Abstract
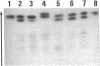
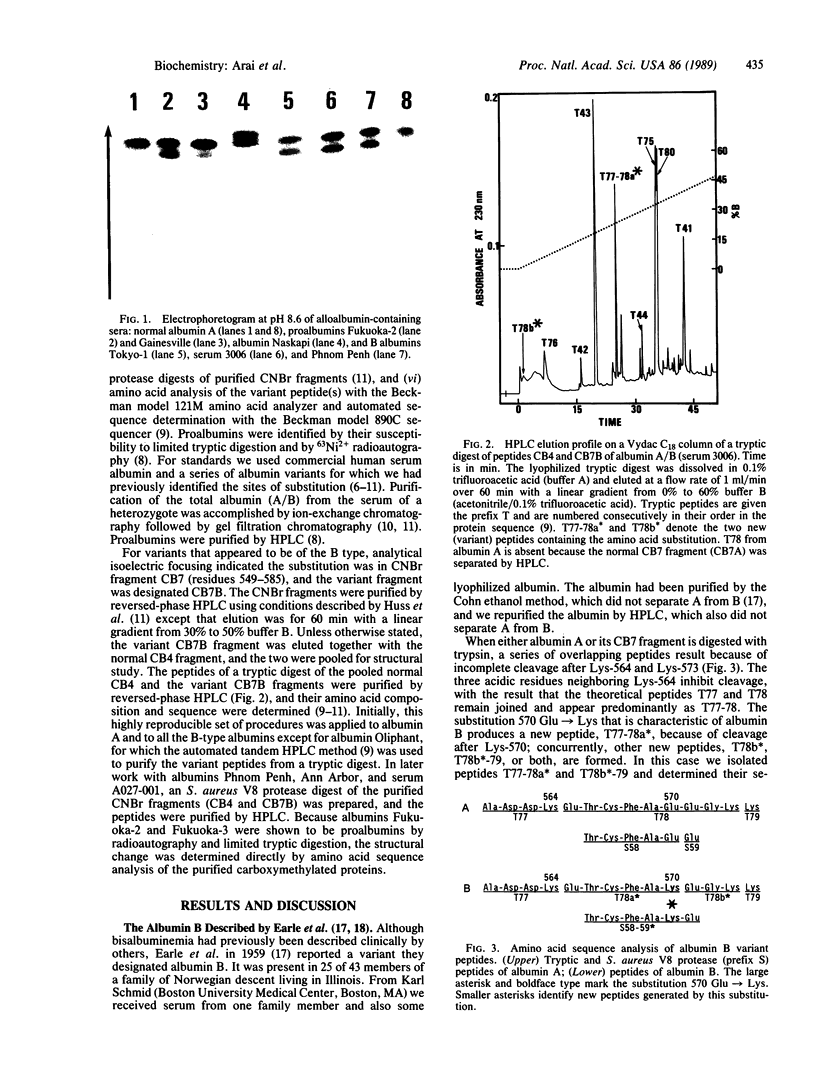
Alloalbuminemia is rare and has a cumulative frequency of only approximately 1 in 3,000 in Europeans and Japanese. The worldwide ethnic and geographic distribution of certain albumin genetic variants appears to be nonrandom. Moreover, we have found that structurally identical variants may occur at different frequencies in ethnically distinct populations, presumably owing to independent mutations. In this study, albumin B and two types of proalbumins, which as a group are the most common European albumin variants, have also been found in Asians. We have identified the amino acid substitution characteristic of albumin B (glutamic acid----lysine at position 570) in alloalbumins from six unrelated individuals of five different European descents and also in two Japanese and one Cambodian. The two types of proalbumins most common in Europe (Lille type, arginine----histidine at position -2; Christchurch type, arginine----glutamic acid at position -1) also occur in Japan. These results provide evidence for independent mutations at single sites in the albumin genome. The clustering of these and of several other amino acid exchanges in certain regions of the albumin molecule suggests two possibilities: that certain sites are hypermutable or that mutants involving certain sites are more subject to selection than mutants involving others.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdo Y., Rousseaux J., Dautrevaux M. Proalbumin Lille, a new variant of human serum albumin. FEBS Lett. 1981 Aug 31;131(2):286–288. doi: 10.1016/0014-5793(81)80386-9. [DOI] [PubMed] [Google Scholar]
- Brennan S. O., Carrell R. W. A circulating variant of human proalbumin. Nature. 1978 Aug 31;274(5674):908–909. doi: 10.1038/274908a0. [DOI] [PubMed] [Google Scholar]
- EARLE D. P., HUTT M. P., SCHMID K., GITLIN D. Observations on double albumin: a genetically transmitted serum protein anomaly. J Clin Invest. 1959 Aug;38(8):1412–1420. doi: 10.1172/JCI103917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine J. M., Marneux M., Rochu D. Human albumin genetic variants: an attempt at a classification of European allotypes. Am J Hum Genet. 1987 Mar;40(3):278–286. [PMC free article] [PubMed] [Google Scholar]
- GITLIN D., SCHMID K., EARLE D. P., GIVERLBER H. Observations on double albumin. II. A peptide difference between two genetically determined human serum albumins. J Clin Invest. 1961 May;40:820–827. doi: 10.1172/JCI104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliano M., Minchiotti L., Iadarola P., Zapponi M. C., Ferri G., Castellani A. A. Structural characterization of a chain termination mutant of human serum albumin. J Biol Chem. 1986 Mar 25;261(9):4283–4287. [PubMed] [Google Scholar]
- Huss K., Madison J., Ishioka N., Takahashi N., Arai K., Putnam F. W. The same substitution, glutamic acid----lysine at position 501, occurs in three alloalbumins of Asiatic origin: albumins Vancouver, Birmingham, and Adana. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6692–6696. doi: 10.1073/pnas.85.18.6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss K., Putnam F. W., Takahashi N., Takahashi Y., Weaver G. A., Peters T., Jr Albumin Cooperstown: a serum albumin variant with the same (313 Lys----Asn) mutation found in albumins in Italy and New Zealand. Clin Chem. 1988 Jan;34(1):183–187. [PubMed] [Google Scholar]
- Iadarola P., Minchiotti L., Galliano M. Localization of the amino acid substitution site in a fast migrating variant of human serum albumin. FEBS Lett. 1985 Jan 21;180(1):85–88. doi: 10.1016/0014-5793(85)80237-4. [DOI] [PubMed] [Google Scholar]
- Matsuda Y., Ogushi F., Ogawa K., Katunuma N. Structure and properties of albumin Tokushima and its proteolytic processing by cathepsin B in vitro. J Biochem. 1986 Aug;100(2):375–379. doi: 10.1093/oxfordjournals.jbchem.a121724. [DOI] [PubMed] [Google Scholar]
- Minchiotti L., Galliano M., Iadarola P., Stoppini M., Ferri G., Castellani A. A. Structural characterization of two genetic variants of human serum albumin. Biochim Biophys Acta. 1987 Dec 18;916(3):411–418. doi: 10.1016/0167-4838(87)90187-7. [DOI] [PubMed] [Google Scholar]
- Minghetti P. P., Ruffner D. E., Kuang W. J., Dennison O. E., Hawkins J. W., Beattie W. G., Dugaiczyk A. Molecular structure of the human albumin gene is revealed by nucleotide sequence within q11-22 of chromosome 4. J Biol Chem. 1986 May 25;261(15):6747–6757. [PubMed] [Google Scholar]
- Naylor D. H., Anhorn C. A., Laschinger C., Males F., Chodirker W. B. Antigenic differences between normal human albumin and a genetic variant. Transfusion. 1982 Mar-Apr;22(2):128–133. doi: 10.1046/j.1537-2995.1982.22282177119.x. [DOI] [PubMed] [Google Scholar]
- Neel J. V. Rare variants, private polymorphisms, and locus heterozygosity in Amerindian populations. Am J Hum Genet. 1978 Sep;30(5):465–490. [PMC free article] [PubMed] [Google Scholar]
- Neel J. V., Satoh C., Goriki K., Asakawa J., Fujita M., Takahashi N., Kageoka T., Hazama R. Search for mutations altering protein charge and/or function in children of atomic bomb survivors: final report. Am J Hum Genet. 1988 May;42(5):663–676. [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Takahashi Y., Blumberg B. S., Putnam F. W. Amino acid substitutions in genetic variants of human serum albumin and in sequences inferred from molecular cloning. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4413–4417. doi: 10.1073/pnas.84.13.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Takahashi Y., Ishioka N., Blumberg B. S., Putnam F. W. Application of an automated tandem high-performance liquid chromatographic system to peptide mapping of genetic variants of human serum albumin. J Chromatogr. 1986 May 30;359:181–191. doi: 10.1016/0021-9673(86)80072-3. [DOI] [PubMed] [Google Scholar]
- Takahashi N., Takahashi Y., Isobe T., Putnam F. W., Fujita M., Satoh C., Neel J. V. Amino acid substitutions in inherited albumin variants from Amerindian and Japanese populations. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8001–8005. doi: 10.1073/pnas.84.22.8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Takahashi Y., Putnam F. W. Structural changes and metal binding by proalbumins and other amino-terminal genetic variants of human serum albumin. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7403–7407. doi: 10.1073/pnas.84.21.7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tárnoky A. L. Genetic and drug-induced variation in serum albumin. Adv Clin Chem. 1980;21:101–146. doi: 10.1016/s0065-2423(08)60087-6. [DOI] [PubMed] [Google Scholar]
- Weitkamp L. R., Franglen G., Rokala D. A., Polesky H. F., Simpson N. E., Sunderman F. W., Jr, Bell H. E., Saave J., Lisker R., Bohls S. W. An electrophoretic comparison of human serum albumin variants: eight distinguishable types. Hum Hered. 1969;19(2):159–169. doi: 10.1159/000152212. [DOI] [PubMed] [Google Scholar]
- Weitkamp L. R., McDermid E. M., Neel J. V., Fine J. M., Petrini C., Bonazzi L., Ortali V., Porta F., Tanis R., Harris D. J. Additional data on the population distribution of human serum albumin genes; three new variants. Ann Hum Genet. 1973 Oct;37(2):219–226. doi: 10.1111/j.1469-1809.1973.tb01829.x. [DOI] [PubMed] [Google Scholar]
- Winter W. P., Weitkamp L. R., Rucknagel D. L. Amino acid substitution in two identical inherited human serum albumin variants: albumin Oliphant and albumin Ann Arbor. Biochemistry. 1972 Feb 29;11(5):889–896. doi: 10.1021/bi00755a031. [DOI] [PubMed] [Google Scholar]