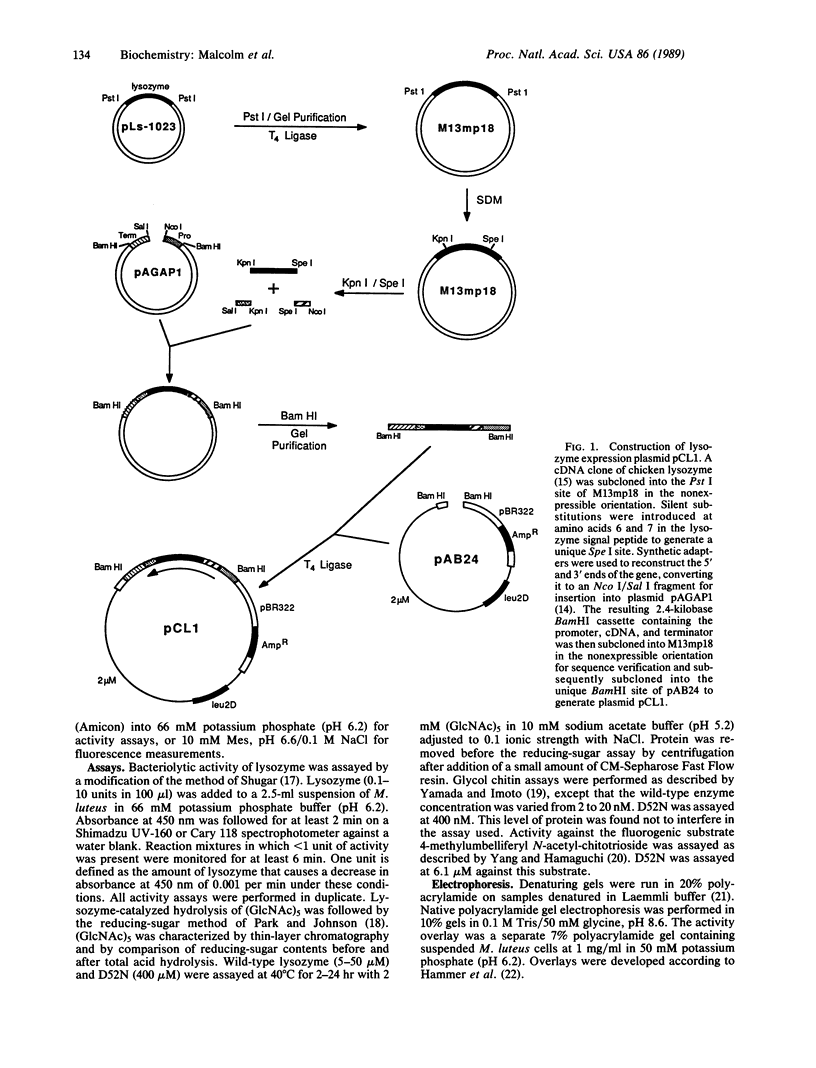
Abstract
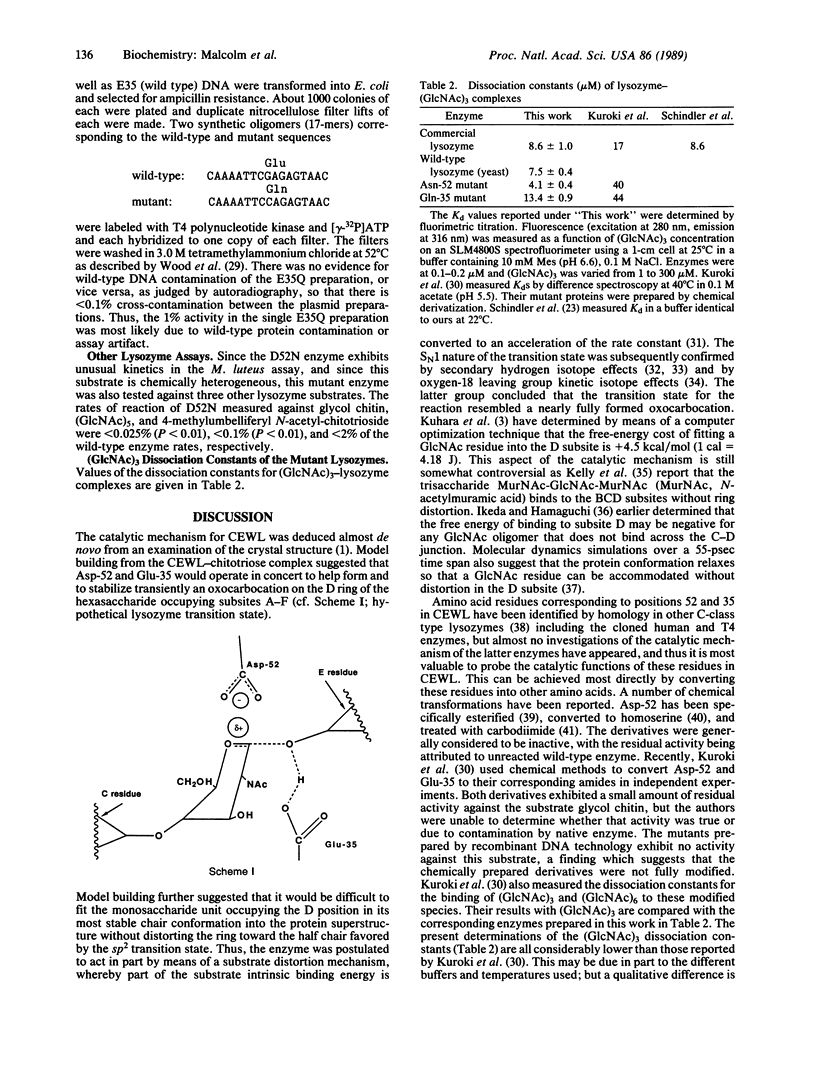
The roles of the catalytic active-site residues aspartic acid-52 and glutamic acid-35 of chicken lysozyme (EC 3.2.1.17) have been investigated by separate in vitro mutagenesis of each residue to its corresponding amide (denoted as D52N and E35Q, respectively). The mutant enzyme D52N exhibits approximately 5% of the wild-type lytic activity against Micrococcus luteus cell walls, while there is no measurable activity associated with E35Q (0.1% +/- 0.1%). The measured dissociation constants for the chitotriose-enzyme complexes were 4.1 microM (D52N) and 13.4 microM (E35Q) vs. 8.6 microM for wild type, indicating that the alterations in catalytic properties may be due in part to binding effects as well as to direct catalytic participation of these residues. The mutant lysozymes have been expressed in and secreted from yeast and obtained at a level of approximately 5 mg per liter of culture by high-salt elution from the cell walls.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barr P. J., Gibson H. L., Enea V., Arnot D. E., Hollingdale M. R., Nussenzweig V. Expression in yeast of a Plasmodium vivax antigen of potential use in a human malaria vaccine. J Exp Med. 1987 Apr 1;165(4):1160–1171. doi: 10.1084/jem.165.4.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Blake C. C., Koenig D. F., Mair G. A., North A. C., Phillips D. C., Sarma V. R. Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution. Nature. 1965 May 22;206(4986):757–761. doi: 10.1038/206757a0. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Brake A. J., Merryweather J. P., Coit D. G., Heberlein U. A., Masiarz F. R., Mullenbach G. T., Urdea M. S., Valenzuela P., Barr P. J. Alpha-factor-directed synthesis and secretion of mature foreign proteins in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4642–4646. doi: 10.1073/pnas.81.15.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlquist F. W., Rand-Meir T., Raftery M. A. Application of secondary alpha-deuterium kinetic isotope effects to studies of enzyme catalysis. Glycoside hydrolysis by lysozyme and beta-glucosidase. Biochemistry. 1969 Oct;8(10):4214–4221. doi: 10.1021/bi00838a045. [DOI] [PubMed] [Google Scholar]
- Douzou P., Hoa G. H., Petsko G. A. Protein crystallography at sub-zero temperatures: lysozyme-substrate complexes in cooled mixed solvents. J Mol Biol. 1975 Aug 15;96(3):367–380. doi: 10.1016/0022-2836(75)90166-7. [DOI] [PubMed] [Google Scholar]
- Erhart E., Hollenberg C. P. The presence of a defective LEU2 gene on 2 mu DNA recombinant plasmids of Saccharomyces cerevisiae is responsible for curing and high copy number. J Bacteriol. 1983 Nov;156(2):625–635. doi: 10.1128/jb.156.2.625-635.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshdat Y., Dunn A., Sharon N. Chemical conversion of aspartic acid 52, a catalytic residue in hen egg-white lysozyme, to homoserine. Proc Natl Acad Sci U S A. 1974 May;71(5):1658–1662. doi: 10.1073/pnas.71.5.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer M. F., Schilling J. W., Prager E. M., Wilson A. C. Recruitment of lysozyme as a major enzyme in the mouse gut: duplication, divergence, and regulatory evolution. J Mol Evol. 1987;24(3):272–279. doi: 10.1007/BF02111240. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler E., Rupley J. A., Hess G. P. Productive and unproductive lysozyme-chitosaccharide complexes. Equilibrium measurements. Biochemistry. 1975 Mar 11;14(5):1088–1094. doi: 10.1021/bi00676a032. [DOI] [PubMed] [Google Scholar]
- Ibrahimi I. M., Prager E. M., White T. J., Wilson A. C. Amino acid sequence of California quail lysozyme. Effect of evolutionary substitutions on the antigenic structure of lysozyme. Biochemistry. 1979 Jun 26;18(13):2736–2744. doi: 10.1021/bi00580a008. [DOI] [PubMed] [Google Scholar]
- Ikeda K., Hamaguchi K. Binding of substrate analogues to subsites D, E, and F of hen egg-white lysozyme. J Biochem. 1976 Feb;79(2):237–247. doi: 10.1093/oxfordjournals.jbchem.a131063. [DOI] [PubMed] [Google Scholar]
- Jencks W. P. Binding energy, specificity, and enzymic catalysis: the circe effect. Adv Enzymol Relat Areas Mol Biol. 1975;43:219–410. doi: 10.1002/9780470122884.ch4. [DOI] [PubMed] [Google Scholar]
- Jollès P., Jollès J. What's new in lysozyme research? Always a model system, today as yesterday. Mol Cell Biochem. 1984 Sep;63(2):165–189. doi: 10.1007/BF00285225. [DOI] [PubMed] [Google Scholar]
- Jung A., Sippel A. E., Grez M., Schütz G. Exons encode functional and structural units of chicken lysozyme. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5759–5763. doi: 10.1073/pnas.77.10.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. A., Sielecki A. R., Sykes B. D., James M. N., Phillips D. C. X-ray crystallography of the binding of the bacterial cell wall trisaccharide NAM-NAG-NAM to lysozyme. Nature. 1979 Dec 20;282(5741):875–878. doi: 10.1038/282875a0. [DOI] [PubMed] [Google Scholar]
- Knowles J. R. Tinkering with enzymes: what are we learning? Science. 1987 Jun 5;236(4806):1252–1258. doi: 10.1126/science.3296192. [DOI] [PubMed] [Google Scholar]
- Kuhara S., Ezaki E., Fukamizo T., Hayashi K. Estimation of the free energy change of substrate binding lysozyme-catalyzed reactions. J Biochem. 1982 Jul;92(1):121–127. doi: 10.1093/oxfordjournals.jbchem.a133908. [DOI] [PubMed] [Google Scholar]
- Kumagai I., Kojima S., Tamaki E., Miura K. Conversion of Trp 62 of hen egg-white lysozyme to Tyr by site-directed mutagenesis. J Biochem. 1987 Oct;102(4):733–740. doi: 10.1093/oxfordjournals.jbchem.a122111. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki R., Yamada H., Moriyama T., Imoto T. Chemical mutations of the catalytic carboxyl groups in lysozyme to the corresponding amides. J Biol Chem. 1986 Oct 15;261(29):13571–13574. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lin T. Y., Koshland D. E., Jr Carboxyl group modification and the activity of lysozyme. J Biol Chem. 1969 Jan 25;244(2):505–508. [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Matthews B. W., Remington S. J., Grütter M. G., Anderson W. F. Relation between hen egg white lysozyme and bacteriophage T4 lysozyme: evolutionary implications. J Mol Biol. 1981 Apr 25;147(4):545–558. doi: 10.1016/0022-2836(81)90399-5. [DOI] [PubMed] [Google Scholar]
- Maurel P., Douzou P. Catalytic implications of electrostatic potentials: the lytic activity of lysozymes as a model. J Mol Biol. 1976 Apr 5;102(2):253–264. doi: 10.1016/s0022-2836(76)80052-6. [DOI] [PubMed] [Google Scholar]
- PARK J. T., JOHNSON M. J. A submicrodetermination of glucose. J Biol Chem. 1949 Nov;181(1):149–151. [PubMed] [Google Scholar]
- Parsons S. M., Raftery M. A. The identification of aspartic acid residue 52 as being critical to lysozyme activity. Biochemistry. 1969 Oct;8(10):4199–4205. doi: 10.1021/bi00838a043. [DOI] [PubMed] [Google Scholar]
- Perry L. J., Heyneker H. L., Wetzel R. Non-toxic expression in Escherichia coli of a plasmid-encoded gene for phage T4 lysozyme. Gene. 1985;38(1-3):259–264. doi: 10.1016/0378-1119(85)90226-4. [DOI] [PubMed] [Google Scholar]
- Prager E. M., Wilson A. C. The dependence of immunological cross-reactivity upon sequence resemblance among lysozymes. I. Micro-complement fixation studies. J Biol Chem. 1971 Oct 10;246(19):5978–5989. [PubMed] [Google Scholar]
- Rosenberg S., Kirsch J. F. Oxygen-18 leaving group kinetic isotope effects on the hydrolysis of nitrophenyl glycosides. 2. Lysozyme and beta-glucosidase: acid and alkaline hydrolysis. Biochemistry. 1981 May 26;20(11):3196–3204. doi: 10.1021/bi00514a032. [DOI] [PubMed] [Google Scholar]
- SHUGAR D. The measurement of lysozyme activity and the ultra-violet inactivation of lysozyme. Biochim Biophys Acta. 1952 Mar;8(3):302–309. doi: 10.1016/0006-3002(52)90045-0. [DOI] [PubMed] [Google Scholar]
- Schindler M., Assaf Y., Sharon N., Chipman D. M. Mechanism of lysozyme catalysis: role of ground-state strain in subsite D in hen egg-white and human lysozymes. Biochemistry. 1977 Feb 8;16(3):423–431. doi: 10.1021/bi00622a013. [DOI] [PubMed] [Google Scholar]
- Smith-Gill S. J., Lavoie T. B., Mainhart C. R. Antigenic regions defined by monoclonal antibodies correspond to structural domains of avian lysozyme. J Immunol. 1984 Jul;133(1):384–393. [PubMed] [Google Scholar]
- Smith L. E., Mohr L. H., Raftery M. A. Mechanism for lysozyme-catalyzed hydrolysis. J Am Chem Soc. 1973 Oct 31;95(22):7497–7500. doi: 10.1021/ja00803a046. [DOI] [PubMed] [Google Scholar]
- Travis J., Owen M., George P., Carrell R., Rosenberg S., Hallewell R. A., Barr P. J. Isolation and properties of recombinant DNA produced variants of human alpha 1-proteinase inhibitor. J Biol Chem. 1985 Apr 10;260(7):4384–4389. [PubMed] [Google Scholar]
- Wood W. I., Gitschier J., Lasky L. A., Lawn R. M. Base composition-independent hybridization in tetramethylammonium chloride: a method for oligonucleotide screening of highly complex gene libraries. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1585–1588. doi: 10.1073/pnas.82.6.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H., Imoto T. A convenient synthesis of glycolchitin, a substrate of lysozyme. Carbohydr Res. 1981 May 18;92(1):160–162. doi: 10.1016/s0008-6215(00)85993-5. [DOI] [PubMed] [Google Scholar]
- Yang Y., Hamaguchi K. Hydrolysis of 4-methylumbelliferyl N-acetyl-chitotrioside catalyzed by hen and turkey lysozymes. pH dependence of the kinetics constants. J Biochem. 1980 Apr;87(4):1003–1014. [PubMed] [Google Scholar]