Abstract
The contribution of histone-DNA interactions to nucleosome positioning in vivo is currently a matter of debate. We argue here that certain nucleosome positions, often in promoter regions, in yeast may be, at least in part, specified by the DNA sequence. In contrast other positions may be poorly specified. Positioning thus has both statistical and DNA-determined components. We further argue that the relative affinity of the octamer for different DNA sequences can vary and therefore the interaction of histones with the DNA is a ‘tunable’ property.
Introduction
Both in vitro and in vivo the histone octamer can form nucleosomes on a wide spectrum of DNA sequences, independent of base composition. It thus lacks the base-specific sequence selectivity typical of transcription factors. Yet both in vitro and in vivo the octamer adopt a rather precise position on a given DNA sequence (1-10). Importantly in vivo histones are essential participants in gene regulation (11, 12). The in vitro data argue strongly that precise positioning is a consequence of selection by histone-DNA interactions but defining the role, if any, of these interactions in vivo has proved elusive. A particular problem is the finding that in vivo the same DNA sequence can accommodate different nucleosome spacings, and hence different positions, not only in different tissues of the same organism (13, 14) but also when DNA from one organism is transferred to another (15, 16). These observations indicate that the DNA sequence by itself need not specify a unique nucleosome array and that in vivo the organisation of nucleosomes may be determined by mechanisms other than direct histone-DNA interactions.
The Structure of the Nucleosome Core Particle
If histone-DNA interactions are strong determinants of positioning the nature of these determinants should be apparent in the structure of the nucleosome core particle and in the octamer itself. The detailed crystal structures of a nucleosome core particle assembled in vitro on a DNA palindrome derived from α-satellite DNA revealed that the DNA was wrapped around octamer in ~1.63 left-handed superhelical turns (17, 18), similar to the value deduced from a low-resolution structure of core particles isolated from chicken erythrocyte chromatin (19). The tight wrapping is confined to the central 12 double-helical turns in which, on average, the DNA is bent by ~49° /double helical turn. Put another way, ~123 bp of DNA are bent by 590° where, by comparison, the persistence length of mixed-sequence B-form DNA in solution is ~140-150 bp (20-26).
The tight wrapping of DNA around the histone octamer is facilitated by certain physicochemical properties of the polymer. One such is anisotropic flexibility (aka anisotropic bendability or deformability), first proposed by Zhurkin (27) and Trifonov (28). This is essentially a geometric property which favours the adoption of DNA trajectory that not only, for a nucleosome, conforms to the superhelical path of the DNA on the surface of the histone octamer but also disfavours other trajectories. Such anisotropic deformability can be conferred by the occurrence of certain short DNA sequences in helical phase (29-32) or by intrinsic curvature dependent upon phased oligo(dA)(dT) tracts (33, 34). The short sequences facilitate tight bending by enabling the distortion of DNA such that the grooves close to the surface of the octamer are compressed while those facing the solvent are widened. These preferences are reflected in the periodic distributions of the AA/TT and GC dinucleotides in chicken erythrocytes core nucleosomal DNA sequences (30). In this set of sequences the AA/TT base-step occurs preferentially where the minor groove is in contact with the octamer, i.e., on the inside of the DNA bend, and GC and other G/C-containing base steps occurs preferentially where the minor groove points away from the octamer. This pattern is characteristic of at least the outer 6 double-helical turns of the nucleosomal DNA and confers directional bending, i.e., the pattern defines the orientation or rotational positioning of DNA on the histone octamer. Similar sequence preferences are associated with tight bending induced by CRP, FIS, Tn3 resolvase and the BPV E4 protein (35-38). However, around the midpoint of the chicken erythrocyte sequences, corresponding to a position where the minor groove points away from the octamer, the periodic phase of the AA/TT and GC dinucleotides is opposite to that in the outer six turns on each side, leading to the suggestion that this singularity constituted a translational positioning signal (39).
The bending preferences conferred by short nucleotide sequences can be directly related to their preferred conformations. A/T containing sequences, especially those containing sequences such as AA/TT and AAA/TTT can preferentially adopt a conformation with a narrow minor groove (40-43). It is this preferred morphology that determines the preferred orientation of these sequences in bent DNA. Conversely widening of the minor groove is energetically unfavourable. Similarly G/C containing sequences can, in crystal structures preferentially adopt a conformation with a wide minor groove (42-45). Because of the tight curvature of nucleosomal DNA the narrowing of the minor groove to ~3 Å at the contacts with the histone octamer is disfavoured by exocyclic groups, notably the purine 2-amino group, that protrude into the minor groove and likely hinder groove compression (46). The electrostatic potential of the narrow minor groove of A/T containing sequences also favours the binding of arginine residues at inward-facing minor grooves around the histone octamer (43). These observations imply that the periodic modulation of di- and tri-nucleotide frequencies results from the preferential exclusion of these sequences from disfavoured rotational orientations.
Although the sequence dependent bending preferences of DNA are likely important determinants of positional preferences, another determinant of affinity is the flexibility of DNA (47). This parameter is essentially the inverse of the persistence length and is a determinant of the deformation energy required for tight wrapping. While flexibillity per se does not contain positional information it may affect positioning indirectly by distinguishing sequences with similar anisotropic bending preferences. Thus intrinsically curved DNA sequences, such as those conferring phased runs of oligo(dA)(dT), are inherently more rigid than isotropically bendable sequences of high flexibility (48), such as oligo(dAT)(dAT), and consequently occupy a smaller region of configurational space. Consequently the entropic penalty on binding to the octamer would be greater for a highly flexible, isotropically bendable sequence than it would be for an anisotropically deformable sequence. Although intrinsic curvature can undoubtedly influence the affinity of the octamer for DNA (33, 47, 49) the relative contributions of flexibility and anisotropic deformability to octamer binding will be determined by the local thermodynamics of nucleosome organisation.
Variation in Positioning Signals
Although the nature of the sequence-dependence of DNA bending is well established the optimum organization of short sequences conferring anisotropic deformability in octamer binding sites is less clear (50). Variation has been reported in the sequence and structural periodicities in nucleosomal DNA, while sequences associated with well-positioned octamers in yeast differ in character from strong positioning sequences selected by in vitro reconstitution of nucleosomes (3, 51-53). This apparent variation can be attributed to two factors - differences in sampling procedures that result in the analysis of differing sets of nucleosomal DNA sequences and intrinsic differences in the DNA sequences associated with the histone octamer.
Analysis of different sets of budding yeast nucleosomal DNA sequences identified by parallel sequencing and by partial micrococcal nuclease (MNase) cleavage reveal differences in sequence organization. Whereas one set of sequences identified by parallel sequencing is enriched in G/C containing sequences at the midpoint (10) another such set, together with that identified by partial MNase cleavage are enriched in A/T containing sequences at the midpoint (9, 51). Again, while nucleosome core sequences isolated from chicken erythrocytes are enriched in A/T sequences at the centre those isolated from chromatosomes from the same source do not (29, 54). Similarly, preferred octamer sequences identified by reconstitution in vitro also differ. The commonly-used salt dilution protocol at low octamer concentration selects sequences with a G/C rich midpoint and strong periodicities of A/T- and G/C-rich short sequences (3, 52, 53). In contrast, octamer-binding sequences selected on the ovine β-lactoglobulin gene, lack a strong A/T periodicity and instead contain GG/CC and CC/GG (defined with respect to the same DNA strand) periodicities in opposite phases (55). Since there is no reason to doubt that the collated sequences are derived from nucleosomes, the observed differences in preferred sequence organisation could likely arise from differences in sampling procedures and/or from intrinsic differences.
Different sampling procedures can potentially identify different sets of nucleosome-associated DNA sequences. Initially nucleosome positions in chromatin were mapped by a method in which partial micrococcal nuclease (MNase) digestion of chromatin was followed by location of the preferred cleavage sites by indirect end labeling (5-8). A related method uses copper-o-phenanthroline (56), which like MNase preferentially cleaves untwisted DNA but lacks the specificity for A-T base-pairs (57). This method assumes that MNase principally, but not necessarily exclusively, cleaves DNA in chromatin in the linker between nucleosomes. The resulting cleavage pattern is then interpreted as defining a preferred array of nucleosome positions. The major drawbacks of this method are that in general it can only be applied to a limited region (up to 7-10 kb (58)) of DNA and that the nucleosome positions are identified by the length of DNA protected and not, in general, by positive identification of the associated histones.
More recently high-throughput methods have been applied to the organization of genomic positioning patterns (for reviews see refs. 49, 59-62). All these studies first isolate core nucleosome particles by limit MNase digestion followed in some cases by immunoprecipitation of particular populations and then analyse the associated DNA either by tiling arrays (63-65) or by parallel sequencing (9, 10, 53, 66, 67) The data from tiling array analysis provides a quantitative estimate of the probability of a given DNA sequence being contained within a nucleosome. These data are then further processed by Hidden Markov Model methods to identify preferred positions. The resolution of this method depends on the density of the tiling array used. In contrast parallel sequencing directly provides the sequences of DNA molecules associated with the isolated core nucleosomes. By mapping these sequences onto genomic DNA preferred nucleosome positions can be identified. Ideally parallel sequencing in principle provides a means for mapping the relative occupancy of all occupied nucleosome positions on genomic DNA.
The coverage of the genomic position maps obtained by the tiling array and parallel sequencing methods is comparable and for many loci there is impressive agreement between the two methods. But there are also loci where there is significant disagreement both between the high throughput methods and between one or other of these methods and the partial MNase method (50). For example, at some yeast loci different studies identify different nucleosome arrays (for example refs. 9, 10) or different average nucleosome spacings (for example: refs 9, 65, 68 at the URA3 locus). Again mismatches occur between parallel sequencing studies and partial MNase cleavage studies. While some of the mismatches observed may simply result from a lack of resolution resulting from, for example, the averaging of a relatively poorly positioned – or ‘fuzzy” – nucleosome, this consideration is unlikely to be the explanation for different studies identifying differently phased arrays at the same locus.
The disagreements between different methods, although limited, raise the issue of how representative the identified populations of nucleosomes are in relation to the actual population in chromatin. This issue is important because estimates of absolute nucleosome occupancy at a given position require that the analysed population be entirely representative. Another measure of whether different studies isolate equivalent populations is DNA sequence organization. While tiling array studies cannot be assessed for this parameter, both the partial MNase digestion and parallel sequencing methods can. When the populations of nucleosome positions identified in different studies are compared striking differences in the occurrence of A/T containing (AA,AT,TA,TT) and G/C containing (CC,CG,GC,GG) dinucleotides at the sequence midpoint – presumed to correspond approximately to the structural dyad – become apparent (50). Whereas the yeast nucleosomes analysed by the parallel sequencing studies of Field et al., (10) and Kaplan et al., (53) are enriched in G/C containing dinucleotides at the midpoint those analysed by the parallel sequencing studies of Albert et al., (66) and Mavrich et al., (9) are both enriched in A/T containing dinucleotides at the same position (50). By the criterion of sequence organization these two sets of populations are, on average, not equivalent. The yeast nucleosomes identified by partial MNase digestion, albeit a much smaller set, are even more enriched in A/T-containing dinucleotides at the midpoint, the average A/T content at this position being 73% compared with the genomic content of 62% (51).
The differences in average A/T content at the midpoint between sequences obtained by partial and limit MNase digestion could simply result from the genomic limit MNase digestion analyses sampling a far larger set of nucleosomal DNA sequences. However, another possible explanation is that the sequence variations are a consequence of procedural differences. In particular, whereas partial MNase digestion identifies the location of presumed nucleosomes in a preferred array, all the other methods involve limit digestion to core nucleosomes. For the isolated populations to be wholly representative of in vivo chromatin these latter studies all require the implicit assumption that during isolation MNase cleavage either occurs only exterior to core nucleosomal DNA or, that any internal cleavage within core DNA is essentially random. However, since MNase cleavage is highly sequence selective (69-71), with preferred cutting at the TA dinucleotide (10, 71), any internal cleavage by this enzyme would potentially bias the population of nucleosomes isolated (72) and also the population of DNA molecules sequenced. Even if a core nucleosome particle containing internally cleaved DNA were isolated the associated DNA would be shorter than full-length nucleosomal DNA and its sequence would not necessarily be incorporated into a database requiring full-length reads.
Differences in the frequency of A/T and G/C containing dinucleotides at the midpoints of sets of sequences obtained by the parallel sequencing could thus be potentially explained simply by differences in the extent of treatment with MNase during the initial isolation of core nucleosome particles. Indeed an early study of the sequences of chicken erythrocyte core nucleosomal DNA noted that the TA dinucleotide, and not other dinucleotides, was significantly depleted, relative to genomic DNA, in the sequences isolated (30). Although studies routinely control for the sequence selectivity of MNase cleavage at the ends of nucleosomal DNA (9, 10) there is no good experimental measure of the extent of internal cleavage in any population of isolated nucleosomes. A recent study comparing nucleosomal DNA sequences isolated from chromatin with different extents of MNase digestion found that some nucleosomes, notably those in the nucleosome-depleted regions of promoter regions, are particularly sensitive to internal cleavage by MNase while others are relatively resistant (73). This finding accords with the observation that the central region of DNA from promoter nucleosomes identified by partial MNase cleavage is enriched in TA, a preferred site for MNase cleavage (50). A further potential complication is that nucleosomes may change their positions during extensive MNase digestion.
However, not all described differences in the organisation of nucleosomal DNA can be ascribed to differences in sampling. First the nature of the short sequences determining the trajectory is variable. In contrast to periodic repetition of the AA/TT and GC base-steps found in chicken and yeast nucleosomal DNA (10, 30), sequences identified by reconstitution of nucleosomes on ovine DNA do not, on average, exhibit a strong periodic occurrence of AA/TT but instead exhibit periodic occurrences of GG/CC and CC/GG in opposite phases (55). A major difference between AA/TT and GG/CC is that whereas the AA/TT step in protein-DNA complexes has a small or negative roll angle, GG/CC can adopt both positive roll/negative slide and negative roll/positive slide configurations (42). These different geometries are important since when AA/TT is placed where the minor groove points in towards the octamer it will, in conjunction with GC in the opposite rotational orientation, confer a planar bend on the DNA. In contrast the preferred configuration for the roll/slide combination is a superhelix (74). This property was first noted by Tolstorukov et al., (74) for the TA base-step which is preferentially located where the minor groove faces the octamer in high-affinity octamer binding sequences selected in vitro (52). The finding that geometrically distinct types of sequence can direct binding to the octamer raises the issue as to whether the DNA can adopt different trajectories under different assembly conditions. On present evidence this cannot be excluded. Nevertheless it should be noted that the short sequences with a preferred high slide geometry are more typically observed when sequences are selected in vitro using octamers from higher eukaryotes.
Another intrinsic variable is the structural periodicity of nucleosomal DNA which has been directly determined from crystal structures (17, 75) and also from the pattern of pyrimidine dimers formed from pyrimidine-pyrimidine base-steps by UVirradiation of chromatin or isolated NCPs (76). The DNA periodicities in crystal structures of the core nucleosome containing 145, 146 and 147 bp of DNA are respectively 10.15, 10.23 and 10.3 bp (17, 75, 77). This variation is accomplished by keeping the number of double-helical turns constant while incorporating additional DNA base-pairs into ‘stretched’ double-helical turns containing 11 instead of 10 base-pairs. These values are close to the average value of 10.3±0.1 bp for the periodicity of pyrimidine dimers.
In contrast to crystal structures the periodicity values obtained from compilations of sequences associated with both core nucleosomes and chromatosomes are average values for the whole population and do not necessarily reflect that in any particular sequence. These sequence periodicities are generally, but not always, characterised by regular modulations in the frequency of occurrence of A/T and G/C containing di- or tri-nucleotides in opposite phases. The periodicity is usually interpreted as the surface helical repeat of the wrapped DNA (29, 30, 78) and, in principle, is equivalent to the structural repeat measured from crystal structures. For nucleosomal DNA sequences from chicken erythrocyte core particles the average sequence periodicity is ~10.2-10.25 bp (30), again in good agreement with the crystal structures and also with the DNase I digestion profile of core particles of similar provenance (79). However this average value conceals variation of periodicities for different short sequences. For example while the AAA/TTT trinucleotide associated with an inward-facing minor groove has an average periodicity of 10.31 bp that of the GGC/GCC trinucleotide has an average periodicity of 10.15 bp. The correlation of periodicity with base-composition is also observed in other compilations with A/T-containing di- and tri-nucleotides invariably exhibiting a higher periodicity than the corresponding G/C-containing dinucleotides (Table). This effect is apparently independent of the rotational orientation of the short sequences since in core particles reconstituted on ovine DNA the dinucleotides GG/CC and CC/GG have the same periodicity but opposite rotational orientations (54). In general only a small number of the 26-28 optimal rotational positions in a given natural octamer-associated sequence contain DNA-bending signals. Thus a possible explanation for a difference in AA/TT and GC periodicities would be that because A/T-rich sequences are more readily untwisted than G/C-rich sequences there would be a slightly greater frequency of ‘stretched’ double-helical turns between tight histone-DNA contacts. This would result in a higher periodicity. In this situation the sequence periodicities of individual sequences could differ from each other. The variation in observed periodicities also suggests that octamer-bound DNA sequences from chromatin preparations containing linker histone may have a slightly lower periodicity and consequently the DNA exit and entry trajectories could differ from those of core particles. While two analyses agree that the periodic occurrence of the AA/TT and TT/AA base-steps exhibit the same phase (80, 81), other studies, based initially on a library of nucleosomal DNA sequences from different sources and subsequently on the analysis of genomic nucleosome positions in Caenorhabditis elegans, find that the periodic occurrences of AA/TT and TT/AA have opposite phases with a periodicity of 10.4 bp (82, 83). This finding was coupled to the identification of a consensus sequence repeat, YYYYYRRRR, in nucleosomal DNA where the YR step is positioned, on average, where the minor groove points away from the histone octamer. Thus, in this analysis, the phases of AA/TT and TT/AA differ by ±90° from the common phase determined in other analyses of nucleosomal DNA sequences.
Table.
Reported periodicities of nucleosomal DNA
| Reference | Dominant periodicity values | Material and/or region of nucleosomal DNA considered |
|---|---|---|
| Sequence periodicities: | ||
| Satchwell et al., 1986 (30) | 10.15 (GGC) - 10.31 (AAA) bp | chicken NCP, isolation involves exposure to 0.6 M NaCl |
| Satchwell & Travers,1989 (119) | 9.80 (GC) - 10.15 (AA/TT) bp | chicken dinucleosomes with linker histone - low salt isolation |
| Muyldermans & Travers, 1994 (54) | 9.90 (GGC) - 10.20 (AAA) bp | chicken chromatosomes - low salt isolation |
| Fraser et al,, 2009 (55) | 9.90 (GG/CC, CC/GG) bp | salt dilution in vitro reconstitution on ovine DNA |
| Thastrom et al., 2004 (52) | 10.14 (TA) bp | SELEX using salt dilution in vitro reconstitution (periodicity value for central 7 turns) |
| Structural periodicities: | ||
| Klug & Lutter, 1981 (79) | ~10.3 bp | DNase I cleavage periodicity of chicken NCP |
| Gale et al., 1986 (76) | 10.3±0.1 bp | soluble chromatin – with and without linker histone (mean minor groove out periodicity) |
| Davey et al., 2002 (75) | 10.15-10.30 bp | crystal structures for NCPs containining 145-147 bp |
| Richmond & Davey, 2003 (18) | 10.28 bp | central 7 turns from high resolution structure of NCP |
Dominant periodicity values refer to the di- or tri-nucleotides with the highest amplitudes of periodic modulation. NCP, nucleosome core particle(s). If no DNA region is specified the calculated periodicity includes all the wrapped DNA.
Is Positioning in vivo Determined by Histone-DNA Sequence Preferences?
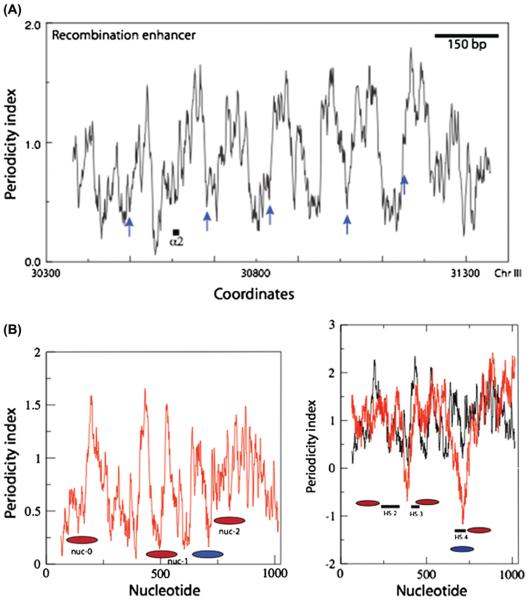
This issue is currently a matter of lively debate with some authors suggesting that DNA sequences play a dominant role in positioning (10) while others reject this view (83). In general parallel sequencing studies identify an, albeit weak, rotational positioning signal of periodic modulations of AA/TT frequency in the population of sequenced nucleosome core DNA molecules (9, 10, 67). In addition a DNA signature characteristic of positioned nucleosomes, particularly those in promoter regions, was char-acterised in a small set of octamer-associated sequences identified in budding yeast by partial MNase digestion (51). This ‘signature’ is based on an analysis of sequence periodicity signals in DNA corresponding to the patterns observed in chicken erythrocyte nucleosome core particle DNA. It has the form of a low periodicity signal at the midpoint of the nucleosomal DNA, flanked on one or both sides by a higher average periodicity (Figure 1). Although initially observed in budding yeast this signature is also found in particular promoter regions of the HIV 5′ LTR (Figure 1). Equally importantly, while there is a preferred sequence organization for DNA wrapped around the histone octamer certain sequences by virtue of their low anisotropic deformability or high average stiffness may have a lower affinity for the histone octamer and may exclude octamer binding under in vivo conditions. Thus in vitro poly(dA)(dT) is a poor ligand for the octamer (85) and oligo(dA)(dT) tracts create barriers for octamer binding (86). In budding yeast such tracts are found preferentially in nucleosome-depleted regions (87, 88). Similarly, some yeast UAS regions contain potentially stiff sequences with a high average stacking energy (51). Taking both positive and negative factors into account the organization of nucleosomes in budding yeast and in Drosophila can be accurately predicted in terms of general location by using an algorithm that considers only the physicochemical properties of DNA (89). Together these observations suggest that histone-DNA interactions are a determinant, and possibly a major one, of positioning in vivo but not necessarily the only one.
Figure 1.
DNA periodicity profiles. The periodicity index Is a measure of coherence of periodicity patterns of e.g., AA/TT and GC over a given length of DNA, usually empirically 50 bp. A helical periodicity of ~10 bp is assumed. If direction of bending changes, the phase of the periodic sequence patterns will change and will interfere with phase of adjacent sequences. A high value of the index indicates the sequence periodicities are strong and coherent over the window chosen. (A) Saccharomyces cerevisiae recombination enhancer (reproduced with permission from ref. 51). Blue arrows indicate centres of positions mapped by partial MNase digestion. Coordinates refer to the nucleotide positions in the appropriate chromosome. (B) 5′ LTR of HIV. In vivo (red ellipses) and in vitro (blue ellipse) nucleosome positions for HIV are taken from ref. 120. Nucleotide indicates the numbers of nucleotides from the 5′ end of the LTR. Lh panel: periodicity profile; rh panel: stacking energy superimposed on periodicity profile. Note that regions of high stacking energy (high negative ΔG) fall within or flank nuclease hypersensitive sites (HS).
By comparing nucleosome positions in yeast chromatin mapped by parallel sequencing with those preferentially occupied on yeast genomic DNA using in vitro reconstitution Kaplan et al., (53) concluded that histone-DNA interactions were a dominant determinant of positioning in vivo. In contrast using similar technique Zhang et al., (84) came to the opposite conclusion. These conclusions relate primarily to translational positioning. The conclusions drawn from the comparisons of in vivo and in vitro positions are subject to two main caveats: the influence of MNase on the sampling of the population, as discussed above, and the assumption that the relative affinities of different DNA sequences for the histone octamer are the same under the chosen in vitro reconstitution conditions as in vivo.
The nature of the problem can be simply defined. The histone octamer, or tetramer, should be regarded as a DNA-binding entity that lacks the capacity for base-specific recognition (17). Instead the selectivity of octamer binding in vitro, and most likely also in vivo, is determined in part, not by a particular defined sequence, but by the overall physicochemical properties of the DNA sequence bound (42, 80). These physicochemical properties comprise not only the anisotropic or directional deformability (27, 28), and the flexibility, but also the torsional flexibility. In practice this implies, at least in vitro, that the affinity of the octamer for different DNA sequences, should spread over a wide range, as indeed is observed (for example, refs. 52, 90). Given a sufficiently large number of different DNA sequences this range of affinities would become a continuum. This phenomenon is also apparent in the DNA-binding properties of certain abundant bacterial proteins that organise the DNA in the bacterial nucleoid and which possess little, if any, capacity for direct base-specific recognition. For example, H-NS, which can repress transcription by stabilising DNA plectonemes (91, 92) and was long considered to bind non-specifically to DNA, has recently been shown, like the histone octamer (3), to bind with high affinity in vitro to sites that nucleate cooperative binding. Another such DNA-bending protein, FIS, with likely only limited base-specific recognition, again binds to different DNA sequences over a wide range of affinity (36).
An additional, and largely neglected, consequence of indirect recognition in DNA-protein interactions is that since the physicochemical properties of DNA are directly influenced by local changes in its environment, so also will be the interaction of DNA with, for example, the histone octamer. Thus both temperature and DNA supercoiling influence the torsional properties of DNA, and hence the elastic constants for unwinding and bending, while water activity and temperature can influence anisotropic deformability by affecting the structure of the relatively rigid oligo(dA)(dT) tracts (93-95). More pertinently contact with histone octamer will likely lower the elastic constants for DNA bending and also possibly for torsion (47). In the context of nucleosome positioning this means that the relative affinities of different DNA sequences for the octamer are not fixed but instead will depend on the precise conditions of nucleosome formation. Indeed, the sequence binding preferences of the octamer in vitro have been shown to be dependent on both octamer concentration and temperature (48).
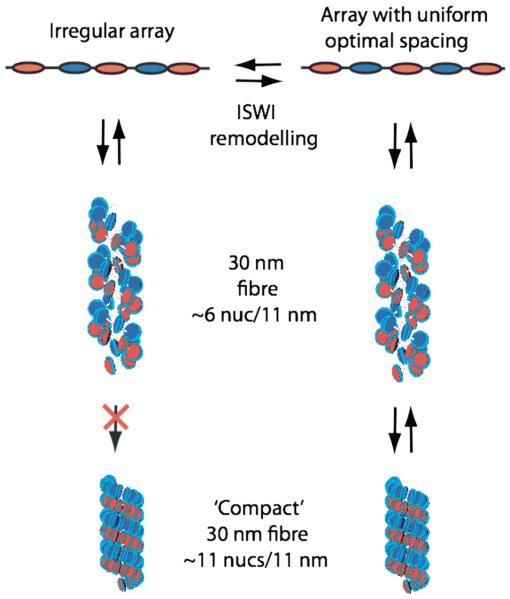
A more realistic way of considering the nucleosome positioning problem might be to view the octamer/DNA interaction in terms of a tunable energy landscape that is sensitive to variables such as temperature and applied torque. In principle these variables could act on the DNA, or the octamer, or both. Discrepancies between positions mapped in vitro and in vivo on the same DNA sequences (96, 97) could thus simply arise because the energy landscapes for nucleosome formation are different in the two cases. Indeed a much closer correspondence between in vivo and in vitro positions has been achieved by reconstituting budding yeast chromatin in the presence of ATP and a crude extract from yeast cells (97). This contrasts with in vitro reconstitution methods including the classic salt dilution protocol (86, 96) or the ACF remodelling complex (84). This is particularly pertinent in the context of positioning in vivo where positioning is maintained by chromatin remodelling complexes and RNA polymerase II activity. Removal of remodelling activities in vivo in yeast results in changes in nucleosome positions (98, 99) while the packaging of promoter DNA in nucleosomes can be dependent on transcriptional activity (70, 100). One remodelling complex, RSC, cradles a nucleosome core particle in a pocket (101-103), creating a topologically constrained DNA microdomain (104). The DNA translocase activity of the remodeling complex applies torque to the core particle which can be manifest as local changes in DNA superhelicity (105, 106). Such torque could affect the energy landscape of both the wrapped DNA and that in its immediate vicinity, potentially changing the preferred position of the octamer by altering the local thermodynamic equilibrium, as well as kinetically facilitating octamer sliding. In principle different remodellers could apply different amounts of torque and thereby stabilise different positions and in some cases, for example the ACF and certain other ISWI-containing complexes, could by constraining the inter-nucleosome distance relocate octamers to positions that are not necessarily energetically favoured by the physicochemical properties of DNA (106-109). In particular, in a repeated array of a strong positioning sequence positioning may be dominated by cation-dependent spacing as well as by the effects of DNA topology (110). In these examples the regular spacing generated by the remodelling complexes might stabilise a more regular nucleosome packing in the form of a compact 30 nm fibre (Figure 2).
Figure 2.
Coupling of nucleosome positioning and chromatin compaction. An array with irregular nucleosome spacing can, in principle, fold to form a 30 nm fibre with a packing density of ~6 nucs/11 nm. When the nucleosomes are regularly spaced at ‘optimal’ distances (121) tight stacking of adjacent nucleosomes can occur. With irregular spacing tight stacking is precluded because the nucleosomes would be inclined at differing orientations to the superhelical axis. The 30 nm fibres are represented as 2-start structures with the two helical stacks of nucleosomes coloured in red and blue.
The Organisation of Nucleosomes in vivo
The elucidation of genome-wide positioning maps - for the budding yeast Saccharomyces cerevisiae, Drosophila melanogaster, Caenorhabditis elegans and Homo sapiens - has revealed some strong similarities in nucleosome organization (9, 10, 53, 63-67, 111, 112). Close to the transcription site there is a nucleosome-depleted or nucleosome-free region flanked downstream, and often upstream, by a well-positioned nucleosome. In many cases these nucleosomes constitute the start of regular arrays in which positioning becomes less distinct or ‘fuzzier’ as the distance from the well-positioned nucleosome increases (9). Another complication is the arrays may not be unique in that alternative overlapping arrays on the same DNA sequence have been observed, particularly in Caenorhabditis elegans and at the CUP1 and HIS3 loci in budding yeast (111, 113, 114).
These patterns address the question raised by Kornberg (115, 116) as to whether nucleosome positioning in vivo is ‘statistical’ or specified entirely by the physicochemical properties of the DNA sequence. Statistical positioning postulates the presence of a ‘boundary’ nucleosome which specifies one end of an array which is not determined by the physicochemical properties of DNA sequence. The boundary nucleosome could be positioned either by a protein or by a strong intrinsic positioning sequence or by both. This model is strongly supported by the observed organisation patterns in vivo (9, 64). In budding yeast nucleosomes can be ordered by a protein - such as the budding yeast α2 repressor (7). Similarly in vitro reconstitution argues that energetically unfavourable DNA barriers can order nucleosomes (86) - again in agreement with the statistical positioning model. Additionally the ‘DNA signature’, based on sequence periodicity patterns, occurs preferentially in promoter-associated nucleosomes in a small set of nucleosomal DNA sequences identified by partial MNase cleavage (51). This signature is often less prominent downstream of the promoter but in some cases it is found within nucleosome arrays - for example at the recombination enhancer locus and at the RVS167/SAC7 locus (51). Because the length of the DNA signature associated with a nucleosome is variable and often quite short such a motif can, and in one well characterised example is, compatible with the positioning of overlapping arrays (117). Nucleosome organization in budding yeast can thus broadly be described in terms of a statistical model but containing additional elements associated with a defined nucleosomal sequence organization. The extent to which these ‘natural’ positioning elements tune, or even merely fine tune, nucleosome organization remains to be established.
This research was reported by the author in part at Albany 2009: The 16th Conversation (118).
References and Footnotes
- 1.Simpson RT, Stafford DW. Proc Natl Acad Sci USA. 1983;80:51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Piña B, Bruggemeier U, Beato M. Cell. 1990;60:719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- 3.Lowary PT, Widom J. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 4.Flaus A, Luger K, Tan S, Richmond TJ. Proc Natl Acad Sci USA. 1996;93:1370–1375. doi: 10.1073/pnas.93.4.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Almer A, Hörz W. Embo J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richard-Foy H, Hager GL. Embo J. 1987;6:2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimizu M, Roth SY, Szent-Gyorgyi C, Simpson RT. Embo J. 1991;10:3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verdone L, Camilloni G, Di Mauro E, Caserta M. Mol Cell Biol. 1996;16:1978–1988. doi: 10.1128/mcb.16.5.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. PLoS Comp Biol. 2008;4:1–25. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han M, Grunstein M. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 12.Schlissel MS, Brown DD. Cell. 1984;37:903–913. doi: 10.1016/0092-8674(84)90425-2. [DOI] [PubMed] [Google Scholar]
- 13.Compton JL, Bellard M, Chambon P. Proc Natl Acad Sci USA. 1976;73:4382–4386. doi: 10.1073/pnas.73.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas JO, Thompson RJ. Cell. 1977;10:633–640. doi: 10.1016/0092-8674(77)90096-4. [DOI] [PubMed] [Google Scholar]
- 15.Bernardi F, Zatchej M, Thoma F. Embo J. 1992;11:1177–1185. doi: 10.1002/j.1460-2075.1992.tb05158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McManus J, Perry P, Sumner AT, Wright DM, Thomson EJ, Allshire RC, Hastie ND, Bickmore WA. J Cell Sci. 1994;107:469–486. doi: 10.1242/jcs.107.3.469. [DOI] [PubMed] [Google Scholar]
- 17.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 18.Richmond TJ, Davey CA. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 19.Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A. Nature. 1984;311:532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- 20.Hagerman PJ. Biopolymers. 1981;20:1503–1535. doi: 10.1002/bip.1981.360200710. [DOI] [PubMed] [Google Scholar]
- 21.Levene SD, Wu H-M, Crothers DM. Biochemistry. 1986;25:3988–3995. doi: 10.1021/bi00362a003. [DOI] [PubMed] [Google Scholar]
- 22.Sobel ES, Harpst JA. Biopolymers. 1991;31:1559–1564. doi: 10.1002/bip.360311311. [DOI] [PubMed] [Google Scholar]
- 23.Pörschke D. Biophys Chem. 1991;40:169–179. doi: 10.1016/0301-4622(91)87006-q. [DOI] [PubMed] [Google Scholar]
- 24.Crothers DM, Drak J, Kahn JD, Levene SD. Methods Enzymol. 1992;212:3–29. doi: 10.1016/0076-6879(92)12003-9. [DOI] [PubMed] [Google Scholar]
- 25.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Proc Natl Acad Sci USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang MD, Yin H, Landick R, Gelles J, Block SM. Biophys J. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhurkin VB, Lysov YP, Ivanov V. Nucleic Acids Res. 1979;6:1081–1096. doi: 10.1093/nar/6.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trifonov EN. Nucleic Acids Res. 1980;8:4041–4053. doi: 10.1093/nar/8.17.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drew HR, Travers AA. J Mol Biol. 1985;186:773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- 30.Satchwell SC, Drew HR, Travers AA. J Mol Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 31.Shrader TE, Crothers DM. Proc Natl Acad Sci USA. 1989;86:7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, Wang JZ, Widom J. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wada-Kiyama Y, Kuwabara K, Sakuma Y, Onishi Y, Trifonov EM, Kiyama R. FEBS Lett. 1999;444:117–124. doi: 10.1016/s0014-5793(99)00041-1. [DOI] [PubMed] [Google Scholar]
- 34.Wolffe AP, Drew HR. Proc Natl Acad Sci USA. 1989;86:9817–9821. doi: 10.1073/pnas.86.24.9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Travers AA, Klug A. Nature. 1987;324:280–281. doi: 10.1038/327280a0. [DOI] [PubMed] [Google Scholar]
- 36.Lazarus LR, Travers AA. Embo J. 1993;12:2483–2494. doi: 10.1002/j.1460-2075.1993.tb05903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blake DG, Boocock MR, Sherratt DJ, Stark WM. Curr Biol. 1995;5:1036–1046. doi: 10.1016/s0960-9822(95)00208-9. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Xi Z, Hegde RS, Shakked Z, Crothers DM. Proc Natl Acad Sci USA. 2004;101:8337–8341. doi: 10.1073/pnas.0402319101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Travers AA. Trends Biochem Sci. 1987;12:108–112. [Google Scholar]
- 40.Dickerson RE, Drew HR. J Mol Biol. 1981;149:761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- 41.Nelson HCM, Finch JT, Luisi BF, Klug A. Nature. 1987;330:221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- 42.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. Proc Natl Acad Sci USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohs R, West SM, Sosinsky A, Liu P, Mannand RS, Honig B. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCall M, Brown T, Kennard O. J Mol Biol. 1985;183:385–396. doi: 10.1016/0022-2836(85)90009-9. [DOI] [PubMed] [Google Scholar]
- 45.Goodsell DS, Kopka ML, Cascio D, Dickerson RE. Proc Natl Acad Sci USA. 1993;90:2930–2934. doi: 10.1073/pnas.90.7.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buttinelli M, Minnock A, Panetta G, Waring M, Travers A. Proc Natl Acad Sci USA. 1998;95:8544–8549. doi: 10.1073/pnas.95.15.8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Virstedt J, Berge T, Henderson RM, Waring MJ, Travers AA. J Struct Biol. 2004;148:66–85. doi: 10.1016/j.jsb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 48.Wu C, Travers A. Biochemistry. 2005;44:14329–14334. doi: 10.1021/bi050915w. [DOI] [PubMed] [Google Scholar]
- 49.Laundon CH, Griffith JD. Cell. 1988;52:545–549. doi: 10.1016/0092-8674(88)90467-9. [DOI] [PubMed] [Google Scholar]
- 50.Travers A, Caserta M, Churcher M, Hiriart E, Di Mauro E. Mol Biosystems. 2009;5:1582–1592. doi: 10.1039/b907227f. [DOI] [PubMed] [Google Scholar]
- 51.Caserta M, Agricola E, Churcher M, Hiriart E, Verdone L, Di Mauro E, Travers A. Nucleic Acids Res. 2009;37:5309–5321. doi: 10.1093/nar/gkp574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thåström A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. J Mol Biol. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 53.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muyldermans S, Travers AA. J Mol Biol. 1994;191:855–870. doi: 10.1006/jmbi.1994.1044. [DOI] [PubMed] [Google Scholar]
- 55.Fraser RM, Keszenman-Pereyra D, Simmen MW, Allan J. J Mol Biol. 2009;390:292–305. doi: 10.1016/j.jmb.2009.04.079. [DOI] [PubMed] [Google Scholar]
- 56.Gencheva M, Boa S, Fraser R, Simmen MW, Whitelaw CBA, Allan J. J Mol Biol. 2006;361:216–230. doi: 10.1016/j.jmb.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 57.Schaeffer F, Rimsky S, Spassky A. J Mol Biol. 1996;260:523–539. doi: 10.1006/jmbi.1996.0419. [DOI] [PubMed] [Google Scholar]
- 58.Fleming AB, Pennings S. Embo J. 2001;20:5219–5231. doi: 10.1093/emboj/20.18.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang C, Pugh BF. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rando OJ, Chang HY. Annu Rev Biochem. 2009;78:245–271. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Segal E, Widom J. Nat Rev Genet. 2009;10:443–456. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stein A, Takasuka TE, Collings CK. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp1043. doi:10.1093/nar/gkp1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guillemette B, Bataille AR, Gévry N, Adam M, Blanchette M, Robert F, Gaudreau L. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan G, Liu Y, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 65.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. Nature Gen. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 66.Albert I, Mavrich TN, Tomsho LP, Zanton SJ, Schuster SC, Pugh BF. Nature. 2007;446:572–578. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 67.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, Gilmour DS, Albert I, Pugh BF. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanaka S, Livingstone-Zatchej M, Thoma F. J Mol Biol. 1996;257:919–934. doi: 10.1006/jmbi.1996.0212. [DOI] [PubMed] [Google Scholar]
- 69.Hörz W, Altenburger W. Nucleic Acids Res. 1981;9:2643–2568. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dingwall C, Lomonossof GP, Laskey RA. Nucleic Acids Res. 1981;9:2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Flick JT, Eissenberg JC, Elgin SCR. J Mol Biol. 1986;190:619–633. doi: 10.1016/0022-2836(86)90247-0. [DOI] [PubMed] [Google Scholar]
- 72.McGhee JD, Felsenfeld G. Cell. 1983;32:1205–1215. doi: 10.1016/0092-8674(83)90303-3. [DOI] [PubMed] [Google Scholar]
- 73.Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tolstorukov MY, Colasanti AV, McCandlish DM, Olson WK, Zhurkin VB. J Mol Biol. 2007;371:725–738. doi: 10.1016/j.jmb.2007.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davey CA, Sargent DF, Luger K, Mäder AW, Richmond TJ. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 76.Gale JM, Nissen KA, Smerdon MJ. Proc Natl Acad Sci USA. 1987;84:6644–6648. doi: 10.1073/pnas.84.19.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Davey CA, Richmond TJ. Proc Natl Acad Sci USA. 2002;99:11169–11174. doi: 10.1073/pnas.172271399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.White JH, Cozzarelli NR, Bauer WR. Science. 1988;241:323–327. doi: 10.1126/science.3388041. [DOI] [PubMed] [Google Scholar]
- 79.Klug A, Lutter LC. Nucleic Acids Res. 1981;9:4267–4283. doi: 10.1093/nar/9.17.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Travers AA, Klug A. Phil Trans Roy Soc Lond B. 1987;317:537–561. doi: 10.1098/rstb.1987.0080. [DOI] [PubMed] [Google Scholar]
- 81.Wang J-PZ, Widom J. Nucleic Acids Res. 2005;22:6743–6755. doi: 10.1093/nar/gki977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Salih F, Salih B, Trifonov EN. J Biomol Struct Dyn. 2008;26:273–281. doi: 10.1080/07391102.2008.10531241. [DOI] [PubMed] [Google Scholar]
- 83.Gadbank I, Barash D, Trifonov EN. J Biomol Struct Dyn. 2009;26:403–412. doi: 10.1080/07391102.2009.10507255. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Struhl K. Nat Struct Mol Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rhodes D. Nucleic Acids Res. 1979;6:1805–16. doi: 10.1093/nar/6.5.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Milani P, Chevereau G, Vaillant C, Audit B, Haftek-Terreau Z, Marilley M, Bouvet P, Argoul F, Arneodo A. Proc Natl Acad Sci USA. 2009;108:22257–22262. doi: 10.1073/pnas.0909511106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iyer V, Struhl K. Embo J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Segal E, Widom J. Curr Opin Struct Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miele C, Vaillant Y, d'Aubenton-Carafa C, Thermes C, Grange T. Nucleic Acids Res. 2008;36:3746–3756. doi: 10.1093/nar/gkn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Widlund HR, Cao H, Simonsson S, Magnusson E, Simonsson T, Nielsen PE, Kahn JD, Crothers DM, Kubista M. J Mol Biol. 1997;267:807–817. doi: 10.1006/jmbi.1997.0916. [DOI] [PubMed] [Google Scholar]
- 91.Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. Nat Struct Mol Biol. 2007;14:441–448. doi: 10.1038/nsmb1233. [DOI] [PubMed] [Google Scholar]
- 92.Lang B, Blot N, Bouffartigues E, Buckle M, Geertz M, Gualerzi CO, Mavathur R, Muskhelishvili G, Pon CL, Rimsky S, Stella S, Babu MM, Travers A. Nucleic Acids Res. 2007;35:6330–6337. doi: 10.1093/nar/gkm712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Diekmann S. Nucleic Acids Res. 1987;15:247–265. doi: 10.1093/nar/15.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koo HS, Crothers DM. Biochemistry. 1987;26:3745–3748. doi: 10.1021/bi00386a070. [DOI] [PubMed] [Google Scholar]
- 95.Drew HR, Travers AA. Cell. 1984;37:491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- 96.Wippo CJ, Krstulovic BS, Ertel F, Musladin S, Blaschke D, Stürzl S, Yuan GC, Hörz W, Korber P, Barbaric S. Mol Cell Biol. 2009;29:2960–2981. doi: 10.1128/MCB.01054-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Korber P, Hörz W. J Biol Chem. 2004;279:35113–35120. doi: 10.1074/jbc.M405446200. [DOI] [PubMed] [Google Scholar]
- 98.Xella B, Goding C, Agricola E, Di Mauro E, Caserta M. Mol Microbiol. 2006;59:1531–1541. doi: 10.1111/j.1365-2958.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- 99.Whitehouse I, Tsukiyama T. Nat Struct Mol Biol. 2006;13:633–640. doi: 10.1038/nsmb1111. [DOI] [PubMed] [Google Scholar]
- 100.Venters BJ, Pugh BF. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Asturias FJ, Chung WH, Kornberg RD, Lorch Y. Proc Natl Acad Sci USA. 2002;99:13477–13480. doi: 10.1073/pnas.162504299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chaban Y, Ezeokonkwo C, Chung WH, Zhang F, Kornberg RD, Maier-Davis B, Lorch Y, Asturias FJ. Nat Struct Mol Biol. 2008;15:1272–1277. doi: 10.1038/nsmb.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Leschziner AE, Saha A, Wittmeyer J, Zhang Y, Bustamante C, Cairns BR, Nogales E. Proc Natl Acad Sci USA. 2007;104:4913–4918. doi: 10.1073/pnas.0700706104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Muskhelishvili G, Travers A. Nucleic Acids Mol Biol. 1997;11:179–190. [Google Scholar]
- 105.Lia G, Praly E, Ferreira H, Stockdale C, Tse-Dinh YC, Dunlap D, Croquette V, Bensimon D, Owen-Hughes T. Mol Cell. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C. Mol Cell. 2006;24:559–68. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT. Genes Dev. 2004;18:170–183. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ. Nat Struct Mol Biol. 2006;13:1078–1083. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]
- 109.Racki LR, Yang JG, Naber N, Partensky PD, Acevodo A, Purcell TJ, Cooke R, Cheng Y, Narlikar GJ. Nature. 2009;460:1016–1021. doi: 10.1038/nature08621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Blank T, Becker PB. J Mol Biol. 1995;260:1–6. doi: 10.1006/jmbi.1996.0377. [DOI] [PubMed] [Google Scholar]
- 111.Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, Sidow A, Fire A, Johnson SM. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shen CH, Clark DJ. J Biol Chem. 2001;276:35209–35216. doi: 10.1074/jbc.M104733200. [DOI] [PubMed] [Google Scholar]
- 114.Kim Y, McLaughlin N, Lindstrom K, Tsukiyama T, Clark DJ. Mol Cell Biol. 2006;26:8607–8622. doi: 10.1128/MCB.00678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kornberg R. Nature. 1981;292:279–280. doi: 10.1038/292579a0. [DOI] [PubMed] [Google Scholar]
- 116.Kornberg RD, Stryer L. Nucleic Acids Res. 1988;16:6677–6690. doi: 10.1093/nar/16.14.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Costanzo G, di Mauro E, Negri R, Pereira G, Hollenberg C. J Biol Chem. 1995;270:11091–11097. doi: 10.1074/jbc.270.19.11091. [DOI] [PubMed] [Google Scholar]
- 118.Abstracts of Albany 2009: 16th Conversation. June 16-20, Albany, New York, USA. Caserta M, Agricola E, Churcher M, Hiriart E, Verdone L, Di Mauro E, Travers A. A Translational Signature for Nucleosome Positioning In vivo, Abstract #179. J Biomol Struct Dyn. 2009;26:787–927. [Google Scholar]
- 119.Satchwell SC, Travers AA. Embo J. 1989;8:229–238. doi: 10.1002/j.1460-2075.1989.tb03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pazin MJ, Sheridan PL, Cannon K, Cao Z, Keck JG, Kadonaga JT, Jones KA. Genes Dev. 1996;10:37–49. doi: 10.1101/gad.10.1.37. [DOI] [PubMed] [Google Scholar]
- 121.Widom J. Proc Natl Acad Sci USA. 1992;89:1095–1099. doi: 10.1073/pnas.89.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]