Abstract
Intrauterine growth restriction (IUGR) increases susceptibility to age-related diseases, including type 2 diabetes (T2DM), and is associated with permanent and progressive changes in gene expression. Our study was designed to test whether epigenomic dysregulation mediates the cellular memory of this intrauterine event. To test this hypothesis, we isolated pancreatic islets from control and IUGR (induced by bilateral uterine artery ligation at day 18 of fetal life) animals at 7 weeks of age. Using the HELP (HpaII tiny fragment enrichment by ligation-mediated PCR) assay, we generated the first DNA methylation map at almost 1 million unique sites throughout the rat genome in normal pancreatic islet cells, allowing us to identify the changes that occur as a consequence of IUGR. We validated candidate dysregulated loci with quantitative assays of cytosine methylation and gene expression. IUGR changes cytosine methylation at ∼1,400 loci (false discovery rate of 4.2%) in male rats at 7 weeks of age, preceding the development of diabetes and thus representing candidate loci for mediating the pathogenesis of metabolic disease that occurs later in life. Epigenetic dysregulation occurred preferentially at conserved intergenic sequences, frequently near genes regulating processes known to be abnormal in IUGR islets, such as vascularization, β-cell proliferation, insulin secretion, and cell death, associated with concordant changes in mRNA expression. These results demonstrate that epigenetic dysregulation is a strong candidate for propagating the cellular memory of intrauterine events, causing changes in expression of nearby genes and long term susceptibility to type 2 diabetes.
Keywords: Diabetes, DNA Methylation, Epigenetics, Gene Regulation, Pancreatic Islet, Rat, Intrauterine Growth Restriction
Introduction
Perturbations in the intrauterine environment resulting in poor fetal growth increase the susceptibility for age-related diseases, particularly type 2 diabetes mellitus (T2DM)4 and cardiovascular disease (1, 2). Moreover, intrauterine growth restriction (IUGR) can cause permanent and progressive changes in gene expression, affecting important metabolically active tissues such as pancreatic islets (3–6). These changes are often present at birth or during early life, and can precede the development of overt disease in rodents by months and in humans by decades. Although the mechanisms that may mediate the “fetal origin of adult disease” (7) remain unclear, dysregulation of the epigenome could play an important role and may explain the changes in gene expression that are heritably maintained through cell divisions in IUGR animals throughout life (8).
We have developed an animal model of IUGR caused by uteroplacental insufficiency, which limits the supply of critical substrates (e.g. nutrients, oxygen, growth factors, and hormones) to the fetus (9). This deficient intrauterine environment affects fetal development through permanent and progressive dysregulation of gene expression and function of susceptible cells (e.g. pancreatic β-cells) and leads to the development of T2DM in adulthood (3, 6, 9). In particular, expression of pancreatic and duodenal homeobox 1 (Pdx1), a transcription factor critical for the regulation of pancreatic development and β-cell differentiation, shows progressive decline in IUGR offspring, with a complete absence of Pdx1 levels in adult rats (3). Moreover, this decline in gene expression is associated with progressive changes in epigenetic regulation at the Pdx1 proximal promoter, through early and progressive alterations in histone post-translational modifications. DNA methylation at this locus is not acquired until much later in life (10).
Previous studies have focused primarily on the identification of epigenetic dysregulation at specific candidate sites, often at promoters of genes of interest (10). In this report, we use the genome-wide HELP assay (11), to define patterns of cytosine methylation at almost 1 million loci in IUGR and control rats. Both groups of animals were studied at the age of 7 weeks, prior to the onset of hyperglycemia in the IUGR animals (12), thus avoiding the confounding effects of overt diabetes. Dysregulation of pancreatic islet β-cell function and development plays a central role in the pathogenesis of diabetes in humans and animal models of IUGR (13). We therefore chose to study isolated pancreatic islets, consisting of ∼95% β-cells by cellular mass (14), as candidates for mediating the functional consequences of IUGR-induced epigenetic changes. Comparison of IUGR with normal rats revealed changes in DNA methylation at a number of novel loci, not limited to canonical CpG islands or promoters. The specific loci affected are in proximity to genes with important roles in β cell function and development. Epigenomic dysregulation may therefore mediate the cellular memory of intrauterine events that are causally associated with adult disease susceptibility.
EXPERIMENTAL PROCEDURES
Bilateral Uterine Artery Ligation and Pancreatic Islet Isolation
Surgical methods have been described previously (9). On day 18 of gestation, time-dated Sprague-Dawley pregnant rats underwent bilateral uterine artery ligation. Controls underwent sham surgery. Pregnant rats were allowed to deliver spontaneously, and the litter size was randomly reduced to 8 at birth to assure uniformity of litter size between IUGR and controls. IUGR and control pups were fostered to unoperated normal female rats and remained with their foster mothers until they were weaned. Only male animals were studied to avoid the potentially confounding influence of hormonal variability. Pancreatic islets were isolated using the Ficoll gradient method (Scharp (48)) from 8 litters each of 7-week-old IUGR and control animals, with islets pooled from a litter as previously described (9). Freshly isolated islets were used for all studies. To remove the confounding factors contributed by even slight differences in purity between islet preparations, we use quantitative RT-PCR to determine the relative levels of endocrine (insulin) and exocrine (amylase) gene expression levels in each sample as previously described (15). This approach allowed us to determine that there was negligible exocrine tissue contamination of isolated pancreatic islets. Using total pancreas RNA for comparison and quantitative RT-PCR, we calculated the enrichment of endocrine mRNA for each of our samples, which averaged 99.1%. Similarly, β-cell fold enrichment (using insulin and glucagon) of islet preps averaged 94.6%. These studies were approved by the Animal Care Committee of the Children's Hospital of Philadelphia and the University of Pennsylvania.
HELP Assay
To define genome-wide patterns of DNA methylation in isolated pancreatic islets of IUGR offspring and controls, we used a high resolution microarray-based approach (HELP (11, 16)) designed to test almost a million sites throughout the rat genome, representing promoters, CpG islands, CG clusters (17), genes, and intergenic regions. Genomic DNA was isolated, digested to completion by either HpaII or MspI separately, and ligated to a mixture of two adapters with overhanging ends complementary to those generated by the restriction digestion. The adapters were used for ligation-mediated PCR to generate products predominantly in the 50- to 2000-bp size range (11). Following PCR, the HpaII and MspI representations were labeled with different fluorophores using random priming and were then cohybridized on a customized genomic microarray representing 917,301 HpaII/MspI fragments of 50–2000 bp in the rat genome.
Microarray Data Analysis
Microarray data were pre-processed and subject to quality control and quantile normalization as we have previously described (15). HpaII/MspI ratio values were then compared between groups for all loci throughout the rat genome using a standard t test for IUGR (n = 4) and controls (n = 4). Changes in methylation state were defined using a HpaII/MspI ratio threshold of zero, where heavily methylated and relatively hypomethylated loci had ratio values less than and greater than zero, respectively. An ordered list of differences was generated using the formula: −log(p) × |IUGR − control|. This method, similar to a modified t test (18), places more weight on the -fold change relative to the within-group variability. However, unlike p values based on the modified t test, computed values are not interpreted in these studies as a probabilistic quantity, and instead are used exclusively to rank and isolate important loci for further validation.
Ingenuity Pathway Analysis
The top 100 loci, identified by means of the ranking system described above, were mapped to RefSeq gene identifiers by location <100 kb upstream of the transcription start site or overlapping the gene body. The list of RefSeq identifiers was then uploaded to the Ingenuity Pathway Analysis program (Redwood City, CA) and mapped to corresponding gene objects (focus genes) in the Ingenuity Pathways Knowledge Base, enabling exploration of ontology and molecular interaction networks. Core networks were constructed for both direct and indirect interactions using default parameters. The ranking score for each network was then computed by a right-tailed Fisher's exact test as the negative log of the probability that the number of focus genes in the network is not due to random chance. Similarly, significances for functional enrichment of specific genes were also determined by the right-tailed Fisher's exact test, using all genes on the microarray as a reference set.
Bisulfite MassArray Validation
Target regions were amplified by PCR using the primers and cycling conditions described in supplemental Table 1. Primers were selected with MethPrimer (available on-line) using parameters as follows: 250–450 bp amplicon size, 56–60 °C Tm, 24–30 bp length, and ≥1 CG in product. PCR reactions were carried out using the Roche FastStart High Fidelity Kit. In cases where products showed primer-dimer or nonspecific amplification, expected bands were excised from 2% agarose gels, purified by the Qiagen Gel Extraction Kit, and eluted with 1× Roche FastStart High Fidelity Reaction Buffer (+MgCl2). Sequenom MassArray assays were performed using the company's standard protocol through Einstein's Genomics Shared Facility. Matched peak data were exported using EpiTYPER software and analyzed for quality and single nucleotide polymorphisms using analytical tools that we have developed (19).
Gene Expression by RT-PCR
mRNA expression levels were determined for eight genes by quantitative RT-PCR. These genes were chosen for having either no changes in methylation in their vicinity (Hint3, Dnmt3a, Dnmt3b, and Gadd45a) or, from the top 100 loci with methylation changes, those with multiple reproducible changes nearby (Fgfr1, Gch1, Vgf, and Pcsk5). Total RNA was extracted from islets of 7-week-old IUGR and control animals (n = 5 from each group) using TRIzol (Invitrogen). Integrity was confirmed on an Agilent Bioanalyzer. cDNA was generated using the SuperScript II Reverse Transcriptase kit (Invitrogen), and real-time quantitative PCR was carried out in triplicate using an ABI 7900HT Real Time PCR system with SYBR Green Master Mix (Applied Biosystems). Results were normalized to β-actin expression and expressed as -fold change compared with control rats. Intron-spanning primers were designed to target exons conserved across the maximal number of expressed mRNA transcripts in the rat genome (primers are listed in supplemental Table 2).
RESULTS
Distinct Patterns of DNA Methylation in IUGR and Control Rats
We performed a whole genome cytosine methylation assay on pancreatic islets isolated at 7 weeks from Sprague-Dawley rats in each of two groups: IUGR offspring (n = 4) and their sham-operated controls (n = 4), and confirmed overall data quality for each sample as previously described (15). To detect global patterns of epigenomic change distinguishing IUGR and control animals, we compared one thousand randomly selected loci to generate a representative heat map. This unsupervised clustering approach showed consistent patterns of methylation across all samples, without any apparent global shift toward hypo- or hypermethylation in the IUGR group (supplemental Fig. 1). Global Pearson correlation coefficients were calculated for the methylation patterns for each pair of samples, confirming a high degree of inter-sample consistency (R = 0.90–0.96, supplemental Fig. 2).
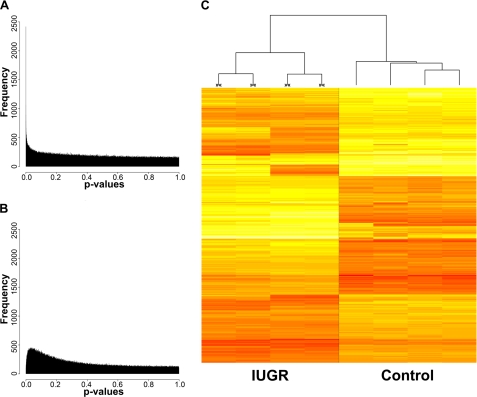
To detect locus-specific epigenetic changes, t tests comparing IUGR and control animals were applied to all individual loci represented on our arrays. This analysis yielded a subset of potentially informative sites with statistically significant differences in methylation. Fig. 1A shows the overall distribution of p values observed for IUGR compared with controls, with a comparable p value distribution calculated for grouping by random permutation (Fig. 1B), reflective of what we would see under the null distribution of no group effect. The IUGR and control group comparisons identify consistently and distinctively methylated loci, with 237 candidate loci enriched for highly significant group differences (p < 1.0 × 10−6, chosen because no iterations of random permutation had a p value below 1.08 × 10−6), and an additional 1675 loci with moderate group differences (p < 0.0001). Additionally, we applied significance analysis of microarrays (18) to the data and show a resulting Q–Q plot (supplemental Fig. 3), which demonstrates ∼1,400 changes in methylation with an estimated false discovery rate of 4.2%. We created a heat map of the top 1000 differentially methylated loci and found that this subset of loci readily distinguishes between IUGR and controls, without an overall tendency toward decreased or increased methylation in either group (Fig. 1C).
FIGURE 1.
Significant differences in cytosine methylation between IUGR and control rats. A, histogram distribution of p values calculated from an unpaired t test of IUGR (n = 4) in one group and controls (n = 4) in another, with p values along the x axis and frequency along the y axis. B, the analogous comparison with data obtained from a representative random grouping (n = 4 per group). C, heat map of the top 1000 loci that distinguish IUGR from control rats. Each row in this heat map corresponds to data from a single locus, whereas columns correspond to individual samples, IUGR (≫≪) and controls. The branching dendrogram corresponds to the relationships among samples, as determined by clustering using these 1000 sites. IUGR and controls show similar numbers of loci becoming relatively hyper- and hypomethylated (red to yellow, respectively).
Genomic Distributions of Cytosine Methylation
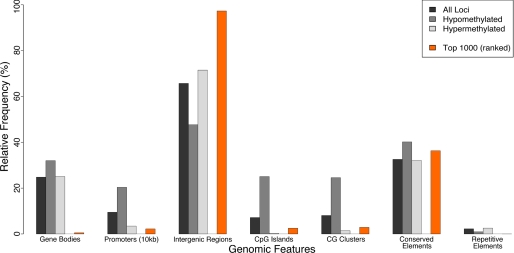
The genomic distributions of methylation were determined for both normal and IUGR pancreatic islets. The data were partitioned into three non-overlapping subsets: (i) consistently hypomethylated sites in both normal and IUGR tissues (average log2(HpaII/MspI) > 2); (ii) consistently hypermethylated sites (average log2(HpaII/MspI) < −1); and (iii) the top 1000 differentially methylated loci. The observed number and associated probabilities of whole and partial sequence overlaps with CpG islands, CG clusters (17), conserved elements, repeat-masked sequences, gene bodies, and promoters were determined for each of these subsets of loci (supplemental Table 3) using UCSC Genome Browser annotation data (20).
Because there has never been a report of genome-wide methylation patterns in rat, nor in normal pancreatic islets, we first investigated the distribution of cytosine methylation in the control animals, focusing on sites that are constitutively hypo- or hypermethylated in all samples. The distributions we observed for genomic sequence compartments in normal islets were consistent with prior studies in other mammals, with hypomethylated loci highly enriched at CpG islands and CG clusters (Fig. 2), as observed in human and mouse (17, 21). Additionally, hypomethylated loci were highly enriched at promoters, and to a lesser extent in gene bodies and conserved sequences (Fig. 2). Repetitive sequences tended to be constitutively hypermethylated (Fig. 2), as has been previously observed in other species (22). Normal pancreatic islets therefore have overall patterns of methylation typical of primary cells from Eutherian mammals. This reference cytosine methylation map is the first published for the rat genome and the first for pancreatic islets in any species, and is available both from the Gene Expression Omnibus (GEO) (accession number: GSE16839) and as an open-access University of California Santa Cruz genome browser-compatible resource available on-line.
FIGURE 2.
Genomic feature distributions of HELP data. Frequencies were calculated for the overlap between four sets of loci (all loci, constitutively hypomethylated, constitutively hypermethylated, and top 1000 differentially methylated) with 7 genomic features (gene body, 10 kb upstream of promoter, intergenic region, conserved element, repetitive element, CpG island, and CG cluster). Associated probabilities that observed differences are non-random (tailed hypergeometric distribution) are shown in supplemental Table 2. Most differential methylation occurs at intergenic and conserved sequences.
We next investigated how these normal patterns of cytosine methylation are dysregulated in IUGR islets. The 1000 most differentially methylated loci were under-represented at gene bodies and promoters as well as CpG islands and CG clusters (Fig. 2). However, they were highly enriched at conserved non-coding intergenic sequences, which may represent important cis-regulatory sites influencing local gene expression (23). The cytosine methylation data from the IUGR animals are also provided as a University of California Santa Cruz genome browser track on-line.
Quantitative Validation of DNA Methylation States
We confirmed our HELP results with a quantitative validation approach, bisulfite MassArray (24), focusing initially on four loci, two each representing constitutively hypomethylated and constitutively methylated sites identified by the HELP assay. Supplemental Fig. 4 shows a strong inverse correlation between methylation values determined independently for these loci by HELP and MassArray, confirming our ability to quantify the methylation status in these samples.
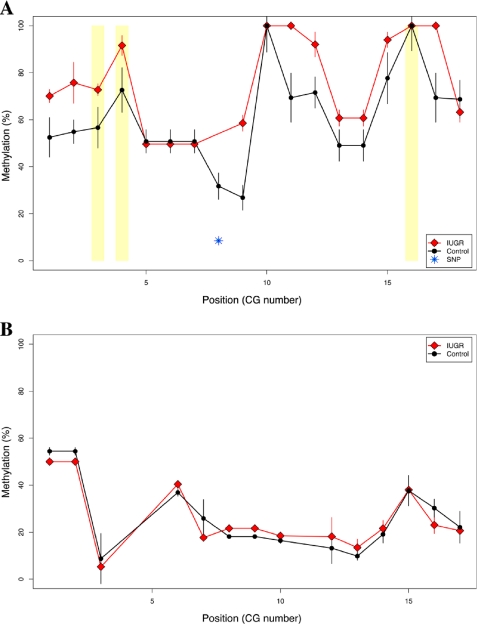
We then tested whether the cytosine methylation differences found by HELP in IUGR rats compared with controls could be validated by bisulfite MassArray. Upstream of the GTP cyclohydrolase 1 (Gch1) gene, the HELP assay identified multiple neighboring changes, with increased methylation in IUGR compared with control rats (supplemental Fig. 5). Among these sites, we identified a region located near a highly conserved non-coding element (present in all mammalian species) ∼45 kb upstream of the Gch1 transcription start site. Differences in methylation at this site were confirmed by bisulfite MassArray, with hypermethylation in IUGR compared with controls at an informative HpaII site (86.3% and 53.1% methylation, respectively; p = 0.003) and multiple neighboring CG dinucleotides (72.8% and 55.7% average overall methylation, respectively; p = 0.01) (Fig. 3A), consistent with the HELP data (log2(HpaII/MspI) = −0.27 ± 0.27 and 2.69 ± 0.59, respectively; p = 4.3 × 10−6). This degree of difference of cytosine methylation is of the magnitude we observe for tissue-specific differences in methylation at other loci (supplemental Fig. 6). Using our recently published analytical pipeline for bisulfite MassArray data (19), we identified one single nucleotide polymorphism in this region that confounded interpretation of methylation status at a single CG dinucleotide (Fig. 3A); therefore, this site was removed from consideration.
FIGURE 3.
Bisulfite MassArray validation confirms IUGR-specific hypermethylation of the Gch1 locus. MassArray was performed for two genomic regions as described by the PCR conditions in supplemental Table 1. Three replicate assays were performed for each of three IUGR offspring and controls. A, MassArray validation results are shown as group-median percent methylation values with standard error bars (y axis) for each individual CG site (in sequential order along the x axis). IUGR is depicted with red lines and diamonds, whereas control data are shown in black lines and filled circles. HpaII sites are highlighted by yellow rectangles. One single nucleotide polymorphism (SNP) was identified in the underlying genomic sequence and was removed from affected samples in the dataset (blue, starred symbol). B, MassArray data are similarly shown for a second genomic region at the Pdx1 gene. No differences in DNA methylation were observed at this age, with relative hypomethylation in all samples.
We tested the cytosine methylation status for the Pdx1 proximal-promoter region. Experimental IUGR causes a permanent and progressive decrease in expression of Pdx1, which is associated with progressive epigenetic changes from a hypomethylated state at 2 weeks of age and culminating in DNA hypermethylation in the adult rat at age 6 months (10). In our 7-week-old rats we found the Pdx1 proximal-promoter region to be hypomethylated in islets and indistinguishable from control animals (Fig. 3B). These findings are consistent with our previous studies in islets from young adult IUGR animals (10), narrowing the window in which the methylation is acquired to between 7 weeks and 6–8 months.
Molecular Interaction Networks of Genes Associated with Epigenetic Dysregulation
We next investigated whether genes associated by proximity to the loci manifesting epigenetic dysregulation in IUGR shared any common functional roles or relationships. We performed ingenuity pathway analysis on the top 53 of 100 loci, which were linked to RefSeq-annotated genes (within 100 kb upstream of the transcription start site or within the gene body). Almost half of the 53 genes tested were associated together in a single functional network centered on a collection of important metabolic and cellular regulators, including extracellular signal-regulated kinase, protein kinase B, and nuclear factor kappa-B (NFκB) (with an associated chance of ∼10−49% that this network might arise randomly) (supplemental Fig. 7). This network also showed interaction of multiple genes directly relevant to β-islet cells, including integrin β5 (Itgb5), glycogen synthase kinase 3 β (Gsk3b), and vascular growth factor nerve growth factor inducible (Vgf). Ingenuity pathway analysis further revealed that the top 53 genes dysregulated in IUGR were significantly enriched for functions in endocrine development (p = 3.1 × 10−3), cellular proliferation and growth (p = 2.7 × 10−3), and cell death (p = 2.1 × 10−3), consistent with our previous observations of impaired β-cell proliferation and increased cell death in this model (3, 6). Cytosine methylation changes in islets from IUGR rats are therefore enriched near RefSeq genes that are candidates for mediating the later T2DM phenotype.
Gene Expression Analysis
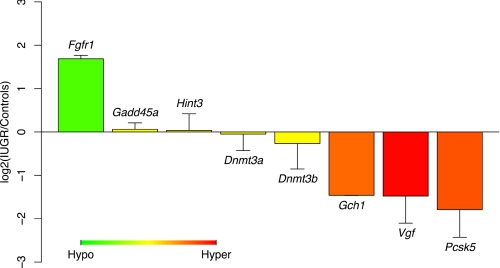
To test whether these mostly non-promoter cytosine methylation changes were having functional effects on gene expression, quantitative RT-PCR was used to determine mRNA expression levels for eight genes, four of which had methylation changes identified by HELP located an average of ∼45 kb upstream of their transcription start sites (fibroblast growth factor receptor 1 (Fgfr1), Gch1, histidine triad nucleotide-binding protein 3 (Hint3), and Vgf) and one of which was differentially methylated within 750 bp of its transcription start site (proprotein convertase subtilisin/kexin type 5 (Pcsk5)). The remaining three (DNA methyltransferases 3a and 3b (Dnmt3a and Dnmt3b) and growth arrest and DNA-damage-inducible 45 alpha (Gadd45a)) are known or candidate regulators of global DNA methylation patterns (25, 26) but showed no appreciable change in cytosine methylation by our HELP analysis. We detected significant changes in mRNA expression levels for IUGR compared with control animals at Fgfr1 (increased 3.2 ± 0.2-fold), Gch1 (decreased 2.8 ± 0.01-fold), Pcsk5 (decreased 3.2 ± 1.0-fold), and Vgf (decreased 2.6 ± 0.8-fold), with p < 0.01 for all these loci (Fig. 4). These changes correlated with differences in DNA methylation observed, with hypomethylation in IUGR rats compared with controls at Fgfr1 and hypermethylation at Vgf, Gch1, and Pcsk5 (Fig. 4; supplemental Table 4). Although the HELP assay initially identified differences in methylation at Hint3, it was subsequently confirmed that these changes were artifacts due to multiple single nucleotide polymorphisms (Fig. 3A). Correspondingly, we observed no differences in gene expression at the Hint3 locus (Fig. 4). Moreover, Dnmt3a, Dnmt3b, and Gadd45a are all similarly expressed in IUGR offspring and controls, indicating that the differences in DNA methylation we observed throughout the genome are independent of transcription levels for these regulators of cytosine methylation.
FIGURE 4.
Quantitative RT-PCR shows changes in mRNA expression concordant with the cytosine methylation changes. Primers were designed to target mRNA transcripts for eight loci, four of which were genes associated with differential methylation (Fgfr1, Vgf, Gch1, and Pcsk5), and four with no changes in methylation, including Hint3 and three genes regulating cytosine methylation (Gadd45a, Dnmt3a, and Dnmt3b). Average log2 ratio (± S.D.) of IUGR expression data relative to controls was calculated for each gene. The color scheme indicates corresponding methylation data (green for hypomethylation, red for hypermethylation) in IUGR samples compared with controls. All methylation values displayed are from HELP data except Hint3, for which MassArray data proved more reliable because of a local sequence polymorphism.
DISCUSSION
Our work contributes the first genome-wide study of DNA methylation in rats and in normal pancreatic islets and uses this foundation to define abnormal patterns in islets from a well characterized model of T2DM. We found that experimentally induced IUGR in the rat causes consistent and non-random changes in cytosine methylation, affecting <1% of HpaII sites in the genome. The majority of these changes take place not at promoters but at intergenic sequences, many of which are evolutionarily conserved. Furthermore, some of these loci are in proximity to genes manifesting concordant changes in gene expression and are enriched near genes that regulate processes that are markedly impaired in IUGR islets (e.g. vascularization, proliferation, insulin secretion, and cell death) (3, 6, 9, 27). This epigenomic dysregulation precedes the development of diabetes and is therefore a potential mediator of the pathogenesis of the disease, preserving cellular memory of the long antecedent intrauterine event. Finally, our identification of differential methylation at conserved intergenic and potentially cis-regulatory sites emphasizes the limitations of study designs that focus solely on promoters and/or CpG islands. We show that changes in methylation, even when not located at promoters, are correlated with transcriptional changes at adjacent genes, consistent with suggestions from previous studies (28).
One gene in particular, GTP cyclohydrolase 1 (Gch1), showed almost a 3-fold reduction in mRNA expression, associated with hypermethylation at a conserved intergenic site ∼45 kb upstream of the gene itself. Additionally, down-regulation of Gch1 is associated with hyperglycemia secondary to progressive β-cell dysfunction (impaired glucose-stimulated insulin secretion) and β-cell loss (reduced number and proliferation) in Tcf1 (hepatocyte nuclear factor 1-α) knock-out mice (data from Ref. 29 and associated Gene Expression Omnibus (GEO) dataset GSE3544). Gch1 is a crucial (rate-limiting) component of the tetrahydrobiopterin synthetic pathway and has been directly implicated in diabetes-associated endothelial dysfunction through its critical role in nitric oxide (NO) synthesis (30, 31). Interestingly, folate supplementation of protein-restricted dams, known to buffer against changes in DNA methylation (32), prevented the development of hypertension and reduced NO synthesis and endothelial dysfunction that was otherwise observed in male offspring (33, 34). We hypothesize that the Gch1 down-regulation we observed with IUGR could play a role in the progressive β-cell decline that we have previously observed (9), through direct effects on the β-cells or islet vascularization (27).
At all four of the candidate genes with cytosine methylation changes we were able to measure changes in transcription, indicating that the genome-wide methylation changes are likely to affect transcription at many loci. In general, these changes occurred at genes involved in cellular growth and development, some implicated in pancreatic function. For instance, the fibroblast growth factor receptor 1 (Fgfr1) is expressed in β-cells, dependent upon Pdx1 activity, and is directly involved in β-cell function and glucose homeostasis (35). Furthermore, Fgfr1 signaling is modulated by the β-cell microenvironment, in particular the extracellular matrix deposited by apposing endothelial cells (36–39). Interestingly, depletion of laminin from the extracellular matrix, which may indicate immature, impaired, or damaged vascular endothelial cells (40, 41), increases Fgfr1 levels and activity (36). We hypothesize that the up-regulation of Fgfr1 we observed could indicate dysregulation of the β-cell and vascular endothelial microenvironment, either in addition to or associated with the depletion of Gch1. The Pcsk5 gene, where we found differential methylation at the promoter region, is a subtilisin/kexin-like proprotein convertase that may impair β-cell activity through Igf1 receptor (42)- and bone morphogenetic protein-4 (43)-mediated pathways (44, 45). Moreover, Pcsk5, which is the major regulator of αv integrin activity (46), interacts in pancreatic islets with β5 integrin to maintain β-cell adhesion to the extracellular matrix (47). Of note is that the gene for β5 integrin is also among the top 100 differentially methylated loci identified in this study, with hypermethylation in IUGR compared with control islets at a site ∼48 kb upstream of the transcription start site (supplemental Fig. 8). Taken together, these changes and functional associations are suggestive of a deficient β-cell microenvironment in IUGR and may explain some of the β-cell phenotype observed in IUGR islets (3, 6, 27).
We tested animals at 7 weeks, prior to the onset of mild fasting hyperglycemia (9). At this age, β-cell remodeling is complete and the animal has gone through puberty, thus avoiding confounding variables due to either of these physiologic states. This 7-week time point also enabled us to study the epigenome in the absence of overt hyperglycemia, which subsequently begins to develop in IUGR animals. On the other hand, we note that animals at 7 weeks already exhibit altered metabolism (e.g. redox state and insulin levels (6)), which may contribute to the altered epigenetic states that we have observed in IUGR islets. As such, we cannot determine whether the IUGR-inducing environment contributes to the altered epigenetic states that we have observed in IUGR islets by direct or indirect mechanisms. It is in fact possible that the epigenome may be influenced by multiple such mechanisms, both direct and indirect. Irrespective of the exact etiology, it is clear that IUGR induces both altered epigenetic patterns and gene expression programs, observable in IUGR animals as early as 7 weeks.
In conclusion, our study has found evidence of early epigenetic dysregulation in pancreatic islets that may mediate, in whole or in part, the long term consequences of a deficient intrauterine environment. Epigenetic dysregulation is therefore a strong candidate for mediating the cellular memory that allows these fetal events to confer susceptibility to adult disease. By using a genome-wide approach, we have uncovered reproducible and validated changes in cytosine methylation that occur predominantly at candidate cis-regulatory elements and are associated with local transcriptional dysregulation. Although the direct causality and subsequent physiologic consequences of these altered epigenetic states remain unclear, this genome-wide approach to studying cytosine methylation provides a novel means of gaining insights into the pathogenesis of T2DM.
This work was supported, in whole or in part, by National Institutes of Health Grants HG004401 and HD044078 (to J. M. G.), DK55704 and DK062965 (to R. A. S.), and AG21654 and AG18381 (to N. B.). This work was also supported by Einstein's Center for Epigenomics and by the Core laboratories of the Albert Einstein Diabetes Research and Training Center (Grant DK 20541).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–8, Tables 1–4, and additional references.
- T2DM
- type 2 diabetes mellitus
- IUGR
- intrauterine growth restriction
- HELP
- HpaII tiny fragment enrichment by ligation-mediated PCR
- CpG/CG
- cytosine-guanine dinucleotide
- RT
- reverse transcription.
REFERENCES
- 1.Barker D. J. (2006) Clin. Obstet. Gynecol. 49, 270–283 [DOI] [PubMed] [Google Scholar]
- 2.Martin-Gronert M. S., Ozanne S. E. (2007) J. Intern. Med. 261, 437–452 [DOI] [PubMed] [Google Scholar]
- 3.Stoffers D. A., Desai B. M., DeLeon D. D., Simmons R. A. (2003) Diabetes 52, 734–740 [DOI] [PubMed] [Google Scholar]
- 4.Arantes V. C., Teixeira V. P., Reis M. A., Latorraca M. Q., Leite A. R., Carneiro E. M., Yamada A. T., Boschero A. C. (2002) J. Nutr. 132, 3030–3035 [DOI] [PubMed] [Google Scholar]
- 5.Dahri S., Snoeck A., Reusens-Billen B., Remacle C., Hoet J. J. (1991) Diabetes 40, Suppl. 2, 115–120 [DOI] [PubMed] [Google Scholar]
- 6.Simmons R. A., Suponitsky-Kroyter I., Selak M. A. (2005) J. Biol. Chem. 280, 28785–28791 [DOI] [PubMed] [Google Scholar]
- 7.Barker D. J. (2007) J. Intern. Med. 261, 412–417 [DOI] [PubMed] [Google Scholar]
- 8.Simmons R. A. (2007) Pediatr. Res. 61, 64R–67R [DOI] [PubMed] [Google Scholar]
- 9.Simmons R. A., Templeton L. J., Gertz S. J. (2001) Diabetes 50, 2279–2286 [DOI] [PubMed] [Google Scholar]
- 10.Park J. H., Stoffers D. A., Nicholls R. D., Simmons R. A. (2008) J. Clin. Invest. 118, 2316–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oda M., Glass J. L., Thompson R. F., Mo Y., Olivier E. N., Figueroa M. E., Selzer R. R., Richmond T. A., Zhang X., Dannenberg L., Green R. D., Melnick A., Hatchwell E., Bouhassira E. E., Verma A., Suzuki M., Greally J. M. (2009) Nucleic Acids Res. 37, 3829–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vuguin P., Raab E., Liu B., Barzilai N., Simmons R. (2004) Diabetes 53, 2617–2622 [DOI] [PubMed] [Google Scholar]
- 13.Muoio D. M., Newgard C. B. (2008) Nat. Rev. Mol. Cell Biol. 9, 193–205 [DOI] [PubMed] [Google Scholar]
- 14.Finegood D. T., Scaglia L., Bonner-Weir S. (1995) Diabetes 44, 249–256 [DOI] [PubMed] [Google Scholar]
- 15.Thompson R. F., Reimers M., Khulan B., Gissot M., Richmond T. A., Chen Q., Zheng X., Kim K., Greally J. M. (2008) Bioinformatics 24, 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khulan B., Thompson R. F., Ye K., Fazzari M. J., Suzuki M., Stasiek E., Figueroa M. E., Glass J. L., Chen Q., Montagna C., Hatchwell E., Selzer R. R., Richmond T. A., Green R. D., Melnick A., Greally J. M. (2006) Genome Res. 16, 1046–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glass J. L., Thompson R. F., Khulan B., Figueroa M. E., Olivier E. N., Oakley E. J., Van Zant G., Bouhassira E. E., Melnick A., Golden A., Fazzari M. J., Greally J. M. (2007) Nucleic Acids Res. 35, 6798–6807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tusher V. G., Tibshirani R., Chu G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson R. F., Suzuki M., Lau K. W., Greally J. M. (2009) Bioinformatics 25, 2164–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D. (2002) Genome Res. 12, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bird A., Taggart M., Frommer M., Miller O. J., Macleod D. (1985) Cell 40, 91–99 [DOI] [PubMed] [Google Scholar]
- 22.Yang A. S., Estécio M. R., Doshi K., Kondo Y., Tajara E. H., Issa J. P. (2004) Nucleic Acids Res. 32, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pennacchio L. A., Ahituv N., Moses A. M., Prabhakar S., Nobrega M. A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K. D., Plajzer-Frick I., Akiyama J., De Val S., Afzal V., Black B. L., Couronne O., Eisen M. B., Visel A., Rubin E. M. (2006) Nature 444, 499–502 [DOI] [PubMed] [Google Scholar]
- 24.Ehrich M., Nelson M. R., Stanssens P., Zabeau M., Liloglou T., Xinarianos G., Cantor C. R., Field J. K., van den Boom D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15785–15790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barreto G., Schäfer A., Marhold J., Stach D., Swaminathan S. K., Handa V., Döderlein G., Maltry N., Wu W., Lyko F., Niehrs C. (2007) Nature 445, 671–675 [DOI] [PubMed] [Google Scholar]
- 26.Okano M., Bell D. W., Haber D. A., Li E. (1999) Cell 99, 247–257 [DOI] [PubMed] [Google Scholar]
- 27.Ham J. N., Crutchlow M. F., Desai B. M., Simmons R. A., Stoffers D. A. (2009) Pediatr. Res. 66, 42–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irizarry R. A., Ladd-Acosta C., Wen B., Wu Z., Montano C., Onyango P., Cui H., Gabo K., Rongione M., Webster M., Ji H., Potash J. B., Sabunciyan S., Feinberg A. P. (2009) Nat. Genet. 41, 178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akpinar P., Kuwajima S., Krützfeldt J., Stoffel M. (2005) Cell Metab. 2, 385–397 [DOI] [PubMed] [Google Scholar]
- 30.Meininger C. J., Cai S., Parker J. L., Channon K. M., Kelly K. A., Becker E. J., Wood M. K., Wade L. A., Wu G. (2004) FASEB J. 18, 1900–1902 [DOI] [PubMed] [Google Scholar]
- 31.Antoniades C., Shirodaria C., Van Assche T., Cunnington C., Tegeder I., Lötsch J., Guzik T. J., Leeson P., Diesch J., Tousoulis D., Stefanadis C., Costigan M., Woolf C. J., Alp N. J., Channon K. M. (2008) J. Am. Coll. Cardiol. 52, 158–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lillycrop K. A., Phillips E. S., Jackson A. A., Hanson M. A., Burdge G. C. (2005) J. Nutr. 135, 1382–1386 [DOI] [PubMed] [Google Scholar]
- 33.Brawley L., Itoh S., Torrens C., Barker A., Bertram C., Poston L., Hanson M. (2003) Pediatr. Res. 54, 83–90 [DOI] [PubMed] [Google Scholar]
- 34.Torrens C., Brawley L., Anthony F. W., Dance C. S., Dunn R., Jackson A. A., Poston L., Hanson M. A. (2006) Hypertension 47, 982–987 [DOI] [PubMed] [Google Scholar]
- 35.Hart A. W., Baeza N., Apelqvist A., Edlund H. (2000) Nature 408, 864–868 [DOI] [PubMed] [Google Scholar]
- 36.Kilkenny D. M., Rocheleau J. V. (2008) Mol. Endocrinol. 22, 196–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parnaud G., Hammar E., Rouiller D. G., Armanet M., Halban P. A., Bosco D. (2006) Diabetes 55, 1413–1420 [DOI] [PubMed] [Google Scholar]
- 38.Kaido T., Yebra M., Cirulli V., Rhodes C., Diaferia G., Montgomery A. M. (2006) Diabetes 55, 2723–2729 [DOI] [PubMed] [Google Scholar]
- 39.Nikolova G., Jabs N., Konstantinova I., Domogatskaya A., Tryggvason K., Sorokin L., Fässler R., Gu G., Gerber H. P., Ferrara N., Melton D. A., Lammert E. (2006) Dev. Cell 10, 397–405 [DOI] [PubMed] [Google Scholar]
- 40.Davis G. E., Senger D. R. (2005) Circ. Res. 97, 1093–1107 [DOI] [PubMed] [Google Scholar]
- 41.Goldin A., Beckman J. A., Schmidt A. M., Creager M. A. (2006) Circulation 114, 597–605 [DOI] [PubMed] [Google Scholar]
- 42.Khatib A. M., Siegfried G., Prat A., Luis J., Chrétien M., Metrakos P., Seidah N. G. (2001) J. Biol. Chem. 276, 30686–30693 [DOI] [PubMed] [Google Scholar]
- 43.Cui Y., Jean F., Thomas G., Christian J. L. (1998) EMBO J. 17, 4735–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kulkarni R. N., Holzenberger M., Shih D. Q., Ozcan U., Stoffel M., Magnuson M. A., Kahn C. R. (2002) Nat. Genet. 31, 111–115 [DOI] [PubMed] [Google Scholar]
- 45.Goulley J., Dahl U., Baeza N., Mishina Y., Edlund H. (2007) Cell Metab. 5, 207–219 [DOI] [PubMed] [Google Scholar]
- 46.Stawowy P., Kallisch H., Veinot J. P., Kilimnik A., Prichett W., Goetze S., Seidah N. G., Chrétien M., Fleck E., Graf K. (2004) Circulation 109, 770–776 [DOI] [PubMed] [Google Scholar]
- 47.Kaido T., Perez B., Yebra M., Hill J., Cirulli V., Hayek A., Montgomery A. M. (2004) J. Biol. Chem. 279, 17731–17737 [DOI] [PubMed] [Google Scholar]