Abstract
Animal models carrying mutations in the hairless (Hr) gene provide a rich resource for study of hair follicle biology. A spontaneous mouse mutant with a phenotype strikingly similar to rhino mutants of Hr arose spontaneously in the mouse facility at Oak Ridge National Laboratory. Sequence analysis of Hr in these mutants uncovered a nonsense mutation in exon 12, designated as Hrrh-R (rhino, Oak Ridge). The mutation led to significant reduction in Hr mRNA levels, predicted to be due to nonsense-mediated decay. Histological analysis indicated dilated hair follicle infundibula at 14 days of age that rapidly became filled with cornified material. Microarray analyses revealed that expression levels of many genes involved in keratinocyte differentiation, epidermal regeneration, and wound healing were significantly upregulated before morphological detection of the phenotype, suggesting their role in onset of the Hrrh-R phenotype. Identification of this new Hr allele and the underlying molecular alterations allows further understanding of the role of Hr in hair follicle biology.
Keywords: alopecia, hair follicle, hairless mice, microarray analysis
Existing allelic mutations in the hairless (Hr) gene compose a set of mouse models that cause hair loss through partial or complete loss of Hr function across a variety of mammalian species.1,5,10,14,45,62,73
Hr encodes a 127-kD protein that includes a single zinc finger and a Jumonji-like C terminus domain, which is a metalloenzyme-like domain implicated in chromatin remodeling.9,20 A predicted role for Hr in control of gene expression is borne out by its demonstrated ability to function as a transcriptional corepressor through heterodimerization with the thyroid hormone receptor, vitamin D receptor, and retinoic acid–like orphan receptor alpha, possibly mediated through association with histone deacetylases.15,29,42,47,65,70 Multiple allelic mutations with various degrees of severity have arisen in Hr, all of which manifest in mice as an inability to regrow a normal coat of hair after the initial catagen stage, resulting in a progressive loss of hair from head-to-tail beginning in the third week of life.18,28 Gaskoin first described a mutant mouse with hair loss and wrinkled skin as “rhinoceros” in 1856.26 More recently, several Hr mutations causing a phenotype more severe than hairless (Hrhr) were characterized as rhino alleles of Hr (Hrrh), most of which were due to nonsense mutations.3,4,20,72 The rhinocerotic appearance and excessive skin wrinkling of rhino mutants at Hr are due to large utricles and deep dermal cysts that form from the remnants of hair follicles.39,44,58,60 The difference in severity between Hrhr and Hrrh alleles is attributed to the degree to which the mutation abolishes wild-type Hr expression.6 Nonsense mutations in Hr are also described in human patients with atrichia with papular lesions (OMIM No. 209500), but the excessive skin wrinkling is not present in affected human patients.2,8,56
A mouse with a hair loss phenotype similar to Hr mutants arose spontaneously in the Oak Ridge National Laboratory mouse colony. We report here that this new mouse model results from a novel nonsense mutation: Hrrh-R (Oak Ridge rhino allele of Hr). We describe in detail the phenotype of Hrrh-R mice, including the changes in gene expression that underlie hair loss in this model.
Materials and Methods
Animals and Tissue Collection
All mice were bred at Oak Ridge National Laboratory, and experiments were conducted under approved Institutional Animal Care and Use Committee protocols. The Hrrh-R mutation was maintained on an inbred, mixed-stock genetic background by mating Hrrh-R/Hrrh-R male with Hrrh-R/+ females. Details of the genetic background are available from the Mouse Genome Informatics database under accession No. MGI:3580645. Coisogenic +/+ mice for sequence comparison were produced by intercross mating of Hrrh-R/+ mice. At various ages, mice were euthanized by cervical dislocation. Tails were harvested from littermates of each genotype for DNA isolation, and dorsal skin was collected for RNA and for histological analyses of hematoxylin and eosin–stained sections.48
DNA Sequencing and Genotyping
Genomic DNA was extracted from tail biopsies (0.2-cm tail snips) using the HotSHOT method.66 All Hr exons including intron–exon boundaries were sequenced according to protocols described previously.37 Sequencing results were analyzed with BLAST (Basic Local Alignment Search Tool).7 A pair of specific primers spanning exon 12 and covering the DNA mutation was used for genotyping of the mice (forward primer: 5′-CCAAGAACCTGAGGACCAGA-3′; reverse primer: 5′-GCCAGTGTTCCTGGAAGAGA-3′).
Gene Expression Microarray Construction, Labeling, Hybridization, and Data Analysis
Total RNA was isolated from dorsal skin of 10-day old mice according to protocols standard for fibrous tissues.37 Microarrays representing about 15,000 unique mouse genes were printed by the Center for Applied Genomics (PHRI, Newark, NJ) using the Compugen Mouse OligoLibrary 2.0. Hybridization experiments were performed as previously described.37
A total of 3 biological replicates (RNA from a pair of Hrrh-R/+ and Hrrh-R/Hrrh-R littermates at 10 days old after birth) were included in this analysis. Data were normalized using Lowess to adjust for intensity-dependent dye bias after removing spots of poor quality or low expression and subtracting local background.54,55 The Significance Analysis of Microarrays software package was used to analyze data for statistically significant differences in gene expression, with a false discovery rate of about 3%.67 Analysis of overrepresented Gene Ontology (GO) categories within the biological process ontology across the set of differentially expressed genes was conducted using the Database for Annotation, Visualization and Integrated Discovery 2006.23 The detailed protocols and primary data from this study are available at the Gene Expression Omnibus database under accession Nos. GSE4055 and GSE4059. A 95% confidence interval was used for all statistical analyses. The oPOSSUM database of transcription factor binding sites (TFBS) identified by phylogenetic footprinting was used to identify binding sites overrepresented among differentially expressed genes. When supplied with a list of genes of interest, oPOSSUM retrieves the TFBS counts for each gene in the list and computes 2 statistics (Z score, Fisher exact test) to measure overrepresentation of TFBS in the gene set relative to a background comprising all genes in the oPOSSUM database.
Northern Blot
Two probes, one corresponding to exon 12 and one to exons 5–11, were amplified from a skin cDNA pool created from wild type mice. Total RNA from dorsal skin of 1-, 7-, 14-, and 35-day old mice was extracted, electrophoresed in agarose gels, transferred to nylon membranes, and cross-linked to the membranes using ultraviolet light according to the standard protocols. Probes were randomly labeled with α-32P-dCTP and hybridized at 42°C overnight. Following washing, autoradiography was used to obtain blot images.
Results
Phenotype
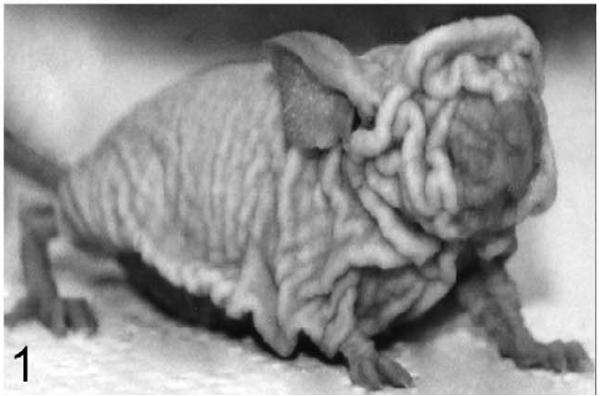
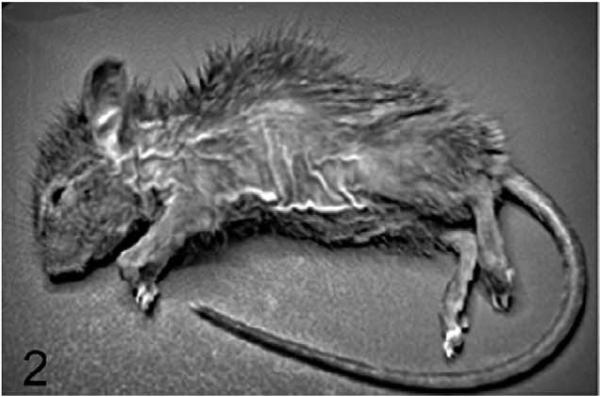
Compared to the well-characterized rhino mutants of Hr, homozygous Hrrh-R/Hrrh-R mice exhibited a similar phenotype whereas Hrrh-R/+ mice were phenotypically normal. Similar to other Hrrh alleles,3,4,17,25,44,52,58 skin wrinkling appeared before the majority of hair was lost, and it progressed with age (Fig. 1). Hair loss in Hrrh-R/Hrrh-R was first detectable on the ventrum at approximately 2.5 weeks of age and proceeded in a diffuse manner across the body, a pattern of loss that differed from other Hr mutants (Fig. 2). Vibrissae were normal but became sparse with age (Fig. 2). Nail length increased, and older adult mice (about 6 months of age) displayed overgrown nails that curled under the foot pad (onychogryposis) (Fig. 3). This is a common feature in all the Hr allelic mutations.59,61 Both sexes of Hrrh-R/Hrrh-R mice were fertile and viable, but females had difficulty nursing owing to a propensity for skin lesions. Thymus weight was significantly reduced in Hrrh-R/Hrrh-R mice compared to Hrrh-R/+ (P < .05).
Figure 1.
Skin; Hrrh-R/Hrrh-R mouse, 5 months old. Skin wrinkling is characteristic of rhino mutants of Hr.
Figure 2.
Skin; Hrrh-R/Hrrh-R mouse, 4 weeks old. Scattered hair loss is readily visible at 4 weeks of age, occurring in a diffuse manner across the trunk.
Figure 3.
Nail; Hrrh-R/Hrrh-R mouse, 6 months old. Nails are increasingly long and curved in Hrrh-R/Hrrh-R mice.
Utricle and Dermal Cysts Formation
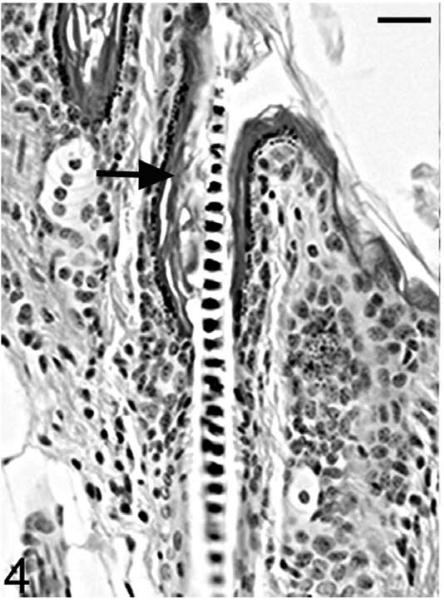
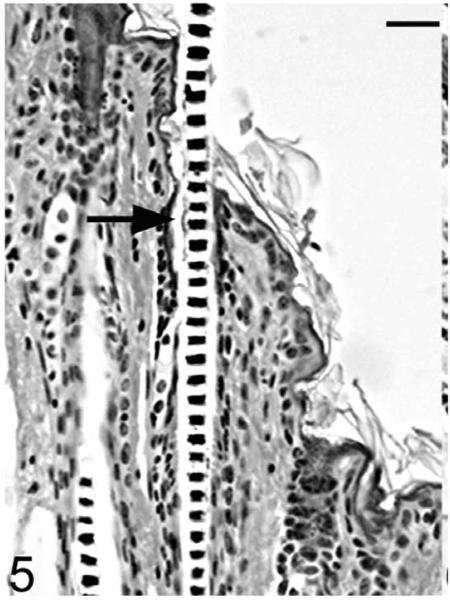
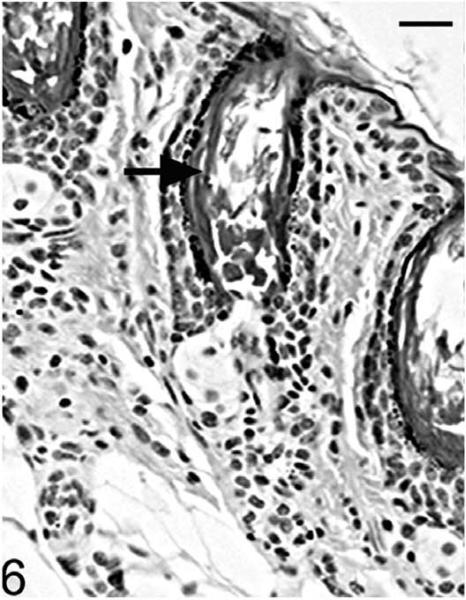
Histological examinations in skin from the dorsal interscapular region suggested grossly evident changes. Hair follicles completed postpartum development normally, and at 5 days of age mutants (Hrrh-R/Hrrh-R) could not be differentiated from littermate controls (Hrrh-R/+ or +/+). Dorsal interscapular skin was examined histologically from replicate mice at 7, 10, 14, 18, 21, and 35 days of age. All mice were in late anagen stage from 7 to 14 days, at which point all entered catagen regardless of genotype. Early changes were observed in Hrrh-R/Hrrh-R mice at 14 days of age (Fig. 4). Infundibula were mildly ectatic and filled with laminated cornified cells. The stratum granulosum extended from the epidermis to the entrance of the sebaceous gland duct. The stratum granulosum was mildly hyperplastic (hypergranulosis) with prominent large basophilic keratohyalin granules. This finding was restricted to the layers producing the cornified material that filled the dilated infundibulum. By contrast, the influndibulae of the normal mice was lined by tight, compact squames that covered a thin-stratum granulosum with fine, dark basophilic granules (Fig. 5). By 18 days of age, the influndibular changes were similar, but the dilation was larger in the mutant mouse (Fig. 6) compared to the normal mice (Fig. 7).
Figure 4.
Skin; Hrrh-R/Hrrh-R mouse, 14 days old. Mouse dorsal skin from the interscapular region was sectioned and stained with HE. Infundibula were mildly ectatic and filled with laminated cornified cells (black arrow). The stratum granulosum extended from the epidermis to the entrance of the sebaceous gland duct. Bar: 20 μm.
Figure 5.
Skin; Hrrh-R/+ mouse, 14 days old. Dorsal skin from the interscapular region was sectioned and stained with HE. The influndibulae (black arrow) were normal and lined by tight, compact squames that covered a thin-stratum granulosum with fine, dark basophilic granules. Bar: 20 μm.
Figure 6.
Skin; Hrrh-R/Hrrh-R mouse, 18 days old. Dorsal skin from the interscapular region was sectioned and stained with HE. The influndibulae were ectatic and filled with more laminated cornified cells (black arrow) when compared to 14-day-old Hrrh-R/Hrrh-R mouse. The dilation also became larger. Bar: 20 μm.
Figure 7.
Skin; Hrrh-R/+ mouse, 18 days old. Dorsal skin from the interscapular region was sectioned and stained with HE. The influndibulae appeared normal (black arrow). Bar: 20 μm.
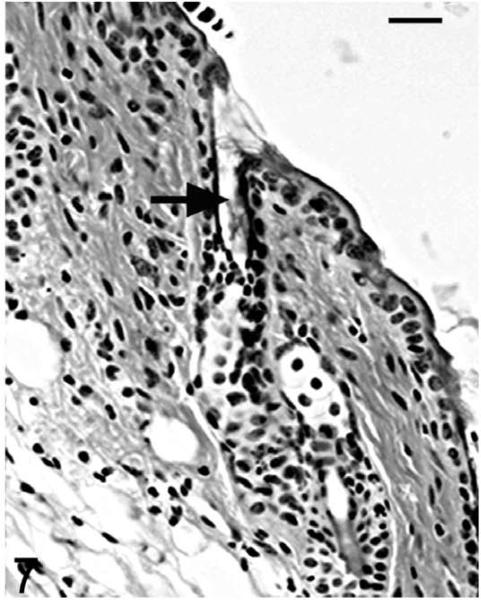
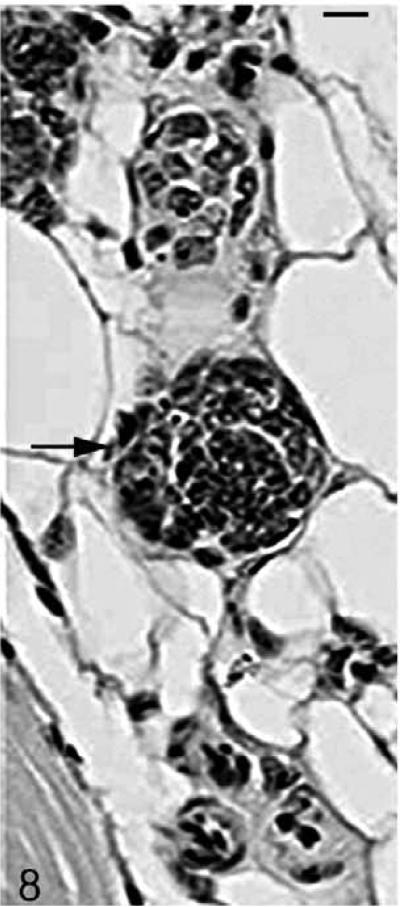
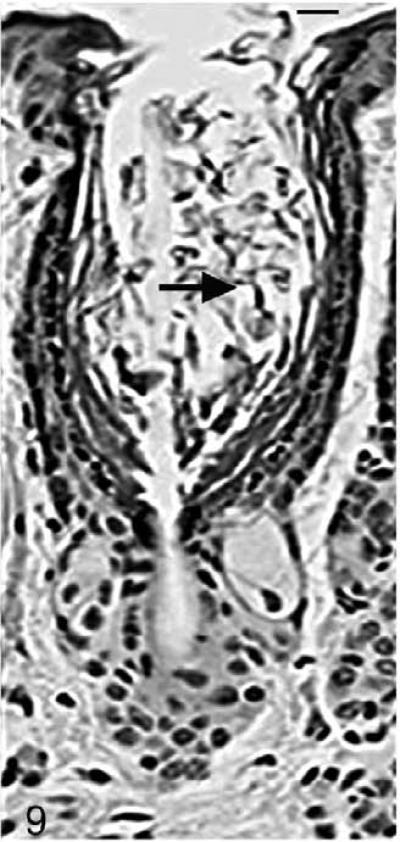
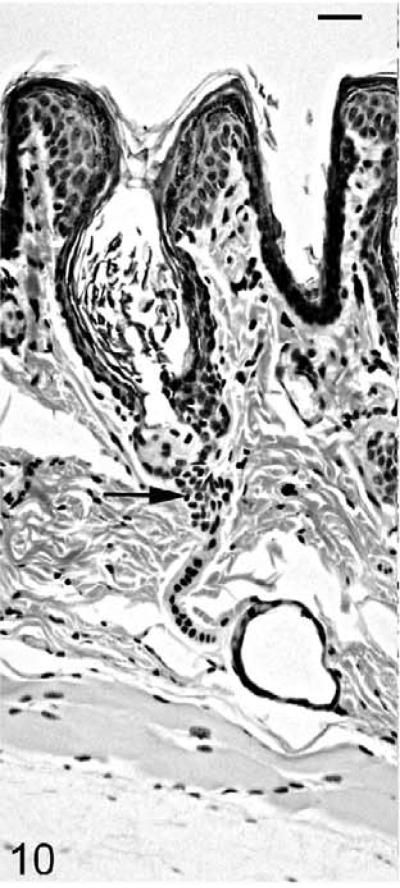
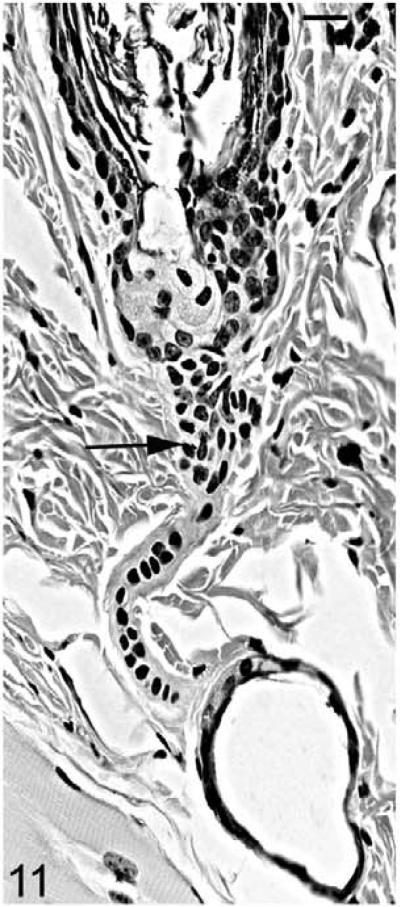
As the mutant hair follicle entered catagen, a rim of keratinocytes surrounded the dermal papilla (Fig. 8). At 21 days of age, when hair follicles were in telogen or early anagen, the remaining club hair was still present in many follicles in which the infundibulum was ectatic and filled with laminated cornified material (Fig. 9). No club hairs remained at 5 weeks of age. As follicles entered catagen at 5 weeks of age, a thin serpentine-shaped band of cells, surrounded by a thick hyalin membrane (glassy membrane), extended from the abnormal ectatic infundibulum to the dilated deep dermal cyst, which developed from keratinocyte remnants that surrounded the dermal papilla (Figs. 10, 11). Some of the cells surrounding these cysts differentiated into sebocytes.63
Figure 8.
Skin; Hrrh-R/Hrrh-R mouse, 21 days old. HE-stained section indicates a rim of keratinocytes surrounding the stranded dermal papilla (black arrow). Bar: 20 μm.
Figure 9.
Skin; Hrrh-R/Hrrh-R mouse, 21 days old. HE-stained section indicates the ectatic infundibula filled with laminated cornified material (black arrow). Bar: 20 μm.
Figure 10.
Skin; Hrrh-R/Hrrh-R mouse, 5 weeks old. HE-stained section indicates the epithelial connection (black arrow) between abnormal infundibulum and deep dermal cyst. Bar: 80 μm.
Figure 11.
Skin; Hrrh-R/Hrrh-R mouse, 5 weeks old. Magnified HE-stained section indicates the epithelial connection (black arrow) between abnormal infundibulum and deep dermal cyst. Bar: 20 μm.
Novel Nonsense Mutation
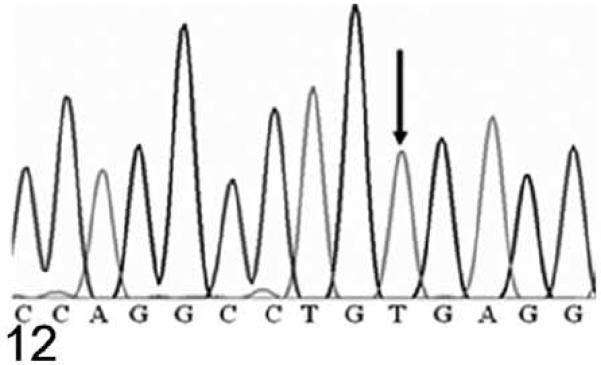
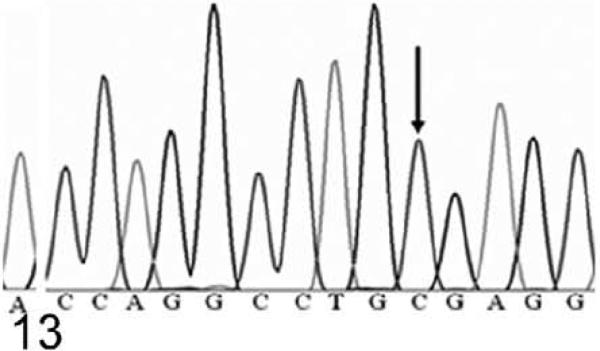
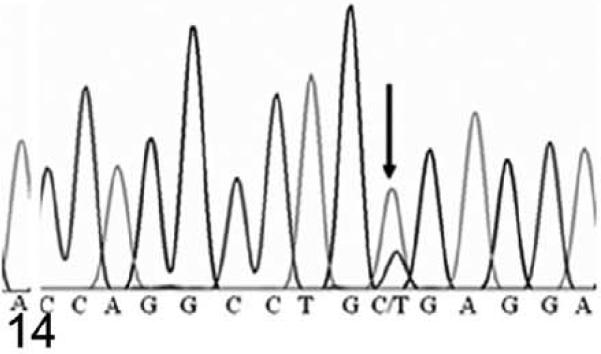
Because of the marked similarity between Hrrh-R/Hrrh-R mice and rhino mutants of Hr, we identified the underlying Hrrh-R mutation by sequencing the exons of Hr in Hrrh-R/Hrrh-R mice. A single nucleotide change (from C to T) was identified in position 3134 of Hr mRNA (NM_021877) in Hrrh-R/Hrrh-R mice (Fig. 12) whereas only C is identified at this position in the wild-type mice (Fig. 13). Hrrh-R/+ mice were found to have both C and T at position 3134 (Fig. 14), consistent with the heterozygosity. This change in exon 12 was predicted to cause a premature stop codon at arginine 814 (R814X). Samples from a total of 100 Hrrh-R/Hrrh-R, Hrrh-R/+, and +/+ mice were sequenced to confirm the mutation, and there were no deviations from the expected genotypes.
Figure 12.
DNA sequence; Hrrh-R/Hrrh-R mouse, 4 weeks old. The electropherogram of DNA sequence around Hrrh-R mutation site in a homozygous mutant mouse (Hrrh-R/Hrrh-R). The arrow indicates the base mutanted in the mutant mice from C to T (3134C to T of NM_021877).
Figure 13.
DNA sequence; wild-type mouse, 4 weeks old. The electropherogram of DNA sequence around Hrrh-R mutation site in a wild-type mouse (+/+). Arrow indicates that the base here is C (3134C in NM_021877).
Figure 14.
DNA sequence; Hrrh-R/+ mouse, 4 weeks old. The electropherogram of DNA sequence around Hrrh-R mutation site in a heterozygous mouse (Hrrh-R/+). Arrow indicates that the base is changed from C to C/T in the heterozygotes (3134C in NM_021877).
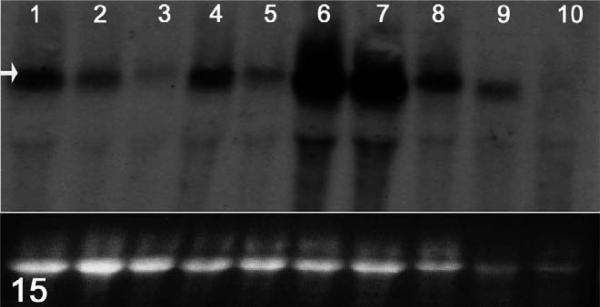
Owing to its distance from the terminal exon, the mutant Hrrh-R mRNA should be degraded through nonsense-mediated decay.68 We used Northern blotting to test this prediction, hybridizing with a probe covering exon 12. Exon 12 is a consensus exon present in the majority of mRNAs reported for Hr and is not known to be alternatively spliced in skin. As shown in Fig. 15, one major band was present in all samples, corresponding to the known size of the Hr transcript. Hr expression was dramatically reduced in Hrrh-R/Hrrh-R mice at all ages examined (1, 7, 14, and 35 days), compared to Hrrh-R/+ littermates. Similar results were obtained with a probe spanning exons 5-11 (data not shown). Therefore, the Hrrh-R mutation is expected to result in hair loss and skin wrinkling through marked loss of HR protein, similar to other rhino alleles of Hr.
Figure 15.
RNA expression; effects of the mutation on Hr mRNA levels. Total RNA was extracted from dorsal skin of each genotype at different ages (1, 7, 14, 35 days after birth). A probe designed to span exon 12 of the Hr cDNA was amplified from wild-type skin cDNA using polymerase chain reaction. From the left side, lanes and samples are as follows: lane 1, 1 day, +/+; lane 2, 1 day, Hrrh-R/+; lane 3, 1 day, Hrrh-R/Hrrh-R; lane 4, 7 days, Hrrh-R/+; lane 5, 7 days, Hrrh-R/Hrrh-R; lane 6, 14 days, +/+; lane 7, 14 days, Hrrh-R/+; lane 8, 14 days, Hrrh-R/Hrrh-R; lane 9, 35 days, Hrrh-R/+; lane 10, 35 days, Hrrh-R/Hrrh-R. Arrow indicates the position of Hr transcript. The bottom panel represents the intensity of 28S rRNA from the ethidium bromide–stained gel as a loading control.
Gene Expression Profiling
Microarrays were used to identify changes in gene expression that preceded onset of the hair follicle phenotype, as a means to profile early molecular events consequent to reduced expression of Hr. At 10 days of age, a total of 120 genes were differentially expressed in skin of Hrrh-R/Hrrh-R mice compared to Hrrh-R/+ littermates (P < .05). Remarkably, only 1 gene exhibited decreased expression; 119 of 120 genes were significantly upregulated. These results are consistent with loss of the transcriptional corepressor function of Hr.71 Results are shown in Table S1.
Within the set of differentially expressed genes, 63 of 120 were annotated with a descriptive gene symbol; 13 of the 63 have established roles in some aspect of skin and hair follicle function. This subset includes epidermal keratins 1 and 10 (Krt1 and Krt10), both of which show increased expression in Hrrh-R/Hrrh-R mutants. Krt 1 and Krt10 are coexpressed in the suprabasal layers of differentiating epidermis and are widely used as markers of keratinocyte differentiation.24,60 Several additional genes, either involved in keratinocyte differentiation or expressed specifically in the differentiated layer of the epidermis, were upregulated, including neuroblastoma myc-related oncogene 1 (Mycn), homeo box D13 (Hoxd13), fos-related antigen 1 (Fos11), and ephrin A3 (Efna3).19,32,40,51,53 Six genes activated in wound healing and epidermal regeneration—stefins A1 and A3 (Stfa1 and Stfa3), integrin beta 4 (Itgb4), inhibin beta E (Inhbe), neuropsin (Prss19), and trefoil factor 1 (Tff1)—also exhibited significantly increased expression in skin from Hrrh-R/Hrrh-R versus Hrrh-R/+ controls.11,22,31,33,34,38,43 In addition, 57 of the 120 differentially expressed genes (47%) encode proteins that play some role in the immune response, including 36 genes or expressed sequence tags that encode components of T-cell receptors and 10 that encode segments of immunoglobulins.
Differentially expressed genes annotated with Entrez GeneIDs (76 of 120) were analyzed for functional enrichment based on GO representation within the biological process ontology using DAVID 2006, with P < .05 for significance.23 The most highly enriched GO category was that of organelle organization and biogenesis (P = .0012); other significantly enriched categories included those of skeletal development, regulation of metabolism, and nucleobase, nucleoside, nucleotide, and nucleic acid metabolism (Table 1).
Table 1.
Significantly Gene Ontology Categories
| Biological Process Gene Ontology Term | No. | P |
|---|---|---|
| Organelle organization and biogenesis | 11 | .0012 |
| Skeletal development | 4 | .0105 |
| Cytoskeleton organization and biogenesis | 6 | .0166 |
| Cortical actin cytoskeleton organization and biogenesis | 2 | .0268 |
| Cortical cytoskeleton organization and biogenesis | 2 | .0301 |
| Regulation of metabolism | 14 | .0427 |
| Nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 17 | .0439 |
Hr acts as a transcriptional regulator that in turn interacts with other transcription factors to control hair follicle development and cycling.12 In silico promoter analysis was used to determine if genes differentially expressed as a result of reduced Hr expression shared one or more regulatory elements, which might further illuminate the molecular pathways through which Hr acts in skin. The oPOSSUM database identifies statistically overrepresented TFBS in a gene's putative promoter regions.27 Like other TFBS identification programs, oPOSSUM is not yet comprehensive across the entire genome; however, it uses phylogenetic footprinting to restrict the search space to regions conserved between mouse and human, making it conservative and markedly reducing the number of false-positive detections.27 As shown in Table 2, a total of 12 TFBS were significantly enriched among the set of differentially expressed genes (Fisher exact test, P < .05). The most significant was the binding site for MAX (P = .00026), a transcription factor that heterodimerizes with members of the Myc superfamily of transcription factors, such as MYCN. The binding site for MYCN was also overrepresented, and the Mycn gene was upregulated in Hrrh-R/Hrrh-R skin. Other enriched TFBS include those for aryl-hydrocarbon receptor nuclear translocator protein (ARNT) and upstream stimulatory factor 1 (USF1). ARNT executes nuclear translocation of the Ah receptor, which has been hypothesized to interact directly with HR.35,64 USF1 is a transcriptional regulator of genes involved in wound repair, a function shared by several genes differentially expressed in Hrrh-R/Hrrh-R skin.6,49
Table 2.
Transcription Factor Binding Sites Significantly Overrepresented Among Genes Differentially Expressed in Hrrh-R/Hrrh,-R
| Transcription Factor | No. Genes | Fisher P |
|---|---|---|
| Max | 13 | .00026 |
| USF | 14 | .0025 |
| Mzf1-4 | 29 | .0028 |
| Spz1 | 6 | .0086 |
| Arnt | 10 | .014 |
| NF-kappaB | 10 | .019 |
| N-Myc | 19 | .026 |
| bZIP910 | 20 | .027 |
| Irf-1 | 6 | .029 |
| Sox-9 | 13 | .042 |
| Yin-Yang | 26 | .049 |
| Aml-1 | 14 | .049 |
Discussion
Mutations in Hr represent the richest set of allelic mutations known to cause hair loss in multiple species. Despite this wealth of models, the mechanisms through which these mutations lead to hair loss remain poorly defined at the molecular level. We identified a novel nonsense mutation in Hr gene that leads to reduced expression, presumably through nonsense-mediated decay. Hrrh-R represents the 15th cloned or putative allele of Hr reported to cause hair loss in mice. One explanation for this abundance of mutant alleles is that Hr represents a gene for which mutations cause an obvious phenotype but not lethal or decreased fecundity, facilitating their recognition and propagation in mouse colonies worldwide.
The phenotype of Hrrh-R/Hrrh-R mice resembles that of other rhino alleles of Hr with respect to lesions within the hair follicle and the eventual wrinkling of the skin as hair follicle remnants become filled with cornified material.45 The main difference between Hrrh-R and other Hr mutants is that hair loss develops in a diffuse manner across the body, first noticeable on the ventrum, rather than in a rostral-caudal pattern coincident with hair cycle progression.61 The reason for this difference remains unknown but might result from modifier alleles that affect retention of club hairs and are unique to the mixed-stock genetic background on which Hrrh-R is maintained. It is worth noting that the pattern of hair follicle lesions in histological sections appeared uniform, as opposed to the diffuse pattern in hair loss of this mouse mutant.
Northern blots indicate that little Hr mRNA remains in the skin of Hrrh-R/Hrrh-R mice. Therefore, Hrrh-R represents an additional member of the allelic series of Hr mutations that impair Hr expression and cause hair loss owing to dramatic reduction in HR protein.
Loss of Hr expression led to upregulation of a number of genes involved in epidermal regeneration and differentiation, before the point at which obvious morphological lesions were detected in sections of dorsal skin. These changes suggest that when HR is lost or significantly reduced, as predicted in Hrrh-R/Hrrh-R mice owing to the marked reduction in Hr mRNA, the coordinated pattern of gene expression necessary for progression of hair follicle differentiation through the hair cycle is significantly altered. Genes upregulated relative to heterozygous controls include several early markers of keratinocyte differentiation such as keratins 1 and 10 and Fos11.24,50,51 Also in this set are genes linked more specifically to the hair follicle, including Mycn, Hoxd13, Efna3, and Bmi1.32,41 Bmi1 has been implicated as a self-renewal gene expressed in hair follicle stem cells,21,46 a population proposed to be controlled in part by Hr.71 Six genes upregulated in Hrrh-R/Hrrh-R skin are also activated during wound healing (Stfa1, Stfa3, Itgb4, Inhbe, Prss19, and Tff1).* The single gene (RIKEN cDNA 1500031M22) that was downregulated in Hrrh-R/Hrrh-R skin is a putative mouse homologue of a wound-healing gene found in Xenopus laevis.57,69
At the same time, about 47% of upregulated genes are involved in some component of immune function. These changes may be reflective of Hr involvement in the immune system, which has been suggested by premature thymic atrophy in both Hrrh-R/Hrrh-R mice and other Hr mutants.45,59
In silico promoter analysis of differentially expressed genes suggests that HR interacts with pathways utilizing the Myc/Max superfamily of transcriptional regulators. Max and Mycn are coexpressed in all epithelial layers of the epidermis and hair follicle and in the follicle bulb, bulge, outer root shealth, and sebaceous gland. Together they play key roles in directing the fate of hair follicle stem cells, a process proposed to be controlled in part by HR.19,71 It cannot be determined from these results if enrichment of Myc and Max binding sites among genes differentially expressed in Hrrh-R/Hrrh-R skin is a primary event of HR loss or secondary to changes in the hair follicles. However, that this enrichment is present in genes differentially expressed before dramatic structural alterations suggests cross-talk between HR and Myc/Max signaling as a potential molecular mechanism for HR regulation of cell populations and differentiation in the hair follicle. Further experiments will be necessary to explore this possibility.
In summary, we have cloned and characterized a new allelic mutation of Hr that produces a rhino phenotype through loss of Hr expression. Upregulation of a number of genes involved in keratinocyte differentiation precedes manifestation of the phenotype and supports a proposed role for Hr in control of keratinocyte fate. Additional study will be necessary to identify the genes that are direct targets of HR and that initiate the cascade of events that lead to hair loss in Hr mutants.
Acknowledgements
We would like to thank Dr L. Webb (Christopher Newport University) for her critical reading of this manuscript. We also thank Dr L. E. King (Vanderbilt University) for insightful comments on the pathogenesis of hair loss in Hrrh-R mice. This work was supported by the Office of Biological and Environmental Research, US Department of Energy, under contract No. DE-AC05-00OR22725 with UT-Battelle and by grants from the National Institutes of Health (RR00173).
Financial Disclosure/Funding
Funding for this research was provided by the Office of Biological and Environmental Research, US Department of Energy, under contract DE-AC05-00OR22725 with UT-Battelle LLC, the managing organization of ORNL for the U.S. DOE.
Footnotes
References
- 1.Ahmad W, Haque MF, Brancolini V, Tsou HC, Haque S, Lam HM, Aita VM, Owen J, deBlaquiere M, Frank J, Cserhalmi-Friedman PB, Leask A, McGrath JA, Peacocke M, Ahmad M, Ott J, Christiano AM. Alopecia universalis associated with a mutation in the human hairless gene. Science. 1998;279:720–724. doi: 10.1126/science.279.5351.720. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad W, Nomura K, McGrath JA, Hashimoto I, Christiano AM. A homozygous nonsense mutation in the zinc-finger domain of the human hairless gene underlies congenital atrichia. J Invest Dermatol. 1999;113:281–283. doi: 10.1046/j.1523-1747.1999.00686.x. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad W, Panteleyev AA, Henson-Apollonio V, Sundberg JP, Christiano AM. Molecular basis of a novel rhino (hrrhChr) pheno-type: a nonsense mutation in the mouse hairless gene. Exp Dermatol. 1998;7:298–301. doi: 10.1111/j.1600-0625.1998.tb00300.x-i1. [DOI] [PubMed] [Google Scholar]
- 4.Ahmad W, Panteleyev AA, Sundberg JP, Christiano AM. Molecular basis for the rhino (hrrh-8J) phenotype: a nonsense mutation in the mouse hairless gene. Genomics. 1998;53:383–386. doi: 10.1006/geno.1998.5495. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad W, Ratterree MS, Panteleyev AA, Aita VM, Sundberg JP, Christiano AM. Atrichia with papular lesions resulting from mutations in the rhesus macaque (Macaca mulatta) hairless gene. Lab Anim. 2002;36:61–67. doi: 10.1258/0023677021911777. [DOI] [PubMed] [Google Scholar]
- 6.Allen RR, Qi L, Higgins PJ. Upstream stimulatory factor regulates E box-dependent PAI-1 transcription in human epidermal keratinocytes. J Cell Physiol. 2005;203:156–165. doi: 10.1002/jcp.20211. [DOI] [PubMed] [Google Scholar]
- 7.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 8.Ashoor GG, Greenstein RM, Lam H, Martinez-Mir A, Zlotogorski A, Christiano AM. Novel compound heterozygous nonsense mutations in the hairless gene causing atrichia with papular lesions. J Dermatol Sci. 2005;40:29–33. doi: 10.1016/j.jdermsci.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Balciunas D, Ronne H. Evidence of domain swapping within the jumonji family of transcription factors. Trends Biochem Sci. 2000;25:274–276. doi: 10.1016/s0968-0004(00)01593-0. [DOI] [PubMed] [Google Scholar]
- 10.Bale SJ. Of hairless mice and men: the genetic basis of congenital alopecia universalis/congenital atrichia. J Cutan Med Surg. 1999;3:309–311. doi: 10.1177/120347549900300607. [DOI] [PubMed] [Google Scholar]
- 11.Bamberger C, Scharer A, Antsiferova M, Tychsen B, Pankow S, Muller M, Rülicke T, Paus R, Werner S. Activin controls skin morphogenesis and wound repair predominantly via stromal cells and in a concentration-dependent manner via keratinocytes. Am J Pathol. 2005;167:733–747. doi: 10.1016/S0002-9440(10)62047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaudoin GM, III, Sisk JM, Coulombe PA, Thompson CC. Hairless triggers reactivation of hair growth by promoting Wnt signaling. Proc Natl Acad Sci U S A. 2005;102:14653–14658. doi: 10.1073/pnas.0507609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beer HD, Fassler R, Werner S. Glucocorticoid-regulated gene expression during cutaneous wound repair. Vitam Horm. 2000;59:217–239. doi: 10.1016/s0083-6729(00)59008-6. [DOI] [PubMed] [Google Scholar]
- 14.Bergman R, Schein-Goldshmid R, Hochberg Z, Ben Izhak O, Sprecher E. The alopecias associated with vitamin D-dependent rickets type IIA and with hairless gene mutations: a comparative clinical, histologic, and immunohistochemical study. Arch Dermatol. 2005;141:343–351. doi: 10.1001/archderm.141.3.343. [DOI] [PubMed] [Google Scholar]
- 15.Bikle DD, Elalieh H, Chang S, Xie Z, Sundberg JP. Development and progression of alopecia in the vitamin D receptor null mouse. J Cell Physiol. 2006;207:340–353. doi: 10.1002/jcp.20578. [DOI] [PubMed] [Google Scholar]
- 16.Bossenmeyer-Pourie C, Kannan R, Ribieras S, Wendling C, Stoll I, Thim L, Tomasetto C, Rio MC. The trefoil factor 1 participates in gastrointestinal cell differentiation by delaying G1-S phase transition and reducing apoptosis. J Cell Biol. 2002;157:761–770. doi: 10.1083/jcb200108056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brancaz MV, Iratni R, Morrison A, Mancini SJ, Marche P, Sundberg J, Nonchev S. A new allele of the mouse hairless gene interferes with Hox/LacZ transgene regulation in hair follicle primordia. Exp Mol Pathol. 2004;76:173–181. doi: 10.1016/j.yexmp.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Brooke HC. Hairless mice. J Hered. 1926;17:173–174. [Google Scholar]
- 19.Bull JJ, Muller-Rover S, Patel SV, Chronnell CM, McKay IA, Philpott MP. Contrasting localization of c-Myc with other Myc superfamily transcription factors in the human hair follicle and during the hair growth cycle. J Invest Dermatol. 2001;116:617–622. doi: 10.1046/j.1523-1747.2001.12771234.x. [DOI] [PubMed] [Google Scholar]
- 20.Cachon-Gonzalez MB, Fenner S, Coffin JM, Moran C, Best S, Stoye JP. Structure and expression of the hairless gene of mice. Proc Natl Acad Sci U S A. 1994;91:7717–7721. doi: 10.1073/pnas.91.16.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claudinot S, Nicolas M, Oshima H, Rochat A, Barrandon Y. Long-term renewal of hair follicles from clonogenic multipo-tent stem cells. Proc Natl Acad Sci U S A. 2005;102:14677–14682. doi: 10.1073/pnas.0507250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dang H, Trempus C, Malarkey DE, Wei SJ, Humble M, Morris RJ, Tennant RW. Identification of genes and gene ontology processes critical to skin papilloma development in Tg.AC transgenic mice. Mol Carcinog. 2006;45:126–140. doi: 10.1002/mc.20154. [DOI] [PubMed] [Google Scholar]
- 23.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 24.Fuchs E. Keratins as biochemical markers of epithelial differentiation. Trends Genet. 1988;4:277–281. doi: 10.1016/0168-9525(88)90169-2. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Atares N, San J,I, Cabo R, Vega JA, Represa J. Changes in the cerebellar cortex of hairless Rhino-J mice (hrrh-J). Neurosci Lett. 1998;256:13–16. doi: 10.1016/s0304-3940(98)00757-5. [DOI] [PubMed] [Google Scholar]
- 26.Gaskoin JS. On a peculiar variety of Mus musculus. Proc Zool Soc London. 1856;24:38. [Google Scholar]
- 27.Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howard A. “Rhino,” an allele of hairless in the house mouse. J Hered. 1940;31:467–470. [Google Scholar]
- 29.Hsieh JC, Sisk JM, Jurutka PW, Haussler CA, Slater SA, Haussler MR, Thompson CC. Physical and functional interaction between the vitamin D receptor and hairless corepressor, two proteins required for hair cycling. J Biol Chem. 2003;278:38665–38674. doi: 10.1074/jbc.M304886200. [DOI] [PubMed] [Google Scholar]
- 30.Inoue N, Kuwae K, Ishida-Yamamoto A, Iizuka H, Shibata M, Yoshida S, Kato K, Shiosaka S. Expression of neuropsin in the keratinizing epithelial tissue-immunohistochemical analysis of wild-type and nude mice. J Invest Dermatol. 1998;110:923–931. doi: 10.1046/j.1523-1747.1998.00212.x. [DOI] [PubMed] [Google Scholar]
- 31.Kanai M, Mullen C, Podolsky DK. Intestinal trefoil factor induces inactivation of extracellular signal-regulated protein kinase in intestinal epithelial cells. Proc Natl Acad Sci U S A. 1998;95:178–182. doi: 10.1073/pnas.95.1.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanzler B, Viallet JP, Le Mouellic H, Boncinelli E, Duboule D, Dhouailly D. Differential expression of two different homeobox gene families during mouse tegument morphogenesis. Int J Dev Biol. 1994;38:633–640. [PubMed] [Google Scholar]
- 33.Kirihara T, Matsumoto-Miyai K, Nakamura Y, Sadayama T, Yoshida S, Shiosaka S. Prolonged recovery of ultraviolet B-irradiated skin in neuropsin (KLK8)-deficient mice. Br J Dermatol. 2003;149:700–706. doi: 10.1046/j.1365-2133.2003.05484.x. [DOI] [PubMed] [Google Scholar]
- 34.Kitayoshi H, Inoue N, Kuwae K, Chen ZL, Sato H, Ohta T, Hosokawa K, Itami S, Yoshikawa K, Yoshida S, Shiosaka S. Effect of 12-O-tetradecanoyl-phorbol ester and incisional wounding on neuropsin mRNA and its protein expression in murine skin. Arch Dermatol Res. 1999;291:333–338. doi: 10.1007/s004030050418. [DOI] [PubMed] [Google Scholar]
- 35.Knutson JC, Poland A. Response of murine epidermis to 2,3,7,8-tetrachlorodibenzo-p-dioxin: interaction of the ah and hr loci. Cell. 1982;30:225–234. doi: 10.1016/0092-8674(82)90028-9. [DOI] [PubMed] [Google Scholar]
- 36.Kuwae K, Matsumoto-Miyai K, Yoshida S, Sadayama T, Yoshikawa K, Hosokawa K, Shiosaka S. Epidermal expression of serine protease, neuropsin (KLK8) in normal and pathological skin samples. Mol Pathol. 2002;55:235–241. doi: 10.1136/mp.55.4.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Das S, Olszewski RE, Carpenter DA, Culiat CT, Sundberg JP, Soteropoulos P, Liu X, Doktycz MJ, Michaud EJ, Voy BH. The near-naked hairless (HrN) mutation disrupts hair formation but is not due to a mutation in the hairless coding region. J Invest Dermatol. 2007;127(7):1605–1614. doi: 10.1038/sj.jid.5700755. [DOI] [PubMed] [Google Scholar]
- 38.Madsen P, Rasmussen HH, Leffers H, Honore B, Celis JE. Molecular cloning and expression of a novel keratinocyte protein (psoriasis-associated fatty acid-binding protein [PA-FABP]) that is highly up-regulated in psoriatic skin and that shares similarity to fatty acid-binding proteins. J Invest Dermatol. 1992;99:299–305. doi: 10.1111/1523-1747.ep12616641. [DOI] [PubMed] [Google Scholar]
- 39.Mann SJ. Hair loss and cyst formation in hairless and rhino mutant mice. Anat Rec. 1971;170:485–499. doi: 10.1002/ar.1091700409. [DOI] [PubMed] [Google Scholar]
- 40.Midorikawa T, Chikazawa T, Yoshino T, Takada K, Arase S. Different gene expression profile observed in dermal papilla cells related to androgenic alopecia by DNA macroarray analysis. J Dermatol Sci. 2004;36:25–32. doi: 10.1016/j.jdermsci.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Mill P, Mo R, Hu MC, Dagnino L, Rosenblum ND, Hui CC. Shh controls epithelial proliferation via independent pathways that converge on N-Myc. Dev Cell. 2005;9:293–303. doi: 10.1016/j.devcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Moraitis AN, Giguere V, Thompson CC. Novel mechanism of nuclear receptor corepressor interaction dictated by activation function 2 helix determinants. Mol Cell Biol. 2002;22:6831–6841. doi: 10.1128/MCB.22.19.6831-6841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nikolopoulos SN, Blaikie P, Yoshioka T, Guo W, Puri C, Tacchetti C, Giancotti FG. Targeted deletion of the integrin beta4 signaling domain suppresses laminin-5-dependent nuclear entry of mitogen-activated protein kinases and NF-kappaB, causing defects in epidermal growth and migration. Mol Cell Biol. 2005;25:6090–6102. doi: 10.1128/MCB.25.14.6090-6102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panteleyev AA, Ahmad W, Malashenko AM, Ignatieva EL, Paus R, Sundberg JP, Christiano AM. Molecular basis for the rhino Yurlovo (hrrhY) phenotype: severe skin abnormalities and female reproductive defects associated with an insertion in the hairless gene. Exp Dermatol. 1998;7:281–288. doi: 10.1111/j.1600-0625.1998.tb00298.x-i1. [DOI] [PubMed] [Google Scholar]
- 45.Panteleyev AA, Paus R, Ahmad W, Sundberg JP, Christiano AM. Molecular and functional aspects of the hairless (hr)genein laboratory rodents and humans. Exp Dermatol. 1998;7:249–267. doi: 10.1111/j.1600-0625.1998.tb00295.x-i1. [DOI] [PubMed] [Google Scholar]
- 46.Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 47.Potter GB, Zarach JM, Sisk JM, Thompson CC. The thyroid hormone-regulated corepressor hairless associates with histone deacetylases in neonatal rat brain. Mol Endocrinol. 2002;16:2547–2560. doi: 10.1210/me.2002-0115. [DOI] [PubMed] [Google Scholar]
- 48.Prophet E, Mills B, Arrington J, Sobin L. AFIP Laboratory Methods in Histotechnology. American Registry of Pathology; Washinton, DC: 1992. [Google Scholar]
- 49.Providence KM, White LA, Tang J, Gonclaves J, Staiano-Coico L, Higgins PJ. Epithelial monolayer wounding stimulates binding of USF-1 to an E-box motif in the plasminogen activator inhibitor type 1 gene. J Cell Sci. 2002;115:3767–3777. doi: 10.1242/jcs.00051. [DOI] [PubMed] [Google Scholar]
- 50.Rundhaug JE, Hawkins KA, Pavone A, Gaddis S, Kil H, Klein RD, Berton TR, McCauley E, Johnson DG, Lubet RA, Fischer SM, Aldaz CM. SAGE profiling of UV-induced mouse skin squamous cell carcinomas, comparison with acute UV irradiation effects. Mol Carcinog. 2005;42:40–52. doi: 10.1002/mc.20064. [DOI] [PubMed] [Google Scholar]
- 51.Rutberg SE, Saez E, Glick A, Dlugosz AA, Spiegelman BM, Yuspa SH. Differentiation of mouse keratinocytes is accompanied by PKC-dependent changes in AP-1 proteins. Oncogene. 1996;13:167–176. [PubMed] [Google Scholar]
- 52.San Jose I, Garcia-Suarez O, Hannestad J, Cabo R, Gauna L, Represa J, Vega JA. The thymus of the hairless rhino-j (hrrh-J) mice. J Anat. 2001;198:399–406. doi: 10.1046/j.1469-7580.2001.19840399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmuth M, Elias PM, Hanley K, Lau P, Moser A, Willson TM, Bikle DD, Feingold KR. The effect of LXR activators on AP-1 proteins in keratinocytes. J Invest Dermatol. 2004;123:41–48. doi: 10.1111/j.0022-202X.2004.22707.x. [DOI] [PubMed] [Google Scholar]
- 54.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 55.Smyth GK, Yang YH, Speed T. Statistical issues in cDNA micro-array data analysis. Methods Mol Biol. 2003;224:111–136. doi: 10.1385/1-59259-364-X:111. [DOI] [PubMed] [Google Scholar]
- 56.Sprecher E, Lestringant GG, Szargel R, Bergman R, Labay V, Frossard PM, Friedman-Birnbaum R, Cohen N. Atrichia with papular lesions resulting from a nonsense mutation within the human hairless gene. J Invest Dermatol. 1999;113:687–690. doi: 10.1046/j.1523-1747.1999.00723.x. [DOI] [PubMed] [Google Scholar]
- 57.Stanchi F, Bertocco E, Toppo S, Dioguardi R, Simionati B, Cannata N, Zimbello R, Lanfranchi G, Valle G. Characterization of 16 novel human genes showing high similarity to yeast sequences. Yeast. 2001;18:69–80. doi: 10.1002/1097-0061(200101)18:1<69::AID-YEA647>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 58.Sundberg JP, Boggess D. Rhino-9J (hrrh9J): a new allele at the hairless locus. Vet Pathol. 1998;35:297–299. doi: 10.1177/030098589803500409. [DOI] [PubMed] [Google Scholar]
- 59.Sundberg JP, Dunstan RW, Compton JG. Hairless mouse, HRS/J hr/hr. In: Jones TC, Mohr U, Hunt RD, editors. Integument and Mammary Glands: Monographs on Pathology of Laboratory Animals. Springer-Verlag; Heidelberg, Germany: 1989. pp. 192–197. [Google Scholar]
- 60.Sundberg JP, Erickson AA, Roop DR, Binder RL. Ornithine decarboxylase expression in cutaneous papillomas in SENCAR mice is associated with altered expression of keratins 1 and 10. Cancer Res. 1994;54:1344–1351. [PubMed] [Google Scholar]
- 61.Sundberg JP, King LE. Skin and its appendages: normal anatomy and pathology of spontaneous, transgenic, and targeted mouse mutations. In: Ward J, Mahler JF, Maronpot RR, Sundberg JP, editors. Pathology of Genetically Engineered Mice. Iowa State University Press; Ames, IA: 2000. pp. 181–213. [Google Scholar]
- 62.Sundberg JP, Price VH, King LE., Jr The ‘‘hairless’’ gene in mouse and man. Arch Dermatol. 1999;135:718–720. doi: 10.1001/archderm.135.6.718. [DOI] [PubMed] [Google Scholar]
- 63.Sundberg JP, Roop DR, Dunstan R, Lavker R, Sun TT. Interaction between dermal papilla and bulge: the rhino mouse mutation as a model system. Ann N Y Acad Sci. 1991;642:496–499. doi: 10.1111/j.1749-6632.1991.tb24430.x. [DOI] [PubMed] [Google Scholar]
- 64.Swanson HI, Chan WK, Bradfield CA. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J Biol Chem. 1995;270:26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- 65.Thompson CC, Bottcher MC. The product of a thyroid hormone-responsive gene interacts with thyroid hormone receptors. Proc Natl Acad Sci U S A. 1997;94:8527–8532. doi: 10.1073/pnas.94.16.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques. 2000;29:52, 54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- 67.Tusher VG, Tibshirani R, Chu G. Significance analysis of micro-arrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wagner E, Lykke-Andersen J. mRNA surveillance: the perfect persist. J Cell Sci. 2002;115:3033–3038. doi: 10.1242/jcs.115.15.3033. [DOI] [PubMed] [Google Scholar]
- 69.Wolfe AD, Crimmins G, Cameron JA, Henry JJ. Early regeneration genes: building a molecular profile for shared expression in cornea-lens transdifferentiation and hindlimb regeneration in Xenopus laevis. Dev Dyn. 2004;230:615–629. doi: 10.1002/dvdy.20089. [DOI] [PubMed] [Google Scholar]
- 70.Xie Z, Chang S, Oda Y, Bikle DD. Hairless suppresses vitamin D receptor transactivation in human keratinocytes. Endocrinology. 2006;147:314–323. doi: 10.1210/en.2005-1111. [DOI] [PubMed] [Google Scholar]
- 71.Zarach JM, Beaudoin GM, III, Coulombe PA, Thompson CC. The co-repressor hairless has a role in epithelial cell differentiation in the skin. Development. 2004;131:4189–4200. doi: 10.1242/dev.01303. [DOI] [PubMed] [Google Scholar]
- 72.Zhang JT, Fang SG, Wang CY. A novel nonsense mutation and polymorphisms in the mouse hairless gene. J Invest Dermatol. 2005;124:1200–1205. doi: 10.1111/j.0022-202X.2005.23744.x. [DOI] [PubMed] [Google Scholar]
- 73.Zlotogorski A, Hochberg Z, Mirmirani P, Metzker A, Ben-Amitai D, Martinez-Mir A, Panteleyev AA, Christiano AM. Clinical and pathologic correlations in genetically distinct forms of atrichia. Arch Dermatol. 2003;139:1591–1596. doi: 10.1001/archderm.139.12.1591. [DOI] [PubMed] [Google Scholar]