Abstract
Drosophila melanogaster is today one of the three foremost models in olfactory research, paralleled only by the mouse and the nematode. In the last years, immense progress has been achieved by combining neurogenetic tools with neurophysiology, anatomy, chemistry, and behavioral assays. One of the most important tasks for a fruit fly is to find a substrate for eating and laying eggs. To perform this task the fly is dependent on olfactory cues emitted by suitable substrates as e.g. decaying fruit. In addition, in this area, considerable progress has been made during the last years, and more and more natural and behaviorally active ligands have been identified. The future challenge is to tie the progress in different fields together to give us a better understanding of how a fly really behaves. Not in a test tube, but in nature. Here, we review our present state of knowledge regarding Drosophila plant-odor-related olfactory neuroethology to provide a basis for new progress.
Keywords: Drosophila melanogaster, Olfaction, Sensory neuron, Behavior, Transduction, Imaging
Introduction
Odor information is known to be of vital importance for most animals, and especially for insects. To achieve optimal fitness insects need to survive and reproduce in an optimal way. Odors are known to play vital roles as cues for both these functions. To survive, optimally an insect needs to eat (in most cases) and to avoid becoming victim to predators or parasites. To reproduce optimally it needs to find a good mate and a suitable oviposition site. For one of the world’s foremost model organisms, the fruit fly Drosophila melanogaster (Dm), odor information is known to be heavily involved in location and quality determination of mates, food and oviposition sites.
Drosophila first appeared as a study organism for genetics when Castle 1906 investigated effects of inbreeding. In the following two decades it was further established as a real model organism by Morgan (1910) who used crossing experiments in order to map genes onto different chromosomes. Due to its small size, short generation time, and easy rearing, Dm quickly became a valuable model. Today, Dm is the most well-developed model animal when it comes to genetic manipulations and presence of mutants for more or less any trait. Here, we describe our present state of knowledge regarding plant-odor-dependent neuroethology in Dm. We discuss which odors are important, how different odor molecules are detected and transduced, how peripheral information is processed and integrated in the antennal lobe, and, finally, which types of behavior that have been studied.
In reviewing scientific progress in a single model organism, it is inevitable that decisive information from other systems is omitted. To get an impression of all the important studies performed in insect olfaction outside Drosophila, we refer the reader to the following books and reviews: Stengl et al. (1992), Hildebrand and Shepherd (1997), Hansson (1999), Hallem et al. (2006), de Bruyne and Baker (2008), Galizia (2008), and Spehr and Munger (2009).
Which plant-related odors are important to Drosophila melanogaster?
The preferred food and oviposition substrate of Dm is decaying fruit (Lachaise and Tsacas 1983) and most key ligands identified can be connected either directly to the fruit, or to microorganisms living in and of the fruit (Stensmyr et al. 2003a). Typical examples of directly fruit-related odors are a number of esters like ethyl hexanoate, geranyl acetate, and pentyl acetate. In addition, more “green” odors like E-2-hexenal and hexanol are detected. Of the bacteria-associated odors 2,3-butanedione seems to play an important role. In addition, flower odors like phenylethyl alcohol and phenylacetaldehyde are detected. A full account of the key stimuli identified so far is provided in Table 1.
Table 1.
Odorant receptors (ORs) of the Drosophila olfactory organs, their sensillumassociation, their central nervous target glomeruli, and their, so far, identified key ligands
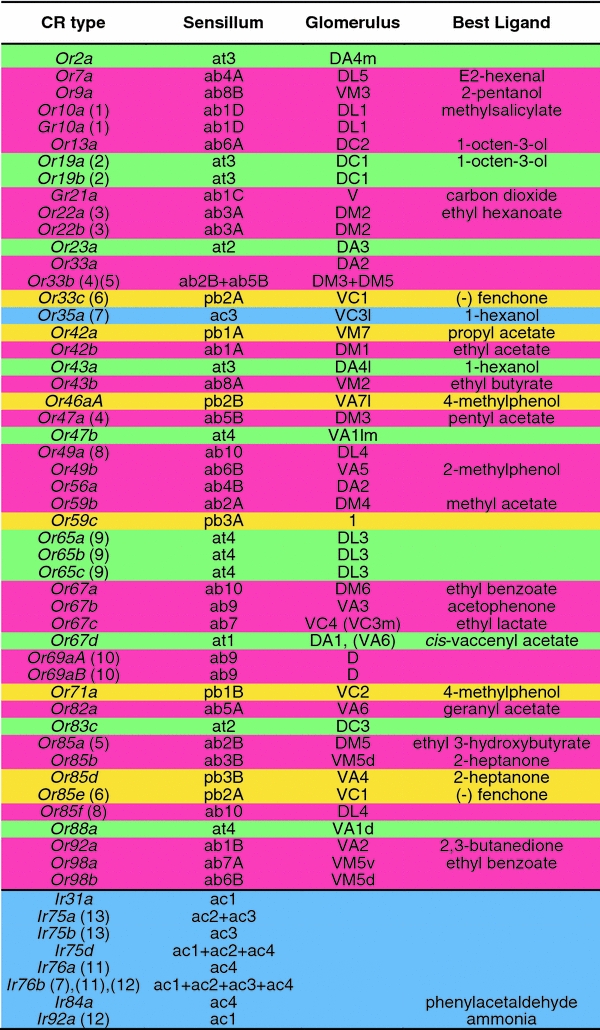
Color labeling represents different sensillum types [red basiconic (antenna), yellow basiconic (maxillary palp), green trichoid, blue coeloconic]
ORs with the same number in brackets are expressed in the same sensillum
Detecting the molecules
In the fruit fly, volatile chemical information is detected by two pairs of olfactory organs; the third antennal segment and the maxillary palps. More specifically, odorants are detected by Odorant Receptors (OR), which are localized in the dendritic membrane of Olfactory Sensory Neurons (OSN). Each antenna carries ca 1200 OSNs, which are housed in about 420 olfactory sensilla, whereas the maxillary palp has about 120 OSNs and 60 olfactory sensilla. The olfactory sensilla come in three morphologically distinct types, namely sensilla basiconica, trichodea, and coeloconica that in turn can be divided into further subtypes (Shanbhag et al. 1999). Each OSN typically expresses only a single OR (beyond the ubiquitous Or83b, see below), which confers a unique odorant response profile on the OSN. In Dm, the OR gene family comprises 62 genes (Clyne et al. 1999; Gao and Chess 1999; Vosshall et al. 1999, 2000; Robertson et al. 2003). Apart from the ORs, a number of Gustatory Receptors (GR) is expressed in the antennae, e.g. Gr21a and Gr63a that detect carbon dioxide (Kwon et al. 2007; Jones et al. 2007). The insect OR genes are quite peculiar; they show an inverted membrane insertion (Benton et al. 2006; Lundin et al. 2007), form heteromultimers (Neuhaus et al. 2005, Benton et al. 2006) and show no sequence homology to any other GPCRs. In addition, the family is highly divergent, even between closely related species. Another peculiarity is that, contrary to what has been found in vertebrates, most insect OSNs express two ORs. One is unique for each OSN type, while one is expressed ubiquitously in all basiconic and trichoid-associated OSNs. This second OR is in Dm named Or83b. Contrary to the OSN-type-specific ORs, Or83b is highly conserved among most insects (Jones et al. 2005). The function of Or83b is discussed in the following section.
The initial coding of volatile chemical information is a consequence of the tuning spectra of the expressed ORs. How the peripheral olfactory system of the fly decodes odors has been rather well studied (e.g. Clyne et al. 1997; De Bruyne et al. 1999, 2001; Stensmyr et al. 2003a, b; Hallem et al. 2004, 2006). That the gene family encoding ORs indeed act as chemosensors was first shown by Störtkuhl et al. (2001). Taking a cue from work done in the mouse (Araneda et al. 2000), Störtkuhl et al. misexpressed one OR gene in most OSNs. Subsequent electroantennogram recordings from the transgenic flies revealed an increased response to the ligand of the misexpressed OR versus control flies. Further evidence showing that the OR family encodes bona fide odor detectors was provided by Dobritsa et al. (2003), who elegantly exploited the genotype of the ΔHalo mutant, which lack (among other things) the Or22a gene. Re-expression of Or22a in the “empty” neuron created by the ΔHalo mutation fully rescued the response to ethyl butyrate, one of the ligands for Or22a. The “empty” neuron system of ΔHalo flies has since been used extensively to examine the molecular receptive range of a large variety of ORs (e.g. Hallem et al. 2004, 2006). The strength of this approach is that it allows for deorphanization in a more natural setting as compared to the alien environments provided by e.g. Xenopus oocytes or human embryonic kidney cells, two frequently used systems for heterologous analysis of protein function. The drawback to the ΔHalo approach is that it is both time and labor intensive compared to heterologous systems, which can be incorporated into high throughput screening pipelines, and that the environment surrounding the OSNs (e.g. OBPs, see below) remains the same, even if the ORs are exchanged.
The fly ORs display a varied degree of selectivity in their receptive range. Some ORs are highly specific whereas others are more broadly tuned. However, those ORs that respond to multiple compounds, typically respond to chemicals of structural proximity (e.g. Stensmyr et al. 2003a; Hallem et al. 2004; Hallem et al. 2006). Available data accordingly suggest that insect ORs are optimally configured for specific chemical properties, or even specific chemicals. A problem though with any data regarding receptive range of ORs is that the obtained tuning spectra are always dependent on the number of odor ligands screened, their relevance to the system as well as the concentrations tested. Most ORs are capable of detecting almost any chemical as long as this compound is tested in high enough concentrations, a feat that extends even to notoriously selective pheromone receptors. Likewise, perceived notions of, for example, extreme selectivity might simply be a consequence of missing key stimuli from the test panel. Thus, although a wide battery of odorants were used to deorphan the fly ORs in the above-mentioned studies, the chosen stimuli only represent a minute fraction of all chemical volatiles that a fly might encounter in its daily life (flies living in the “wild” that is). For example, a banana alone contains well over 500 volatile compounds, most fruits contain similar numbers of volatiles (Stensmyr et al. 2001, 2003a, b). Hence, it is quite possible that for many of the ORs there exist better ligand matches that would differentiate the system further. The use of gas chromatography linked with electrophysiology and/or optical imaging is a way of identifying potent new ligands for the system, which would increase our understanding of how odorants are decoded. In the case of Dm, odor sources should preferably be looked for among afrotropical fruit from the ancestral home range.
The OR and GR families are not the only chemosensory-specific gene families expressed in the peripheral olfactory system. The odorant binding protein family, first discovered in the moth (Vogt and Riddiford 1981) comprises another large gene family [51 members in Dm (Hekmat-Scafe et al. 2002)] encoding olfactory specific proteins. The OBPs are produced and secreted by auxillary cells surrounding the OSNs. Within the sensillium lymph, The OBPs bind the hydrophobic odor molecules and presumably guide them through the aqueous sensillum lymph to the ORs in the dendritic membrane of the OSNs (Vogt and Riddiford 1981; Vogt 2003; Xu et al. 2005). The OBP family member LUSH nicely illustrates the importance of OBPs. LUSH mutants fail to respond to the fly pheromone cis-vaccenyl acetate, both physiologically as well as behaviorally (Xu et al. 2005). Although the importance of OBPs for pheromone detection has been clearly demonstrated, their role in the detection of plant odors remains unclear. ORs misexpressed in the “empty” neurons of the ΔHalo mutant retain their odorant specificity in their new environment, in spite of a differing OBP set-up. OBPs undergo a conformational change upon binding of an odor ligand. Recent work done on LUSH actually suggests that what the OR is detecting is the conformationally altered LUSH molecule, rather than the pheromone itself (Laughlin et al. 2008). If this is a unique feature of the pheromone system or also holds true for the part of the system detecting, e.g. food-related volatiles remains to be elucidated. Detection of pheromones is furthermore dependent on the presence of another class of molecules, namely Sensory Neuron Membrane Proteins (SNMP) (Rogers et al. 1997). SNMPs constitute an insect-specific sub-group of the CD36 family, that comprises transmembrane receptors involved in lipid binding and transport. The SNMPs act as co-receptors with ORs in the pheromone detection subsystem of Dm (Benton et al. 2007; Jin et al. 2008). Whether SNMPs are also of importance outside pheromone detection remains to be investigated.
The OSNs found in coeloconica sensilla do not conform to the norm, as they do not express the ubiquitous Or83b (with the exception of coeloconica sensilla type 3, that express Or35a as well as Or83b), suggesting that odorants in these sensilla are detected and/or transduced differently. The identity of the coeloconica ORs was recently revealed to be divergent members of the ionotropic glutamate receptor family (Benton et al. 2009). The receptors, called Ionotropic Receptors (IRs) form a large and divergent gene family comprising 63 members (61 putatively functional), of which 15 are expressed in the antennae. Contrary to the ORs, the IRs seem to be expressed in a combinatorial manner and also display a slightly different ligand specificity compared to the ORs, responding to nitrogen-containing volatiles, such as ammonia and 1,4-diaminobutane.
Chemical to electrical
In vertebrate olfactory ORNs the stimulated ORs couple to Golf proteins that stimulate—like Gs proteins—adenylyl cyclase and raise the level of cAMP. These messenger molecules diffuse to and bind on ion channels, the cyclic-nucleotide-gated (CNG) channels, which they activate. CNG channels are non-selective cation channels that conduct Na+, K+, and Ca2+. The rise in intracellular Ca2+ activates Ca2+-dependent Cl− channels that cause a robust depolarization and is the step in the signal transduction cascade providing the strongest amplification of the odor signal (Kaupp et al. 2008).
The present knowledge regarding how odorant molecule binding to its OR is converted into an electrical signal in insects arises from two sources. First, from manipulating parts of putative signal transduction cascades and observing the effect on odor response, and second, from studies of OR function in heterologous expression systems.
Dm mutants with disturbed cAMP signaling (dunce and rutabaga) show abnormal olfactory behavior (Martin et al. 2001). This was especially the case when the phosphodiesterase dunce was overexpressed in ORN subsets, and thereby decreasing the cAMP level in these neurons (Gomez-Diaz et al. 2004). Thus, proper function of olfactory signaling relies on an intact cAMP signaling cascade.
There is also evidence that DAG/IP3 signaling may play a role in odorant signal transduction (Krieger and Breer 1999). Dm norpA mutants show impaired olfaction, which indicates that PLC activity is required for appropriate processing of odor information (Riesgo-Escovar et al. 1995). Furthermore, mutations in the dgq gene, which encodes the Gq alpha subunit result in reduced responses to odor stimulation (Kain et al. 2008b). The responses were further attenuated by additional mutations in plc21C, a gene encoding for a PLCb. Interestingly, the dgq mutant phenotype was rescued by mutation of the DAG kinase rdgA (Kain et al. 2008b). This finding indicates that the efficiency of olfactory signal transduction is controlled by a phospholipid messenger. Support for this hypothesis comes from a study on mutants in stmbhA, a gene encoding a putative PIP2-DAG lipase. These mutants show a markedly reduced electroantennogram response to odor stimulation (Kain et al. 2008a). Taken together, any disturbance of the PIP2 cleavage and regeneration cycle seems to impair an appropriate processing of the olfactory signal in insect ORNs. Thus, both a Gs- and a Gq-mediated pathway seem to be involved in Dm olfactory transduction.
Expression of ORs in heterologous systems allows us either to perform functional tests by using endogeneous parts of the signal transduction machinery or to coexpress useful reporters. To identify ligands for vertebrate ORs a variety of assays has been developed, such as detection of cAMP production and Ca2+ imaging (Katada et al. 2003). For example, by coexpression of Drosophila Or43a with the promiscous G protein Gaa15 in Xenopus ooytes receptor activation could be monitored by measuring Cl− currents activated by intracellular Ca2+ release (Wetzel et al. 2001). Elevation of free Ca2+ could also be obtained in HEK293 cells when Or22a was coexpressed with the ubiquitous receptor protein Or83b (Neuhaus et al. 2005). Further investigations of the Or22a/Or83b complex in HEK293 cells (Wicher et al. 2008) or Or47a/Or83b in HELA cells, HEK293T cells and Xenopus oocytes (Sato et al. 2008) revealed that these heterodimers form non-selective cation channels permeable to Na+, K+, and Ca2+. Even in the absence of any stimulation, these channels show some constitutive activity leading to elevated levels of free Ca2+. Short odor stimulation was seen to produce an immediate transient response that was independent of G proteins, i.e. the ORs act as ionotropic receptors. This type of response requires a sufficient strength of stimulation (Sato et al. 2008; Wicher et al. 2008). The fast ionotropic response is followed by a slower developing, but more sensitive, metabotropic response (Wicher et al. 2008, Fig. 1). It was shown that stimulation of Or22a, either expressed alone or coexpressed with Or83b, lead to cAMP production. On the other hand, Or83b, either expressed alone or coexpressed with Or22a, responded to cAMP elevation with current production. Moreover, it could also be activated by cGMP. Mutagenesis in the Or83b protein caused a change in the permeability ratio of ions, which demonstrates that Or83b is responsible for the channel activity of Dm ORs (Wicher et al. 2008). Taken together, the odorant-specific part of the OR dimere seems to couple to stimulatory G proteins to stimulate cAMP production. This G-protein-dependent pathway provides a highly sensitive odor detection, while at higher odor concentrations there is a very fast, ionotropic mechanism of OR activation. It remains to be seen what role Gq signaling may play (Kain et al. 2008a, b).
Fig. 1.
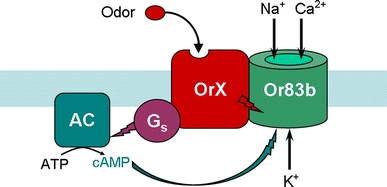
Scheme of odorant signal transduction. The insect odorant receptor complex is composed of an odorant-specific receptor protein OrX and the non-selective cation channel Or83 that conducts Na+, K+ and Ca2+. Odor-stimulation of OrX activates Or83 by an ionotropic and a metabotropic pathway. The direct activation of Or83b by OrX (red flash) leads to a fast and transient cation flow. The metabotropic pathway stimulates—via activation of Gs proteins by OrX—the adenylyl cyclase (AC) activity (violet flash) and thus the cAMP production. cAMP in turn activates Or83b (blue-green flash). The ionotropic pathway ensures a very rapid recognition of high odor concentrations while the metabotropic pathway allows highly sensitive odor detection
Primary information processing—the antennal lobe
The neural components, mediating olfactory information in the brain of the fruit fly, have been intensively studied and are well described (Stocker 1994). The olfactory system of the Dm adult is organized according to similar principles as the vertebrate olfactory system, but with vastly reduced numerical complexity (Hildebrand and Shepherd 1997; Hallem and Carlson 2004) (Fig. 2a). As mentioned in the two previous sections, odors are recognized by OSNs (not shown in Fig. 2a). The OSNs send their axons to the first olfactory neuropil in the insect brain, the antennal lobe (AL), which processes the olfactory information. The AL is a structure common to all insects except some anosmic species that have lost it secondarily (Strausfeld and Hildebrand 1999). The insect AL represents an analogous structure to the olfactory bulb in vertebrates but has evolved independently (Strausfeld and Hildebrand 1999). Both structures consist of discrete neuropil structures, so-called olfactory glomeruli. These represent structural as well as functional units involved in olfactory processing. The Drososphila AL comprises 49 olfactory glomeruli, which can be identified individually due to their specific position and size (Laissue et al. 1999). Each OSN axon innervates a single ipsilateral glomerulus in the AL in most species. However, in Dm, OSNs form an axonal commissure between the two ALs and individual axons innervate homologous glomeruli in the two lobes (Stocker 1994). Each bilaterally innervated glomerulus receives equal input from both antennae (Vosshall et al. 2000). Interestingly, a few glomeruli (as V, VL1, VP1, VP2 and VP3) are innervated only unilaterally (Stocker et al. 1983; Stocker 2001). As mentioned above, each OSN expresses only a single OR gene (besides OR83b). All the OSNs expressing the same OR converge onto a common glomerulus, which thus collects OSN input from only one functional type. The glomerulus acts as a “collecting basket” of OSNs with identical odor response profiles. This correspondence was first shown morphologically for some moth species (Hansson et al. 1992, 1995; Ochieng’ et al. 1995; Todd et al. 1995), but was finally proven by molecular techniques in Dm (Gao et al. 2000; Vosshall et al. 2000). This condition is also assumed to hold true for other insect species, but has not yet been experimentally proven. However, a few aberrant cases of 1 OSN type to 2 glomeruli and of 2 to 1 innervation ratios in Dm have been described as well (Couto et al. 2005; Fishilevich and Vosshall 2005).
Fig. 2.
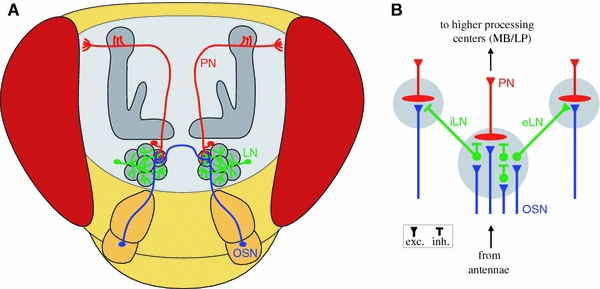
Schematic of the Drosophila olfactory pathway. a Antennal olfactory sensory neurons (OSN, blue) converge in specific glomeruli of the antennal lobe. Some of them send an axonal branch through the antennal commissure to the other hemisphere. Local interneurons (LN, green) branch in all glomeruli and interconnect these with each other. Projections neurons (PN, red) collect the olfactory information within the antennal lobe and send their axons to higher processing centers as the calyx and the lateral protocerebrum. b Circuit diagram of the antennal lobe. The three principal populations of neurons and their synaptic connections within the glomeruli (gray circles) are represented. The diagram summarizes anatomical data from several insect species. Excitatory synapses are symbolised by triangles, inhibitory synapses by bars. In Drosophila, the existence of both inhibitory local interneurons (iLN) as well as excitatory local interneurons (eLN) has been shown
A glomerulus receives not only the input from OSNs, but contains a highly ordered synaptic organization including two types of interneurons (Fig. 2b): the local interneurons (LNs; approximately 100, probably analogous to periglomerular cells in mammals) and projection neurons (PNs; approximately 150, analogous to mitral/tufted cells in mammals). LNs of the AL represent an enigmatic element of early olfactory coding in the CNS of insects. They have all their projections restricted to the AL, where most form wide-field arborizations within most glomeruli. The majority of LNs appears to be inhibitory (iLNs) and release γ-aminobutyric acid (GABA) as a neurotransmitter (Wilson and Laurent 2005), indicating that they form a lateral multi-level inhibitory network modulating the olfactory signal within the AL. However, a recently described population of excitatory cholinergic LNs (eLNs) forms a dense network of lateral excitatory connections between different glomeruli assumed to boost the AL output (Olsen et al. 2007; Shang et al. 2007). PNs have dendritic arborizations within the glomeruli and convey the olfactory signals through axonal projections to higher olfactory processing centers such as the lateral protocerebrum and the mushroom bodies. A great majority of the PNs is uniglomerular with dendrites confined to a single glomerulus, but a few have multiglomerular dendritic fields.
How are odors neuronally represented in the first olfactory neuropil? Optical recording techniques allow visualization of odor-evoked patterns in the AL. Several imaging studies in different insect species using either calcium-sensitive dyes (e.g. Joerges et al. 1997; Galizia et al. 1999; Sachse et al. 1999; Carlsson et al. 2002), genetically encoded reporters to measure intracellular calcium (Fiala et al. 2002; Wang et al. 2003; Silbering et al. 2008) or synaptic vesicle release (Ng et al. 2002; Yu et al. 2004) have shown that odors are encoded as specific spatio-temporal “across-glomeruli” patterns. Each odor evokes activity in several glomeruli, whereas each glomerulus participates in the patterns of several odors (Fig. 3). These activity patterns are species-specific and conserved between different individuals. The olfactory system has in this way developed a strategy to encode a huge number of odors with a limited number of coding units. Although the different studies agree on how odors are encoded, different publications report contradicting results regarding the transformation of odor representations at the different processing levels within the AL (i.e. OSN versus PN responses, Fig. 3). These results reach from sharpening and contrast-enhancement of the olfactory input due to inhibitory processing in honeybees (Sachse and Galizia 2002) to either not processing at all (Ng et al. 2002; Wang et al. 2003) or a broadening of the output pattern in comparison to the input pattern in Dm (Wilson et al. 2004). Adding to the already existing complexity of the AL network, a recently published study in Dm shows that presynaptic inhibition onto OSN terminals leads to an inhibitory network activity (Olsen and Wilson 2008). Odor information processing mechanisms underlying olfactory coding are thus highly diverse and appear to be specific for particular glomerulus-odor combinations (Silbering et al. 2008).
Fig. 3.
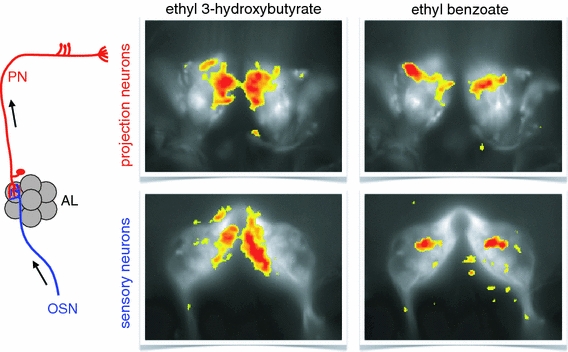
Odors evoke specific patterns of glomerular activity in the Drosophila antennal lobe. The calcium-sensitive protein G-CaMP has been genetically expressed in either projection neurons (above) or sensory neurons (below). Calcium signals to two different odors have been superimposed onto the morphological image of the antennal lobe. Both odors lead to a specific, but different pattern of activated glomeruli. The activities are bilaterally symmetric between the left and the right antennal lobe. Comparison of the activity patterns between the sensory and the projection neurons to the same odor reveals similar but not identical responses
The AL output responses are further processed on their way to higher processing centers. There is a massive divergence between PNs and the intrinsic neurons of the mushroom body, the Kenyon cells. While most PNs have a relatively broad response profile with various levels of activity, a large number of units encode that same information with very few spikes, in an almost binary fashion, with only few units active at any time among Kenyon cells (Perez-Orive et al. 2002). This coding strategy is in strong contrast to the one found in the AL, where each PN covers a wide dynamic range. Deciphering the coding of odors on all processing levels is crucial for understanding the remarkable capacity of the olfactory system in recognizing and discriminating the vast array of odor molecules.
The result—behavior
When investigating the sense of smell the first question that comes up is, do flies have to smell? In an elegant set of experiments, Asahina et al. (2008) demonstrated that the sense of smell is important at least for the survival of fruit fly larvae: when being reared under food ad libitum, anosmic OR83b mutants developed as successfully as mutants whose OR83b system had been rescued. However, when food was limited, i.e. when larvae had to search for new food sources, the anosmic flies were less successful compared to the rescued flies. In a direct competition experiment anosmic mutants were selectively eliminated in competition with fly larvae whose OR83b had been rescued. Interestingly, partial rescue of the olfactory system (only one of 61 receptors was functioning) led to survival rates that were intermediate between the anosmic and the fully rescued mutants. Although a simple sense of smell (partially rescued mutants) already increased the fly larvae’s survival rates, a more powerful smell (fully rescued mutants) was even more efficient.
The efficiency of the adults’ sense of smell can be demonstrated easily by putting about 50 flies in an enclosed arena that is equipped with two traps. When one trap is empty and the other is filled, e.g. with rotten banana, one of Drosophila’s favorite food sources, the first flies will enter the smelling trap already after a few minutes and after 24 h between 90 and 100% of the total number is usually captured. This simple bioassay was introduced by Larsson et al. (2004) and was recently used, e.g. to demonstrate that different Dm strains like Wildtype Berlin, Wildtype Canton S, etc. differ substantially in their odor preferences and in their response latencies (Ruebenbauer et al. 2008).
However, as a Dm fly can live for more than 2 weeks, it has ample time to experience diverse and varying qualities of different food sources. Although the flies are equipped with some innate odor preferences, i.e. every inexperienced Drosophila will head for a plume of rotten banana or balsamico vinegar, they can learn associations of odors with good or bad experiences. A substantial amount of work has been done on olfactory learning, mainly using Pavlovian conditioning procedures, where one odor (conditioned stimulus) is temporally paired with a positive (i.e. sugar water) or negative (i.e. electric shock) reinforcement, the so-called unconditioned stimulus (Tully and Quinn 1985). This procedure provided knowledge regarding the formation and capacity of short-term and long-term memory in fruit flies. Furthermore, by performing the conditioning procedure with transgene flies Krashes et al. (2007) could map which cells in the brain are necessary for olfactory memory retrieval (Kenyon cells in the mushroom body whose axons form the α/β-lobes), and which are needed for acquisition and consolidation of memory (α′/β′-Kenyon cells). However, apart from helping to understand learning in flies, conditioning experiments provide a solution to another problem: only few odors provoke strong innate behavioral responses in Dm, e.g. the main component of banana odor isoamylacetate is highly attractive, while benzaldehyde—the typical smell of bitter almond—is a repellent. In order to check the flies’ olfactory threshold for innately neutral odors these have to be provided with a meaning by positive or negative reinforcement. After conditioning, it is possible to test, for example, the minimum concentration the flies react to.
However, to tell the truth, for an ethologist interested in the sense of smell Drosophila would not have been the first choice. While trap assays and mass learning experiments often yield valuable results, the investigation of individual flies is often frustrating. In order to study the sense of smell, one would like to make predictions on how a specific fly responds before and after some kind of manipulation. However, to make any predictions about the fly’s response to an odor you need to know the fly’s motivational state. Contrary to, for example, workers of eusocial insects that have no sex but are dedicated just to foraging, fruit flies are sometimes in the mood for sex, sleeping, ovipositioning or foraging. Each motivational stage can provoke different responses to different stimuli, which makes the prediction how a fly will respond rather difficult. Despite these problems, experiments using individual flies are necessary in order to investigate, for example, the speed of odor processing or the response to minute differences in an odor’s concentration. To illustrate the difficulties in establishing good bioassays for single Dm, we will describe the story of the “Knaden olfactometer”. One of us invested considerable time in establishing an experiment where individual flies were exposed to a continuous airflow within 1 cm-wide glass tubes. The flies were tracked automatically and an elaborate stimulus controller allowed exposure of the flies to odor pulses well-defined in timing, blend composition, and concentration. We expected the flies to show upwind runs whenever facing a positive odor, e.g. banana or aceto balsamico. By coupling the stimulus controller to the tracking device, we were able to compare the upwind speed directly before odor contact with the one afterwards. However, despite a comprehensive analysis until now we never found any consistent responses of the flies to different kinds of stimuli (Fig. 4). The picture changes when flies are not tested while sitting or running but during flying. Budick and Dickinson (2006) successfully investigated individual Drosophila flies in a free-flight wind tunnel. Under these conditions, the contact to an odor plume provoked directed upwind flights, while the loss lead to a so-called casting behavior, where the trajectory of the flies becomes more and more perpendicular to the wind direction (a strategy that is well known from moths and that under natural conditions increases the chance to re-enter the odor plume, Fig. 4). Like in our walking assay, the use of a tracking device allowed the authors to measure the response latencies after the flies entered (<250 ms) and lost the plume (ca. 330 ms). This experiment thus provides the tool to investigate fine scale temporal dynamics of the flies’ sense of smell. Meanwhile additional experiments, using individual flies, have been established. These experiments use innate responses of tethered, flying individuals to odor stimuli provided in a constant airflow (Duistermars and Frye 2008). The development of admittedly rather complicated experimental procedures gives a vantage point to unravel more details about the fine structure of the Dm’s sense of smell.
Fig. 4.
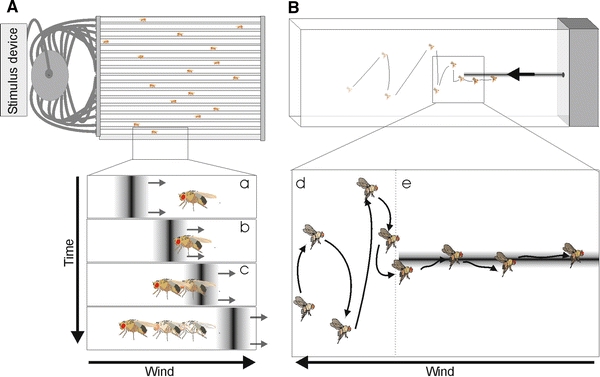
Steck-Knaden assay. A 16 individually tracked flies are exposed to a clean-air flow. a–c A computer-controlled stimulus device generates odor stimuli, that meet the flies at a predictable time. When encountering an attractive odor, flies walk upwind. The tracking system calculates upwind speed before and after the stimulus reaches the fly. B Wind tunnel. d Flies show crosswind flights when exposed to clean-air flow. e When getting in contact with an odor plume, flies start upwind flights. A 3-D tracking system detects the time of plume contact and plume loss and calculates the flight’s directionality and upwind speed before and afterwards
Conclusion
Considering the progress reported above, we are presently in an extremely interesting time regarding Dm olfactory neuroethology. So much background data have been produced on neural and molecular function at different levels that we are approaching stages where behavioral questions can be attacked with a full arsenal of neurogenetic tools. By using the knowledge regarding receptors, transduction mechanisms, and primary and higher integration we can dissect the system, but only when we have relevant and preferably nature-based bioassays and stimuli will we start to really understand the olfactory neuroethology of Drosophila melanogaster. This is one of the major challenges ahead in this fascinating research field.
Acknowledgments
The writing of this review was supported by funding from the Max Planck Society to BSH and from the Bundesministerium für Bildung und Forschung (BMBF) to SS. We are also grateful to M. Jean-Marie Delwart for his continued support and interest in the chemical senses and chemical communication.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Asahina K, Pavlenkovich V, Vosshall LB. The survivial advantage of olfaction in a competitive environment. Curr Biol. 2008;18:1153–1155. doi: 10.1016/j.cub.2008.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Vosshall LB. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. 2007;450:289–293. doi: 10.1038/nature06328. [DOI] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budick SA, Dickinson MH. Free-flight responses of Drosophila melanogaster to attractive odors. J Exp Biol. 2006;209:3001–3017. doi: 10.1242/jeb.02305. [DOI] [PubMed] [Google Scholar]
- Carlsson MA, Galizia CG, Hansson BS. Spatial Representation of Odours in the Antennal Lobe of the Moth Spodoptera littoralis (Lepidoptera: Noctuidae) Chem Senses. 2002;27:231–244. doi: 10.1093/chemse/27.3.231. [DOI] [PubMed] [Google Scholar]
- Castle WE. Inbreeding, crossbreeding and sterility in Drosophila. Science. 1906;23:153. doi: 10.1126/science.23.578.153. [DOI] [PubMed] [Google Scholar]
- Clyne P, Grant A, O’Connell R, Carlson JR (1997) Odorant response of individual sensilla on the Drosophila antenna. Invert Neurosci (2–3):127–135 [DOI] [PubMed]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR (1999) A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22:327–338 [DOI] [PubMed]
- Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15:1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- de Bruyne M, Baker TC. Odor detection in insects: volatile codes. J Chem Ecol. 2008;34:882–897. doi: 10.1007/s10886-008-9485-4. [DOI] [PubMed] [Google Scholar]
- de Bruyne M, Clyne PJ, Carlson JR. Odor coding in a model olfactory organ: the Drosophila maxillary palp. J Neurosci. 1999;19:4520–4532. doi: 10.1523/JNEUROSCI.19-11-04520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyne M, Foster K, Carlson JR. Odor coding in the Drosophila antenna. Neuron. 2001;30:537–552. doi: 10.1016/S0896-6273(01)00289-6. [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, van Naters W, Warr CG, Steinbrecht RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37:827–841. doi: 10.1016/S0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- Duistermars BJ, Frye MA. Crossmodal visual input for odor tracking during fly flight. Curr Biol. 2008;18:270–275. doi: 10.1016/j.cub.2008.01.027. [DOI] [PubMed] [Google Scholar]
- Fiala A, Spall T, Diegelmann S, Eisermann B, Sachse S, Devaud JM, Buchner E, Galizia CG. Genetically expressed cameleon in Drosophila melanogaster is used to visualize olfactory information in projection neurons. Curr Biol. 2002;12:1877–1884. doi: 10.1016/S0960-9822(02)01239-3. [DOI] [PubMed] [Google Scholar]
- Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- Galizia G (2008) Insect olfaction. In: Basbaum AI, Kaneko A, Shepherd GM, Westheimer G (eds) The senses: a comprehensive reference, vol 4 [Olfaction & Taste, Firestein S and Beauchamp GK (eds)]. Academic Press, San Diego, pp 725–770
- Galizia CG, Sachse S, Rappert A, Menzel R. The glomerular code for odor representation is species specific in the honeybee Apis mellifera. Nat Neurosci. 1999;2:473–478. doi: 10.1038/8144. [DOI] [PubMed] [Google Scholar]
- Gao Q, Chess A. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics. 1999;60:31–39. doi: 10.1006/geno.1999.5894. [DOI] [PubMed] [Google Scholar]
- Gao Q, Yuan B, Chess A. Convergent projections of Drosophila olfactory neurons to specific glomeruli in the antennal lobe. Nat Neurosci. 2000;3:780–785. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]
- Gomez-Diaz C, Martin F, Alcorta E. The cAMP transduction cascade mediates olfactory reception in Drosophila melanogaster. Behav Genet. 2004;34:395–406. doi: 10.1023/B:BEGE.0000023645.02710.fe. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. The odor coding system of Drosophila. Trends Genet. 2004;20:453–459. doi: 10.1016/j.tig.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Dahanukar A, Carlson JR. Insect odor and taste receptors. Annu Rev Entomol. 2006;51:113–135. doi: 10.1146/annurev.ento.51.051705.113646. [DOI] [PubMed] [Google Scholar]
- Hansson BS. Insect olfaction. Berlin: Springer; 1999. [Google Scholar]
- Hansson BS, Ljungberg H, Hallberg E, Lofstedt C. Functional specialization of olfactory glomeruli in a moth. Science. 1992;256:1313–1315. doi: 10.1126/science.1598574. [DOI] [PubMed] [Google Scholar]
- Hansson BS, Almaas TJ, Anton S. Chemical communication in heliothine moths. V. Antennal lobe projection patterns of pheromone-detecting receptor neurons in the male Heliothis virescens (Lepidoptera: Noctuidae) J Comp Physiol A. 1995;177:535–543. doi: 10.1007/BF00207183. [DOI] [Google Scholar]
- Hekmat-Scafe DS, Scafe CR, McKinney A, Tanouye MA. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002;12:1357–1369. doi: 10.1101/gr.239402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand JG, Shepherd GM. Mechanisms of olfactory discrimination: converging evidence for common principles across phyla. Ann Rev Neurosci. 1997;20:595–631. doi: 10.1146/annurev.neuro.20.1.595. [DOI] [PubMed] [Google Scholar]
- Jin X, Ha TS, Smith DP. SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc Natl Acad Sci USA. 2008;105:10996–11001. doi: 10.1073/pnas.0803309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerges J, Küttner A, Galizia CG, Menzel R. Representation of odours and odour mixtures visualized in the honeybee brain. Nature. 1997;387:285–288. doi: 10.1038/387285a0. [DOI] [Google Scholar]
- Jones WD, Nguyen TA, Kloss B, Lee KJ, Vosshall LB (2005) Functional conservation of an insect odorant receptor gene across 250 million years of evolution. Curr Biol 22;15(4):R119–R121 [DOI] [PubMed]
- Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445:86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- Kain P, Chandrashekaran S, Rodrigues V, Hasan G. Drosophila mutants in phospholipid signaling have reduced olfactory responses as adults and larvae. J Neurogenet. 2008;17:1–10. doi: 10.1080/01677060802372494. [DOI] [PubMed] [Google Scholar]
- Kain P, Chakraborty TS, Sundaram S, Siddiqi O, Rodrigues V, Hasan G. Reduced odor responses from antennal neurons of G(q)alpha, phospholipase Cbeta, and rdgA mutants in Drosophila support a role for a phospholipid intermediate in insect olfactory transduction. J Neurosci. 2008;28:4745–4755. doi: 10.1523/JNEUROSCI.5306-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Nakagawa T, Kataoka H, Touhara K. Odorant response assays for a heterologously expressed olfactory receptor. Biochem Biophys Res Commun. 2003;305:964–969. doi: 10.1016/S0006-291X(03)00863-5. [DOI] [PubMed] [Google Scholar]
- Kaupp UB, Kashikar ND, Weyand I. Mechanisms of sperm chemotaxis. Annu Rev Physiol. 2008;70:93–117. doi: 10.1146/annurev.physiol.70.113006.100654. [DOI] [PubMed] [Google Scholar]
- Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S. Sequential use of mushroom body neuron subsets during Drosophila odor memory processing. Neuron. 2007;53:103–115. doi: 10.1016/j.neuron.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger J, Breer H. Olfactory reception in invertebrates. Science. 1999;286:720–723. doi: 10.1126/science.286.5440.720. [DOI] [PubMed] [Google Scholar]
- Kwon JY, Dahanukar A, Weiss LA, Carlson JR. The molecular basis of CO2 reception in Drosophila. Proc Natl Acad Sci USA. 2007;104:3574–3578. doi: 10.1073/pnas.0700079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D, Tsacas L (1983) Breeding-sites in tropical African Drosophilids. In: Ashburner M, Carson HL, Thompson JN (eds) The genetics and biology of Drosophila vol 3d, pp 221–332
- Laissue PP, Reiter C, Hiesinger PR, Halter S, Fischbach KF, Stocker RF. Three-dimensional reconstruction of the antennal lobe in Drosophila melanogaster. J Comp Neurol. 1999;405:543–552. doi: 10.1002/(SICI)1096-9861(19990322)405:4<543::AID-CNE7>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Laughlin JD, Ha TS, Jones DN, Smith DP. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell. 2008;33(7):1255–1265. doi: 10.1016/j.cell.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin C, Käll L, Kreher SA, Kapp K, Sonnhammer EL, Carlson JR, Heijne G, Nilsson I. Membrane topology of the DrosophilaOr83b odorant receptor. FEBS Lett. 2007;581(29):5601–5604. doi: 10.1016/j.febslet.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Charro MJ, Alcorta E. Mutations affecting the cAMP transduction pathway modify olfaction in Drosophila. J Comp Physiol A. 2001;187:359–370. doi: 10.1007/s003590100208. [DOI] [PubMed] [Google Scholar]
- Morgan TH. Sex-limited inheritance in Drosophila. Science. 1910;32:120–122. doi: 10.1126/science.32.812.120. [DOI] [PubMed] [Google Scholar]
- Neuhaus EM, Gisselmann G, Zhang W, Dooley R, Stortkuhl K, Hatt H. Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat Neurosci. 2005;8:15–17. doi: 10.1038/nn1371. [DOI] [PubMed] [Google Scholar]
- Ng M, Roorda RD, Lima SQ, Zemelman BV, Morcillo P, Miesenbock G. Transmission of olfactory information between three populations of neurons in the antennal lobe of the fly. Neuron. 2002;36:463–474. doi: 10.1016/S0896-6273(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Ochieng’ SA, Anderson P, Hansson BS. Antennal lobe projection patterns of olfactory receptor neurons involved in sex pheromone detection in Spodoptera littoralis (Lepidoptera: Noctuidae) Tiss Cell. 1995;27(2):221–232. doi: 10.1016/S0040-8166(95)80024-7. [DOI] [PubMed] [Google Scholar]
- Olsen S, Wilson R. Lateral presynaptic inhibition mediates gain control in an olfactory circuit. Nature. 2008;452:956–960. doi: 10.1038/nature06864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SR, Bhandawat V, Wilson RI. Excitatory interactions between olfactory processing channels in the Drosophila antennal lobe. Neuron. 2007;54:89–103. doi: 10.1016/j.neuron.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Orive J, Mazor O, Turner GC, Cassenaer S, Wilson RI, Laurent G. Oscillations and sparsening of odor representations in the mushroom body. Science. 2002;297:359–365. doi: 10.1126/science.1070502. [DOI] [PubMed] [Google Scholar]
- Riesgo-Escovar J, Raha D, Carlson JR. Requirement for a phospholipase C in odor response: overlap between olfaction and vision in Drosophila. Proc Natl Acad Sci USA. 1995;92:2864–2868. doi: 10.1073/pnas.92.7.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Wanner KW. The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 2006;16:1395–1403. doi: 10.1101/gr.5057506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Warr CG, Carlson JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci USA. 2003;100(Suppl. 2):14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers ME, Sun M, Lerner MR, Vogt RG. Snmp-1, a novel membrane protein of olfactory neurons of the silk moth Antheraea polyphemus with homology to the CD36 family of membrane proteins. J Biol Chem. 1997;272:14792–14799. doi: 10.1074/jbc.272.23.14792. [DOI] [PubMed] [Google Scholar]
- Ruebenbauer A, Schlyter F, Hansson BS, Löfstedt C, Larsson MC. Genetic variability and robustness of host odor preference in Drosphila melanogaster. Curr Biol. 2008;18:1438–1443. doi: 10.1016/j.cub.2008.08.062. [DOI] [PubMed] [Google Scholar]
- Sachse S, Galizia CG. Role of inhibition for temporal and spatial odor representation in olfactory output neurons: a calcium imaging study. J Neurophysiol. 2002;87:1106–1117. doi: 10.1152/jn.00325.2001. [DOI] [PubMed] [Google Scholar]
- Sachse S, Rappert A, Galizia CG. The spatial representation of chemical structures in the antennal lobes of honeybees: steps towards the olfactory code. Eur J Neurosci. 1999;11:3970–3982. doi: 10.1046/j.1460-9568.1999.00826.x. [DOI] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Shanbhag SR, Muller B, Steinbrecht RA. Atlas of olfactory organs of Drosophila melanogaster. 1. Types, external organization, innervation and distribution of olfactory sensilla. Int J Insect Morphol Embryol. 1999;28:377–397. doi: 10.1016/S0020-7322(99)00039-2. [DOI] [Google Scholar]
- Shang Y, Claridge-Chang A, Sjulson L, Pypaert M, Miesenböck G. Excitatory local circuits and their implications for olfactory processing in the fly antennal lobe. Cell. 2007;128:601–612. doi: 10.1016/j.cell.2006.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbering AF, Okada R, Ito K, Galizia CG. Olfactory information processing in the drosophila antennal lobe: anything goes? J Neurosci. 2008;28:13075–13087. doi: 10.1523/JNEUROSCI.2973-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spehr M, Munger SD (2009) Olfactory receptors: G protein-coupled receptors and beyond. J Neurochem. doi:10.1111/j.1471-4159.2009.06085 [DOI] [PMC free article] [PubMed]
- Stengl M, Hatt H, Breer H. Peripheral processes in insect olfaction. Annu Rev Physiol. 1992;54:665–681. doi: 10.1146/annurev.ph.54.030192.003313. [DOI] [PubMed] [Google Scholar]
- Stensmyr MC, Larsson MC, Bice S, Hansson BS. Detection of fruit- and flower-emitted volatiles by olfactory receptor neurons in the polyphagous fruit chafer Pachnoda marginata (Coleoptera: Cetoniinae) J Comp Physiol [A] 2001;187:509–519. doi: 10.1007/s003590100222. [DOI] [PubMed] [Google Scholar]
- Stensmyr MC, Giordano E, Balloi A, Angioy AM, Hansson BS. Novel natural ligands for Drosophila olfactory receptor neurones. J Exp Biol. 2003;206:715–724. doi: 10.1242/jeb.00143. [DOI] [PubMed] [Google Scholar]
- Stensmyr MC, Dekker T, Hansson BS. Evolution of the olfactory code in the Drosophila melanogaster subgroup. Proc Biol Sci. 2003;270:2333–2340. doi: 10.1098/rspb.2003.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker RF. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. 1994;275:3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- Stocker RF. Drosophila as a focus in olfactory research: mapping of olfactory sensilla by fine structure, odor specificity, odorant receptor expression, and central connectivity. Microsc Res Tech. 2001;55:284–296. doi: 10.1002/jemt.1178. [DOI] [PubMed] [Google Scholar]
- Stocker R, Singh R, Schorderet M, Siddiqi O. Projection patterns of different types of antennal sensilla in the antennal glomeruli of Drosophila melanogaster. Cell Tissue Res. 1983;232:237–248. doi: 10.1007/BF00213783. [DOI] [PubMed] [Google Scholar]
- Störtkuhl KF, Kettler R. Functional analysis of an olfactory receptor in Drosophila melanogaster. Proc Natl Acad Sci USA. 2001;98:9381–9385. doi: 10.1073/pnas.151105698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld NJ, Hildebrand JG. Olfactory systems: common design, uncommon origins? Curr Opin Neurobiol. 1999;9:634–639. doi: 10.1016/S0959-4388(99)00019-7. [DOI] [PubMed] [Google Scholar]
- Todd JT, Anton S, Hansson BS, Baker TC. Functional organization of the macroglomerular complex related to behaviorally expressed olfactory redundancy in male cabbage looper moths. Physiol Entomol. 1995;20:349–361. doi: 10.1111/j.1365-3032.1995.tb00826.x. [DOI] [Google Scholar]
- Tully T, Quinn WG. Classical Conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol. 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- Vogt RG. Insect pheromone biochemistry and molecular biology, the biosynthesis and detection of pheromones and plant volatiles. London: Elsevier; 2003. [Google Scholar]
- Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293:161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/S0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/S0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/S0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Wetzel CH, Behrendt HJ, Gisselmann G, Stortkuhl KF, Hovemann B, Hatt H. Functional expression and characterization of a Drosophila odorant receptor in a heterologous cell system. Proc Natl Acad Sci USA. 2001;98:9377–9380. doi: 10.1073/pnas.151103998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- Wilson RI, Laurent G. Role of GABAergic inhibition in shaping odor-evoked spatiotemporal patterns in the Drosophila antennal lobe. J Neurosci. 2005;25:9069–9079. doi: 10.1523/JNEUROSCI.2070-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RI, Turner GC, Laurent G. Transformation of olfactory representations in the Drosophila antennal lobe. Science. 2004;303:366–370. doi: 10.1126/science.1090782. [DOI] [PubMed] [Google Scholar]
- Xu P-X, Atkinson R, Jones DNM, Smith DP. Drosophila OBP LUSH is required for activity of Pheromone-sensitive neurons. Neuron. 2005;45:193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Yu D, Ponomarev A, Davis RL. Altered representation of the spatial code for odors after olfactory classical conditioning; memory trace formation by synaptic recruitment. Neuron. 2004;42:437–449. doi: 10.1016/S0896-6273(04)00217-X. [DOI] [PubMed] [Google Scholar]


