL. Using the quantitative real-time PCR assay, we did not find a statistically significant difference (P > 0.05) in the accumulation of alternatively spliced mRNA with increasing thresholds in C57BL/6J, C57BR/cdJ, NOD/LtJ, DBA/2J and CBy-Atp2b2dfw-2J (Fig. 2b).
Figure 2.
Cdh23753A affects splicing of exon 7. Cdh23-specific primers located in exon 6 and exon 8 amplify wild-type (389 bp) and alternatively spliced transcripts (260 bp) from cochlea cDNA (see Supplementary Note online). Gapd was included as reference. (a) RT-PCR analyses of exon 7 in 23 common inbred strains. Strains, allele status at Cdh23753 and amplified PCR fragments are shown. Strains within genealogical subgroups of mice (Swiss mice, Castle's mice) have different allele status and alternative splicing; compare NOD/LtJ with NON/LtJ and 129P1/ReJ with 129T2SvEmsJ. (b) ABR analysis was used to assess hearing function in the indicated strains (at the indicated ages), and the averaged response (dB SPL) to a click stimulus is plotted. ΔN = NGapd − NCdh23, where N is the cycle number at which a significant increase of fluorescence signal above background (usually 0.01 units) was observed. The mean ± s.d. (n ≥ 10 assays) and statistical significance (P value) are given. Both Cby-Atp2b2+/+ and Cby-Atp2b2+/dfw-2J are homozygous with respect to mdfw.
To test the functionality of the Cdh23753A allele, we studied its trans effect on the frame-shift allele Cdh23834–835insG (Cdh23v). We determined by SNP marker analysis that Cdh23v arose on an ancestral Mus musculus molossinus chromosome and that the retained congenic interval contains both ahl and mdfw loci (Fig. 1a). Because MOLF/Ei mice have normal hearing and are resistant to NIHL, we assumed that if Cdh23 and ahl were different genes, then the Cdh23v allele would be in coupling phase with the protecting allele of ahl in the V/Le strain. If so, hybrid mice derived from matings between AHL-susceptible strains and V/Le would be protected from AHL by the dominant V/Le-derived resistance allele at ahl. If Cdh23753A underlies the hearing loss associated with ahl, however, then such hybrid mice would have AHL because the Cdh23753A allele from the AHL-susceptible strain would be combined with the Cdh23v null allele from the V/Le strain. Cdh23753A/Cdh23v compound heterozygotes had significantly higher auditory-brainstem response (ABR) thresholds to a series of acoustic stimuli (Supplementary Table 3 online). In comparison, Cdh23753G/Cdh23v had normal waveforms and thresholds.
The stereocilia hair bundle has a highly organized staircase-like architecture, which is central to the function of cochlea and vestibular hair cells. Mice deficient in Cdh23 develop a structurally disorganized hair bundle9. Recent data provide evidence that cadherin 23 forms a complex with harmonin b and myosin 7A that localizes to stereocilia and is a component of interciliary links10,11. The peptide of 43 amino acids encoded by exon 7 is part of the second and third ectodomain and lies in the potential homodimerization site of cadherin 23. The CDH235712A mutation in humans is associated with Usher syndrome type 1, and its mechanism of action is similar to that of the Cdh23753A allele12. Together, the data suggest that Cdh23753A is a pathological, hypomorphic allele; predisposition to AHL and NIHL may be conferred through altered adhesion or intracellular targeting of misfolded protein.
Homozygosity with respect to Cdh23753A significantly increases susceptibility to AHL but is not the only cause of its phenotypic manifestation. Predisposition to early-onset AHL conferred by Cdh23753A depends on the effects of several strain-specific genetic factors, including the mitochondrial mutation mt-Tr9827ins8 (as in A/J; ref. 13), ahl2 (as in NOD/LtJ; ref. 14) and ahl3 (K.R.J and Q.Y.Z, unpublished data). Combination of any of these `accelerating alleles' with Cdh23753A is sufficient to induce AHL expression (Supplementary Fig. 1 online). An additional genetic factor is the null allele of Atp2b2, which is an important regulator of intrastereocilia Ca2+ levels15. Haploinsufficiency at Atp2b2 and homozygosity with respect to Cdh23753A together, but neither alone, cause early-onset hearing loss in mdfw mice (Atp2b2+/dfw-2J mdfw/mdfw; Fig. 2b). The heterogeneity of secondary factors suggests additive or stochastic interactions with Cdh23753A. The genetic architecture of AHL and NIHL may provide a paradigm for predisposition to AHL and NIHL in human and defines a presbyacusis model to explore therapeutic avenues, such as stem cell therapy.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Pellegrino, M. Irby, A. Calderon, A. Derr and J. Fiallos for technical assistance, K. Steel for the V/Le strain and C. Kubisch, N. Fischel-Ghodsian and D. Drayna for discussions and their comments on the manuscript.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
References
- 1.Erway LC, Willott JF, Archer JR, Harrison DE. Hear. Res. 1993;65:125–132. doi: 10.1016/0378-5955(93)90207-h. [DOI] [PubMed] [Google Scholar]
- 2.Willott JF, et al. Hear. Res. 1998;115:162–174. doi: 10.1016/s0378-5955(97)00189-5. [DOI] [PubMed] [Google Scholar]
- 3.Zheng QY, Johnson KR, Erway LC. Hear. Res. 1999;130:94–107. doi: 10.1016/s0378-5955(99)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis RR, et al. Hear. Res. 2001;155:82–90. doi: 10.1016/s0378-5955(01)00250-7. [DOI] [PubMed] [Google Scholar]
- 5.Johnson KR, Zheng QY, Erway LC. Genomics. 2000;70:171–180. doi: 10.1006/geno.2000.6377. [DOI] [PubMed] [Google Scholar]
- 6.Noben-Trauth K, Zheng QY, Johnson KR, Nishina PM. Genomics. 1997;44:266–272. doi: 10.1006/geno.1997.4869. [DOI] [PubMed] [Google Scholar]
- 7.Zheng QY, Johnson KR. Hear. Res. 2001;154:45–53. doi: 10.1016/s0378-5955(01)00215-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petit C, Levilliers J, Hardelin JP. Annu. Rev. Genet. 2001;35:589–646. doi: 10.1146/annurev.genet.35.102401.091224. [DOI] [PubMed] [Google Scholar]
- 9.Di Palma F, et al. Nat. Genet. 2001;27:103–107. doi: 10.1038/83660. [DOI] [PubMed] [Google Scholar]
- 10.Boeda B, et al. EMBO J. 2002;21:6689–6699. doi: 10.1093/emboj/cdf689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siemens J, et al. Proc. Natl. Acad. Sci. USA. 2002;99:14946–14951. doi: 10.1073/pnas.232579599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Brederlow B, et al. Hum. Mutat. 2002;19:268–273. doi: 10.1002/humu.10049. [DOI] [PubMed] [Google Scholar]
- 13.Johnson KR, Zheng QY, Bykhovskaya Y, Spirina O, Fischel-Ghodsian N. Nat. Genet. 2001;27:191–194. doi: 10.1038/84831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson KR, Zheng QY. Genomics. 2002;80:461–464. [PMC free article] [PubMed] [Google Scholar]
- 15.Yamoah EN, et al. J. Neurosci. 1998;18:610–624. doi: 10.1523/JNEUROSCI.18-02-00610.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.