Abstract
We have previously reported hepatitis C virus (HCV) replication using a novel binary expression system in which mammalian cells were transfected with a T7 polymerase-driven full-length genotype 1a HCV cDNA plasmid (pT7-flHCV-Rz) and infected with vaccinia-T7 polymerase. We hypothesized that the use of replication-defective adenoviral vectors expressing T7 (Ad-T7pol) or cell lines stably transfected with T7 (Huh-T7) would alleviate cell toxicity and allow for more sustained HCV replication.
CV-1, Huh7, and Huh-T7 cells were transfected with pT7-flHCV-Rz and treated with Ad-T7pol (CV-1 and Huh7 only). Protein and RNA were harvested from cells on days 1, 2, 3, 5, 7, and 9 post-infection. No cytotoxicity was observed at 9 days post-infection in any cell type. HCV positive- and negative-strand RNA expression were strongest during days 1–3 post-infection; however, HCV RNA remained detectable throughout the 9-day observation period. Furthermore, transfection with a replication-incompetent plasmid suggested that efficient HCV replication is dependent upon NS5B gene expression. Finally, after 1–2 days of IFN treatment, HCV positive-strand levels decreased significantly compared to HCV-infected but untreated samples (p < 0.05).
In conclusion, these refined binary systems offer more durable and authentic models for identification of host cellular processes critical to HCV replication and will permit longer-term analysis of virus–host interactions critical to HCV pathogenesis and the treatment of genotype 1 infections.
Keywords: Hepatitis C virus, HCV, Replication, Genotype 1, Adenovirus vector, Huh-T7
1. Introduction
Hepatitis C virus (HCV) is a leading cause of chronic liver disease, including hepatitis, cirrhosis, and hepatocellular carcinoma (Alter et al., 1999). The combination of interferon (IFN) and ribavirin (RBV) is the standard treatment for chronic HCV infection; however, their effectiveness remains limited (McHutchison and Poynard, 1999). The lack of a full-length HCV tissue culture model has limited not only the ability to screen novel antiviral agents but also the ability to precisely characterize the antiviral effect of IFN, particularly against genotype 1 infections.
We recently reported successful cell-based HCV replication using a novel binary expression system in which mammalian cells were transfected with a T7 polymerase-driven full-length genotype 1a HCV cDNA plasmid and infected with a recombinant vaccinia vector encoding T7 polymerase (Chung et al., 2001). However, HCV replication driven by vaccinia-based vectors is restricted to short-term studies due to the cytotoxic effects of vaccinia. Moreover, vaccinia encodes two proteins, E3L (Chang et al., 1992; Watson et al., 1991) and K3L (Carroll et al., 1993; Gale et al., 1996), that act as potent inhibitors of the IFN-induced double-stranded RNA-activated protein kinase (PKR). Due to these limitations, we sought to further refine our HCV replication model using alternative, less disruptive modes of T7 polymerase delivery.
We hypothesized that the use of replication-defective adenoviral vectors expressing T7 or cell lines stably expressing T7 would alleviate cell toxicity and allow for more sustained HCV replication.
Recombinant replication-defective adenoviral vectors have comparable infectivity to vaccinia vectors. These adenoviral vectors cannot replicate inside infected cells, because they lack the E1A and E1B proteins necessary for viral vector replication. Moreover, these vectors lack the E3 gene that inhibits immune responses by interacting with cytoplasmic MHC class-I molecules (Wold and Gooding, 1989). Thus, adenoviral vectors maintain infectivity and protein delivery with minimal cytotoxicity. By transfecting the HCV cDNA construct into Huh7 cell lines stably expressing T7 polymerase (Huh-T7) (Schultz et al., 1996), the need for viral delivery systems was removed altogether. Using these alternative delivery methods, we have established a refined HCV replication model that produces more sustained viral RNA replication, leads to less perturbation of host genes, and represents a more authentic system for studying virus–host interactions relevant to HCV pathogenesis. These refined models were also utilized to characterize the antiviral kinetics of IFN on HCV replication.
2. Materials and methods
2.1. Cell lines
CV-1 cells (American Type Culture Collection, Manassas, VA) and Huh7 (Dr. Robert Lanford, Southwest Foundation for Biomedical Research) and Huh-T7 (Dr. Stanley Lemon, University of Texas) (Schultz et al., 1996) were maintained in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum.
2.2. Plasmids and transfection-infection
The binary replication system has been described previously and is capable of successful positive-strand and negative-strand HCV RNA synthesis, efficient HCV protein production, and quasispecies generation (Chung et al., 2001; Contreras et al., 2002). Briefly, a plasmid containing the infectious full-length genotype 1 cDNA sequence corresponding to the H77 prototype strain (Yanagi et al., 1997) was adapted at its 5′ and 3′ termini with the T7 promoter and a hepatitis delta virus ribozyme sequence, respectively, to yield pT7-flHCV-Rz (hereafter referred to as H77). As a negative control, a mutant plasmid in which the GDD active site of the NS5B RNA-dependent RNA polymerase (RdRp) was mutated to AAG (hereafter referred to as H77GDD→AAG) was generated by site directed mutagenesis (Quick Change; Stratagene; La Jolla, CA). This substitution is associated with replication-incompetence in replicon models (Blight et al., 2000). H77 and H77GDD→AAG were used to transfect CV-1, Huh7, or Huh-T7 cells at 70% confluency on 6-well plates with Lipofectamine (Invitrogen, Carlsbad, CA). Plasmids were transfected at concentrations of 1µg/well for CV1 cells and 3µg/well for Huh7 and Huh-T7 cells. Transfection efficiency was assessed by co-transfection with 0.1 µg/well of phRL-TK (Int−) (Promega, Madison, WI) and luciferase activity quantified using the Dual-Luciferase reporter assay system (Promega). For CV-1 and Huh7 cells, T7 polymerase was delivered using a recombinant vaccinia virus vector (vTF7-3) (Fuerst et al., 1986) or a recombinant adenovirus vector (Ad-T7pol) 24 h after H77 transfection. In control experiments, a replication-defective adenovirus vector lacking the T7 polymerase gene (Ad-Psi5) was used. Adenoviral vectors were provided by the Harvard Gene Therapy Initiative’s Viral Vector core (Boston, MA).
2.3. X-gal staining of pOS8-transfected cells
To compare the transfection and infection efficiency of the vaccinia and adenovirus vectors, the pOS8 plasmid, which contains a T7 promoter flanking the β-galactosidase gene, was cotransfected into cells. After 48 h, cultured cells were washed with PBS, fixed with 0.25% glutaraldehyde for 1 h at 4 °C, and stained with 0.1% 5-bromo-5-chloro-3-indolyl-β-d-galactopyranoside (X-gal) as described previously (Hiasa et al., 1998; Miyake et al., 1996).
2.4. Interferon experiments
Interferon alpha 2b was obtained from Schering Plough (Kenilworth, NJ). For CV-1 and Huh7 cells, 100–1000 IU/mL of IFN was added 5 h after infection with adenovirus vector. For Huh-T7 cells, 100–1000 IU/mL of IFNwas added 5 h after transfection with H77. Medium with or without IFN was changed at day 1 post-infection and every 2 days thereafter.
2.5. Cellular RNA extraction and qualitative strand-specific rTth RT-PCR
Cells were washed three times with phosphate-buffered saline. RNA was extracted using TRIzol (Invitrogen; Carlsbad, CA), and treated two times for 4 h with DNase I using the DNA-free kit (Ambion; Austin, TX) following the manufacturer’s protocol. RNA was quantified by UV spectrum analysis, and adjusted to 0.3 µg/µL. HCV RNA was detected utilizing a previously described qualitative strand-specific rTth reverse transcription PCR (RT-PCR) assay (Castet et al., 2002; Lanford et al., 1995). For detection of negative-strand HCV RNA, 1 µg of RNA in 10 µL of dietyl pyrocarbonate-treated water was layered with mineral oil and heated at 95 °C for 1 min, and lowered to 70 °C. A 20 µ L mixture containing 10 pM of HCV-II sense primer (5′-CAC TCC CCT GTG AGG AAC T-3′, nucleotides [nt] 38–56 of the 5′UTR) (Laskus et al., 1997), 1 × RT buffer (Applied Biosystems; Foster City, CA), 1mM MnCl2, 200 µM (each) deoxynucleoside triphosphate, and 5U of rTth enzyme (Applied Biosystems) was then added. The temperature was dropped to 60 °C for 2 min for annealing and then raised to 70 °C for 20 min for the cDNA reaction. To inactivate the RT activity of rTth, chelating buffer (Applied Biosystems) was added. Forty microliters of the prewarmed PCR mixture containing 10 pM of HCV-I antisense primer (5′-TGG ATG CAC GGT CTA CGA GAC CTC-3′, nt 342–320 of the 5′UTR) (Laskus et al., 1997) and 3.75 mM MgCl2 was added. Twenty-five cycles of PCR (94 °C 15s, 58 °C 30s, 72 °C 30 s) were performed.
For GAPDH measurements, RT was carried out using an oligo d(T)16 primer under standard conditions (Hiasa et al., 2003). The cDNA product was subjected to 25 cycles of PCR (95 °C 1 min, 60 °C 2 min, 73 °C 2 min), using 50 pM of the GAPDH sense and antisense primers (forward primer 5′-GAA GGT GAA GGT CGG AGT-3′, reverse primer 5′-GAA GAT GGT GAT GGG ATT TC-3′), 0.1mM of each dNTP, 2.5mM MgCl2, and 0.5U Taq polymerase. Reaction products were separated on 1.5 % agarose gels.
To ensure efficient removal of plasmid DNA after DNase I treatment, a qualitative PCR was performed. The plasmid DNA was completely digested as no PCR products were observed using this approach.
2.6. RNase protection assay
Antigenomic HCV RNA was detected as described previously (Chung et al., 2001). Briefly, utilizing the sense-oriented [α-32P] UTP-labeled probe (corresponding to 98 nucleotides of the 3′ terminal HCV genome), antigenomic RNA was generated by in vitro transcription using T7 polymerase from the vector pHCV-3′T (Chung and Kaplan, 1999). Transcripts were generated using the RPA III kit according to the manufacturer’s instructions (Ambion).
2.7. Real-time quantification of HCV positive- and negative-strand RNA
Positive- and negative-strand HCV RNAs were quantified by real-time PCR using LightCycler technology (Roche Diagnostics, Mannheim, Germany) and SYBR green I dye as described previously (Blackard et al., 2005). One microgram of RNA was used for cDNA synthesis in a mixture containing 5U of rTth and 10 pM of the appropriate RT primer (HCV-I for positive-strand HCV RNA or HCV-II for negative-strand HCV RNA). cDNA was purified with the High Pure PCR product purification kit (Roche Diagnostics).
Positive- and negative-strand HCV PCR amplifications were performed with 2 µL of purified cDNA in a reaction mixture containing 1 µL of LightCycler Fast Start DNA Master SYBR Green I, 4mM of MgCl2, and 5 pM of antisense primer KY78 (5′-CTC GCA AGC ACC CTA TCA GGC AGT-3′, nt 311–288 of the 5′UTR) and 5 pM of sense KY80 (5′-GCA GAA AGC GTC TAG CCA TGG CGT-3′, nt 68–91 of the 5′UTR). The PCR consisted of an initial denaturation step of 10 min at 95 °C, followed by 40 cycles of the following thermal conditions: 15 s at 95 °C, 5 s at 70 °C, and 15 s at 72 °C. All samples were analyzed in triplicate. The sensitivity of the PCR for HCV was previously determined to be approximately 230 copies HCV/µL.
For quantification of GAPDH mRNA, RT was performed with the same amount of RNA used for HCV positive- and negative-strand analysis, using the oligo d(T)16 primer under standard conditions. For real-time PCR amplification of GAPDH, a commercial GAPDH primer set (Roche Search LC, Mannheim, Germany) was used under the recommended conditions. For real-time PCR amplification of LacZ, sense (5′-GCC TGC GAT GTC GGT TTC CGC GAG G-3′) and antisense primers (5′-GCC AGC GCG GAT CAT CGG TCA GAC G- 3′) were utilized under the following conditions: 10 s at 95 °C, 10s at 68 °C, 16 s at 72 °C (Dobson et al., 1990). The sensitivity of detection was approximately 210 copies/µL.
DNA was quantified measuring SYBR green I dye incorporation into PCR products at 530 nm following manufacturer’s instructions. An HCV standard curve was generated using a PCR product corresponding to nucleotides 38–342 of the 5′UTR. At the end of each run, a DNA melting curve was performed to control for sample homogeneity and quality. In a subset of samples, electroporation and sequencing were performed to confirm the correct identity of the amplified PCR product. Data were expressed as the copy number of HCV positive-strand (or negative-strand) RNA per molecule of GAPDH. This analysis was done in quadruplicate for each sample and presented as the mean and standard deviation. Each value was analyzed statistically using the SPSS 10.0 software (SPSS, Chicago, IL). Differences in mean values were compared using the Mann–Whitney U-test.
2.8. Western blotting analysis
Cells were washed twice with PBS, and lysed with 100 µL of Nonidet P-40 buffer (0.5% Nonidet P-40, 10mM Tris, pH 7.4, 150 mM NaCl, 1% SDS). Protein lysate concentrations were measured using the DC protein assay Kit (Bio-Rad, Hercules, CA). Forty microliters of protein lysate were utilized. Separated products were then blotted onto Immobilon-P membranes, and each membrane was incubated with the relevant antibody. The ECL Kit (Amersham Pharmacia, Buckinghamshire, UK) was used for detection. Monoclonal antibody to HCV core protein (515s) (Kashiwakuma et al., 1996) was provided by Dr. M. Kohara, Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan; monoclonal antibody to β-galactosidase was purchased from Promega. Appropriate species-specific conjugated secondary antibodies were obtained commercially (Amersham Pharmacia).
2.9. ELISA for HCV core antigen
Cell culture lysates were adjusted to 0.2 mg/mL. HCV core antigen concentrations were quantified using the HCV core protein ELISA kit (Ortho-Clinical Diagnostics, Raritan, NJ) following the manufacturer’s instructions (Bouvier-Alias et al., 2002). Core ELISA data were expressed as fmol of HCV core antigen per µg of total protein. The lower level of detection for this assay was less than 1.5 pg/mL.
3. Results
3.1. Replication-defective adenoviral vectors successfully replicate HCV RNA without cytotoxicity
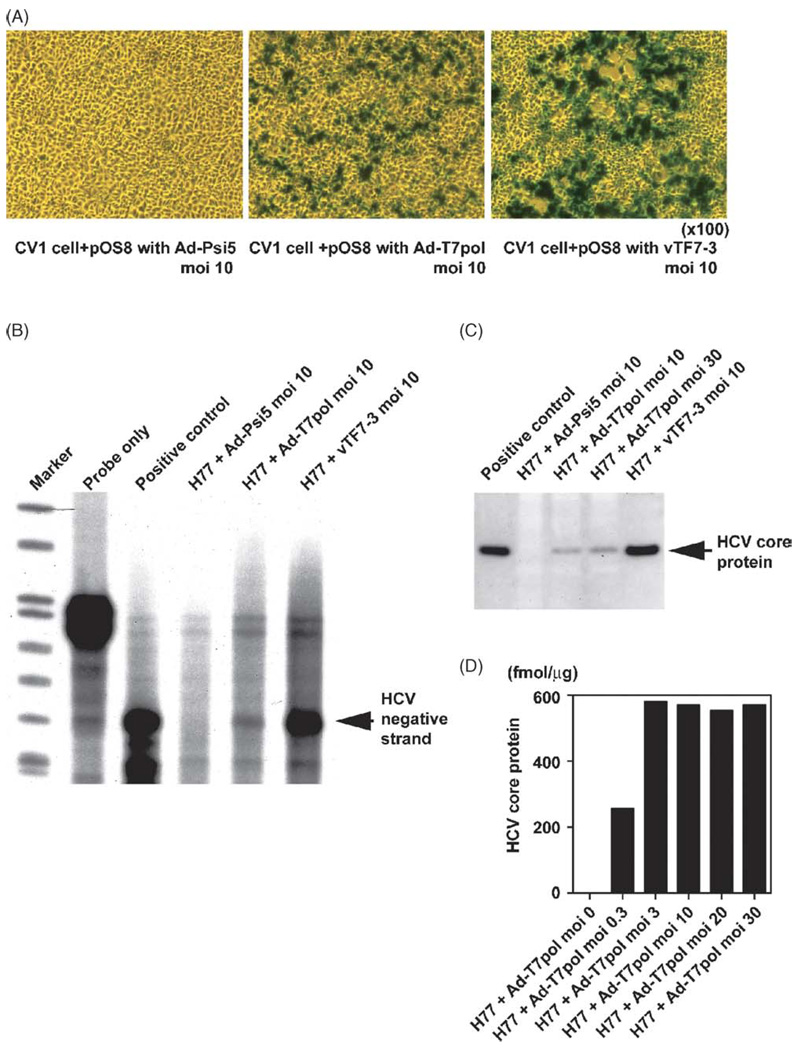
To compare the transfection and infection efficiency of the vaccinia and adenovirus vectors, the pOS8 plasmid, which contains aT7 promoter flanking the β-galactosidase gene, was transfected into cells that were then infected with either vaccinia-T7 (vTF7-3) or Ad-T7pol at a multiplicity of infection (MOI) of 10. The parental replication-incompetent vector Ad-Psi5 was used as an adenoviral vector control. At an MOI of 10, each viral vector efficiently expressed β-galactosidase in approximately 50% of cells 24 h after infection (Fig. 1A). Using trypan-blue staining, cell injury was observed in cells transfected with vaccinia-T7 but not in cells transfected with the Ad-T7pol or Ad-Psi5 vectors (data not shown).
Fig. 1.
Comparison of the transfection and infection efficiency of HCV replication system using vaccinia-T7 and adeno-T7 vectors: (A) after transfection with the pOS8 plasmid, cells were infected with virus vectors using control adenovirus (Ad-Psi5), recombinant adeno-T7 polymerase (Ad-T7pol), or vaccinia-T7 polymerase (vTF7-3) at an MOI of 10. (B) RPA for negative-strand HCV RNA was performed with H77 plasmid as a positive control. (C)Western blotting for HCV core protein was performed on CV-1 cell lysates 24 h after infection. (D) Quantitative HCV core ELISA results indicated that an MOI of 10 was optimal for adenoviral-driven HCV protein production.
Ribonuclease protection assay for negative-strand HCV RNA (Fig. 1B) and Western blotting for HCV core protein (Fig. 1C) were performed 24 h after infection. Expression of negative-strand HCV RNA was lower after Ad-T7pol infection compared to vTF7-3 infection, yet was clearly detectable. Similarly, HCV core protein production was less robust after Ad-T7pol infection; nonetheless, it was clearly detectable. Negative-strand HCV RNA and HCV core protein were not detected when the control Ad-Psi5 vector was used. Quantitative HCV core ELISA results suggested that an MOI of 10 was optimal for adenoviral-driven HCV protein production (Fig. 1D); therefore, an MOI of 10 was selected for all subsequent experiments.
In contrast to increased HCV RNA synthesis and protein production in transfected/infected cells, LacZ mRNA levels decreased rapidly after day 1 and were not detectable after day 7 (data not shown).
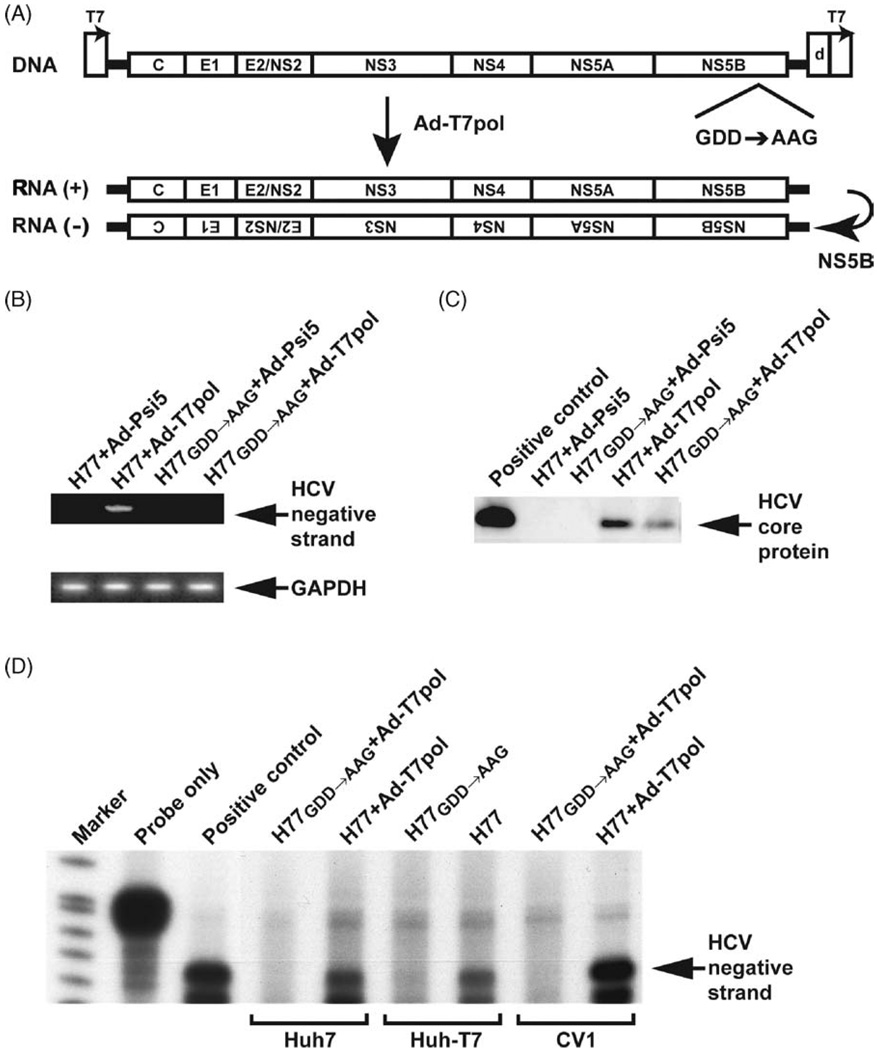
3.2. Adenoviral-T7-driven HCV replication is dependent on an intact HCV polymerase gene
The H77GDD→AAG mutant (Fig. 2A) was used to assess whether the HCV RNA polymerase gene (NS5B) was necessary for viral replication. By qualitative RT-PCR of the 5′UTR, HCV negative-strand synthesis was detectable only in the presence of both H77 and Ad-T7pol (Fig. 2B) in CV-1 cells. The absence of detectable negative-strand HCV RNA upon transfection of the mutant plasmid (H77GDD→AAG) indicates that replication was dependent on an intact polymerase sequence. H77 + Ad-T7pol expressed significantly higher core protein levels compared to H77GDD→AAG + Ad-T7pol (Fig. 2C), further suggesting an intact polymerase sequence is necessary for robust HCV protein production. Ribonuclease protection assay demonstrated the presence of HCV negative-strand in CV-1, Huh7, and Huh-T7 cell lines on day 2 (Fig. 2D). However, negative-strand HCV RNA was not detected when the H77GDD→AAG mutant was transfected, indicating that an intact NS5B sequence was necessary for negative-strand HCV RNA synthesis.
Fig. 2.
(A) A control plasmid was prepared by mutating the active site motif from GDD to AAG in the NS5B RNA-dependent RNA polymerase sequence (H77GDD→AAG). (B) A qualitative strand-specific RT-PCR for negative-strand HCV RNA was performed as previously described (Lanford et al., 1995). (C)Western blotting analysis demonstrated that transfection/infection with H77 + Ad-T7-pol also resulted in HCV core protein production. (D) Ribonuclease protection assay demonstrated detectable negative-strand HCV RNA in CV-1, Huh7, and Huh-T7 cell lines on day 2.
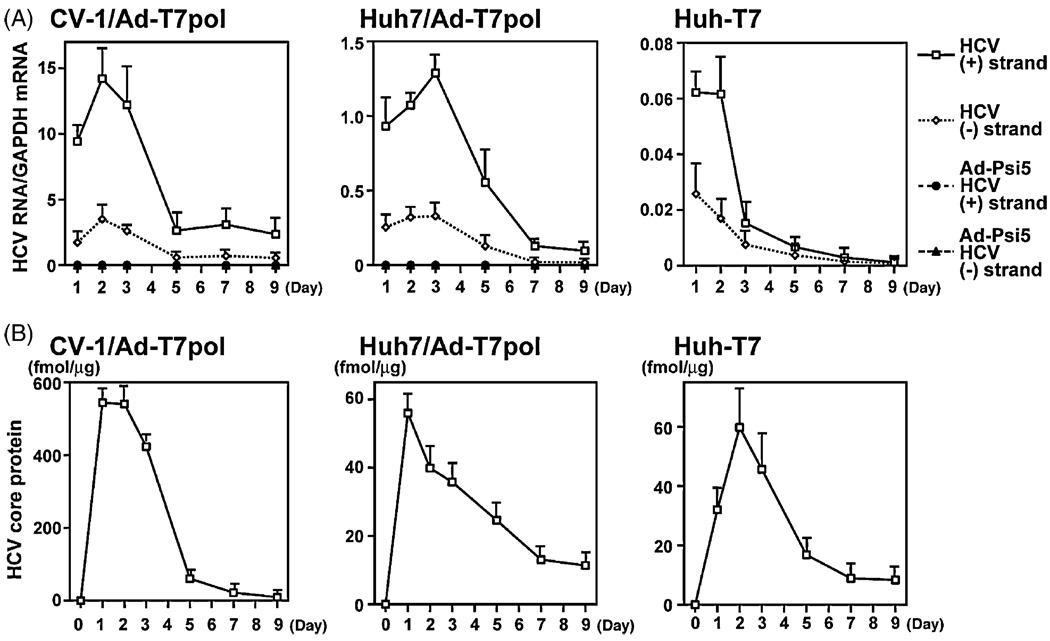
3.3. Kinetic analysis of HCV RNA synthesis and core protein production
The data described above suggest that the Ad-T7pol replication system results in efficient HCV RNA and protein expression without inducing cell toxicity. Thus, the time course of strand-specific HCV RNA synthesis (Fig. 3A) and HCV protein production (Fig. 3B) was examined further in CV-1 and Huh7 cells (transfected with H77 and infected with Ad-T7pol), as well as Huh-T7 cells (transfected with H77). Both positive- and negative-strand HCV RNA were detectable for the entire 9-day experiment in each cell line. Positive-strand HCV RNA levels increased significantly after infection and continued to be expressed at high levels for 3 days and diminished thereafter. Negative-strand HCV RNA synthesis paralleled that of positive-strand throughout the time course; however, the quantity of negative-strand HCV RNA was approximately 10% of positive-strand levels. This is consistent with positive-/negative-strand ratios reported from infected human liver samples (Komurian-Pradel et al., 2004; Laskus et al., 1998). As expected, HCV RNA was not detected in CV-1 or Huh7 cells infected with the Ad-psi5 control vector.
Fig. 3.
(A) Real-time PCR of HCV positive- and negative-strand RNA was performed as described. Data are expressed as the strand-specific HCV copy number per molecule of GAPDH. Error bars indicate the mean ± standard error (S.E.) of four replicates. (B) A quantitative core ELISA measured HCV protein production in CV-1, Huh7, and Huh-T7 cell lines. Data are expressed as fmol (mean ± S.E.) of HCV core per µg of total protein for three replicates.
Using an identical experimental approach, HCV core protein production was expressed strongly during days 1–3 in all cell lines examined, and diminished with similar kinetics as HCV RNA (Fig. 3B). Similar to HCV RNA, HCV core protein was detectable for the entire 9-day experiment in each cell line.
To circumvent potential perturbations in the cellular environment due to transfection/infection with viral vectors, experiments in a Huh7 cell line stably transfected with T7 polymerase (Huh-T7) were performed. After transfection of H77 into these cells, positive- and negative-strand HCV RNA were detected (Fig. 3A), as well as HCV core protein (Fig. 3B), throughout the entire 9-day experiment. Interestingly, HCV RNA levels were lower in Huh-T7 cells than in CV-1 and Huh7 cells, although core levels were not appreciably different between Huh7 and Huh-T7 cells.
3.4. IFN efficiently inhibits HCV expression
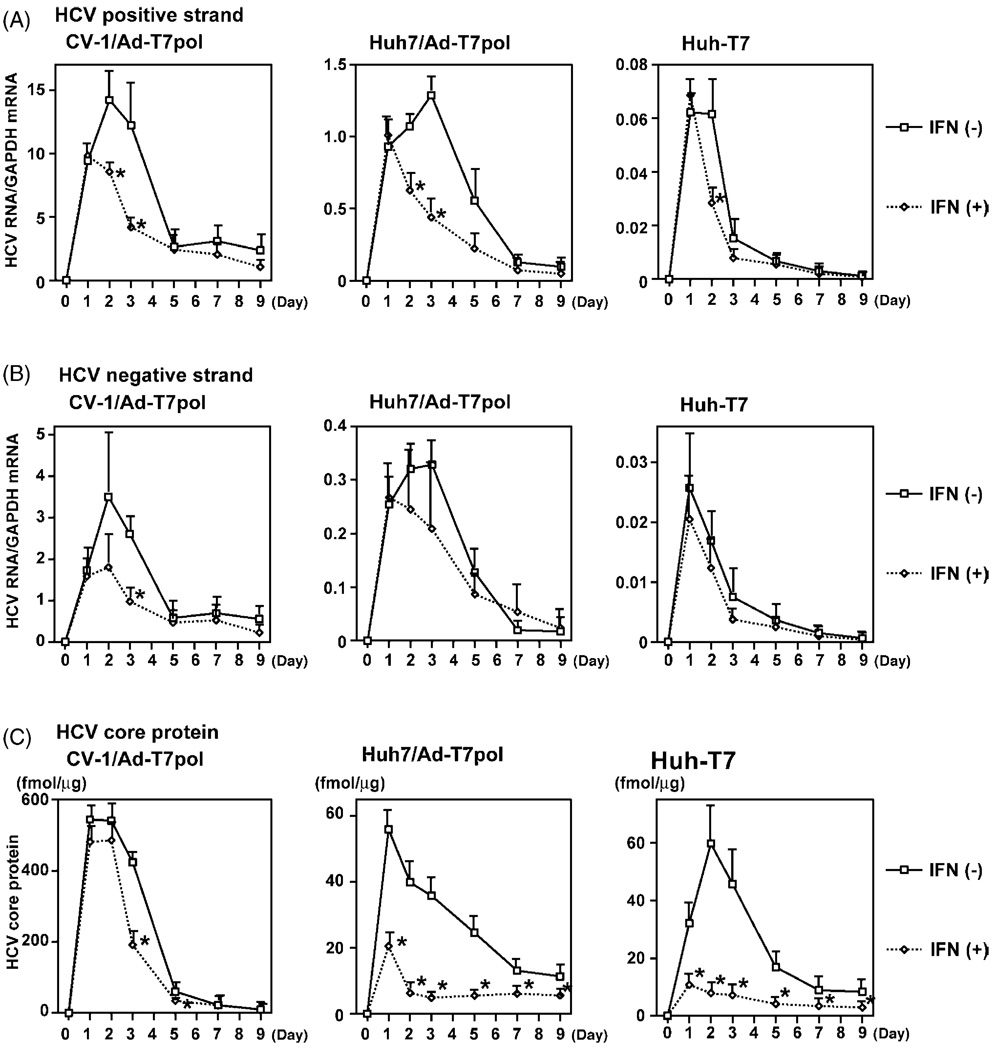
Utilizing these refined models of HCV replication, the inhibitory effects of IFN on HCV expression were examined. To determine the potential effects of IFN on cellular gene translation, the plasmid OS8 was transfected, and LacZ mRNA levels were measured in the presence of several doses of IFN. LacZ mRNA expression was slightly reduced; however, no significant toxicity in cells exposed to IFN was observed using trypan-blue staining (data not shown).
In CV-1 cells (Fig. 4A), HCV positive-strand RNA levels were significantly decreased in the presence of 1000 IU/mL IFN at day 2 (14.21 ± 3.95 versus 8.55 ± 0.61 p < 0.05). A trend toward reduced HCV RNA was also observed on day 3 (12.23 ± 5.43 versus 4.13 ± 0.74, p < 0.10). In Huh7 cells, a significant decrease of HCV positive-strand was observed on days 2 (1.07 ± 0.07 versus 0.63 ± 0.14, p < 0.05) and 3 (1.29 ± 0.13 versus 0.44 ± 0.15, p < 0.05). In Huh-T7 cells, a significant decrease was also observed on day 2 (0.06 ± 0.03 versus 0.03 ± 0.007, p < 0.05). For HCV negative-strand RNA (Fig. 4B), only day 3 IFN-treated CV-1 cells had significantly decreased levels compared to untreated cells (2.60 ± 0.41 versus 0.97 ± 0.31, p < 0.05).
Fig. 4.
(A) Real-time PCR of positive-strand HCV RNA was performed in the presence of 1000 IU/mL IFN. Error bar indicates mean ± S.E. for four replicates (*p < 0.05; **p < 0.10). (B) Real-time PCR of negative-strand HCV RNA was also performed in the presence of 1000 IU/mL IFN. (C) Quantitative core ELISA also demonstrated decreased HCV protein production in IFN-treated cells.
HCV core protein expression was approximately 10-fold higher in CV-1 cells compared to either Huh7 or Huh-T7 cells (Fig. 4C). IFN treatment of CV-1 cells did not appear to have a large effect on HCV core protein levels; however, HCV core levels were decreased in IFN-treated CV-1 cells compared to untreated cells at days 3 (423.51 ± 25.73 fmol/µg versus 190.92 ± 35.25 fmol/µg, p < 0.05) and 5 (60.24 ± 12.89 fmol/µg versus 34.15 ± 0.76 fmol/µg, p < 0.05). For Huh7 and Huh-T7 cells, HCV core expression was significantly reduced when treated with IFN compared to untreated cells at all time points (p < 0.05).
4. Discussion
Because of the cytotoxic nature of vaccinia virus, long-term evaluation of HCV RNA synthesis and protein production, as well as characterization of the inhibitory effects of antiviral agents, such as IFN and RBV, was not possible using our previous replication model. By using adenovirus-derived T7 vectors, vaccinia-induced cytotoxicity was removed, allowing sustained detection of HCV replication and protein production for 9 days in multiple cell types. The refined binary HCV replication system efficiently synthesized HCV negative-strand RNA, an important indicator of ongoing, active viral replication, in an NS5B-dependent manner, as no negative-strand HCV RNA was detected upon transfection of an NS5B mutant. Using Huh7 cell lines stably expressing T7 polymerase (Huh-T7), dependence of the replication models on any viral vectors was removed. Quantities of HCV RNA synthesis and protein production in Huh-T7 cells were not as robust as in CV-1 or Huh7 cells transfected with Ad-T7. However, sustained HCV replication in Huh-T7 cells, with no obvious signs of cytotoxicity, suggests that this viral vector-independent replication model will be useful for future studies of virus–host interactions and the development of antiviral agents with activity against HCV genotype 1.
These binary systems offer several advantages over currently available HCV replication systems. First, these replication models do not require continuous antibiotic selection as do current replicon systems (Blight et al., 2000; Frese et al., 2001; Guo et al., 2001; Lohmann et al., 1999). Second, the requirement of highly adaptive viral mutations for efficient replicon activities that are not necessarily viable in vivo (Bukh et al., 2002) may limit the interpretability of certain findings obtained from replicon systems. Because the refined replication models do not require continuous selection and do not possess highly ‘adaptive’ viral mutations, they are more authentic for characterization of antiviral agents, virus–host interactions, and viral fitness. Third, these replication systems can be used to study HCV replication in a variety of hepatocyte- and non-hepatocyte-derived cell types; in contrast, replicon systems only replicate efficiently in Huh7 cells. Most importantly, these replication systems use a full-length infectious genotype 1a cDNA construct that yields an authentic dual-function template in vivo that is both translated and transcribed. Moreover, transfected cells in our replication systems are able to express all HCV structure and non-structural proteins (Lin et al., 2005). Thus, they are more likely to carry out authentic HCV RNA replication than replicon systems based on sub-genomic constructs.
Several significant differences exist between the vaccinia and adeno-T7 replication systems (Table 1). Both are capable of positive- and negative-strand HCV RNA synthesis, protein production, and quasispecies generation without the need for cell culture adaptive mutations (Chung et al., 2001; Contreras et al., 2002; Blackard and Hiasa et al., unpublished data). HCV replication is inhibited significantly by IFN in both systems. However, the vaccinia-based system replicates at much higher levels than the adeno-T7-based system; yet, HCV RNA synthesis occurs for at least 9 days in the former due to the lack of vector-induced cytotoxicity. Nonetheless, the decrease of HCV RNA synthesis and protein production after 3 days suggests an inhibitory effect exerted by key host cells proteins, such as protein kinase R (PKR), since adenoviruses do not inhibit PKR function as does vaccinia virus (Chang et al., 1992; Watson et al., 1991). Further examination of host antiviral pathways that limit robust long-term viral replication in culture is necessary.
Table 1.
Several similarities and differences between the vaccinia and adenovirus systems exist
| Vaccinia | Ad-T7 | |
|---|---|---|
| T7 polymerase delivery | Vaccinia virus | Adenovirus |
| Cytotoxicity | Yes | No |
| HCV replication | (+) and (−) strand synthesis; high levels |
(+) and (−) strand synthesis; low but effective levels |
| Duration of replication | 24 h | 9 days |
| HCV protein production | Yes | Yes |
| Quasispecies generation | Yes | Yes |
| IFN inhibits replication | Yes | Yes |
We used these refined replication systems to explore the kinetics of HCV RNA synthesis and protein production in the presence of IFN. When 1000 IU/mL of IFN was added to the culture medium of HCV-expressing cells, there was no difference in HCV positive- or negative-strand quantity compared to untreated cells at day 1. Despite this lack of short-term antiviral activity, HCV RNA was significantly decreased in IFN-treated cells at days 2 and 3, suggesting that the full effects of IFN may require at least 24 h.
This cell-based HCV replication system has already been used to examine the interaction between HCV protein expression and host type I IFN signaling components in the Jak-STAT kinase pathway (Lin et al., 2005). Recently, in vitro systems that support infectious HCV production have been reported. However, these systems are based on HCV genotype 2a (Wakita et al., 2005; Zhong et al., 2005) and do not support replication in cells other than the highly permissive Huh-7 cell line and its derivatives. Importantly, the replication systems described here are based on genotype 1a isolate and replicate in several hepatocyte- and non-hepatocyte-derived cell lines. Thus, these refined replication models provide the opportunity to explore HCV molecular biology and the interactions between antiviral agents and specific HCV and/or host proteins that are relevant to genotype 1 infection.
Acknowledgements
We thank Drs. Michinori Kohara and Bryan R.G. Williams for anti-HCV core and anti-PKR antibodies, respectively. We thank Dr. Stanley M. Lemon for the Huh-T7 cell line. This work supported by a Postdoctoral Research Fellowship Award from the American Liver Foundation (to Y.H.), a Grant-in-Aid for Scientific Research Grant (JSPS KAKENHI 15790350 and 17590650) from the Japanese Ministry of Education, Culture, Sports, Science and Technology (to Y.H.), and NIH Grants RO1 DK57857 (to R.T.C.) and RO1 AI43478 (to E.V.S.).
Contributor Information
Yoichi Hiasa, Email: hiasa@m.ehime-u.ac.jp.
Jason T. Blackard, Email: jblackard@partners.org.
Wenyu Lin, Email: wlin1@partners.org.
Yoshitaka Kamegaya, Email: ykamegaya@partners.org.
Norio Horiike, Email: horiike@m.ehime-u.ac.jp.
Morikazu Onji, Email: onjimori@m.ehime-u.ac.jp.
Emmett V. Schmidt, Email: eschmidt@partners.org.
Raymond T. Chung, Email: rtchung@partners.org.
References
- Alter MJ, Kruszon-Moran D, Nainan OV, McQuillan GM, Gao F, Moyer LA, Kaslow RA, Margolis HS. The prevalence of hepatitis C virus infection in the United States, 1988 through. N. Engl. J. Med. 1994;341:556–562. doi: 10.1056/NEJM199908193410802. [DOI] [PubMed] [Google Scholar]
- Blackard JT, Smeaton L, Hiasa Y, Horiike N, Onji M, Jamieson DJ, Rodriguez I, Mayer KH, Chung RT. Detection of hepatitis C virus (HCV) in serum and peripheral blood mononuclear cells of HCV-monoinfected and HIV/HCV-coinfected persons. J. Infect. Dis. 2005;192:258–265. doi: 10.1086/430949. [DOI] [PubMed] [Google Scholar]
- Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- Bouvier-Alias M, Patel K, Dahari H, Beaucourt S, Larderie P, Blatt L, Hezode C, Picchio G, Dhumeaux D, Neumann A, McHutchison JG, Pawlotsky JM. Clinical utility of total HCV core antigen quantification: a new indirect marker of HCV replication. Hepatology. 2002;36:211–218. doi: 10.1053/jhep.2002.34130. [DOI] [PubMed] [Google Scholar]
- Bukh J, Pietschmann T, Lohmann V, Krieger N, Faulk K, Engle RE, Govindarajan S, Shapiro M, St. Claire M, Bartenschlager R. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14416–14421. doi: 10.1073/pnas.212532699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll K, Elroy-Stein O, Moss B, Jagus R. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2 alpha-specific protein kinase. J. Biol. Chem. 1993;268:12837–12842. [PubMed] [Google Scholar]
- Castet V, Fournier C, Soulier A, Brillet R, Coste J, Larrey D, Dhumeaux D, Maurel P, Pawlotsky JM. Alpha interferon inhibits hepatitis C virus replication in primary human hepatocytes infected in vitro. J. Virol. 2002;76:8189–8199. doi: 10.1128/JVI.76.16.8189-8199.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H-W, Watson JC, Jacobs BL. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung RT, He W, Saquib A, Contreras AM, Xavier RJ, Chawla A, Wang TC, Schmidt EV. Hepatitis C virus replication is directly inhibited by INF-a in a full-length binary expression system. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9847–9852. doi: 10.1073/pnas.171319698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung RT, Kaplan LM. Heterogenous nuclear ribonucleoprotein 1 (hnRNP-1/PTB) selectively binds the conserved 3′ terminus of hepatitis C viral RNA. Biochem. Biophys. Res. Commun. 1999;254:351–362. doi: 10.1006/bbrc.1998.9949. [DOI] [PubMed] [Google Scholar]
- Contreras AM, Hiasa Y, He W, Terella A, Schmidt EV, Chung RT. Viral RNA mutations are region specific and increased by ribavirin in a full-length hepatitis C virus replication system. J. Virol. 2002;76:8505–8517. doi: 10.1128/JVI.76.17.8505-8517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson AT, Margolis TP, Sedarati F, Stevens JG, Feldman LT. A latent, nonpathogenic HSV-1-derived vector stably expresses b-galactosidase in mouse neurons. Neuron. 1990;5:353–360. doi: 10.1016/0896-6273(90)90171-b. [DOI] [PubMed] [Google Scholar]
- Frese M, Pietschmann T, Moradopour D, Haller O, Bartenschlager R. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 2001;82:723–733. doi: 10.1099/0022-1317-82-4-723. [DOI] [PubMed] [Google Scholar]
- Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M, Jr, Tan S-L, Wambach M, Katze MG. Interaction of the interferon-induced PKR protein kinase with inhibitory proteins p58IPK and vaccinia virus K3L is mediated by unique domains. Mol. Cell Biol. 1996;16:4172–4181. doi: 10.1128/mcb.16.8.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JT, Bichko VV, Seeger C. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 2001;75:8516–8523. doi: 10.1128/JVI.75.18.8516-8523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiasa Y, Horiike N, Akbar SM, Saito I, Miyamura T, Matsuura Y, Onji M. Low stimulatory capacity of lymphoid dendritic cells expressing hepatitis C virus genes. Biochem. Biophys. Res. Commun. 1998;249:90–95. doi: 10.1006/bbrc.1998.9089. [DOI] [PubMed] [Google Scholar]
- Hiasa Y, Kamegaya Y, Nuriya H, Onji M, Kohara M, Schmidt EV, Chung RT. PKR is increased and is functional in hepatitis C virus-related hepatocellular carcinoma. Am. J. Gastroenterol. 2003;98:2528–2534. doi: 10.1111/j.1572-0241.2003.08663.x. [DOI] [PubMed] [Google Scholar]
- Kashiwakuma T, Hasegawa A, Kajita T, Tanaka A, Mori H, Ohta Y, Tanaka E, Kiyosawa K, Tanaka T, Tanaka S, Hattori N, Kohara M. Detection of hepatitis C virus specific core protein in serum of patients by a sensitive fluorescence enzyme immunoassay (FEIA) J. Immunol. Methods. 1996;190:79–89. doi: 10.1016/0022-1759(95)00261-8. [DOI] [PubMed] [Google Scholar]
- Komurian-Pradel F, Perret M, Deiman B, Sodoyer M, Lotteau V, Paranhos-Baccala G, Andre P. Strand specific quantitative real-time PCR to study replication of hepatitis C virus genome. J. Virol. Methods. 2004;116:103–106. doi: 10.1016/j.jviromet.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Chavez D, Chisari FV, Sureau C. Lack of detection of negative-strand hepatitis C virus RNA in peripheral blood mononuclear cells and other extrahepatic tissues by the highly strand-specific rTth reverse transcriptase PCR. J. Virol. 1995;69:8079–8083. doi: 10.1128/jvi.69.12.8079-8083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskus T, Radkowski M, Wang LF, Cianciara J, Vargas H, Rakela J. Hepatitis C virus negative strand RNA is not detected in peripheral blood mononuclear cells and viral sequences are identical to those in serum: a case against extrahepatic replication. J. Gen. Virol. 1997;78:2747–2750. doi: 10.1099/0022-1317-78-11-2747. [DOI] [PubMed] [Google Scholar]
- Laskus T, Radkowski M, Wang LF, Vargas H, Rakela J. Search for hepatitis C virus extrahepatic replication sites in patients with acquired immunodeficiency syndrome: specific detection of negative-strand viral RNA in various tissues. Hepatology. 1998;28:1398–1401. doi: 10.1002/hep.510280531. [DOI] [PubMed] [Google Scholar]
- Lin W, Choe WH, Hiasa Y, Kamegaya Y, Blackard J, Schmidt EV, Chung RT. Hepatitis C virus expression suppresses interferon signaling by degrading STAT1. Gastroenterology. 2005;128:1034–1041. doi: 10.1053/j.gastro.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- McHutchison JG, Poynard T. Combination therapy with interferon plus ribavirin for the initial treatment of chronic hepatitis C. Semin. Liver Dis. 1999;19:57–65. [PubMed] [Google Scholar]
- Miyake S, Makimura M, Kanegae Y, Harada S, Sato Y, Takamori K, Tokuda C, Saito I. Efficient generation of recombinant adenoviruses using adenovirus DNA-terminal protein complex and a cosmid bearing the full-length virus genome. Proc. Natl. Acad. Sci. U.S.A. 1996;93:1320–1324. doi: 10.1073/pnas.93.3.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DE, Honda M, Whetter LE, McKnight KL, Lemon SM. Mutations within the 5′ nontranslated RNA of cell culture-adapted hepatitis A virus which enhance cap-independent translation in cultured African green monkey cells. J. Virol. 1996;70:1041–1049. doi: 10.1128/jvi.70.2.1041-1049.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JC, Chang H-W, Jacobs BL. Characterization of a vaccinia virus-encoded double-stranded RNA-binding protein that may be involved in inhibition of the double-stranded RNA-dependent protein kinase. Virology. 1991;185:206–216. doi: 10.1016/0042-6822(91)90768-7. [DOI] [PubMed] [Google Scholar]
- Wold WS, Gooding LR. Adenovirus E3 proteins that prevent cytolysis by cytotoxic T cells and tumor necrotic factor. Mol. Biol. Med. 1989;6:433–452. [PubMed] [Google Scholar]
- Yanagi M, Purcell RH, Emerson SU, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U.S.A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]