Abstract
Nuclear DBF-2-related (NDR) kinases are essential regulators of cell cycle progression, growth, and development in many organisms and are activated by the binding of an Mps One Binder (MOB) protein partner, autophosphorylation, and phosphorylation by an upstream STE20 family kinase. In the protozoan parasite, Trypanosoma brucei, the causative agent of human African trypanosomiasis, the NDR kinase, PK50, is expressed in proliferative life cycle stages and was shown to complement a yeast NDR kinase mutant cell line. However, the function of PK50 and a second NDR kinase, PK53, in T. brucei has not been determined to date, although trypanosome MOB1 is known to be essential for cytokinesis, suggesting the NDR kinases may also be involved in this process. Here, we show that specific depletion of PK50 or PK53 from bloodstream stage trypanosomes resulted in the rapid accumulation of cells with two nuclei and two kinetoplasts, indicating that cytokinesis was specifically inhibited. This led to a deregulation of the cell cycle and cell death and provides genetic validation of these kinases as potential novel drug targets for human African trypanosomiasis. Recombinant active PK50 and PK53 were produced and biochemically characterized. Both enzymes autophosphorylated, were able to trans-phosphorylate generic kinase substrates in vitro, and were active in the absence of phosphorylation by an upstream kinase. Additionally, both enzymes were active in the absence of MOB1 binding, which was also demonstrated to likely be a feature of the kinases in vivo. Biochemical characterization of recombinant PK50 and PK53 has revealed key kinetic differences between them, and the identification of in vitro peptide substrates in this study paves the way for high throughput inhibitor screening of these kinases.
Keywords: Phosphorylation/Kinases/Serine-Threonine, Signal Transduction/Protein Kinases/Serine/Threonine, Cell Division, Enzyme Kinetics, Parasitology, RNA Interference (RNAi), Trypanosoma brucei, Cytokinesis, Drug Target, NDR Kinase
Introduction
Nuclear DBF2-related (NDR)3 kinases are important regulators of a variety of cellular processes, including centrosome duplication, mitotic exit, cytokinesis, morphogenesis, and cell growth and development. The founding member of this family, DBF2, is part of the mitotic exit network in Saccharomyces cerevisiae (1), although its counterpart in fission yeast, Sid2, is a component of the septation initiation network (2). Both kinases were recently shown to phosphorylate CDC14 phosphatase, promoting its retention in the cytoplasm and allowing it to inactivate mitotic cyclin/cyclin-dependent kinase and promote cell cycle progression (3, 4). Other NDR kinases in these yeasts (CBK1 in S. cerevisiae and Orb6 in Schizosaccharomyces pombe) regulate cellular morphogenesis (5, 6). In contrast, the fungal pathogen, Candida albicans, expresses only a single DBF2 orthologue, which is essential for viability and appears to have evolved multiple functions, being required for mitotic spindle organization, actomyosin ring contraction, and hyphal growth (7). In Drosophila, Tricornered/NDR1 (Trc/NDR1) regulates dendritic tiling and branching (8) as well as morphogenesis of epidermal outgrowths (9). A second NDR kinase, Warts/Lats (Wts/Lts), in addition to a role in dendrite maintenance (10), acts as a tumor suppressor, coordinating cell proliferation and apoptosis (11), and phosphorylates the transcriptional co-activator, Yorkie, which stimulates transcription of cyclin E and the apoptosis inhibitor, Diap1. NDR phosphorylation of Yorkie promotes its binding to 14-3-3 proteins in the cytoplasm and hence inhibits its nuclear localization and function (12). Human cells express four NDR kinases, NDR1/2 and LATS1/2. NDR1/2 regulate centrosome duplication (13), and LATS1/2 phosphorylate the Yorkie homologue, Yes-associated protein (YAP) (14), regulate organ size (15), and have tumor suppressor functions, regulating progression through mitosis (16).
NDR kinases are members of the AGC group of protein kinases and are known to be activated by the binding of a Mps One Binder (MOB) protein partner and also via phosphorylation of the kinase domain activation loop and a C-terminal hydrophobic motif. In most AGC kinases, an upstream kinase phosphorylates both sites. However, NDR kinases autophosphorylate on the activation loop residue (Ser-281 in human NDR1) (17). They also possess a basic insert sequence just before the activation loop, which negatively regulates activity. MOB protein binding at the N terminus of the kinase relieves this inhibition, stimulates autophosphorylation, and activates the kinase (18). It has also been suggested that in S. pombe, MOB protein binding reduces an inhibitory self-association of the Sid2 NDR kinase (2). STE20-like kinases further activate NDR kinases by phosphorylating the hydrophobic motif (Thr-444 in human NDR1) and also indirectly by phosphorylating MOB, increasing its binding affinity for the NDR kinase (18–20). Scaffolding proteins are also known to enhance the NDR/MOB interaction (21, 22), and MOB binding has been shown to target the NDR kinase to the plasma membrane, bringing it into close proximity with its STE20 kinase activator (23, 24). Recently, phosphorylation of MOB1 by CDK1 in S. cerevisiae was reported to inhibit DBF2 kinase activity without interfering with the MOB1/DBF2 interaction (25). Regulation of NDR kinase activity is therefore complex and not yet fully understood.
The protozoan parasite, Trypanosoma brucei, causes devastating diseases of humans (human African trypanosomiasis) and animals (N′gana) in sub-Saharan Africa. It has a digenetic life cycle, split between mammalian and tsetse fly hosts. In the mammalian bloodstream, the surface coat of the parasite undergoes antigenic variation, and therefore a vaccine against the parasite has so far proved elusive (26). Chemotherapies are available to treat the disease but can cause undesirable and even dangerous side effects, and only two drugs, melarsoprol and difluoromethylornithine (DFMO or eflornithine), both parenterally administered, are effective against late stage disease, characterized by the parasite crossing the blood-brain barrier (27). The recently approved nifurtimox-eflornithine combination therapy (28) offers improved safety over DFMO monotherapy, but new drugs are still urgently needed.
The trypanosome cell cycle displays several unusual features compared with mammalian cells, and it is likely that novel signal transduction pathways regulate these cellular processes. Elucidation of these pathways could identify novel drug targets. In particular, cytokinesis occurs via a distinctive physical mechanism (29, 30). Whereas mammalian cells divide using a contractile actomyosin ring, in the parasite, a furrow ingresses unidirectionally from the anterior to the posterior along the helical axis of the cell to divide the microtubule cytoskeleton and cell contents. Parasite cell division is highly precise; a number of single copy organelles must be replicated, segregated, and positioned accurately so that the furrow can pass between them to generate two daughter cells with equivalent complements of organelles. Any perturbation of cytokinesis results in disastrous consequences for the parasite (30), suggesting that inhibiting a parasite cytokinesis regulator might have therapeutic potential for human African trypanosomiasis.
T. brucei expresses two MOB1 proteins (MOB1A and MOB1B) and two putative NDR kinases, PK50 and PK53 (31, 32). The MOB1 proteins play essential roles during furrow ingression in bloodstream stage trypanosomes, because their depletion leads to an accumulation of post-mitotic cells with partially ingressed furrows, resulting in deregulation of the cell cycle (32). Previous data also suggested that MOB1A and PK50 can interact in procyclic (insect) form T. brucei (32). The functions of the NDR kinases in trypanosomes have not been studied previously, although it was reported that PK50 is expressed in dividing parasite life cycle stages and is able to complement an Orb6 fission yeast mutant (31). We therefore sought to determine whether the trypanosome NDR kinases were, like the MOB1 proteins, also key cell cycle regulators, to investigate the interactions between the NDR kinases and MOB1 proteins and to evaluate the potential of the NDR kinases as novel drug targets for human African trypanosomiasis.
EXPERIMENTAL PROCEDURES
Gene Sequences
In this study, gene fragments for cloning were amplified from DNA from either EATRO795 or 427 strains. For PK53, DNA sequencing revealed only one nonsynonymous mutation, resulting in a single predicted amino acid change (E440D) compared with the 927 genome sequence (GeneDB accession number Tb927.7.5770), present in both EATRO795 (EMBL accession number FN550379) and 427 (FN550380) strains. For PK50, however, despite the corresponding gene sequences from EATRO795 (FN550382) and 427 (FN550381) strains being almost identical, there were more extensive changes compared with the 927 sequence (Tb927.10.4940) and the AnTat1.1 sequence previously published (supplemental Fig. S1) (31). The start codon of PK50 in bloodstream T. brucei of strain 427 was confirmed by 5′-rapid amplification of cDNA ends to be equivalent to that previously identified in AnTat1.1 (31), despite the presence of another in-frame start codon upstream (supplemental Fig. S1A). Additionally, EATRO795, 427, and 927 PK50 sequences lack three amino acids (Glu-111, His-112, and Arg-113) compared with the AnTat1.1 sequence, and a temporary frameshift is also present in AnTat1.1 compared with the other strains (supplemental Fig. S1, B and C). Amino acids of PK50 referred to in this study are numbered according to the 427 sequence (437 amino acids in length).
Culturing and Transfection of T. brucei
Bloodstream stage T. brucei brucei were cultured and transfected as described previously (33, 34). For RNA interference (RNAi) of PK50 or PK53, the cell line 427 pLew13 pLew90 (35) was transfected with pGL1709 or pGL1708, respectively, and dsRNA production was induced by the addition of 1 μg ml−1 tetracycline to the medium. pGL1709 was generated by cloning a 425-bp fragment of the PK50 open reading frame amplified using the PCR with oligonucleotides OL2518 and OL2519 (supplemental Table S1), into p2T7ti:GFP (36), in place of the GFP-coding sequence. pGL1708 was generated similarly, using a 427-bp fragment of PK53 amplified with oligonucleotides OL2516 and OL2517 (supplemental Table S1).
Real Time PCR Analysis
RNA preparation and cDNA synthesis were carried out as described previously (33). Real time PCR was performed using a Prism7500 real time PCR machine (Applied Biosystems). Oligonucleotides (PR23–PR26, supplemental Table S1) amplified an ∼70-bp fragment of the gene of interest distinct from the RNAi vector insert. Oligonucleotides OL2272 and OL2273 (supplemental Table S1), recognizing a fragment of GPI8 (37), were used as an endogenous control. PCRs were set up in triplicate, with each reaction containing 12.5 μl of SYBR Green master mix (Applied Biosystems), 2.5 μl each of oligonucleotide (3 μm), 1 μl of cDNA, and 6.5 μl of PCR-grade H2O. PCR conditions were as follows: 1 cycle of 50 °C for 2 min, 1 cycle of 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Resultant data were analyzed using Applied Biosystems 7500 system software.
Generation of Recombinant Proteins and Antisera
The open reading frames encoding MOB1A, MOB1B, PK50, and PK53 were PCR-amplified using the oligonucleotide pairs detailed in supplemental Table S1, sequenced, and cloned into pGEX-5X-1 (GE Healthcare), generating pGL1671 (GST:MOB1A), pGL1672 (GST:MOB1B), pGL1674 (GST:PK50), and pGL1673 (GST:PK53). Plasmids allowing the expression of C-terminally His6-tagged NDR kinases were also constructed by amplifying the PK50 and PK53 open reading frames (see supplemental Table S1 for details of oligonucleotide pairs) and cloning into pET21a (Novagen), generating pHG120 (PK50his) and pHG119 (PK53his).
To allow the expression of NDR kinase dead variants, site-directed mutagenesis of pGL1674, pGL1673, pHG120, and pHG119 was performed using the Stratagene QuikChange XL mutagenesis kit with oligonucleotides OL2531 and OL2532 (PK50) and OL2529 and OL2530 (PK53) (supplemental Table S1) to mutate the invariant catalytic lysines to methionine (K84M and K100M) (38), generating pGL1767, pGL1766, pHG127, and pHG128, respectively. Sequencing confirmed only the desired mutations had been introduced during the PCR.
To express GST-tagged recombinant proteins, each plasmid was transformed separately into Escherichia coli strain BL-21 (F−, ompT, hsdSB(rB− mB−), dcm, gal) (GE Healthcare). Protein expression was induced by the addition of 1 mm isopropyl β-d-thio-galactopyranoside to cultures growing at 30 °C, and recombinant proteins were purified according to the Bulk and RediPack GST purification module protocol (GE Healthcare). For His-tagged protein purification, constructs were transformed into Rosetta2(DE3) pLysS cells (Novagen) (F− ompT hsdSB(rB− mB−) gal dcm (DE3) pLysSRARE2 (CamR)), and protein expression was induced by the addition of 1 mm isopropyl β-d-thio-galactopyranoside to cultures growing at 37 °C. Following cell lysis by sonication, His-tagged proteins were purified on nickel-nitrilotriacetic acid-agarose.
Purified GST:PK50 and GST:PK53 were sent to the Scottish Antibody Production Unit, Penicuik, Scotland, United Kingdom, for immunization of sheep to generate specific antisera against PK50 and PK53, which were affinity-purified using protein A/G and GST columns sequentially and stored in phosphate-buffered saline. Western blotting showed the purified anti-PK50 and anti-PK53 antiserum (used at 1:100 dilution) to recognize proteins of the predicted sizes for PK50 (50.1 kDa) and PK53 (53.0 kDa) in trypanosome cell lysates that were depleted following PK50 or PK53 RNAi (Fig. 1C).
FIGURE 1.
PK50 and PK53 are essential for proliferation in bloodstream stage T. brucei. A, cumulative growth curves for two independent PK50 (left) and PK53 (right) bloodstream stage RNAi clones in the presence and absence of tetracycline (tet). B, real time PCR, 10 h post-induction for PK50 (left) and PK53 (right) RNAi clones with PK50- and PK53-specific oligonucleotides (oligos). RT, reverse transcriptase. Error bars show the standard error over three replicates. C, Western blotting, 15 h post-induction for PK50 (left) and PK53 (right) RNAi clones with anti-PK50 (top panel) and anti-PK53 (middle panel) antibodies. The lower panel shows part of the polyvinylidene difluoride membrane following blotting, stained with Ponceau S, as a loading control. 106 cell eq were loaded per lane.
Anti-MOB1 antiserum (32) was used at a 1:100 dilution for Western blotting. Anti-TY antibody (39), used at a 1:25 dilution for Western blotting, was a gift from Keith Matthews, University of Edinburgh.
Immunofluorescence, 4,6-Diamidino-2-phenylindole (DAPI) Staining, and Flow Cytometry
For immunofluorescence, 427 bloodstream stage cells (unsynchronized or synchronized with hydroxyurea (40)) were fixed in 2.4% paraformaldehyde overnight at 4 °C and processed according to Ref. 41. Anti-PK50 or anti-PK53 antisera were used undiluted as the primary antibodies, while a 1:1000 dilution of AlexaFluor488-donkey anti-sheep IgG (Molecular Probes) was used as secondary antibody. DAPI staining of methanol-fixed T. brucei and flow cytometry of propidium iodide-stained cells were carried out as described previously (33).
Immunoprecipitation of NDR Kinases and MOB Proteins and Kinase Assays
NDR kinases were immunoprecipitated from 427 wild type bloodstream cells or from the procyclic cell line 427 pHD449 (42) transfected with pGL1090 (MOB1Aty) or pGL1165 (MOB1Bty). pGL1090 and pGL1165, based on a pHD675 backbone (42), were generated by replacing the hygromycin resistance markers of pGL850 and pGL1156 (32), respectively, with a neomycin resistance marker. Induction of the tagged MOB1 proteins was induced by the addition of 1 μg ml−1 tetracycline for 16 h. 1 × 108 trypanosome cells were lysed by snap-freezing in dry ice, resuspending in 1 ml of ice-cold lysis buffer (50 mm MOPS, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 10 mm NaF, 1 mm Na3VO4, 1% Triton X-100, 10% glycerol) with protease inhibitors (234 μm leupeptin, 500 μm phenylmethanesulfonyl fluoride, 7.3 μm pepstatin A, 2.5 μm Pefabloc, 250 μm 1,10-phenanthroline, and 10 μm E-64d), and incubating on ice for 15 min. Cell debris was removed by centrifuging at 100,000 × g at 4 °C for 45 min. The clarified supernatant was incubated with the appropriate antibody for 2–3 h at 4 °C. Protein A/G beads (Pierce; 100 μl bed volume) were then added, and the resulting suspension was incubated with end over end mixing for 2 h at 4 °C. The beads were washed four times with lysis buffer, and immunoprecipitated proteins were eluted by boiling in 2× NuPAGE loading buffer (Invitrogen). Eluted proteins were analyzed by SDS-PAGE followed by Western blotting. Immunoprecipitated proteins were also analyzed for kinase activity by mixing 40 μl of beads (4 × 107 cell eq) with 20 μl of kinase assay buffer (50 mm MOPS, pH 7.4, 20 mm MgCl2, 2 mm dithiothreitol, 10 mm EGTA) supplemented with 5 μg of myelin basic protein, 4 μm ATP, and 0.037 MBq [γ-32P]ATP (44). Reactions were incubated at 30 °C for 30 min before being stopped by the addition of 20 μl of 2× NuPAGE loading buffer (Invitrogen) and heating to 70 °C for 10 min. Reaction mixes were electrophoresed on NuPAGE gels (Invitrogen); gels were stained with Coomassie Blue, dried, and analyzed by phosphorimaging.
Kinase Assays Using Recombinant Proteins
Initial assays to test the activity of recombinant protein preparations were performed as described above by incubating 2 μg of recombinant protein(s) with 20 μl of kinase assay buffer containing myelin basic protein (MBP), histone H1, α- or β-casein substrates at 30 °C for 30 min.
Substrate Optimization
80 peptides (30 μm) and 3 proteins (MBP, casein, and histone H1, 0.1 mg ml−1) were tested in duplicate as possible substrates of GST:PK50 or GST:PK53 (50 nm) in a filter plate assay (50 mm MOPS, pH 7.4, 20 mm MgCl2, 25 mm EGTA, 3 mm dithiothreitol, 30 μm ATP, 7.4 kBq/well [γ-33P]ATP, at room temperature for 2 h). 5 nm Leishmania mexicana CRK3/Leishmania major CYC6 with histone H1 was used as a positive control. Reactions were initiated by the addition of GST:PK50 or GST:PK53, incubated at room temperature, and stopped by the addition of 150 mm phosphoric acid. Stopped reactions were then transferred to P-81 phosphocellulose filter plates (Whatman), allowing the capture of peptide substrate. Following three washes with 75 mm phosphoric acid and one wash with MeOH, plates were dried at 40 °C for 20 min, and the retained signal was read on a TopCount NXT HTS counter (PerkinElmer Life Sciences).
Biochemical Characterization of GST:PK50 and GST:PK53
For more detailed characterization of the NDR kinases, optimized kinase assay buffers (GST:PK50: 50 mm Tris-HCl, pH 7.9, 3 mm MnCl2, 0.1 mm EGTA, 3 mm dithiothreitol, 0.02% CHAPS, 3 μm Na3VO4, 0.2 mg/ml bovine serum albumin and 30 μm ATP; GST:PK53: as for GST:PK50 but with 50 mm Tris-HCl, pH 7.0, 0.02% CHAPS and 1 mm MnCl2) with 8.9 kBq/well [γ-33P]ATP and 30 μm of the peptide substrate KKLRRTLSVA (GST:PK50) or KKLNRTLSFAEPG (GST:PK53) were used. Michaelis-Menten constants for the substrate peptide and ATP were determined in a time course-based matrix experiment using the filter plate format. Under the conditions tested, all initial velocity reactions utilized less than 10% of the substrate load.
Staurosporine Inhibition Assays
Staurosporine potency against GST:PK50 and GST:PK53 was tested across 10-point serial dilutions of compound from 10 μm to 5 nm (1% DMSO) using the filter plate format. Potency was determined at the respective Km values for ATP for each kinase with varying concentrations of peptide substrate.
Data Analysis
To determine bireactant kinetic parameters, Equation 1 was used,
 |
where v represents the measured velocity; Vmax is the maximum velocity; [A] and [B] and Ka and Kb are the concentrations and Michaelis-Menten constants, respectively, of substrates A and B, and α is the cooperativity factor between the two substrates (45). The α parameter was fitted either as free or fixed to 1, and the best fit was assessed using the Akaike information criterion (46). Graphs were plotted and analyses performed using SigmaPlot 10.0. IC50 values were determined using the following two parameter Equation 2 by nonlinear regression using Prism 5,
where the experimental data were corrected for background and expressed as a percentage of inhibited enzyme activity. In Equation 2, Y is the percentage of inhibition; X represents the inhibitor concentration, and Hill slope is the slope factor.
Phosphosite Analysis
Phosphosite analysis was performed by the FingerPrints Proteomics Facility, University of Dundee. GST:PK50 and GST:PK53 were digested in-gel with trypsin before being subjected to nano-liquid chromatography-tandem mass spectrometry and precursor ion scanning (Parents of −79 (PO3-)) using a 4000 QTRAP (Applied Biosystems).
RESULTS
PK50 and PK53 Are Essential Enzymes in Bloodstream Stage T. brucei
To determine whether PK50 and PK53 are essential for viability, and therefore have potential as new trypanosomiasis drug targets, their expression in bloodstream stage parasites was knocked down using an inducible RNAi system (35, 36). Stable PK50 and PK53 RNAi bloodstream cell lines were generated, and two independent clonal cell lines for each kinase were selected for analysis. Induction of the RNAi response for either kinase resulted in a rapid growth defect observable from 12 h post-induction in both clones (Fig. 1A), leading to cell death from 48 h post-induction, as evidenced by the decreasing cell numbers. For PK50 RNAi cells, clone 1 died completely following 96 h of RNAi induction, whereas the second clone began to recover at this time, most likely due to the presence of RNAi revertants within the culture as has been observed previously (47). PK53 RNAi cells died more slowly, with only one of the two clones dying completely by around 120 h post-induction. However, for each kinase, the growth defects observed were accompanied by a reduction in specific mRNA levels (60–70% at 10 h post-induction) as demonstrated by real time PCR analysis (Fig. 1B). Down-regulation of the kinase in question was also confirmed at the protein level by Western blotting cell lysates, showing PK50 and PK53 abundance decreased by ∼90% at 15 h post-induction (Fig. 1C). Down-regulation of either NDR kinase did not significantly affect the mRNA or protein levels of the other (Fig. 1, B and C), demonstrating that each NDR kinase was specifically targeted.
Trypanosome NDR Kinases Are Required for Cytokinesis
To determine whether the growth defect upon PK50 or PK53 depletion occurred as a result of a cell cycle defect, and whether these kinases play roles in cytokinesis like MOB1, cell cycle progression following RNAi induction was analyzed by fluorescence microscopy of DAPI-stained cells to monitor the number of nuclei and kinetoplasts per cell and flow cytometry of propidium iodide-stained cells to monitor their ploidy. DAPI staining is a useful tool to monitor cell cycle progression because trypanosomes contain a single mitochondrion whose DNA is organized into a disc termed the kinetoplast; the kinetoplast replicates and divides ahead of the nucleus allowing the classification of cell cycle stage for individual cells (48). The results for both clones for the PK50 and PK53 RNAi cell lines were very similar, and hence only data obtained for clone 1 is presented here. Following depletion of either kinase, DAPI staining revealed an initial increase in cells with two nuclei and two kinetoplasts (2N2K cells), with a concomitant decrease in 1N1K and 1N2K cells (Figs. 2A and 3A). Following PK50 RNAi, 2N2K cells, which constituted around 10% of the population in uninduced cells, rose to over 25% of the population at 9 h post-induction (Fig. 2A). Further examination revealed that nearly 80% of these 2N2K cells had not yet commenced cytokinesis furrowing, around 15% were in the process of furrowing, and only ∼5% were in the final stages of cytokinesis (abscission) (Fig. 2B). In uninduced cultures, the distribution was notably different, with around 60% 2N2K cells yet to commence furrowing, ∼15% with a visible cleavage furrow, and 30% cells having reached abscission (Fig. 2B). This suggests that onset of cytokinesis furrowing is delayed following depletion of PK50. At later time points, cells with >2 nuclei and >2 kinetoplasts were generated (Fig. 2, A, Other cell types, and C), but very few zoids (0N1K cells) or 2N1K cells were formed. Flow cytometry profiles of these cells showed that cells re-replicated their nuclei and kinetoplasts, as demonstrated by the appearance of 6C and 8C peaks following induction (Fig. 2D). These data strongly suggest that PK50 is essential for initiation of cytokinesis, and following its depletion, the delay in entering cytokinesis allows cells to re-replicate their DNA, leading to cell cycle deregulation.
FIGURE 2.
Characterization of bloodstream stage PK50 RNAi cell lines. A, nucleus and kinetoplast configurations of cells over time following induction, as revealed by DAPI staining and fluorescence microscopy (n >200 per time point). N, nucleus; K, kinetoplast. Other cell types predominantly include cells with >2N and >2K. B, characterization of cytokinesis stage of 2N2K cells from RNAi cell lines cultured in the presence or absence of tetracycline (tet) at 9 h post-induction. C, example images of multinucleate and multikinetoplast cells present in induced PK50 RNAi cell lines 15 h post-induction. Left panels, differential interference contrast images; right panels, DAPI images. D, flow cytometry profiles of propidium iodide-stained cells over time after PK50 RNAi induction. The x axis shows fluorescence intensity in the FL2-A channel. Time points and ploidy of peaks are indicated.
FIGURE 3.
Characterization of bloodstream stage PK53 RNAi cell lines. A, nucleus and kinetoplast configurations of cells over time following induction, as revealed by DAPI staining and fluorescence microscopy (n >200 per time point). N, nucleus; K, kinetoplast. Other cell types predominantly include cells with >2N and >2K. B, characterization of cytokinesis stage of 2N2K cells from RNAi cell lines cultured in the presence or absence of tetracycline (tet) at 10 h post-induction. C, example images of multinucleate and multikinetoplast cells present in induced PK53 RNAi cell lines 15 h post-induction. Left panels, differential interference contrast images; right panels, DAPI images. D, flow cytometry profiles of propidium iodide-stained cells over time after PK53 RNAi induction. The x axis shows fluorescence intensity in the FL2-A channel. Time points and ploidy of peaks are indicated.
A slightly different phenotype was observed following PK53 RNAi. 2N2K cells doubled in abundance by 10 h post-RNAi induction (Fig. 3A), and nearly 40% of these cells had a visible cleavage furrow compared with around 10% in uninduced control cells. There was no change in the proportion of 2N2K cells yet to commence furrowing, and very few cells progressed to abscission, strongly suggesting cytokinesis was blocked at the level of furrow ingression. This represents a later cell cycle block than that observed in PK50 RNAi cell lines and could indicate the two kinases have different functions in cell division. Nevertheless, as with PK50 RNAi, PK53 RNAi cells re-initiated multiple rounds of organelle replication and division at later time points as evidenced by DAPI staining (Fig. 3, A, Other cell types, and C, images showing cells with multiple, partially ingressed cleavage furrows) and flow cytometry (Figs. 3D, 6C and 8C peaks). 2N1K and 0N1K cells also appeared in similar proportions after ∼20 h post-induction, suggesting that some 2N2K cells may have been able to divide, but at the expense of accuracy, thus resulting in nonequivalent progeny. A similar phenotype was observed when MOB1 was depleted from the insect procyclic form of T. brucei (32).
FIGURE 6.
Analysis of interactions between NDR kinases and MOB1 proteins in T. brucei 427 bloodstream stage cell lysates. A, PK50 was immunoprecipitated with anti-PK50 antiserum, and immunoprecipitated proteins were detected by Western blotting with anti-PK50 and anti-MOB1 antibodies. As controls, immunoprecipitation (IP) was also performed with an irrelevant antibody (anti-GFP), and cell lysates were also incubated with beads without antibody (no Ab). I, sample input to beads; FT, flow-through from beads; E, eluate. Ratio of loading (immunoprecipitation/flow-through/eluate), 1:1:10. B, as for A, but showing immunoprecipitation of PK53 with anti-PK53 antiserum. C, kinase assays using eluates from immunoprecipitations. Left panel, Coomassie Blue-stained NuPAGE gel; right panel, phosphorimager scan. Lane 1, eluate from control immunoprecipitation (no antibody); lane 2, eluate from immunoprecipitation with irrelevant antibody (anti-GFP); lane 3, eluate from anti-PK50 IP; lane 4, eluate from anti-PK53 immunoprecipitation. The positions of the substrate, MBP, and the 50-kDa marker are indicated. The ∼50-kDa band visible on the autoradiograph may represent autophosphorylation of PK53.
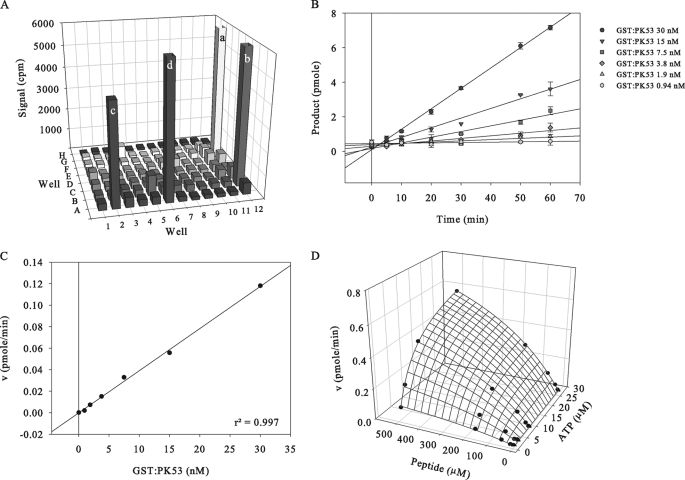
FIGURE 8.
Assay development for GST:PK53. A, substrate optimization. 80 peptides and 3 proteins (concentration 30 μm and 0.1 mg ml−1, respectively) were tested in duplicate as possible substrates of GST:PK53 (50 nm) in a filter plate assay. Mean counts per min (cpm) obtained are shown. a and b, positive controls (5 nm Leishmania mexicana CRK3/L. major CYC6 against histone H1); c, KKLNRTLSFAEPG peptide, d, RARTLSFAEPG peptide. B, linearity of GST:PK53 assay over time at the GST:PK53 concentrations indicated. Assays used 30 μm KKLNRTLSFAEPG peptide substrate and 30 μm ATP. C, linearity of GST:PK53 assay with respect to enzyme concentration. D, matrix experiment to allow the determination of the Km(app) for ATP and KKLNRTLSFAEPG peptide. Reactions contained 5 nm GST:PK53 and the concentrations of peptide and ATP indicated. The initial reaction velocities (v) are shown.
Localization of NDR Kinases in T. brucei
To investigate the localization of the NDR kinases throughout the cell cycle, immunofluorescence of PK50 and PK53 was performed in bloodstream stage cells that had been synchronized with hydroxyurea (40) to enrich for cells in particular cell cycle stages (Fig. 4 and supplemental Fig. S2). Localizations were confirmed by examining unsynchronized cell populations. Similar to a previous study where PK50 was shown to localize to the cytoplasm in long slender bloodstream and procyclic form parasites (31), immunofluorescence of bloodstream trypanosomes (using the anti-PK50 antibody generated in this study) gave a punctate cytoplasmic signal that disappeared in PK50 RNAi cell lines following tetracycline induction (supplemental Fig. S2). No obvious changes were noted in the distribution of the PK50 signal throughout the cell cycle (supplemental Fig. S2A), including during cytokinesis (supplemental Fig. S2B). PK53 also appeared to be localized throughout the cytoplasm for most of the cell cycle (Fig. 4A, 1N2K and 2N2K cells), and the specificity of the antibody signal, as for PK50, was demonstrated by performing immunofluorescence in cells induced for PK53 RNAi. However, in many cells that were at an early cell cycle stage, PK53 showed a more restricted localization, confined predominantly to an area between the nucleus and kinetoplast (Fig. 4A, 1N1K cell). Analysis of cells undergoing cytokinesis revealed a widespread punctate localization of PK53 during furrowing that became more restricted during abscission as cells were about to re-enter into a new cell cycle (Fig. 4B).
FIGURE 4.
Subcellular localization of PK53. A, immunofluorescence of 427 wild type bloodstream stage cells (top three cells) or PK53 RNAi cells (induced for 24 h with 1 μg ml−1 tetracycline (tet), bottom cell) using anti-PK53 antibody. Differential interference contrast, DAPI, PK53, and DAPI/PK53 merged (left to right) channels are shown for cells of different cell cycle stages. N, nucleus; K, kinetoplast. B, PK53 immunofluorescence as in A for 2N2K cells undergoing cytokinesis. Scale bars, 5 μm.
Purification and Activity of Recombinant NDR Kinases
To confirm that PK50 and PK53 are bona fide protein kinases, they were tagged at their N termini with a GST tag and purified from E. coli. The recombinant kinases displayed activity in vitro against the generic kinase substrate, MBP (Fig. 5), as well as against histone H1 and α- and β-casein (data not shown). This activity was abrogated when the invariant catalytic lysine in the kinase domain of each kinase was mutated to methionine (GST:PK50(K84M) and GST:PK53(K100M)) (Fig. 5, A and B, lane 5), indicating that contaminating co-purified protein kinases were not responsible for the kinase activity detected. Autophosphorylation of the NDR kinases was also observed, in the presence or absence of substrate, but not for the kinase-dead mutants. Neither of the kinases was able to phosphorylate GST (data not shown), suggesting that the autophosphorylation occurred on the NDR kinase rather than the GST tag. This was confirmed by tryptic digest and mass spectrometry analysis of GST:PK50 and GST:PK53, which identified a number of phosphorylation sites on the protein kinases (Tables 1 and 2). The PK50 activity data differ from a previous report, where an equivalent GST:PK50 fusion protein autophosphorylated but did not transphosphorylate MBP or histone substrates (31) and could reflect the more optimal kinase assay buffer used here.
FIGURE 5.
Kinase assays with recombinant PK50 and PK53. A, left panel, Coomassie Blue-stained NuPAGE gel; right panel, phosphorimager scan. Lane 1, GST:PK50 only; lane 2, GST:PK50 plus MBP substrate; lane 3, GST:PK50 with GST:MOB1A and MBP; lane 4, GST:PK50 with GST:MOB1B and MBP; lane 5, GST:PK50(K84M) plus MBP substrate. The positions of the different proteins are indicated. *, autophosphorylation of GST:PK50. B, as for A but with GST:PK53 or GST:PK53(K100M) in place of GST:PK50 or GST:PK50(K84M), respectively.
TABLE 1.
Identified phosphosites for GST:PK50
GST:PK50 was in-gel digested with trypsin, and resultant peptides were analyzed by nano-liquid chromatography-tandem mass spectrometry combined with parents of −79 (PO3−) analysis. The phosphosites identified and their mass-to-charge ratios are shown in the table. Phosphorylated residues are highlighted in boldface. The underlined serine or threonine residues indicate putative phosphosites where one of the underlined residues was phosphorylated, but the spectrum did not allow the phosphorylated residue to be discriminated.
| Residue | Peptide | Mascot (m/z) | Pre79 (m/z) |
|---|---|---|---|
| Tyr-25 | YTGEYYVK | 551.7 (2+) | 549.3 (2−) |
| Ser-36 | GMVGDKKSFAIEHSR | 871.5 (2+) | 869.5 (2−) |
| Ser-216, Thr-217, or Ser-218 | RDPDQAESTSVADDSYLTEDVTVDDDVK | 1056.2 (2+) | 1054.6 (2−) |
| Ser-251 or Thr-252 | KVMFFSTVGSPAYIAPEVLIGR | 1231.8 (2+) | 1229.7 (2−) |
| Ser-251 or Thr-252 | VMFFSTVGSPAYIAPEVLIGR | 1167.7 (2+) | 1165.7 (2−) |
| Thr-394 and Ser-402 | QPMTAVSREDQSVFVGFTSK | 1187.6 (2+) | 1185.7 (2−) |
| Ser-402 | EDQSVFVGFTSK | 712.4 (2+) | 711.4 (2−) |
| Ser-402 and Thr-408 or Ser-409 | EDQSVFVGFTSKLCDR | 1024.5 (2+) | 1022.6 (2−) |
| Ser-434 | FHELQNFSDDD | 723.8 (2+) | 721.4 (2−) |
TABLE 2.
Identified phosphosites for GST:PK53
GST:PK53 was in-gel digested with trypsin, and resultant peptides were analyzed by nano-liquid chromatography-tandem mass spectrometry combined with parents of −79 (PO3-) analysis. The phosphosites identified and their mass-to-charge ratios are shown in the table. Phosphorylated residues are highlighted in boldface. The underlined serine or threonine residues indicate putative phosphosites where one of the underlined residues was phosphorylated, but the spectrum did not allow the phosphorylated residue to be discriminated.
| Residue | Peptide | Mascot (m/z) | Pre79 (m/z) |
|---|---|---|---|
| Ser-2 or Thr-3 | GIPEFSTR (GIPEF is C-terminal end of GST sequence) | 493.7 (2+) | |
| Thr-17 or Ser-20 or Ser-23 | GAAYTECSPISDVSHHK | 970.4 (2+) | 967.9 (2−) |
| Ser-31 and Ser-33 | ASASKAFLENHYR | 827.3 (2+) | |
| Ser-33 | ASASKAFLENHYR | 787.4 (2+) | 785.3 (2−) |
| Thr-48 | GMLRDTR | 464.7 (2+) | |
| Ser-50 or Thr-53 | SGNTARPVGPRPR | 482.2 (2+) | |
| Ser-114 or Ser-118 | QISHVRSEK | 582.3 (2+) | |
| Ser-218 | LTDFGLSKR | 558.8 (2+) | |
| Ser-298 | IFESIVGSPGYIAPEILLR | 1077.6 (2+) | 1076.6 (2−) |
| Ser-298 | IFESIVGSPGYI | 702.8 (2+) | 701.4 (2−) |
| Ser-298 | RIFESIVGSPGYIAPEILLR | 770.7 (3+) | 1153.6 (2−) |
| Tyr-458 | GVIFADYKFNLK | 747.8 (2+) | 745.5 (2−) |
Given that previous data suggested PK50 could interact with MOB1 in T. brucei (32), and in other eukaryotes, MOB1 is required to activate its NDR kinase partner, the two trypanosome MOB1 proteins, MOB1A and MOB1B, were also purified as N-terminal GST fusion proteins and added to the kinase assays. However, the activities of the NDR kinases were not increased in the presence of either GST:MOB1 protein (Fig. 5, lanes 3 and 4), and subsequent analyses showed that GST:MOB1A/B did not bind to GST:PK50 or GST:PK53 (data not shown). Attempts to alleviate possible steric hindrance of interactions between these proteins due to the presence of the GST tags by removal or replacement of the tags with smaller His6 tags were confounded by the poor solubility of the resulting MOB1 proteins. NDR kinases tagged at the C terminus with a His6 tag, like the GST-tagged kinases, underwent autophosphorylation and transphosphorylated MBP but were also not bound or further activated by the GST:MOB1 proteins (supplemental Fig. S3), indicating that the N-terminal GST tag on the NDR kinase was not responsible for the lack of GST:MOB1 binding.
Previously, using a procyclic EATRO795 cell line inducibly expressing MOB1Aty, MOB1Aty was immunoprecipitated with anti-TY antibody and a 50-kDa interacting protein detected with a rabbit anti-PK50 antibody (31), suggesting that PK50 and MOB1Aty formed a complex (32). In view of the data above that indicate no such interaction between the NDR kinases and MOB1 proteins in vitro, the interaction of these proteins in vivo was analyzed in more detail. Because the anti-PK50 antibody used in the original study no longer exists, immunoprecipitations were performed with the sheep anti-PK50 and anti-PK53 antibodies generated here, which were demonstrated to be specific (Figs. 1C and 4A and supplemental Fig S2A). Surprisingly, MOB1A/B were not co-immunoprecipitated from bloodstream cell lysates (strain 427) with PK50 or PK53 (Fig. 6, A and B). Furthermore, the immunoprecipitated kinase fractions were active against MBP despite the absence of MOB1 proteins (Fig. 6C). To determine whether this represented a life cycle stage-specific difference in MOB1:NDR kinase association, the immunoprecipitations were also carried out using procyclic T. brucei 427 cell lysates inducibly expressing MOB1Aty or MOB1Bty (supplemental Fig. S4). MOB1A/Bty did not co-purify with immunoprecipitated PK50 or PK53 (supplemental Fig. S4, A and B). Similarly, the tagged MOB1 proteins immunoprecipitated from these cell lines did not pull down either of the NDR kinases (supplemental Fig. S4C). Additionally, MOB1Aty could not be co-immunoprecipitated with PK50 using our sheep anti-PK50 antibody from the EATRO795-derived cell line used previously (data not shown) (32).
Despite being unable to confirm the PK50:MOB1Aty interaction or to detect any interactions between PK50 and MOB1B or between PK53 and MOB1A/B, we cannot completely rule out that an interaction occurs between the MOB1 proteins and PK50 and/or PK53 in vivo. Such an interaction may only occur very transiently or under very specific conditions in the cell cycle. For example, the phosphorylation status of the NDR kinases or MOB1 proteins may be crucial to their interaction. Nevertheless, the data generated here suggest that a significant part of the NDR kinase pool does not interact with MOB1 proteins and that MOB1 association with the NDR kinases is not required for their activity in vivo.
Biochemical Characterization of NDR Kinases
To facilitate subsequent biochemical analyses, NDR kinase assay conditions were optimized. A collection of 80 synthetic peptides and generic protein substrates (obtained from the Division of Signal Transduction Therapy, University of Dundee) was screened with the GST:PK50 and GST:PK53 to identify optimal substrates for each kinase. Two peptides, KKLNRTLSFAEPG and KKLRRTLSVA, were found to be reproducibly phosphorylated by GST:PK50 in vitro (Fig. 7A); KKLNRTLSFAEPG was also found to be a good substrate for GST:PK53, along with RARTLSFAEPG (Fig. 8A). Although these peptides have been reported as relatively nonspecific kinase substrates (49), they do contain an RXXS motif previously reported to be an NDR kinase consensus sequence (3, 50). KKLRRTLSVA and KKLNRTLSFAEPG were used as the PK50 and PK53 substrates, respectively, in all subsequent studies. Buffer optimization matrix experiments were performed, varying pH and MgCl2/MnCl2 concentration, and showed that for GST:PK50, a buffer of pH 7.9 containing 3 mm MnCl2 increased the assay signal 12-fold. For GST:PK53, the optimal buffer was of pH 7.0 with 1 mm MnCl2. All subsequent activity assays used these optimized conditions, under which it was determined that levels of NDR kinase autophosphorylation were negligible compared with exogenous substrate trans-phosphorylation (data not shown).
FIGURE 7.
Assay development for GST:PK50. A, substrate optimization. 80 peptides and 3 proteins (concentration 30 μm and 0.1 mg ml−1, respectively) were tested in duplicate as possible substrates of GST:PK50 (50 nm) in a filter plate assay. Mean counts per min (cpm) obtained are shown. a and b, positive controls (5 nm L. mexicana CRK3/L. major CYC6 against histone H1); c, KKLNRTLSFAEPG peptide, d, KKLRRTLSVA peptide. B, linearity of GST:PK50 assay over time at the GST:PK50 concentrations indicated. Assays used 30 μm KKLRRTLSVA peptide substrate and 30 μm ATP. C, linearity of GST:PK50 assay with respect to enzyme concentration. D, matrix experiment to allow the determination of the Km(app) for ATP and KKLRRTLSVA peptide. Reactions contained 15 nm GST:PK50, and the concentrations of peptide and ATP are indicated. The initial reaction velocities (v) are shown.
GST:PK50 and GST:PK53 were found to trans-phosphorylate their peptide substrates linearly with respect to time and enzyme concentration (Figs. 7, B and C, and 8, B and C). Furthermore, by measuring initial reaction velocities in a matrix experiment of varied ATP and peptide substrate concentrations (Figs. 7D and 8D), the Km values of GST:PK50 and GST:PK53 for ATP were determined to be 25.7 ± 4.8 and 7.0 ± 0.3 μm, respectively, and for substrate 66.9 ± 11.4 and 495.1 ± 26.1 μm, respectively, with no evidence of cooperativity. This was further confirmed by the fact that the IC50 values for the generic kinase inhibitor, staurosporine (216 ± 47 nm for GST:PK50 and 4.6 ± 0.3 nm for GST:PK53 (supplemental Fig. S5C)), did not significantly shift under differing peptide concentrations (supplemental Fig. S5, A and B).
DISCUSSION
This study demonstrates that the two trypanosome NDR kinases are essential enzymes in bloodstream stage T. brucei, thus providing genetic validation for these enzymes as putative novel drug targets. Depletion of either kinase disrupted cytokinesis, leading to cell cycle deregulation and cell death. The early increase in 2N2K cells, observed at 5 h post-induction of PK50 or PK53 RNAi, argues that depletion of either of these kinases has a specific effect on cytokinesis, because a growth arrest and large numbers of abnormal cells in the population were only observed after the appearance of increased numbers of 2N2K cells. The phenotypes observed following depletion of the two kinases were not identical, however. Following PK50 depletion, most of the abnormal cells had multiple nuclei and kinetoplasts, and, like the majority of the 2N2K cells, lacked cleavage furrows. Very few zoids (0N1K cells), cells commonly formed following aberrant cytokinesis, or 2N1K cells, which can form following defects in basal body or kinetoplast replication or segregation, or as a result of inaccurate furrowing, were present following PK50 RNAi. Together, these data indicate that depletion of PK50 prevents bloodstream stage cells from initiating cytokinesis. In comparison, following PK53 depletion, partially furrowed 2N2K cells accumulated. Additionally, at later time points, 2N1K cells and zoids accumulated in equal numbers, suggesting that some 2N2K cells did manage to divide, but with a loss of accuracy of furrow positioning. However, the accumulation of cells with multiple nuclei and kinetoplasts partitioned into multiple cell bodies with multiple partially ingressed cleavage furrows (Fig. 3C) indicates that completion of furrow ingression was blocked in other dividing cells.
The different phenotypes observed for PK50 and PK53 could indicate that these kinases act sequentially in a cytokinesis signaling pathway, most likely with PK50 above PK53 in the cascade. However, neither kinase was able to use the other as a substrate in vitro (data not shown). Alternatively, because both kinases were apparently depleted by similar proportions following RNAi induction (Fig. 1C), the different phenotypes observed may reflect different threshold levels for their function at different stages of cytokinesis. Both kinases could conceivably be required for the initiation and ingression of furrowing, but the amount of residual PK53 protein was sufficient for initiation of cytokinesis, whereas that of PK50 was not.
Immunofluorescence was performed for PK50 and PK53 across the cell cycle. However, despite the RNAi phenotypes obtained, no specific localization to the cytokinesis furrow was observed. One of the problems inherent in performing immunofluorescence for any kinase is that, without a phosphospecific antibody, it is not possible to distinguish the active protein kinase pool from inactive forms. Hence, the total kinase pool may mask a specific localization of the active pool. It is also possible that these kinases do not localize to the cytokinesis furrow themselves but instead phosphorylate a substrate, which as a result then translocates to the furrow to bring about cytokinesis. The more restricted localization of PK53 during abscission and early stages in the cell cycle is intriguing but not easy to explain without further study. However, this finding may suggest additional roles for this kinase that have not been revealed by our analysis to date of the RNAi cell lines.
PK50 and PK53 join a growing list of T. brucei signaling proteins with direct roles in cytokinesis. A chromosomal passenger complex has been reported to regulate the mitosis to cytokinesis transition (51–53) in bloodstream and procyclic trypanosomes. TbRACK1 is required for furrow ingression in procyclic T. brucei, although its depletion in bloodstream trypanosomes appears to prevent cytokinesis initiation (54), and TbRACK1 is thought to facilitate continued translation during cytokinesis (55). The Polo-like kinase, PLK1, is essential for furrow ingression in the bloodstream form (56), as is the F-box protein, CFB2 (57), and MOB1 (32). MOB1 also plays roles in accuracy of furrow ingression in procyclic trypanosomes. RNAi of each of these regulators gives slightly different phenotypes, and given the incomplete nature and varied kinetics of protein knockdown, as well as the relatively few protein/protein interactions that have been determined between these regulators, it is difficult to interpret the temporal order of the signaling events that control trypanosome cytokinesis at present.
This study provides the first biochemical analysis of the trypanosome NDR kinases. Despite MOB1 proteins being known activators of NDR kinases in a wide range of organisms, the recombinant trypanosome NDR kinases were active in the absence of MOB1 activation or phosphorylation by upstream kinases. For PK50, this property was not unduly influenced by the affinity tag, but for PK53, the C-terminal His6 tag led to autophosphorylation being favored over MBP phosphorylation (supplemental Fig. S3), although the reason for this is not clear at present. In mammalian cells, protein phosphatase 2A has been shown to remove the activatory phosphate groups (in the activation loop and C-terminal hydrophobic motif) to inhibit NDR1 (21, 58). It will be interesting to discover whether the activities of the trypanosome kinases are regulated in a similar manner in vivo.
Because of GST:MOB1A/B being unable to bind to recombinant NDR kinases, we were unable to determine whether MOB1 binding might have increased the activity of the kinases still further. It is possible that the GST tag on the MOB1 proteins prevented them from interacting with the NDR kinases, or that GST:MOB1 was not folded correctly, possibilities that we have not been able to rule out at present. However, we were unable to co-immunoprecipitate the NDR kinases and MOB1A/B proteins in vivo either, at least under the conditions used in this study, suggesting the intriguing possibility that the trypanosome NDR kinases are regulated by different mechanisms in the trypanosome compared with other eukaryotes. The reason for the discrepancy between the interaction data generated here and previous data indicating an interaction between PK50 and MOB1Aty in procyclic trypanosomes is unclear and not possible to determine, given that supplies of the original rabbit anti-PK50 antibody have been exhausted. Although technical issues with the original experiment cannot be ruled out, it is also possible that the rabbit anti-PK50 antibody (31) used previously recognized an ∼50-kDa protein in trypanosome cell lysates that was not PK50, since a PK50 RNAi cell line was not available at the time to confirm its in vivo specificity. Because of the more comprehensive analysis of the NDR kinase/MOB1 interactions, we believe that the data set presented here is the more robust of the two analyses.
Given the data presented here, we favor a hypothesis whereby the NDR kinases function largely independently of the MOB1 proteins. However, we cannot completely rule out that interactions between subpopulations of the MOB1 proteins and NDR kinases do occur, perhaps very transiently. A specific phosphorylation event or a scaffold protein could be required for an NDR/MOB1 interaction to occur. Phosphorylation of mammalian MOB1 at Thr-74 by the STE20 kinase, MST2, is essential for its interaction with, and activation of, NDR1 (19), and in Drosophila, phosphorylation of MOBKL1A/B at Thr-12 and Thr-35 by MST1/2 kinases increases their affinity for LATS1 (20). Scaffolding proteins containing multiple HEAT/Armadillo-like repeats can promote the activation of NDR kinases in a variety of organisms (21). In Drosophila, Trc/NDR1 and MOB2 were shown to interact with the Furry scaffolding protein, leading to activation of NDR1 (22, 59). In this system, the MOB2/Furry interaction was dependent on phosphorylation of MOB2. A recent phosphoproteomic study of bloodstream T. brucei (60) revealed that Ser-23 of MOB1A (Ser-40 in MOB1B) was phosphorylated in vivo, but further investigation will be required to determine the significance of this modification.
PK50 and PK53 have been shown to be phosphorylated in vivo on Ser-434 and Ser-298, respectively (60). Ser-298 of PK53 is the activation loop serine, whereas Ser-434 of PK50 corresponds to the most C-terminal serine residue and is not conserved in other NDR kinases. Hence, this could indicate a trypanosome-specific phosphoregulatory event. Phosphorylation of the activation loop serine (Ser-251) or the hydrophobic motif threonine (Thr-408) of PK50 was not detected in the study by Nett et al. (60. No other in vivo phosphosites were detected for PK53, and the C-terminal hydrophobic motif is not conserved in this protein, again suggesting trypanosome-specific regulation of this kinase may occur. In this study, recombinant GST:PK50 was shown to be phosphorylated on Ser-434, mimicking the in vivo situation, and Ser-251 and Thr-408 were identified as putative phosphorylation sites. Additional phosphorylation sites were identified in the recombinant protein at Tyr-25, Ser-36, Ser-216/Thr-217/Ser-218, Thr-394, and Ser-402. GST:PK53 was found to be phosphorylated on Ser-298, mimicking the in vivo data, and additional phosphorylation sites were detected at Ser-2/Thr-3, Thr-17/Ser-20/Ser-23, Ser-31, Ser-33, Thr-48, Ser-50/Thr-53, Ser-114/Ser-118, Ser-218, and Tyr-458. The in vivo significance of these phosphorylations is not clear at present. However, although the presence of these phosphosites suggests that PK50 and PK53 are able to autophosphorylate on these residues in vitro, this does not rule out that these modifications are performed by an upstream kinase in vivo. NDR2 kinase is known to autophosphorylate its hydrophobic motif threonine in vitro, although this event is primarily performed by the STE20 kinase MST3 in vivo (17, 61). Additionally, although it is likely that some of these residues, e.g. Ser-251 (activation loop) and Thr-408 (hydrophobic motif) in PK50, are phosphorylated in vivo, similar to NDR kinases in other organisms, it is not yet clear whether the other phosphosites identified represent bone fide in vivo modifications of the NDR kinases.
NDR kinases are known to phosphorylate substrates with the consensus sequence RXXS (3, 50), although relatively few in vivo substrates have been identified for these kinases. Our substrate screen showed our kinases to favor peptides with the RXXS consensus sequence. Both kinases phosphorylated the peptide KKLNRTLSFAEPG, but GST:PK50 was unable to phosphorylate the GST:PK53 substrate RARTLSFAEPG, whereas the GST:PK50-preferred substrate, KKLRRTLSVA, was relatively poorly phosphorylated by GST:PK53. The in vivo substrate specificities of these enzymes are still unknown, but it is clear from the lethal RNAi phenotypes obtained upon depletion of either kinase that they are not redundant enzymes in bloodstream stage T. brucei. Biochemical characterization also revealed differences between these kinases. Although GST:PK50 and GST:PK53 were shown to have similar cation preferences (for both kinases, the in vitro activity dropped dramatically in presence of Mg2+ and was restored by Mn2+), they differ in their catalytic pocket as demonstrated by their different affinities for ATP and the different potencies of staurosporine. These findings are consistent with the presence of a 36-amino acid insertion in the catalytic pocket of PK53 compared with PK50.
PK50 and PK53 are about as similar to each other at the amino acid level (∼30% identity and ∼45% similarity, rising to ∼35% identity and ∼50% similarity over the kinase domain) as they are to human NDR/LATS kinases. It has been reported that for kinases sharing >60% identity over their catalytic domain, there is a high likelihood that these enzymes will be inhibited by the same group of low molecular weight compounds (62) and suggested that there may therefore be a higher probability of being able to identify selective small molecule inhibitors for kinases sharing <60% identity (43). The difference in affinity of GST:PK50 and GST:PK53 for the generic kinase inhibitor staurosporine provides preliminary evidence that selective inhibition of these kinases is possible. Indeed, using recombinant GST:PK50 and GST:PK53, high throughput screening for small molecule inhibitors has now been performed.4 Inhibitors based on distinct scaffolds with sub-micromolar IC50 values were identified for each recombinant kinase that were selective against the majority of human kinases tested. Work is ongoing to determine the trypanosome activity and mode of action of these inhibitors.
Acknowledgments
We thank Emily Gaunt for generating the GST:PK50 and GST:PK53 kinase-dead expression constructs; Keith Matthews for providing anti-TY antibody; the FingerPrints Proteomic Facility, University of Dundee, for the phosphosite analysis of the NDR kinases; and the Division of Signal Transduction Therapy for providing the peptide substrate collection.
This work was supported by Medical Research Council Career Development Fellowship G120/1001 (to T. C. H.), Medical Research Council New Investigator Research Grant G0900239 (to T. C. H.), Research Councils UK Academic Fellowship GR/T28003/01 (to T. C. H.), a Wellcome Trust Value in People award (to C. B.), and Wellcome Trust Strategic Award WT077705 (Drug Discovery for Tropical Disease Initiative).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
R. Grimaldi, J. Ma, L. Cleghorn, C. Stockdale, C. Benz, A. Woodland, P. Wyatt, T. C. Hammarton, and J. Frearson, manuscript in preparation.
- NDR
- nuclear DBF2-related
- GST
- glutathione S-transferase
- MBP
- myelin basic protein
- MOB
- Mps One Binder
- RACE
- rapid amplification of cDNA ends
- MBP
- myelin basic protein
- RNAi
- RNA interference
- DAPI
- 4,6-diamidino-2-phenylindole
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- MOPS
- 4-morpholinepropanesulfonic acid.
REFERENCES
- 1.Mah A. S., Jang J., Deshaies R. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7325–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hou M. C., Guertin D. A., McCollum D. (2004) Mol. Cell. Biol. 24, 3262–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen C. T., Feoktistova A., Chen J. S., Shim Y. S., Clifford D. M., Gould K. L., McCollum D. (2008) Curr. Biol. 18, 1594–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohl D. A., Huddleston M. J., Collingwood T. S., Annan R. S., Deshaies R. J. (2009) J. Cell Biol. 184, 527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss E. L., Kurischko C., Zhang C., Shokat K., Drubin D. G., Luca F. C. (2002) J. Cell Biol. 158, 885–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou M. C., Wiley D. J., Verde F., McCollum D. (2003) J. Cell Sci. 116, 125–135 [DOI] [PubMed] [Google Scholar]
- 7.González-Novo A., Labrador L., Pablo-Hernando M. E., Correa-Bordes J., Sánchez M., Jiménez J., Vázquez de Aldana C. R. (2009) Mol. Microbiol. 72, 1364–1378 [DOI] [PubMed] [Google Scholar]
- 8.Emoto K., He Y., Ye B., Grueber W. B., Adler P. N., Jan L. Y., Jan Y. N. (2004) Cell 119, 245–256 [DOI] [PubMed] [Google Scholar]
- 9.Geng W., He B., Wang M., Adler P. N. (2000) Genetics 156, 1817–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emoto K., Parrish J. Z., Jan L. Y., Jan Y. N. (2006) Nature 443, 210–213 [DOI] [PubMed] [Google Scholar]
- 11.Huang J., Wu S., Barrera J., Matthews K., Pan D. (2005) Cell 122, 421–434 [DOI] [PubMed] [Google Scholar]
- 12.Oh H., Irvine K. D. (2008) Development 135, 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hergovich A., Lamla S., Nigg E. A., Hemmings B. A. (2007) Mol. Cell 25, 625–634 [DOI] [PubMed] [Google Scholar]
- 14.Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., Zheng P., Ye K., Chinnaiyan A., Halder G., Lai Z. C., Guan K. L. (2007) Genes Dev. 21, 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A., Pan D. (2007) Cell 130, 1120–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hisaoka M., Tanaka A., Hashimoto H. (2002) Lab. Invest. 82, 1427–1435 [DOI] [PubMed] [Google Scholar]
- 17.Stegert M. R., Tamaskovic R., Bichsel S. J., Hergovich A., Hemmings B. A. (2004) J. Biol. Chem. 279, 23806–23812 [DOI] [PubMed] [Google Scholar]
- 18.Bichsel S. J., Tamaskovic R., Stegert M. R., Hemmings B. A. (2004) J. Biol. Chem. 279, 35228–35235 [DOI] [PubMed] [Google Scholar]
- 19.Hirabayashi S., Nakagawa K., Sumita K., Hidaka S., Kawai T., Ikeda M., Kawata A., Ohno K., Hata Y. (2008) Oncogene 27, 4281–4292 [DOI] [PubMed] [Google Scholar]
- 20.Praskova M., Xia F., Avruch J. (2008) Curr. Biol. 18, 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hergovich A., Stegert M. R., Schmitz D., Hemmings B. A. (2006) Nat. Rev. Mol. Cell Biol. 7, 253–264 [DOI] [PubMed] [Google Scholar]
- 22.Chiba S., Ikeda M., Katsunuma K., Ohashi K., Mizuno K. (2009) Curr. Biol. 19, 675–681 [DOI] [PubMed] [Google Scholar]
- 23.Hergovich A., Bichsel S. J., Hemmings B. A. (2005) Mol. Cell. Biol. 25, 8259–8272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hergovich A., Schmitz D., Hemmings B. A. (2006) Biochem. Biophys. Res. Commun. 345, 50–58 [DOI] [PubMed] [Google Scholar]
- 25.König C., Maekawa H., Schiebel E. (2010) J. Cell Biol. 188, 351–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy P. G. (2008) Ann. Neurol. 64, 116–126 [DOI] [PubMed] [Google Scholar]
- 27.Barrett M. P., Boykin D. W., Brun R., Tidwell R. R. (2007) Br. J. Pharmacol. 152, 1155–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Priotto G., Kasparian S., Mutombo W., Ngouama D., Ghorashian S., Arnold U., Ghabri S., Baudin E., Buard V., Kazadi-Kyanza S., Ilunga M., Mutangala W., Pohlig G., Schmid C., Karunakara U., Torreele E., Kande V. (2009) Lancet 374, 56–64 [DOI] [PubMed] [Google Scholar]
- 29.Hammarton T. C. (2007) Mol. Biochem. Parasitol. 153, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammarton T. C., Monnerat S., Mottram J. C. (2007) Curr. Opin. Microbiol. 10, 520–527 [DOI] [PubMed] [Google Scholar]
- 31.García-Salcedo J. A., Nolan D. P., Gijón P., Gómez-Rodriguez J., Pays E. (2002) Mol. Microbiol. 45, 307–319 [DOI] [PubMed] [Google Scholar]
- 32.Hammarton T. C., Lillico S. G., Welburn S. C., Mottram J. C. (2005) Mol. Microbiol. 56, 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammarton T. C., Clark J., Douglas F., Boshart M., Mottram J. C. (2003) J. Biol. Chem. 278, 22877–22886 [DOI] [PubMed] [Google Scholar]
- 34.Burkard G., Fragoso C. M., Roditi I. (2007) Mol. Biochem. Parasitol. 153, 220–223 [DOI] [PubMed] [Google Scholar]
- 35.Wirtz E., Leal S., Ochatt C., Cross G. A. (1999) Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 36.LaCount D. J., Bruse S., Hill K. L., Donelson J. E. (2000) Mol. Biochem. Parasitol. 111, 67–76 [DOI] [PubMed] [Google Scholar]
- 37.Lillico S., Field M. C., Blundell P., Coombs G. H., Mottram J. C. (2003) Mol. Biol. Cell 14, 1182–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snyder M. A., Bishop J. M., McGrath J. P., Levinson A. D. (1985) Mol. Cell. Biol. 5, 1772–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bastin P., Bagherzadeh Z., Matthews K. R., Gull K. (1996) Mol. Biochem. Parasitol. 77, 235–239 [DOI] [PubMed] [Google Scholar]
- 40.Forsythe G. R., McCulloch R., Hammarton T. C. (2009) Mol. Biochem. Parasitol. 164, 131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grünfelder C. G., Engstler M., Weise F., Schwarz H., Stierhof Y. D., Morgan G. W., Field M. C., Overath P. (2003) Mol. Biol. Cell 14, 2029–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biebinger S., Wirtz L. E., Lorenz P., Clayton C. (1997) Mol. Biochem. Parasitol. 85, 99–112 [DOI] [PubMed] [Google Scholar]
- 43.Naula C., Parsons M., Mottram J. C. (2005) Biochim. Biophys. Acta 1754, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Hellemond J. J., Neuville P., Schwarz R. T., Matthews K. R., Mottram J. C. (2000) J. Biol. Chem. 275, 8315–8323 [DOI] [PubMed] [Google Scholar]
- 45.Segel I. H. (1993) Enzyme Kinetics, Wiley Classics Library Ed., pp. 274–275, John Wiley & Sons, Inc., New York [Google Scholar]
- 46.Burnham K. P., Anderson D. R. (2002) Model Selection and Inference: A Practical Information-Theoretic Approach, 2nd Ed., pp. 362–371, Springer-Verlag, New York [Google Scholar]
- 47.Chen Y., Hung C. H., Burderer T., Lee G. S. (2003) Mol. Biochem. Parasitol. 126, 275–279 [DOI] [PubMed] [Google Scholar]
- 48.Woodward R., Gull K. (1990) J. Cell Sci. 95, 49–57 [DOI] [PubMed] [Google Scholar]
- 49.Ross H., Armstrong C. G., Cohen P. (2002) Biochem. J. 366, 977–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mah A. S., Elia A. E., Devgan G., Ptacek J., Schutkowski M., Snyder M., Yaffe M. B., Deshaies R. J. (2005) BMC Biochem. 6, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Z., Lee J. H., Chu F., Burlingame A. L., Günzl A., Wang C. C. (2008) Plos ONE 3, e2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Z., Umeyama T., Wang C. C. (2008) Plos ONE 3, e3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z., Umeyama T., Wang C. C. (2009) Plos Pathogens 5, e1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rothberg K. G., Burdette D. L., Pfannstiel J., Jetton N., Singh R., Ruben L. (2006) J. Biol. Chem. 281, 9781–9790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Regmi S., Rothberg K. G., Hubbard J. G., Ruben L. (2008) Mol. Microbiol. 70, 724–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hammarton T. C., Kramer S., Tetley L., Boshart M., Mottram J. C. (2007) Mol. Microbiol. 65, 1229–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benz C., Clayton C. E. (2007) Mol. Biochem. Parasitol. 156, 217–224 [DOI] [PubMed] [Google Scholar]
- 58.Millward T. A., Hess D., Hemmings B. A. (1999) J. Biol. Chem. 274, 33847–33850 [DOI] [PubMed] [Google Scholar]
- 59.He Y., Fang X., Emoto K., Jan Y. N., Adler P. N. (2005) Mol. Biol. Cell 16, 689–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nett I. R., Martin D. M., Miranda-Saavedra D., Lamont D., Barber J. D., Mehlert A., Ferguson M. A. (2009) Mol. Cell. Proteomics 8, 1527–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stegert M. R., Hergovich A., Tamaskovic R., Bichsel S. J., Hemmings B. A. (2005) Mol. Cell. Biol. 25, 11019–11029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vieth M., Higgs R. E., Robertson D. H., Shapiro M., Gragg E. A., Hemmerle H. (2004) Biochim. Biophys. Acta 1697, 243–257 [DOI] [PubMed] [Google Scholar]