Abstract
Seven transmembrane (7TM) or G protein-coupled receptors constitute a large superfamily of cell surface receptors sharing a structural motif of seven transmembrane spanning alpha helices. Their activation mechanism most likely involves concerted movements of the transmembrane helices, but remains to be completely resolved. Evolutionary Trace (ET) analysis is a computational method, which identifies clusters of functionally important residues by integrating information on evolutionary important residue variations with receptor structure. Combined with known mutational data, ET predicted a patch of residues in the cytoplasmic parts of TM2, TM3, and TM6 to form an activation switch that is common to all family A 7TM receptors. We tested this hypothesis in the rat Angiotensin II (Ang II) type 1 (AT1) receptor. The receptor has important roles in the cardiovascular system, but has also frequently been applied as a model for 7TM receptor activation and signaling. Six mutations: F66A, L67R, L70R, L119R, D125A, and I245F were targeted to the putative switch and assayed for changes in activation state by their ligand binding, signaling, and trafficking properties. All but one receptor mutant (that was not expressed well) displayed phenotypes associated with changed activation state, such as increased agonist affinity or basal activity, promiscuous activation, or constitutive internalization highlighting the importance of testing different signaling pathways. We conclude that this evolutionary important patch mediates interactions important for maintaining the inactive state. More broadly, these observations in the AT1 receptor are consistent with computational predictions of a generic role for this patch in 7TM receptor activation.
Keywords: 7TM receptor, GPCR, Evolutionary Trace, constitutive activity, Angiotensin
1. INTRODUCTION
7TM receptors constitute a large family of cell surface receptors all characterized by a conserved motif of seven transmembrane spanning alpha helices. The receptors are involved in numerous processes in the human body with important roles in many physiological processes [1].
The molecular mechanisms underlying 7TM receptor activation have not been completely resolved, but they most likely involve similar or even conserved concerted rearrangements of the helical bundle with an outward movement of TM6. This is summarized in the global toggle switch activation model [2]. One of the frequently applied methods to identify molecular interactions involved in receptor activation is the study of constitutively activated receptor mutants (CAMs). The elevated basal activity of these mutants is attributed to removal of restraints normally maintaining the inactive state of the receptor [1].
Thus, an extensive amount of information currently exists regarding receptor regions involved in conversion between inactive and active states now exists. Some of these regions have been verified by mutational studies in several family A 7TM receptors and an extensive amount of mutational data is available. Evolutionary Trace (ET) analysis is a computational method that identifies functional sites in protein families by looking at the evolutionary conservation of the residue positions in homologues sequences [3]. The analysis ranks all the residue positions in a protein family according to how well their side chain variations correlate with major evolutionary divergences of the protein family [3]. Top-ranked residues have important proteome-wide biological properties: they cluster structurally [4]; and these clusters predict functional sites [5]. Thus ET may be used to identify functional sites [6] and to guide mutations that selectively alter these sites and hereby separate singular functions [7] or to exchange functions [8, 9]. Specifically, for family A 7TM receptors this analysis has led to the prediction of a universally conserved activation mechanism consistent with mutations in rhodopsin [3, 10] and in the β-adrenergic receptor [11].
The Angiotensin II (Ang II) type 1 (AT1) receptor is the primary effector of the peptide hormone Ang II. The receptor is coupled to Gq/11, leading to inositol phosphate pathway induction, which in turn triggers an increase in intracellular calcium and protein kinase C activation [12]. However, G protein independent, β-arrestin dependent signaling also has been demonstrated for the receptor [13]. The receptor has frequently been applied as a model receptor for 7TM receptor activation. Previous studies on the molecular determinants of AT1 receptor activation support the general model for 7TM receptor activation with special attention to interactions between and movements of TM2, TM3, TM6, and TM7 (for review: Petrel and Clauser 2009 [14]). Studies have shown that constitutive AT1 receptor activity sometimes reveals itself as constitutive internalization of the receptor or promiscuous activation [15–17]. Combined with the reports of G protein-independent signaling from this receptor, this suggests that testing of more than one signaling pathway is advisable when studying AT1 receptor activation.
In this study, we combined Evolutionary Trace analysis with mutational data from the GPCR database to predict a patch of six amino acids in the cytoplasmic core of the rat AT1a receptor involved in conversion between inactive and active state. These residues were mutated individually and mutant receptors were characterized for expression, ligand binding and G protein-dependent as well as G protein-independent signaling outcomes. Five out of six mutant receptors showed signs of a changed activation state in one or more assays hereby supporting a role for the patch in AT1 receptor activation. We find that our analysis supports the general ET model of 7TM receptor function thus displaying the strength of combining an unbiased computational approach with mutational history to guide mutational studies in 7TM receptors and that it highlights the importance of testing multiple signaling endpoints to ensure accurate characterization of mutation phenotypes.
2. EXPERIMENTAL METHODS
2.1 ET analysis
To identify key residues that may form part of an activation switch in the AT1 receptor (rat AT1a), we followed a computational approach first described by Madabushi et al.[10], which is based on the ET method. The underlying hypothesis is that residues predicted to be important among a large group of 7TM receptors most likely take part in functions common to the entire group of 7TM receptors rather than functions specific to their ligands. A comparison with the large body of mutational data described in the literature can further narrow down this set of key functional residues to some more closely associated with ligand sensitivity, conformational switching, or G protein-coupling.
In this study, we have focused on residues involved in conformational switching from the inactive to the active state of the receptor. Thus, we compared the ET data to mutational data where mutations caused constitutive activity. This led to the identification of six amino acids in TM2, TM3 and TM6. To test the role of this patch in the conformational switch mechanism, we then predicted disruptive mutations most likely to induce constitutive activity.
Specifically, a BLAST [18] search retrieved 129 visual opsins, 69 bioamine, 58 olfactory, and 82 chemokine family A receptor sequences from NCBI data base (See supplementary material table 1). An alignment of each receptor family was generated by ClustalW [19]. 7TM receptors are most conserved in the transmembrane helices whereas the loops are highly divergent. Therefore the alignments were performed on seven transmembrane helix residues only. Specifically, the analyzed residues of the rat AT1a sequence are: 27–55 (TM1), 63–88 (TM2), 99–132 (TM3), 145–167 (TM4), 194–222 (TM5), 238–265 (TM6), and 282–307 (TM7). We then performed an Evolutionary Trace on the concatenated gapless TM segments. This produced a relative ranking of evolutionary importance of the transmembrane sequence residues.
To further narrow down our search to residues most likely to have a direct role in the conformational switch mechanism linked to activation, we picked a subset of residues implicated in constitutive activity or misfolding from the top 20th percentile of importance residues. These choices were either based on mutational data (sixteen residues) or on structural proximity (two others). We then selected six top-ranked residues from this group (15th percentile or better) by ET located in TM2, TM3 and TM6 and with a clustering z-score of 2.8 in the rhodopsin structure 1F88, details are listed in Table 1 and below. Four of these, L67, L119, D125, I245 in the rat AT1a receptor (Ballesteros # in order is 2.43, 3.43, 3.49, 6.40), had cognate residues mutations causing constitutive activity in luteinizing hormone receptor, muscarinic acetylcholine, rhodopsin, and anaphylatoxin chemotactic receptors [20–23]. One residue L70 (2.46) was a novel prediction that was found to cause constitutive activity in rhodopsin [10], and F66 (2.42) was not yet tested for constitutive activity to our knowledge.
TABLE 1. Residues of the rat AT1a receptor activation switch patch identified by ET Analysis.
To predict residues involved in 7TM receptor activation, ET analysis was performed in combination with analysis of mutational data on constitutive activity from the literature. Mutations most likely to cause constitutive activity in the AT1 receptor were then predicted based on records of constitutive activity in the literature, natural variance at the sequence loci in Ang II receptors, and lastly more dispruptive substitution were favored. The resulting six amino acids and selected mutations are reported here. Residues are shown both as numbered from the N-terminal of the AT1 receptor and according to the Schwartz and Ballesteros standardized numbering schemes. Percent rank in ET analysis is reported along with mutation records on constitutive activity (CA) in the literature and variations in Ang II receptor sequences.
| Residue in rat AT1a receptor | Residue # (Schwartz/Ballesteros) | Residue # Bovine Rhodopsin | % Rank | # of CA Mutations / Mutations with recorded effect | CA causing target residues (# of observations) | Natural variations in Ang II sequences at loci | Selected mutation |
|---|---|---|---|---|---|---|---|
| F66 | II:02/2.42 | 75 | 15 | 0 | Novel | YF | A |
| L70 | II:06/2.46 | 79 | 5 | 1/1 | A(1), S(1) | L | R |
| L119 | III:19/3.43 | 128 | 5 | 21/28 | R(13) Q(2) H(1) | ILV | R |
| I245 | VI:05/6.40 | 257 | 10 | 7/9 | F(3) A(3) T(1) T(3) R(2) M(2) |
VI | F |
| L67 | II:03/2.43 | 76 | 15 | 9/15 | Q(1) A(8) Q(6) N(4) |
ILV | R |
| D125 | III:25/3.49 | 134 | 5 | 21/54 | D(1) E(1) S(1) T(1) |
D | A |
To pick substitutions likely to induce constitutive activity for each of these residues, we followed several specific criteria. First, we favored substitution that were already implicated in constitutive activation in the literature; second, we rejected substitutions if these naturally occurred in the family of Ang II receptor sequences; and third, we favored substitutions that were thought to be more disruptive substitutions by traditional criterion (i.e. positive charge to negative charge residue – although we recognize that the meaning of disruptive is statistical rather and may not stringently apply at any one specific site). For example, the L67R mutation switches a non polar sequence position into a positively charged. It also mimics two similar Arg susbstitutions at the cognate position that led to constitutive activity in Glucagon and Vasoactive Intestinal peptide 1 receptors [24, 25]. Finally, no Ang II receptor sequences have an Arg at that position. Likewise, for other mutations such as L119R, D125A and I245F: L119 (3.43) is well reported to be a conformational switch residue; D125 (3.49) is part of the TM3 DRY motif frequently reported to have mixed effects on conformational activation, mechanistic effects and G protein uncoupling; and I245 (6.40) has been reported to induce constitutive activation of receptor in rhodopsin and anaphylatoxin chemotactic receptors [20, 22]. We also note that L70R mutation partially goes along with our previous study [10] showing that a mutation to Ala at this residue position causes constitutive activity in rhodopsin. And finally, F66A is a novel change with no previous mutational data on constitutive activity from the literature, but the Phe to Ala mutation is a highly disruptive change. A detailed evolutionary information and mutation recommendation of above residues is listed in Table 1.
2.2 Ligands
Ang II and Sar1- Thr8 (ST) Ang II were from Sigma-Aldrich (St. Louis, MO). Sar1 - Ile4 - Ile8 (SII) Ang II was synthesized at the Cleveland Clinic, Lerner Research Institute (Cleveland, OH, USA). Telmisartan was from Boehringer Ingelheim (Ingelheim am Rhein, Germany)
2.3 Recombinant DNA plasmids
Mutations were generated using the Quickchange mutagenesis protocol (Stratagene, La Jolla, CA) with the rat AT1a or Myc-tagged human AT1 receptor as template. Constructs were subcloned into a modified pCDNA3.1vector containing the M1 Flag-tag inserted after a hemagglutinin signal peptide (rat AT1a constructs; [26]) or into the pSI vector (human AT1 constructs, see Hansen et al, 2004 [27]). Mutations were verified by sequencing at Eurofins MWG Operon (Ebersberg, Germany). The hAT1 N111A construct was described in Hansen et al, 2008 [28]. GFP tagged β-arrestin2 was a gift from Dr. Marc Caron [29].
2.4 Cell culture
Human embryonic kidney (HEK) 293 cells (American Type Culture Collection, Manassas, VA, USA) were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 4.5 g/L glucose and L-Glutamine (Invitrogen, Carlsbad, CA) or supplemented with 0.03% L-Glutamine (Substrate Department at Panum Institute, Copenhagen, Denmark), both supplemented with 10% fetal bovine serum (HyClone, Logan, UT or BioChrome AG, Berlin Germany). N-terminally M1 FLAG-tagged constructs were stably expressed in HEK293 cells. For generation of clonal stable cell lines, single colonies were selected and propagated in the presence of selection-containing media (Zeocin, 0.2ug/mL (Invitrogen, Carlsbad, CA)).
2.5 Whole-cell competitive radioligand binding assay
Binding assay was conducted as described in Hansen et al., 2004 [27] with minor modifications. In brief, HEK293 cells stably expressing AT1 receptor constructs were seeded in poly-L-lysine (Sigma-Aldrich, St. Louis, MO) coated 48-well dishes at 180 000 cells/well. On the following day, cells were kept at 4°C for 30–60 mins., washed once in cold Hanks Balanced Salt Solution (HBSS) with 20 mM Hepes supplemented with 0.9 mM CaCl2 and 1.05 mM MgCl2 (HBSS+). This was followed by 3h of incubation at 4°C with 0.2 mL of HBSS+ containing radioligand ~ 4.5 pM of 125I - Sar1- Ile8 (SI) Ang II (Perkin Elmer, Waltham, MA) and increasing amounts of unlabeled Ang II (triplicate measures for each point). Cells were washed twice in ice cold HBSS+ and lysed in 0.5 mL of lysis buffer (1% TritonX, 50 mM Tris-HCl pH 7.5, 100 mM NaCl, and 5mM EDTA) at room temperature (RT) for 30–60 mins. Well content was transfered to scintillation vials containing 4mL Ultima Gold scintillation liquid (Perkin Elmer, Waltham, MA). Total radioactivity was measured in a TriCarb 2800 TR Liquid Scintillation analyzer (Perkin Elmer, Waltham, MA). Results were analyzed in GraphPad Prism and Excel. Curves were fitted using the non-linear regression analysis (one-site competition) in GraphPad Prism. Kd values for SI Ang II were identified by homologous competition binding using unlabeled SI Ang II as competitor (data not shown), Kd = IC50-[radioligand]. Ki values for Ang II were then calculated using the formula Ki=IC50/[1+([radioligand]/Kd)]. Statistical significance was evaluated by Student’s t-test (two tailed, paired) on pKi values in Excel.
2.6 Inositol phosphate accumulation
Cells were seeded in poly-L-lysine coated 96 well dishes at 100 000/well in 100 μL DMEM without I-inositol with Glutamax (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum and 2 μCi H3myo inositol (GE Healthcare, Buckinghamshire, UK) pr. mL medium. On the following day, cells were preincubated for 25 min at 37°C with either inverse agonist or vehicle (DMSO). After incubation, a wash was performed in Hanks Balanced Salt Solution (HBSS) with) and 20mM Hepes supplemented with 0.9 mM CaCl2, 1.05 mM MgCl2, 10 mM LiCl (HBSS++ vehicle and thereafter incubated 20 mins at 37°C with HBSS++ and inverse agonist for Telmisartan condition to ensure steady state interaction. This was followed by ligand stimulation for 45 mins at 37°C. Hereafter, HBSS++ was removed, and cells were incubated at 4°C with 20 mM formic acid for about 45 mins. 20 μL of the supernatant was transferred to a white 96 well plate (Perkin Elmer, Waltham, MA) and 80 μL SPA beads (GE Healthcare, Buckinghamshire, UK, freshly diluted in MilliQ H20 to final concentration of 1mg/well) was added. Plate was sealed with “Top Seal” cover (Perkin Elmer, Waltham, MA) and incubated for 30 min at constant shaking. Hereafter, SPA beads settled at RT for at least 8h before counted on Topcounter. Data was analyzed in GraphPad Prism and Excel. Statistical analysis was performed in Excel (unpaired Student’s t-test, two-tailed) on un-normalized means.
2.7 ERK1/2 phosphorylation by Western blotting
Stable cell lines were seeded in poly-L-lysine coated 6 well dishes at 400 000/well and grown to confluency. Cells were then serum starved for 4h, stimulated with ligands, washed twice in ice cold PBS and frozen at −20°C. Lysis, SDS-PAGE, and immunoblotting were conducted as described in Theilade et al. with minor modifications [30]. Antibodies were diluted in Tris Buffered Saline with 0.2% Tween20, 0.1% sodium azide was added to primary antibody dilutions. For total ERK1/2 protein: anti p42/44 MAPK antibody (Cell Signaling, Danvers, MA, #9107) 1:2000; HRP conjugated anti-mouse antibody (GE Healthcare, Buckinghamshire, UK) 1:5000. For phosphorylated ERK1/2 protein: anti phospho-p42/44 MAPK antibody (Cell Signaling, Danvers, MA, #9101) 1:1000, HRP conjugated anti-rabbit antibody (GE Healthcare, Buckinghamshire, UK) 1:10 000. Primary antibody incubations were performed either overnight at 4°C or at 1h at RT. Secondary antibody incubation was performed at 1h at RT. Blots were developed using an enhanced chemiluminescence system (GE Healthcare, Buckinghamshire, UK) and visualized at a Fluor Chem HD2 reader (Alpha Innotech, San Leandro, CA). Blots were quantified with densitometric gel analysis in the program Image Gauge. Results were analyzed in Excel and Graphpad Prism. Representative images were rendered in the Irfanview program.
2.8 Cell surface/whole cell ELISA
Cells were seeded for triplicate measures with and without primary antibody in poly-L-lysine coated 96 well dishes at 35 000/well in 100 μL DMEM supplemented with L-glutamine and serum. The following day, cells were preincubated for 90 mins at 37°C with either Telmisartan or vehicle (DMSO). In this assay, we used a concentration of 10μM of the inverse agonist. This was done, because higher concentrations are often needed to detect changes in secondary parameters such as beta-arrestin recruitment and internalization[31, 32]. Cells were subsequently placed on ice. For surface detection of the M1 Flag tag, cells were incubated with cold DMEM and anti M1 Antibody (Sigma-Aldrich, (St. Louis, MO); 1:2000) for 1h at 4°C (cells without primary antibody were control incubated in cold DMEM). Cells were washed first in cold DMEM, then in PBS, and fixed with 4% paraformaldehyde in PBS for 10 mins. Cells were washed twice in PBS and blocked for 30 mins in PBS with 1% Bovine Serum Albumin (blocking buffer). Cells were incubated with secondary antibody (HRP-conjugated anti-mouse IgG, GE Healtcare) diluted 1:3000 in blocking buffer. Cells were washed in PBS and developed using 3,3′,5,5′-tetramethylbenzidine (TMB) liquid substrate system for ELISA (Sigma-Aldrich, St. Louis, MO). Reactions were stopped with 1M hydrochloric acid after development of a blue color and absorbance at 450 nm was measured. For detection of total protein, cells were washed in PBS and fixed as described above. Blocking, primary, and secondary antibody conditions were conducted in blocking buffer with 0.1% Triton X-100 for permeabilization. Results were analyzed in GraphPad Prism and Excel. Triplicate means were normalized firstly by subtraction of values for cells without antibody, and secondly, by substraction of values for empty HEK293 cells.
2.9 Immunocytochemistry and confocal microscopy
Cells stably expressing M1 FLAG-tagged rat AT1 WT and mutant receptors were seeded in 6 cm dishes and grown to 80 – 90 % confluency. Cells were transfected with 3 μg GFP-tagged β-arrestin2 using Fugene 6 (Roche, Basel, Switzerland) according to manufacturer’s protocol. The day after transfection, cells were plated on poly-D-lysine (Sigma-Aldrich, St. Louis, MO) coated coverslips and grown to about 50% confluency. On the assay day, media was changed on cells and 10 μM Telmisartan was added to “Telmisartan” well for 30 mins. Cells were then incubated with M1 antibody (IgG2b, Sigma-Aldrich, St. Louis, MO) directed against the FLAG tag (1:1000, 30 mins) and hereafter treated with 1 μM Ang II or vehicle (PBS) for 30 mins. Cells were fixed with 4% formaldehyde in PBS for 20 mins, then permeabilized in Blotto (50 mM Tris-Cl pH 7.5, incubated with Alexa 594 conjugated 1mM CaCl2, 3% milk, 0.1% Triton X-100) for 20 mins and (red) antibody (1:500, Invitrogen, Carlsbad, CA) for 45 mins. Coverslips were mounted antiIgG2b using mounting media from Vectashield (Vector laboratories, Burlingame, CA) containing 4′,6′-diamidino-2-phenylidole (DAPI). Hence receptor (red), β-arrestin2 (green) and cell nuclei (blue) could be individually visualized. Mounted slides were analyzed on a 510 LSM laser confocal microscope (Zeiss, Thornwood, NY) using the 63x oil objective.
2.10 Receptor Selection and Amplification Technology (R-SAT®)
The R-SAT® assay measures the ability of transiently expressed oncogenes, proto-oncogenes, and many 7TM receptors to confer partial or total transformation of NIH3T3 cells causing a loss of contact inhibition of these cells, hereby allowing them to proliferate when they would otherwise stop (Brauner-Osborne and Brann, 1996; Burstein et al., 2006). In R-SAT®, a reporter gene (in this case β-galactosidase) is co-transfected into the cells with the 7TM receptor of interest to quantify this proliferative response. The β-galactosidase reporter is constitutively expressed in this system and does not participate in driving the biological response but rather works as an indirect measure of proliferation [33, 34]. The R-SAT® assay was performed as described previously [27]. Briefly, NIH3T3 cells at 70–80% confluence were transfected with plasmids containing WT or mutant human AT1 receptor cDNA (25 ng of receptor and 20 ng of β-galactosidase reporter/well of a 96-well plate) using the PolyFect Reagent (QIAGEN, Valencia, CA) as described in the manufacturer’s protocol. One day after transfection, ligands were added in DMEM supplemented with penicillin (100 U/ml), streptomycin (100 g/ml), and 25% Ultraculture synthetic supplement (Cambrex, Walkersville, MD) instead of calf serum to a final volume of 200 μL. After 5 days, the media was aspirated and cells were lysed, O-nitrophenyl-β-D-galactopyranoside was added, and the resulting absorbance was measured spectrophotometrically. Concentration response curves were performed in at least duplicates. Data was processed in GraphPad Prism and Excel. Statistical analysis was performed in Excel (unpaired Student’s t-test, two-tailed).
3. RESULTS
3.1 ET analysis
To identify key residues involved in conformational switching from the inactive to the active state of the AT1 receptor (rat AT1a), we performed an ET analysis and compared this to mutational data where mutations caused constitutive activity (details are described in the methods section 2.1). This analysis led to the identification of six amino acids in TM2, TM3 and TM6. To test the role of this patch in the conformational switch mechanism, we predicted mutations most likely to induce constitutive activity (Figure 1, Table 1). To investigate the effect of the suggested mutations in the patch, we generated N-terminally FLAG tagged constructs of the WT and mutated AT1 receptors and generated HEK293 cell lines stably expressing each construct. These cell lines were characterized for ligand affinity and signaling properties. The cell line expressing L67R did not respond in ligand binding or signaling assays. This receptor shows very low cell surface expression and consequently is included only in select analyses (Figure 4 and in supplementary material Figure S1).
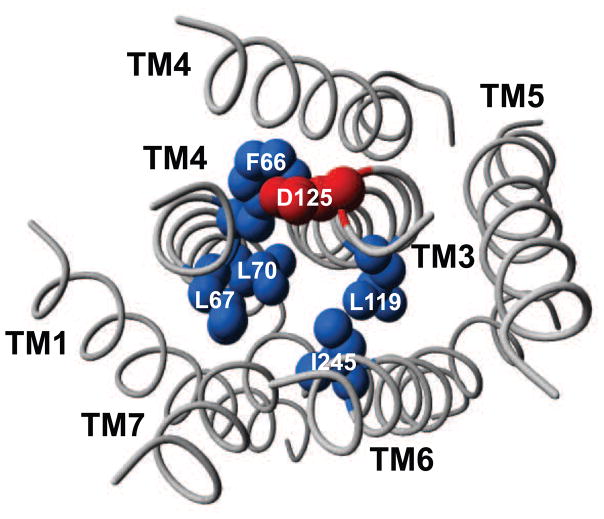
Figure 1. The predicted activation switch patch in a helical model of the rat AT1a receptor.
The residues F66, L67, L70, L119, D125, and I245 are depicted in a model of the helical region of the rAT1a receptor made using Swiss model’s GPCR mode (http://swissmodel.expasy.org//SWISS-MODEL.html) (for details and standard nomenclature, see Table 1). The rhodopsin structure PDB: 1F88 was used for helical alignment. The helical bundle is viewed from the cytoplasmic side. Hydrophobic residues are shown in blue and D125 is shown in red. The figure was made using the Yasara software.
Figure 4. Cell surface expression and trafficking properties of mutant receptors.
A. Cell surface expression levels were determined using an ELISA approach against the FLAG tag of the receptors. To assay total protein, cells were permeabilized during blocking and antibody incubation periods. Graph shows normalized mean values ± S.E.M., n=4–7 for either non-permeabilized cells with or without pre-incubation with 10 μM Telmisartan for 90 mins. (Surface and Surface plus Tel) or for permeabilized cells (Total Protein). Triplicate mean values were normalized firstly by subtraction of values for cells without antibody, and secondly, by subtraction of values for empty HEK293 cells. B (WT) and C (F66A) cell surface expression, internalization, and β-arrestin2 colocalization patterns were analyzed by antibody feeding assay on HEK293 cells stably expressing receptor variants and transiently transfected with GFP tagged β-arrestin2. Cells were pre-incubated with medium containing 10 μM Telmisartan or control treated with regular medium for 30 mins, fed with M1 anti Flag antibody for 30 mins, and then stimulated with 1 μM Ang II or control treated with PBS for 30 mins. Cells were subsequently fixed, blocked, permeabilized and stained with fluorescent secondary antibody. Slides were analyzed by confocal microscopy. For each figure (B and C), left panel shows staining for M1 Flag tag, middle panel shows GFP tagged β-arrestin2, and right panel shows overlay of M1 Flag tag (red) and GFP tagged β-arrestin2 (green). Pictures are representative of at least two independent experiments. Results for remaining mutant receptors can be found in supplementary material Figure S1.
3.2 Agonist binding properties of receptor mutants
To assess the ligand binding properties of the AT1 receptor mutants relative to WT receptor, we performed a whole cell competitive ligand binding assay using the peptide antagonist 125I–SI Ang II as radioligand and unlabeled Ang II as competitor. The analysis showed an increased affinity for the agonist Ang II for all of the mutated receptors compared to WT receptor (Table 2; Figure 2). L119R shows a moderate increase in affinity about 3-fold compared to WT. L70R, D125A, and I245F show a 10–15 fold increase, while the F66A mutant shows a robust increase of about 70-fold compared to WT.
TABLE 2. pKi values for Ang II competition binding.
Whole cell competition binding assay was performed on stable HEK293 cell lines expressing WT or mutated AT1 receptors. 125I - SI Ang II was used as radioligand and competed with unlabeled Ang II. Average pKi ± S.D. for 3 independent experiments are reported. pKi values were calculated based on pKd values from experiments using unlabeled SI Ang II as competitor.
| Receptor | pKi Ang II | Fold compared to WT | n |
|---|---|---|---|
| WT | 6.48 ± 0.06 | - | 3 |
| F66A | 8.34 ± 0.18* | 72 | 3 |
| L67R | N.A. | - | - |
| L70R | 7.69 ± 0.13* | 16 | 3 |
| L119R | 7.02 ± 0.13* | 3 | 3 |
| D125A | 7.50 ± 0.20* | 10 | 3 |
| I245F | 7.65 ± 0.23* | 15 | 3 |
= p < 0.05 determined by two-tailed paired Student’s t-test compared to WT.
Figure 2. Effect of receptor point mutations on Ang II affinity tested by competitive ligand binding.
Whole cell competition binding assay was conducted on stable cell lines as indicated in Table 2. Normalized mean values ± S.E.M. from 3 independent experiments are reported. Statistical analysis is indicated in Table 2.
3.3 Signaling properties of receptor mutants
3.3.1 G protein activation
Gq/11-dependent signaling properties of WT and mutant receptors were analyzed assaying inositol phosphate accumulation in the HEK293 cell lines stably expressing the individual receptors. Partial agonists/neutral antagonists ST and SII Ang II were included to investigate if any of the mutated receptors showed promiscuous activation compared to WT receptor, as has been found for other AT1 CAMs [14]. Compounds were used at high doses to yield maximum responses (Ang II 0.1μM, SII Ang II 18.75 μM, ST Ang II 5 μM, Telmisartan 1 μM). The analysis revealed that receptor mutants L70R, L119R and D125A had moderate, yet significant increases in basal activity compared to WT (~2–3 fold, p<0.05) (Figure 3A). L119R and D125A mutants showed very robust activation by ST and SII Ang II. I245F had an activation profile very similar to WT. The Ang II response was similar to WT for mutants L119R, D125A and I245F (102 ± 8 %, 102 ± 14 %, and 79 ± 4 %, respectively) while reduced for L70R (56 ± 4 %) and very low for the F66A mutant (12 ± 1 %).
Figure 3. Signaling properties of mutant receptors determined by inositol phosphate accumulation and ERK1/2 phosphorylation.
A: Inositol phosphate accumulation in response to full and partial AT1 receptor agonists was determined on stable HEK293 cell lines expressing mutant and WT receptors. Left to right bars represent: Basal, Ang II 0.1 μM, SII Ang II 18.75 μM, ST Ang II 5 μM, Telmisartan 1 μM. Data represent normalized mean values ± S.E.M. from at least 4 independent experiments. *, #, and ¤ indicate p<0.05 compared to WT basal, SII Ang II, and ST Ang II respectively in two tailed unpaired Student’s t-test on un-normalized mean values. B: ERK1/2 phosphorylation was determined by Western blotting of lysates from stable HEK293 cell lines expressing WT or mutant receptors stimulated with 0.1 μM Ang II for 10 mins. or 1 μM Telmisartan for 20 mins. Top: To quantify ERK1/2 phosphorylation response, Phospho ERK1/2:Total ERK1/2 ratios were normalized to max WT response (WT Ang II) for each set of blots. Normalized mean values ± S.E.M. from at least 3 independent experiments are reported. Bottom: Representative blots from the experiments are depicted: B: Basal, A: Ang II, and T: Telmisartan. WT Basal and Ang II were included on every gel for normalization.
3.3.2 ERK1/2 activation
To characterize signaling of the AT1 receptor mutants through other pathways, ERK1/2 phosphorylation was assayed by immuno-blotting for total- and phospho-ERK1/2. To assess basal activity and Ang II induced ERK1/2 activation, HEK293 cell lines stably expressing receptors were stimulated with either 1μM Telmisartan for 20 mins or 0.1μM Ang II for 10 mins. Ang II induced responses in ERK1/2 phosphorylation assays (Figures 3B) were comparable to the inositol phosphate assay (Figure 3A). Based on densitometric quantification of the blots, the F66A and D125A showed a tendency to slight increases basal activity (basal values, percent of max WT: F66A: 13 ± 4, D125A: 16 ± 4 compared to WT: 8 ± 1).
3.4 Receptor surface expression, localization, trafficking, and β-arrestin2 co-localization
Receptor signaling properties, especially the magnitude of the G protein-dependent signaling response, are usually positively correlated to receptor surface expression. Studies have shown that some AT1 CAMs are constitutively internalized. This constitutive internalization can be mediated by constitutive association to β-arrestin2 and inhibited by incubation with inverse agonists [14, 16, 35]. To compare the cell surface expression levels of WT and mutated receptors, we applied ELISA for quantitative evaluation and immunocytochemistry and confocal microscopy for qualitative evaluation of receptor localization and trafficking. In both assays, we also tested the effect of the inverse agonist Telmisartan, because previous studies have shown that incubation with an inverse agonist can increase cell surface expression of CAMs [16, 35].
In the ELISA assay, cell surface expression was determined by incubation with anti the M1 anti-FLAG antibody at 4°C followed by fixation and secondary antibody incubation, while the total amount of protein recognized by the M1 FLAG antibody in the cell was determined by permeabilizing cells prior to and during antibody incubation (Figure 4A). Noticeably, F66A and L67R mutants showed very low cell surface expression, but had total protein levels that were about 80% of WT receptor, suggesting that these mutant receptors were likely located intracellularly. The F66A mutant showed approximately 3 fold increased cell surface expression upon preincubation with 10 μM Telmisartan (Tel/noTel ratio 2.8 vs WT 1.1). Preincubation also yielded a moderate increase in I245F expression (Ratio 1.4). The L119R mutant was expressed at slightly higher levels than WT receptor (0.12 ± 0.02 vs. 0.086 ± 0.01 (WT)), D125A (0.073 ± 0.01) at similar levels as WT, while L70R and I245F showed expression levels at approximately half of WT (0.046 ± 0.01 and 0.047 ± 0.01, respectively).
For visual evaluation of receptor distribution and β-arrestin2 co-localization in basal conditions as well as in the presence of Ang II and Telmisartan, we performed an antibody feeding assay on cells stably expressing AT1 WT or mutant receptors that had been transiently transfected with GFP-tagged β-arrestin2 [29]. For this analysis, cells were incubated (fed) with the M1 anti-FLAG antibody recognizing the extracellular epitope tag while alive thereby labeling a population of receptors that have reached the cell surface. The distribution of these labeled receptors is then monitored in the presence or absence of exogenous ligand. As depicted in Figure 4B, WT receptor is expressed at the cell surface in the absence of ligand and does not colocalize with β-arrestin2. After 30 mins of 1 μM Ang II stimulation, the receptor co-localizes with β-arrestin2 intracellularly, as described previously [36]. Cells expressing WT receptor that were preincubated with 10 μM Telmisartan looked similar to the untreated condition. In agreement with the ELISA data (Figure 4A), the F66A mutant was located intracellularly, even in the absence of agonist ligand, thus, showing constitutive internalization (Figure 4C). Furthermore, Telmisartan preincubation of cells expressing F66A induced a redistribution of the F66A receptor mutant to the cell surface, also consistent with the increased ELISA signal in the presence of Telmisartan (Figure 4A). However, F66A mutants that were either constitutively internalized or internalized in the presence of Ang II, were not colocalized with β-arrestin2 (Figure 4C), unlike the WT receptor (Figure 4B). In this assay, we observed that overexpresssion of β-arrestin2 enhanced internalization of the WT receptor in response to Ang II, possibly because Ang II treatment appeared to be somewhat toxic for the cell lines, particularly WT, L119R, and D125A. The L67R mutant was localized intracellularly suggesting this mutant receptor, like F66A is also constitutively internalized. However, unlike F66A, the localization of L67R was not altered by Telmisartan (supplementary material Figure S1). The remaining mutants, L70R, L119R, D125A and I245F, were all primarily localized to the cell surface in the absence of ligand, and were co-localized intracellularly with β-arrestin2 upon Ang II treatment (Figure S1). As for the WT receptor, internalization of the L119R mutant by Ang II was also enhanced by β-arrestin2 overexpression, while the others (L70R, D125A, and I245F) were internalized in response to Ang II even in the absence of β-arrestin2 overexpression.
3.5 R-SAT® assay
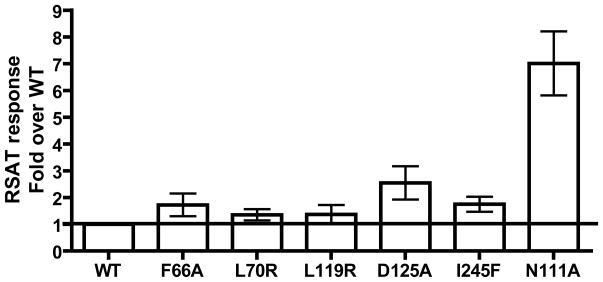
To measure effects on more downstream signaling pathways, the receptor mutants were tested in R-SAT®. R-SAT® assays are typically very sensitive to detecting constitutive activation, possibly due to the duration of the assay [34]. The N111A mutant was included as a positive control since this mutant has been shown previously to be constitutively active in R-SAT® [28]. Figure 5 depicts basal responses normalized to WT response. F66A, D125A and I245F show moderate constitutive activity, while the positive control N111A shows robust constitutive activity. pEC50 and efficacy values from Ang II dose response curves are depicted in Table 3. The F66A mutant had lower efficacy when compared to WT. The remaining mutants show efficacies similar to that of WT, thus, following the tendency of the inositol phosphate and ERK1/2 phosphorylation assays. The positive control N111A shows super efficacy for Ang II as reported previously [28].
Figure 5. R-SAT® assay basal activity, normalized to WT.
on cells transiently transfected with hAT1 WT or mutated receptors. Normalized means and S.E.M. from at least 8 experiments are reported. Basal activities could be inhibited by Telmisartan, data not shown.
Table 3. Ang II dose response curves, carried out in R-SAT® assay.
on cells transiently transfected with human AT1 WT or mutated receptors. pEC50 values ± S.D. are reported,
| Receptor | pEC50 | Efficacy (%) | n |
|---|---|---|---|
| WT | 7.1 ± 0.6 | 100 | 38 |
| F66A | 6.6 ± 1.3 | 35 ± 4 | 8 |
| L67R | N.A. | ||
| L70R | 6.6 ± 0.7* | 87 ± 12 | 10 |
| L119R | 7.6 ± 0.6* | 103 ± 8 | 10 |
| D125A | 7.4 ± 1.0 | 97 ± 9 | 10 |
| I245F | 6.9 ± 0.7 | 113 ± 14 | 9 |
| N111A | 7.5 ± 1.0 | 192 ± 49 | 8 |
= p<0.05 compared to WT in unpaired two-tailed Student’s t-test. Efficacy in percent compared to WT ± S.E.M.
4. DISCUSSION
In this study, we characterize six AT1 receptor mutants predicted computationally to play a role in the conformational switch linked to 7TM receptor activation and which form a patch localized to the cytoplasmic core of the helical bundle of 7TM receptors. Overall, the experimental data support the role for this patch in AT1 receptor conformational switching, since each mutant (except the poorly expressed L67R) shows either moderate constitutive activity or a change of state towards activation, detectable in certain assays. These findings sustain the notion that 7TM receptors, in our case the AT1 receptor, can adopt different conformations and that monitoring several different signaling endpoints is critical for accurate detection of changed activation states. Collectively, the data support a role for this patch in receptor activation and hereby illustrate the benefits of combining computational analysis and mutational data to generate insights to specific functional properties of family A 7TM receptors.
The receptor mutants show different signs of a changed activation state in the different assays. This is consistent with other mutational studies of molecular determinants of AT receptor activation that have also reported mutants that show only slightly elevated basal activity or increased EC50 values through the inositol phosphate pathway as reviewed recently by Petrel et al. [14, 15, 37] and also confirms the advantage of probing different pathways when looking at the AT1 receptor as mentioned in the introduction section. Notably, the WT AT1 receptor also shows no or low constitutive activity compared to untransfected cells indicating that the basal state of the receptor might be more tightly controlled than for certain other 7TM receptors [14, 38].
All of the mutants show increased binding affinity for Ang II in the competition binding assay. This has been associated with a shift towards the active conformation based on the theory that agonists preferably bind and thus stabilize the active conformation of the receptor [1]. Despite this only the L70R, L119R and D125A mutants showed increased basal activity compared to WT in the inositol phosphate assay whereas the I245F mutant showed a tendency towards increased basal activity in the R-SAT® assay. Signaling responses and receptor-surface expression levels are usually positively correlated. Although, this correlation is most likely not completely linear, a normalization of signaling responses to the surface levels of the receptor may provide certain insights about the receptor mutants and suggest tendencies to constitutive activity. If we normalize the inositol phosphate data to the surface expression, there is a tendency towards constitutive activity for all of the receptor mutants in the range of 2–4 fold of the WT receptor. Similarly, if ERK1/2 data are correlated to ELISA data, not only the F66A and D125A, but also the L70R mutant show tendencies towards increased basal activity.
F66A showed very low surface expression, constitutive internalization and responded poorly to Ang II stimulation in all signaling assays, which is most likely a reflection of the low expression levels. If we normalize the inositol phosphate accumulation response of F66A to its surface expression, it shows a basal activity of 42% of WT Ang II response whereas the WT receptor only shows a basal activity of 3% of WT Ang II response. Furthermore, the normalized F66A Ang II induced response is a 178% when compared to the WT response. Similarly, the basal activity in the ERK phosphorylation assay is 203%, and the Ang II response is 456% of WT Ang II for ERK1/2 phosphorylation. Although these calculations may indicate that F66A is substantially more constitutively active than it appears,, a linear correlation between signaling and surface expression is clearly unlikely when the difference in expression levels are this pronounced. Therefore, these calculations should be considered with caution.
Surface expression of the F66A mutant was increased both qualitatively and quantitatively by pre-incubation with the inverse agonist Telmisartan. This could suggest this internalization is dependent on an active receptor conformation, which has been observed for other constitutively activated AT1 receptor mutants [16]. Previous studies have shown that constitutive internalization can be mediated by constitutive association with β-arrestin2 [35]. However, we did not see co-localization with β-arrestin2 in the untreated, nor the Ang II stimulated condition. Importantly, it cannot be excluded that the low levels of receptor expression, validated by ELISA, complicate detection of recruitment by this method. The low cell surface expression of some of the receptor mutants compared to the WT could also result from structural instability of the receptors. Studies of constitutively active receptor mutants of the β2-adrenergic receptor also showed decreased stability compared to the WT receptor possibly reflecting a gain of flexibility due to removal of structural constraints by mutation [39, 40]. The expression of these mutants could be stabilized by the presence of either an inverse agonist or agonist.
The results on the F66A mutant clearly represent the most significant example of the importance of testing several endpoints in characterization of 7TM receptor mutants. Since most mutations reported to be non-signaling have only been tested for G protein dependent signaling endpoints, these mutants could in fact be constitutively active with respect other signaling pathways. Thus, a reevaluation of some of these mutants in more signaling pathways could potentially yield new insights in the molecular mechanisms behind receptor activation.
The identified activation switch patch of residues is located in the cytoplasmic part of TM2 (F66, L67, L70), TM3 (L119, D125) and TM6 (I245, location and standard nomenclature is reported in Table 1 and Figure 1). With the exception of the F66A mutation (2.42), which to our knowledge, has not been reported previously, all the other mutations in the ET patch were previously reported to confer constitutive activity in some 7TM receptors. Based on the general theories of 7TM receptor activation, which involves outward movements of the cytoplasmic part the helical bundle, it seems plausible that the patch mediates interactions that stabilize the inactive conformation [2]. The most extensively studied being the D125 (3.49) residue which is part of the conserved DRY motif. The residue has been proposed to interact with the neighboring Arg (3.50) residue and hereby aiding to keep the receptor in the inactive conformation [2]. The increasing number of available crystal structures of 7TM receptors published during recent years also provides interesting perspectives to the role of the patch in receptor activation. Here, residue positions from the patch have been found to be involved in hydrophobic interactions (2.43, 2.46, 3.43, and 6.40) [41] or participate in a hydrogen bonding network (2.43 and 6.40) [42]. Interestingly, in the structure of the Gα peptide bound opsin, position 2.42 (corresponding to the F66A mutation), which has not been previously characterized) was proposed to indirectly interact with the 3.49 residue through a water molecule (supplementary information to [43]). This suggests a role for this residue position in conversion between inactive and active states, though further studies will be needed to establish the exact nature of these interactions.
In conclusion, we were able to computationally predict a patch of conserved residues in the cytoplasmic core of the rat AT1a receptor important for receptor activation as confirmed by experimental studies. We illustrate how a combination of unbiased computational prediction and the knowledge from published mutational data provides an exciting tool to explore the molecular mechanisms of specific receptor functions. Due to the nature of the analysis, this approach will most likely be applicable to family A 7TM receptors in general. Our study support the current theories of 7TM receptor activation involving shared concerted movements of the transmembrane helices, but also show how the ability of 7TM receptors to adopt multiple conformations requires the need of testing several signaling endpoints to accurately detect and characterize a change in activation state for the individual mutant.
Supplementary Material
Acknowledgments
We thank Christina Lyngsoe and Anne Yaël Nossent for valuable input on the manuscript. Marie Mi Bonde thanks former colleagues in Jennifer L. Whistler’s laboratory at the Ernest Gallo Clinic and Research Center for technical training, scientific input and helpful discussions during her stay. We acknowledge the use of the GPCRDB data base (http://www.gpcr.org/7tm/). This work was sponsored by The Danish Heart Foundation, The Danish National Research Foundation, The Koebmand i Odense Johan og Hanne Weimann f. Seedorffs legat, Aase og Ejnar Danielsens Foundation, the Novo Nordisk Foundation, Danish Cardiovascular Research Academy, and funds provided by the State of California through the University of California San Francisco to J.L.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gether U. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr Rev. 2000;21:90–113. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- 2.Nygaard R, Frimurer TM, Holst B, Rosenkilde MM, Schwartz TW. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol Sci. 2009;30:249–59. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol. 1996;257:342–58. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 4.Madabushi S, Yao H, Marsh M, Kristensen DM, Philippi A, Sowa ME, et al. Structural clusters of evolutionary trace residues are statistically significant and common in proteins. J Mol Biol. 2002;316:139–54. doi: 10.1006/jmbi.2001.5327. [DOI] [PubMed] [Google Scholar]
- 5.Yao H, Kristensen DM, Mihalek I, Sowa ME, Shaw C, Kimmel M, et al. An accurate, sensitive, and scalable method to identify functional sites in protein structures. J Mol Biol. 2003;326:255–61. doi: 10.1016/s0022-2836(02)01336-0. [DOI] [PubMed] [Google Scholar]
- 6.Sowa ME, He W, Wensel TG, Lichtarge O. A regulator of G protein signaling interaction surface linked to effector specificity. Proc Natl Acad Sci U S A. 2000;97:1483–8. doi: 10.1073/pnas.030409597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ribes-Zamora A, Mihalek I, Lichtarge O, Bertuch AA. Distinct faces of the Ku heterodimer mediate DNA repair and telomeric functions. Nat Struct Mol Biol. 2007;14:301–7. doi: 10.1038/nsmb1214. [DOI] [PubMed] [Google Scholar]
- 8.Raviscioni M, Gu P, Sattar M, Cooney AJ, Lichtarge O. Correlated evolutionary pressure at interacting transcription factors and DNA response elements can guide the rational engineering of DNA binding specificity. J Mol Biol. 2005;350:402–15. doi: 10.1016/j.jmb.2005.04.054. [DOI] [PubMed] [Google Scholar]
- 9.Sowa ME, He W, Slep KC, Kercher MA, Lichtarge O, Wensel TG. Prediction and confirmation of a site critical for effector regulation of RGS domain activity. Nat Struct Biol. 2001;8:234–7. doi: 10.1038/84974. [DOI] [PubMed] [Google Scholar]
- 10.Madabushi S, Gross AK, Philippi A, Meng EC, Wensel TG, Lichtarge O. Evolutionary trace of G protein-coupled receptors reveals clusters of residues that determine global and class-specific functions. J Biol Chem. 2004;279:8126–32. doi: 10.1074/jbc.M312671200. [DOI] [PubMed] [Google Scholar]
- 11.Shenoy SK, Drake MT, Nelson CD, Houtz DA, Xiao K, Madabushi S, et al. beta-arrestin-dependent, G protein-independent ERK1/2 activation by the beta2 adrenergic receptor. J Biol Chem. 2006;281:1261–73. doi: 10.1074/jbc.M506576200. [DOI] [PubMed] [Google Scholar]
- 12.Hunyady L, Catt KJ. Pleiotropic AT1 receptor signaling pathways mediating physiological and pathogenic actions of angiotensin II. Mol Endocrinol. 2006;20:953–70. doi: 10.1210/me.2004-0536. [DOI] [PubMed] [Google Scholar]
- 13.Aplin M, Bonde MM, Hansen JL. Molecular determinants of angiotensin II type 1 receptor functional selectivity. J Mol Cell Cardiol. 2009;46:15–24. doi: 10.1016/j.yjmcc.2008.09.123. [DOI] [PubMed] [Google Scholar]
- 14.Petrel C, Clauser E. Angiotensin II AT1 receptor constitutive activation: from molecular mechanisms to pathophysiology. Mol Cell Endocrinol. 2009;302:176–84. doi: 10.1016/j.mce.2008.10.049. [DOI] [PubMed] [Google Scholar]
- 15.Gaborik Z, Jagadeesh G, Zhang M, Spat A, Catt KJ, Hunyady L. The role of a conserved region of the second intracellular loop in AT1 angiotensin receptor activation and signaling. Endocrinology. 2003;144:2220–8. doi: 10.1210/en.2002-0135. [DOI] [PubMed] [Google Scholar]
- 16.Miserey-Lenkei S, Parnot C, Bardin S, Corvol P, Clauser E. Constitutive internalization of constitutively active agiotensin II AT(1A) receptor mutants is blocked by inverse agonists. J Biol Chem. 2002;277:5891–901. doi: 10.1074/jbc.M108398200. [DOI] [PubMed] [Google Scholar]
- 17.Feng YH, Zhou L, Qiu R, Zeng R. Single mutations at Asn295 and Leu305 in the cytoplasmic half of transmembrane alpha-helix domain 7 of the AT1 receptor induce promiscuous agonist specificity for angiotensin II fragments: a pseudo-constitutive activity. Mol Pharmacol. 2005;68:347–55. doi: 10.1124/mol.105.011601. [DOI] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 20.Han M, Lin SW, Minkova M, Smith SO, Sakmar TP. Functional interaction of transmembrane helices 3 and 6 in rhodopsin. Replacement of phenylalanine 261 by alanine causes reversion of phenotype of a glycine 121 replacement mutant. J Biol Chem. 1996;271:32337–42. doi: 10.1074/jbc.271.50.32337. [DOI] [PubMed] [Google Scholar]
- 21.Kraaij R, Post M, Kremer H, Milgrom E, Epping W, Brunner HG, et al. A missense mutation in the second transmembrane segment of the luteinizing hormone receptor causes familial male-limited precocious puberty. J Clin Endocrinol Metab. 1995;80:3168–72. doi: 10.1210/jcem.80.11.7593421. [DOI] [PubMed] [Google Scholar]
- 22.Baranski TJ, Herzmark P, Lichtarge O, Gerber BO, Trueheart J, Meng EC, et al. C5a receptor activation. Genetic identification of critical residues in four transmembrane helices. J Biol Chem. 1999;274:15757–65. doi: 10.1074/jbc.274.22.15757. [DOI] [PubMed] [Google Scholar]
- 23.Lu Z-L, Hulme EC. The Functional Topography of Transmembrane Domain 3 of the M1 Muscarinic Acetylcholine Receptor, Revealed by Scanning Mutagenesis. Journal of Biological Chemistry. 1999;274:7309. doi: 10.1074/jbc.274.11.7309. [DOI] [PubMed] [Google Scholar]
- 24.Hjorth SA, Orskov C, Schwartz TW. Constitutive activity of glucagon receptor mutants. Mol Endocrinol. 1998;12:78–86. doi: 10.1210/mend.12.1.0045. [DOI] [PubMed] [Google Scholar]
- 25.Gaudin P, Maoret JJ, Couvineau A, Rouyer-Fessard C, Laburthe M. Constitutive activation of the human vasoactive intestinal peptide 1 receptor, a member of the new class II family of G protein-coupled receptors. J Biol Chem. 1998;273:4990–6. doi: 10.1074/jbc.273.9.4990. [DOI] [PubMed] [Google Scholar]
- 26.Benned-Jensen T, Rosenkilde MM. Structural motifs of importance for the constitutive activity of the orphan 7TM receptor EBI2: analysis of receptor activation in the absence of an agonist. Mol Pharmacol. 2008;74:1008–21. doi: 10.1124/mol.108.049676. [DOI] [PubMed] [Google Scholar]
- 27.Hansen JL, Haunso S, Brann MR, Sheikh SP, Weiner DM. Loss-of-function polymorphic variants of the human angiotensin II type 1 receptor. Mol Pharmacol. 2004;65:770–7. doi: 10.1124/mol.65.3.770. [DOI] [PubMed] [Google Scholar]
- 28.Hansen JL, Aplin M, Hansen JT, Christensen GL, Bonde MM, Schneider M, et al. The human angiotensin AT(1) receptor supports G protein-independent extracellular signal-regulated kinase 1/2 activation and cellular proliferation. Eur J Pharmacol. 2008;590:255–63. doi: 10.1016/j.ejphar.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Barak LS, Ferguson SS, Zhang J, Caron MG. A beta-arrestin/green fluorescent protein biosensor for detecting G protein-coupled receptor activation. J Biol Chem. 1997;272:27497–500. doi: 10.1074/jbc.272.44.27497. [DOI] [PubMed] [Google Scholar]
- 30.Theilade J, Lerche Hansen J, Haunso S, Sheikh SP. Extracellular signal-regulated kinases control expression of G protein-coupled receptor kinase 2 (GRK2) FEBS Lett. 2002;518:195–9. doi: 10.1016/s0014-5793(02)02701-1. [DOI] [PubMed] [Google Scholar]
- 31.Enquist J, Skroder C, Whistler JL, Leeb-Lundberg LM. Kinins promote B2 receptor endocytosis and delay constitutive B1 receptor endocytosis. Mol Pharmacol. 2007;71:494–507. doi: 10.1124/mol.106.030858. [DOI] [PubMed] [Google Scholar]
- 32.Jorgensen R, Kubale V, Vrecl M, Schwartz TW, Elling CE. Oxyntomodulin differentially affects glucagon-like peptide-1 receptor beta-arrestin recruitment and signaling through Galpha(s) J Pharmacol Exp Ther. 2007;322:148–54. doi: 10.1124/jpet.107.120006. [DOI] [PubMed] [Google Scholar]
- 33.Brauner-Osborne H, Brann MR. Pharmacology of muscarinic acetylcholine receptor subtypes (m1-m5): high throughput assays in mammalian cells. Eur J Pharmacol. 1996;295:93–102. doi: 10.1016/0014-2999(95)00639-7. [DOI] [PubMed] [Google Scholar]
- 34.Burstein ES, Piu F, Ma JN, Weissman JT, Currier EA, Nash NR, et al. Integrative functional assays, chemical genomics and high throughput screening: harnessing signal transduction pathways to a common HTS readout. Curr Pharm Des. 2006;12:1717–29. doi: 10.2174/138161206776873662. [DOI] [PubMed] [Google Scholar]
- 35.Wilbanks AM, Laporte SA, Bohn LM, Barak LS, Caron MG. Apparent loss-of-function mutant GPCRs revealed as constitutively desensitized receptors. Biochemistry. 2002;41:11981–9. doi: 10.1021/bi020275m. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Barak LS, Anborgh PH, Laporte SA, Caron MG, Ferguson SS. Cellular trafficking of G protein-coupled receptor/beta-arrestin endocytic complexes. J Biol Chem. 1999;274:10999–1006. doi: 10.1074/jbc.274.16.10999. [DOI] [PubMed] [Google Scholar]
- 37.Parnot C, Bardin S, Miserey-Lenkei S, Guedin D, Corvol P, Clauser E. Systematic identification of mutations that constitutively activate the angiotensin II type 1A receptor by screening a randomly mutated cDNA library with an original pharmacological bioassay. Proc Natl Acad Sci U S A. 2000;97:7615–20. doi: 10.1073/pnas.110142297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 39.Gether U, Ballesteros JA, Seifert R, Sanders-Bush E, Weinstein H, Kobilka BK. Structural instability of a constitutively active G protein-coupled receptor. Agonist-independent activation due to conformational flexibility. J Biol Chem. 1997;272:2587–90. doi: 10.1074/jbc.272.5.2587. [DOI] [PubMed] [Google Scholar]
- 40.Rasmussen SG, Jensen AD, Liapakis G, Ghanouni P, Javitch JA, Gether U. Mutation of a highly conserved aspartic acid in the beta2 adrenergic receptor: constitutive activation, structural instability, and conformational rearrangement of transmembrane segment 6. Mol Pharmacol. 1999;56:175–84. doi: 10.1124/mol.56.1.175. [DOI] [PubMed] [Google Scholar]
- 41.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–38. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 42.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–73. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 43.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.