Abstract
ErbB family of the receptor protein-tyrosine kinase plays an important role in the progression of human cancers including breast cancer. Finding protein-tyrosine phosphatase (PTPs) that can specifically regulate the function of ErbB should help design novel therapies for treatment. By performing a small interfering RNA screen against 43 human PTPs, we find that knockdown of protein-tyrosine phosphatase PTPN9 significantly increases ErbB2 tyrosyl phosphorylation in the SKBR3 breast cancer cell line. In addition, knockdown of PTPN9 expression also enhances tyrosyl phosphorylation of the ErbB1/epidermal growth factor receptor (EGFR) in the MDA-MB-231 breast cancer cell line. Conversely, increasing expression of PTPN9 wild type (WT) inhibits tyrosyl phosphorylation of ErbB2 and EGFR. To test whether ErbB2 and EGFR are substrates of PTPN9, PTPN9 WT, and a substrate trapping mutant (PTPN9 DA) are overexpressed in SKBR3 and MDA-MB-231 cells. Compared with vector control, expression of PTPN9 WT significantly inhibits whereas expression of PTPN9 DA dramatically enhances tyrosyl phosphorylation of ErbB2 and EGFR, respectively. In contrast, expression of PTPN9 WT or DA mutant does not affect tyrosyl phosphorylation of ErbB3 and Shc. Importantly, coimmunoprecipitation and glutathione S-transferase fusion protein pulldown experiments show that tyrosol-phosphorylated ErbB2 or EGFR is preferentially associated with PTPN9 DA compared with PTPN9 WT, indicating that ErbB2 and EGFR are substrates of PTPN9. Furthermore, PTPN9 WT expression specifically impairs EGF-induced STAT3 and STAT5 activation, and inhibits the cell growth in soft agar. Last, PTPN9 WT expression also reduces invasion and MMP2 expression of MDA-MB-231 cells. Our data suggest PTPN9 as a negative regulator of breast cancer cells by targeting ErbB2 and EGFR and inhibiting STAT activation.
Keywords: Breast Cancer, Cell Motility, Receptor-tyrosine Kinase, siRNA, Tyrosine-protein Phosphatase (Tyrosine Phosphatase), EGFR, ErbB2, PTPN9, STAT3/5
Introduction
The ErbB family of receptor protein-tyrosine kinase (PTK),2 which include EGFR/ErbB1, ErbB2, ErbB3, and ErbB4, plays an important role in the development of various types of cancers, including breast cancer (1). ErbB1/EGFR is usually overexpressed in a significant fraction of triple negative breast tumors that are negative in estrogen and progesterone receptors, and ErbB2/Her2 expression with a worse outcome (2). ErbB2/Her2 is overexpressed in ∼25% of breast cancer patients who usually have a poor prognosis (3). ErbB3 is frequently expressed in breast cancers with ErbB2 overexpression (4) and is required for ErbB2-induced breast cancer cell growth (5). ErbB members are activated upon ligand binding, inducing homodimerization (i.e. EGFR) or heterodimerization of EGFR or ErbB3 with ErbB2. ErbB dimerization initiates phosphorylation on various tyrosine residues in the cytoplasmic tail of ErbB, which serve to recruit and activate multiple signaling pathways including Ras/ERK, phosphatidylinositol 3-kinase/Akt, Src, and STAT that drive the growth, migration, and invasion of cancer cells (1). Although great efforts have been made to develop drugs to down-regulate cell surface expression (by monoclonal antibodies) and kinase activity (by small molecule kinase inhibitors) of EGFR and ErbB2, many breast cancer patients with overexpression of ErbB2 and/or EGFR still do not respond to or develop resistance to these drug treatments. Clearly, further research is needed to uncover new ways to inhibit ErbB initiated signaling in breast cancer cells.
Protein-tyrosine phosphatases (PTPs), which include membrane-associated receptor and cytoplasmic types, are enzymes that remove phosphates from phosphorylated tyrosine residues in proteins (6). Therefore, PTPs are thought to antagonize the action of PTKs that add phosphates on tyrosine residues in proteins. PTP that specifically dephosphorylates tyrosine phosphorylation of the cytoplasmic tails of ErbBs should in principal be able to inhibit oncogenic growth and invasion of EGFR/ErbB2/ErbB3 expressing breast cancer cells. No specific ErbB3 PTP has been identified so far. Published reports indicate that PTP1B (7–9) and PTPN6/Shp-1 (10) can dephosphorylate EGFR. In addition, PTPN13 was reported to negatively regulate ErbB2 signaling through direct dephosphorylation (11). However, the role of these PTPs in breast cancer cells is still not clear. In fact, PTP1B expression promotes ErbB2-evoked breast carcinogenesis both in vitro (12) and in mice (13, 14).
PTPN9, also called PTP-MEG2, is a cytoplasmic PTP. PTPN9 plays an important role in promoting intracellular secretory vesicle fusion in hematopoietic cells (15). It is required for embryonic development (16) and growth and expansion of erythroid cells (17). The role of PTPN9 in receptor PTK signaling is less well known. Only one report shows that PTPN9 can antagonize insulin signaling by reducing insulin receptor phosphorylation and Akt activation in insulin responsive cells (18). However, it is not clear whether PTPN9 inhibits insulin signaling by direct dephosphorylation of the insulin receptor. In this article, we show that PTPN9 inhibits EGF-evoked signaling and STAT3 and STAT5 by direct dephosphorylation of EGFR and ErbB2. Overexpression of PTPN9 impairs oncogenic growth and invasion of breast cancer cells overexpressing ErbB2 and/or EGFR.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
293T cells and human breast cancer cell lines SKBR3 and MDA-MB-231 were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Hyclone), 1 mm sodium pyruvate, 100 units/ml of penicillin, and 100 μg/ml of streptomycin (Hyclone). Human recombinant EGF and β1-heregulin were from PeproTech and R&D Systems, respectively. Non-target control siRNA and On-Target Plus human PTPN9 siRNA oligos were from Dharmacon (Colorado). The sequence of PTPN9 siRNA oligo is 5′-GAAAACAACGCTAGAAATT-3′.
Plasmids and Retrovirus Production
Retroviral MSCV-IRES-GFP (pMIG) plasmid expressing human PTPN9 and its substrate trapping mutant D470A (DA) cDNAs were as described (17). pCMV plasmid expressing N-terminal FLAG-tagged PTPN9 WT and PTPN9 DA were generated by PCR. Details of these constructs are available upon request. pCDNA3 expressing the rat oncogenic (activated) form of ErbB2/NeuNT was as described (19). cDNAs of the human Shp-1 wild type (WT) and phosphatase-dead substrate trapping mutant Cys → Ser (CS) (20) were inserted into the EcoRI site of the pLNCX2 retroviral vector. EGFP-EGFR plasmid, which does not express GFP, was a kind gift from Dr. A. Sorkin (University of Colorado, Denver). For vesicular stomatitis virus (VSV)-G pseudotyped retrovirus production, 293T cells were co-transfected with retroviral plasmids together with the packaging plasmids pCMV-VSV-G, JK3, and pCMV-TAT2 as described (21).
Retroviral Infection of Cells, Transfection of Cells with siRNA Oligo, and Cell Growth Assays
SKBR3 and MDA-MB-231 cells were infected with retroviruses and stable pools were used for experiments. SKBR3 or MDA-MB-231 cells were incubated with pMIG retroviruses or pLNCX2 retroviruses for 24 h. For the pMIG-infected cells, GFP positive cells were isolated by fluorescence-activated cell sorting 2 days later. For the pLNCX2-infected cells, stable pools of cells were selected in the presence of 200 μg/ml of G418 (Invitrogen) for 6 days. For each construct, three different stable pools of cells from different retroviral infections were used in our experiments, which gave the same results. To knockdown PTPN9 expression, SKBR3 and MDA-MB-231 cells were transiently transfected with 100 nm non-target control siRNA oligo or PTPN9 siRNA oligo using transfection reagent 1 (Dharmacon), and lysed for biochemical analyses 3 days after transfection. 293T cells were transiently transfected with the indicated amount of DNA using FuGENE transfection reagent (Roche Applied Science), and lysed for biochemical analyses 3 days post-transfection. Cell proliferation was analyzed by MTS assay according to the manufacturer's instruction (Promega) in 96-well plates. For soft agar assay, cells were resuspended in culture medium containing 0.5% (MDA-MB-231) or 0.25% (SKBR3) agar (Fisher) and added to the top of the 0.5% base agar in 6-well plates. After 2–4 weeks, the colonies were stained with 0.01% crystal violet and counted.
Antibodies, Immunoblotting, and Immunoprecipitation
Anti-PTPN9 rabbit serum was described previously (17). Anti-phospho-ERK (pERK), phospho-Akt (pAkt) (Ser473), phospho-EGFR (pEGFR) (Tyr845, Tyr998, Tyr1068), phospho-ErbB2 (pErbB2) (Tyr877, Tyr1221/Tyr1222), phospho-STAT3 (pSTAT3) (Tyr705), phospho-STAT5 (pSTAT5) (Tyr694), phospho-Shc (pShc) (Tyr239/240), and EGFR antibodies were from Cell Signaling Technology, Inc. Anti-STAT3, STAT5, ErbB2, Shc, Akt, and ERK1/2 antibodies were purchased from Santa Cruz Biotechnology, Inc. Anti-phosphotyrosine antibody (4G10) was from Millipore. For Western blot, cells were directly lysed in 1× SDS sample buffer. For immunoprecipitation, cells were lysed in 1% Triton X-100, 25 mm Tris-HCl, pH 7.4, 2.5 mm EDTA, 150 mm NaCl, 30 mm β-glycerophosphate, 10 mm NaF, 1 mm sodium vanadate, 1 mm phenylmethylsulfonyl fluoride and protease inhibitor mixtures. Lysates were clarified by centrifugation, incubated with primary antibody followed by incubation with protein A-agarose beads (Invitrogen), or directly incubated with anti-FLAG M2 beads (Sigma). The immune complexes were washed, and resuspended in 1× SDS sample buffer. Protein samples were separated by SDS-PAGE, transferred to Immobilon-P membrane (Millipore Inc.), immunoblotted using the appropriate primary and secondary antibodies, and detected by enhanced chemiluminescence. The intensities of bands in Western blots were quantified by densitometry analysis using NIH Image J software.
Expression and Purification of GST-PTPN9 Fusion Proteins
PTPN9 WT and PTPN9 DA cDNAs were cloned into the pGEX4T-2 plasmid, and transformed into BL21 Escherichia coli (kindly provided by Dr. C. K. Kassenbrock). GST and GST-PTPN9 fusion protein expression were induced under room temperature overnight with 10 μm isopropyl β-d-1-thiogalactopyranoside, and purified on glutathione-agarose beads as described previously (20). Glutathione-agarose beads containing equal amounts (15 μg) of GST or GST-PTPN9 fusion proteins were used to incubate with equal amounts of cell lysates from MDA-MB-231 or 293T-transfected cells for 2 h at 4 °C.
Matrigel Invasion Assay
Matrigel invasion assay was performed in Matrigel-coated transwells (BD Biosciences Invasion chambers). Equal numbers (105) of cells resuspended in DMEM only were added to the transwells, which were placed into the 24-well plate containing 0.5 ml of DMEM with 10% FBS. Eighteen hours later, the transwells were washed and fixed with 3.7% formaldehyde, and stained with 0.05% crystal violet. The non-invaded cells on the upper surface were removed with a cotton swab and the invaded cells on the lower surface were counted from 8 random fields under a ×10 objective lens of a phase-contrast microscopy.
Gelatin Zymography
Equal numbers of MDA-MB-231 vector control and PTPN9 WT expressing cells were plated in a 6-well plate and 24 h later changed to DMEM without FBS. Equal volumes of the culture supernatants were harvested 24 h later, mixed with SDS sample buffer without reducing agents, and resolved in 10% SDS-PAGE containing 2 mg/ml of gelatin. The gel was washed with 2.5% Triton X-100, incubated in 15 mm CaCl2 overnight, stained with Coomassie Blue, and destained.
Quantitative Reverse Transcription-PCR
Total RNAs (1 μg) isolated from cells using TRIzol Reagent (Invitrogen) were treated with DNase I and reverse transcribed into cDNA using Superscript III (Invitrogen). Quantitative reverse transcription-PCR was performed on a 7500 fast real time PCR system (Applied Biosystems) using Power SYBR Green PCR Master Mix (Applied Biosystems). PCR primers were: human MMP2 gene forward primer 5′-CGCTCAGATCCGTGGTGAG-3′, reverse primer 5′-TGTCACGTGGCGTCACAGT-3′; human MMP9 gene forward primer 5′-CTGGGCAGATTCCAAACCT-3′, reverse primer 5′-CGGCAAGTCTTCCGAGTAGT-3′; human β-actin gene forward primer 5′-AAATCTGGCACCACACCTTC-3′, reverse primer 5′-CAGAGGCGTACAGGGATAGC-3′. The PCR results were normalized to β-actin and expressed as relative mRNA levels compared with MMP2 vector control sample.
Statistical Analysis
Unpaired one-tailed Student's t test was used for statistical analysis (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
RESULTS
Changing the Expression of PTPN9 Affects Tyrosyl Phosphorylation of ErbB2 and EGFR in Breast Cancer Cells
We performed a siRNA screen against 43 human PTPs using the On-Target Plus pool of siRNA (Dharmacon) in SKBR3 cells, which overexpress ErbB2, to look for protein-tyrosine phosphatases that can specifically dephosphorylate ErbB2. Individual wells, in a 96-well plate, containing a pool of siRNAs against each PTP or scramble control RNA was incubated with cells in the presence of transfection reagent (Dharmacon). Three days later, transfected cells were harvested for immunoblotting with antibody against phospho-ErbB2 followed by reprobing with antibody against ErbB2. From three independent screens, we found that only the siRNA against PTPN9 significantly increases (∼2-fold) ErbB2 tyrosine phosphorylation in SKBR3 cells compared with scramble control RNA (data not shown).
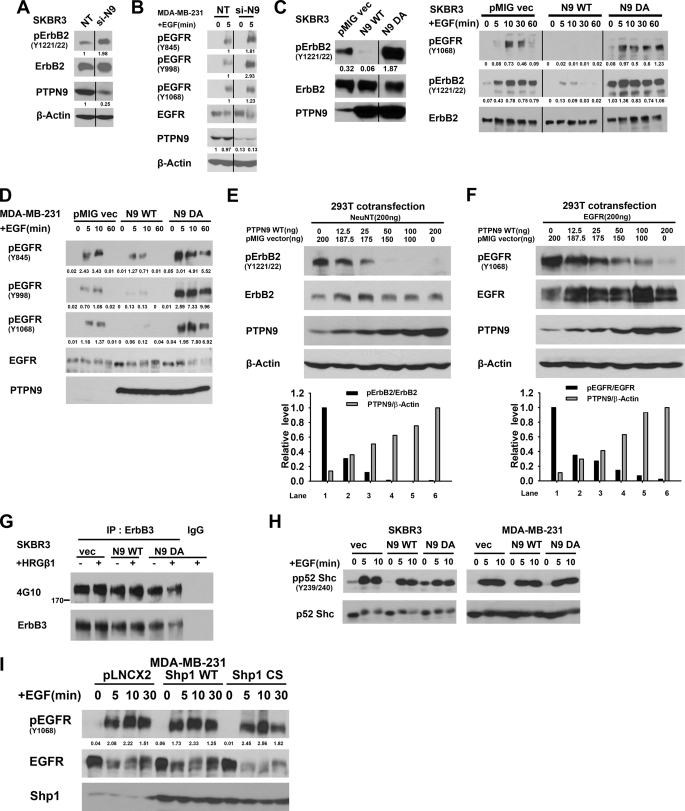
To verify the validity of the siRNA screening result, we knocked down PTPN9 expression by transfecting SKBR3 cells with the Non-Target control siRNA oligo (NT) and an On-Target Plus siRNA oligo against human PTPN9 (Fig. 1A). We decided to use one PTPN9 siRNA oligo (number 5) because we found that this siRNA oligo inhibits 70–80% PTPN9 expression, as shown in Figs. 1, A and B, and 3E, whereas the other three PTPN9 siRNA oligos in the On-Target Plus pool did not reduce PTPN9 expression by more than 50% (supplemental Fig. S1). Compared with cells transfected the NT oligo, cells transfected with the PTPN9 siRNA oligo (number 5) showed a significant increase (∼2-fold) in tyrosyl phosphorylation of ErbB2. Reprobing the same blot with anti-ErbB2 antibody indicated that PTPN9 siRNA does not affect expression of the ErbB2 protein (Fig. 1A). We also examined whether knockdown of PTPN9 affects tyrosyl phosphorylation of EGFR in MDA-MB-231 cells, which resemble triple negative breast cancer cells, expressing a higher level of EGFR but not ErbB2 (Fig. 1B). In response to EGF stimulation, EGFR becomes tyrosyl phosphorylated in cells transfected with the NT oligo. In contrast, EGF-stimulated EGFR tyrosyl phosphorylation is further enhanced in cells transfected with PTPN9 siRNA. Reprobing the same blot with anti-EGFR antibodies indicates that PTPN9 siRNA does not affect the expression level of the EGFR protein (Fig. 1B). We also examined the effects of knocking down PTPN9 expression on tyrosyl phosphorylation of ErbB2 and EGFR using the PTPN9 siRNA oligo (number 5) in BT-474 (ErbB2 positive) and BT-20 (ErbB2 negative and EGFR positive) breast cancer cell lines. Despite various transfection conditions tested, the PTPN9 siRNA (number 5) can only knockdown PTPN9 expression by ∼50% in BT-474 and BT-20 cells, which leads to a smaller increase in ErbB2 (supplemental Fig. S2A) (∼30%) and EGFR (supplemental Fig. S2B) (∼20–30%) phosphorylation, respectively. These data indicate that PTPN9 contributes to tyrosyl dephosphorylation of ErbB2 and EGFR in breast cancer cells.
FIGURE 1.
PTPN9 regulates tyrosyl phosphorylation of ErbB2 and EGFR. A and B, PTPN9 knockdown enhances tyrosyl phosphorylation of ErbB2 and EGFR. SKBR3 (A) and MDA-MB-231 (B) cells were transfected with the non-target control siRNA (NT) and PTPN9 siRNA (si-PTPN9) oligos. Two days later, MDA-MB-231 cells were starved and stimulated with 25 ng/ml of EGF. Equal amounts of cell lysates were immunoblotted with the indicated antibodies (B). C and D, overexpression of PTPN9 inhibits EGF-induced tyrosyl phosphorylation of ErbB2 and EGFR in breast cancer cells. SKBR3 (C) and MDA-MB-231 (D) cells stably expressing pMIG vector control, PTPN9 WT, and PTPN9 DA by retroviral infections were randomly grown (C, left panel) or starved and stimulated with 25 ng/ml of EGF for the indicated times, lysed, and immunoblotted with antibodies against pErbB2 (Y1221/22), pEGFR (Y845, Y998, Y1068) and reprobed with antibodies against ErbB2, EGFR, and PTPN9. E and F, increasing expression of PTPN9 inhibits tyrosyl phosphorylation of NeuNT and EGFR. 293T cells were transiently cotransfected with vector (200 ng) expressing NeuNT (E) or EGFR (F) together with pMIG vector alone, and the indicated increasing amounts (0–200 ng) of pMIG expressing PTPN9 WT. 293T cells cotransfected with EGFR and PTPN9 were stimulated with 25 ng/ml of EGF for 10 min before cell lysis. Equal amounts of lysates were immunoblotted with the indicated antibodies (top panels). Densitometry was used to quantify bands in Western blots (bottom panels). The ratio of the pErbB2/total ErbB2 or pEGFR/total EGFR in pMIG vector control cells is arbitrarily set to 1 (lane 1). The ratio of PTPN9/β-actin in pMIG-PTPN9 (200 ng) of transfected cells is arbitrarily set to 1 (lane 6). G, overexpression of PTPN9 WT does not affect tyrosyl phosphorylation of ErbB3. The same SKBR3 cells used in C were starved, and stimulated with 5 ng/ml of heregulin (HRGβ1). Equal amounts of lysates were immunoprecipitated (IP) with anti-ErbB3 antibodies, immunoblotted with anti-phosphotyrosine antibody (4G10), and reprobed with anti-ErbB3 antibodies. H, overexpression of PTPN9 WT does not affect tyrosyl phosphorylation of Shc. The same SKBR3 cells used in C were starved and stimulated with EGF. Equal amounts of lysates were immunoblotted with anti-pShc (Y239/240) antibodies and reprobed with anti-Shc antibodies. I, overexpression of Shp-1 does not affect tyrosyl phosphorylation of EGFR. MDA-MB-231 cells stably expressing pLNCX2 vector alone, and pLNCX2 expressing Shp-1 WT and CS mutant by retroviral infection were starved and stimulated with 25 ng/ml of EGF. Equal amounts of lysates were immunoblotted with the pEGFR (Y1068) antibodies and reprobed with anti-EGFR antibodies. Densitometry was used to quantify bands in Western blots (Fig. 1, A–D and I). Numbers under the pErbB2 or pEGFR blots show the ratios of pErbB2/total ErbB2 or pEGFR/total EGFR. Numbers under the PTPN9 blots show the ratios of PTPN9/β-actin. Data shown in this figure are representative from at least three independent experiments.
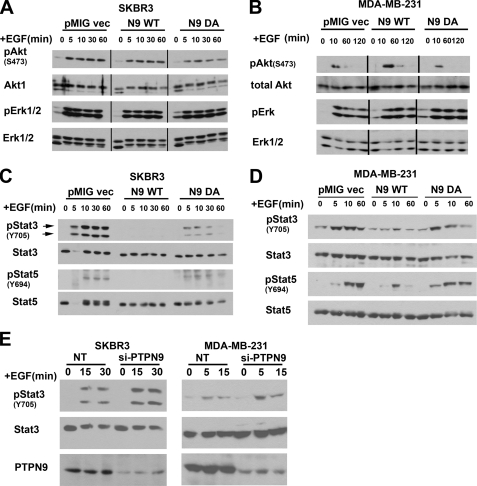
FIGURE 3.
Expression of PTPN9 specifically inhibits EGF-induced STAT3 and STAT5 activation. A and B, expression of PTPN9 WT does not inhibit EGF-induced activation of ERK and Akt. SKBR3 (A) and MDA-231 (B) cells stably expressing pMIG vector, PTPN9 WT, and PTPN9 DA (same as used in Fig. 1, C and D) were starved, stimulated with 25 ng/ml of EGF for the indicated times, lysed, immunoblotted with antibodies against pERK1/2, pAkt (Ser473), and reprobed with antibodies against ERK1/2, Akt, and PTPN9. C and D, expression of PTPN9 WT inhibits EGF-induced STAT3, and STAT5 in SKBR3 and MDA-MB-231 cells. The same SKBR3 (C) and MDA-231 (D) cells used in A and B, respectively, were starved, stimulated with 25 ng/ml of EGF for the indicated times, lysed, and immunoblotted with antibodies against the indicated pSTAT3 (Y705) and pSTAT5 (Y694), and reprobed with antibodies against STAT3 and STAT5. Shown (A–D) are the representative results from three independent experiments. E, PTPN9 knockdown enhances EGF-induced STAT3 activation. SKBR3 (left) and MDA-MB-231 (right) cells were transfected with control (NT) and si-PTPN9 oligos, starved, and stimulated with 25 ng/ml of EGF for the indicated times. Equal amounts of lysates were immunoblotted with anti-pSTAT3 antibodies and reprobed with anti-STAT3 and anti-PTPN9 antibodies. Shown here is the representative result from two independent experiments.
Conversely, we also examined the effects of PTPN9 overexpression on phosphorylation of ErbB2 and EGFR. PTPN9 WT and PTPN9 DA mutant were stably expressed in SKBR3 (Fig. 1C) and MDA-MB-231 cells (Fig. 1D) by retroviral infection. The PTPN9 DA mutant has minimal phosphatase activity due to the Asp to Ala mutation in the WPD loop of the PTP domain, and can still bind (“trap”) its potential protein substrates (9) (17). Thus, expression of the PTPN9 DA mutant in cells results in enhanced tyrosyl phosphorylation of its protein substrates.
The levels of PTPN9 WT and DA mutant overexpression are about 20-fold over the endogenous PTPN9 in the stable pools of SKBR3 (Fig. 1C, left panel). In randomly grown SKBR3 cells, expression of PTPN9 WT inhibits whereas expression of PTPN9 DA enhances ErbB2 tyrosyl phosphorylation (Fig. 1C, left panel). In response to EGF stimulation, tyrosyl phosphorylation of ErbB2 and EGFR was detected robustly in vector control cells but was inhibited in PTPN9 WT-expressing cells. In contrast, EGF-induced tyrosyl phosphorylation of ErbB2 and EGFR were enhanced and prolonged in PTPN9 DA expressing cells compared with vector control cells (Fig. 1C, right panel). Overexpression of PTPN9 WT or the DA mutant does not significantly affect expression of the ErbB2 protein.
The levels of PTPN9 WT and DA mutant overexpression are also about 20-fold over the endogenous PTPN9 in stable pools of MDA-MB-231 cells (Fig. 1D). MDA-MB-231 cells express a high level of EGFR but not ErbB2. Immunoblotting with anti-pEGFR antibodies revealed that EGF-invoked tyrosyl phosphorylation (Tyr845, Tyr998, and Tyr1068) of EGFR was reduced greatly in PTPN9 WT-expressing cells and was enhanced and prolonged significantly in PTPN9 DA-expressing cells compared with vector control cells (Fig. 1D). Expression of PTPN9 WT and DA does not affect protein expression of EGFR (Fig. 1D). These data (Fig. 1, C and D) strongly suggest that ErbB2 and EGFR are potential substrates of PTPN9.
To test whether a lower level of PTPN9 WT overexpression also inhibits ErbB2 and EGFR phosphorylation, 293T cells, which express low levels of endogenous ErbB2 and EGFR, were transiently cotransfected with the activated ErbB2 (NeuNT) or EGFR together with increasing amounts of PTPN9 WT. Although NeuNT (Fig. 1E) or EGF-induced EGFR (Fig. 1F) are robustly tyrosyl phosphorylated in cells cotransfected with vector alone (lane 1), increasing the expression of PTPN9 WT gradually reduces tyrosyl phosphorylation of NeuNT (Fig. 1E) or EGFR (Fig. 1F). Importantly, tyrosyl phosphorylation on NeuNT and EGFR were reduced by more than 95 (Fig. 1E) and 80% (Fig. 1F), respectively, when PTPN9 WT was only overexpressed ∼5-fold over the endogenous PTPN9 (compare lanes 4 and 1), which is similar to the effects of PTPN9 WT overexpression in SKBR3 and MDA-MB-231 cells (Fig. 1, C and D). These results indicate that inhibition of ErbB2 or EGFR phosphorylation by PTPN9 overexpression is not due to the overexpression artifact.
We also examined tyrosyl phosphorylation of ErbB3, a member of the ErbB family, and Shc, an adapter protein that becomes tyrosyl phosphorylated upon ErbB activation, in cells expressing PTPN9 WT and DA mutant. ErbB3 was immunoprecipitated by an ErbB3 antibody from lysates of SKBR3 cells expressing vector alone, PTPN9 WT, and PTPN9 DA mutant followed by immunoblotting with an anti-phosphotyrosine antibody (4G10) (Fig. 1G). Compared with vector control, expression of PTPN9 WT or DA mutant has minimal effects on total tyrosyl phosphorylation of ErbB3 (Fig. 1G) and ErbB3 Tyr1289 phosphorylation (Data not shown). Likewise, EGF-induced tyrosyl phosphorylation of p52 Shc (Tyr239/240) was also not affected by expression of PTPN9 WT or DA mutant in SKBR3 (Fig. 1H, left panel) and MDA-MB-231 cells (Fig. 1H, right panel) compared with vector control cells. The p66 and p46 Shc isoforms are expressed at a low level (<10% of p52 Shc) in these two cell lines, which are not included here. Protein-tyrosine phosphatase Shp-1 is expressed in breast cancer cells (22). However, we found that overexpression of Shp-1 WT and Shp-1 CS mutant (a phosphatase-dead substrate trapping mutant) has minimal effects on EGF-evoked tyrosyl phosphorylation of EGFR (Tyr1068) compared with vector control in MDA-MB-231 cells (Fig. 1I). These results indicate that overexpression of PTPN9 specifically affects tyrosyl phosphorylation of ErbB2 and EGFR, rather than nonspecifically dephosphorylating tyrosyl-phosphorylated proteins in breast cancer cells.
ErbB2 and EGFR Are Potential Substrates of PTPN9
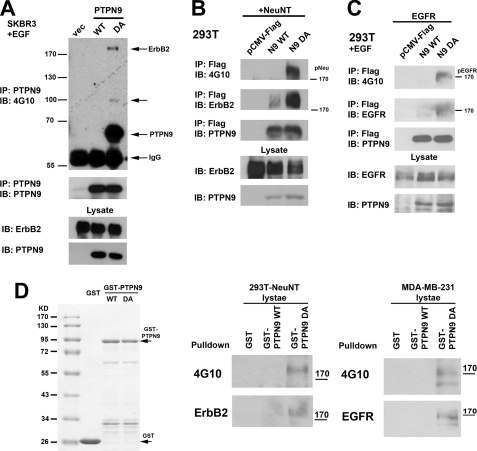
To further address whether PTPN9 expression inhibits ErbB2 and EGFR phosphorylation by directly dephosphorylating tyrosine residues in ErbB2 and EGFR in vivo, we took advantage of the observation that PTPs with a aspartic acid → alanine (DA) mutation in the WPD loop of the PTP catalytic domain have little phosphatase activity, but still maintain the ability to bind (trap) potential substrates (9). Expression of the DA mutant behaves as a biochemical dominant-negative in cells by protecting dephosphorylation of its substrates, resulting in enhanced tyrosyl phosphorylation of the potential protein substrates and association of the DA mutant with its substrates. ErbB2 association with PTPN9 WT and DA mutant in stable pools of SKBR3 cells was investigated by immunoprecipitation (Fig. 2A). In both vector and PTPN9 WT overexpressing cells, no tyrosyl-phosphorylated proteins were detected in the PTPN9 immunoprecipitates. In contrast, tyrosyl-phosphorylated (Tyr(P)) proteins with molecular masses of 175, 100, and 65 kDa were specifically associated with the PTPN9 DA mutant (Fig. 2A, lane 3). Reprobing the PTPN9 immunoprecipitates with anti-PTPN9 antibodies showed that the 65-kDa Tyr(P) band was comigrated with PTPN9 DA, strongly suggesting that the 65-kDa band is the tyrosyl-phosphorylated PTPN9 DA mutant. The same amounts of PTPN9 WT and PTPN9 DA proteins were immunoprecipitated (Fig. 2A). One likely possibility is that PTPN9 DA is hyperphosphorylated. PTPN9 WT is normally phosphorylated by a tyrosine kinase in the cells, and rapidly dephosphorylated by itself. PTPN9 DA trapping mutant is not only phosphatase-dead, but also protects the DA mutant from dephosphorylation by trapping the phosphorylated tyrosine residues in PTPN9 DA, thus resulting in hyperphosphorylation of the PTPN9 DA mutant. Hyperphosphorylation was also seen in the substrate trapping mutants of other PTPs including Shp-1 (20).
FIGURE 2.
PTPN9 substrate trapping mutant DA associates with tyrosyl-phosphorylated ErbB2 and EGFR. A and B, PTPN9 DA co-immunoprecipitates with tyrosyl-phosphorylated ErbB2. Lysates from SKBR3 cells stably expressing vector alone (vec), PTPN9 WT, and PTPN9 DA (A), and 293T cells transiently cotransfected with NeuNT together with pCMV-FLAG vector alone, FLAG-tagged PTPN9 WT, and FLAG-tagged PTPN9 DA (B) were immunoprecipitated (IP) with anti-PTPN9 antibodies (A) and anti-FLAG antibody (B), and immunoblotted (IB) with the indicated antibodies. C, PTPN9 DA co-immunoprecipitates with tyrosine-phosphorylated EGFR. 293T cells cotransfected with the pEGFP-EGFR plasmid together with pCMV-FLAG vector alone, FLAG-tagged PTPN9 WT, and FLAG-tagged PTPN9 DA were starved, stimulated with 25 ng/ml of EGF for 5 min, lysed, immunoprecipitated with anti-FLAG antibody, and immunoblotted with the indicated antibodies. D, GST-PTPN9 DA fusion protein pulls down tyrosyl-phosphorylated ErbB2 or EGFR. Glutathione-agarose beads containing equal amounts of GST alone, GST-PTPN9 WT, and GST-PTPN9 DA proteins were resolved by SDS-PAGE and stained with Coomassie Blue (left panel), or incubated with equal amounts of cells lysates from 293T cells transiently transfected with NeuNT (middle panel) or MDA-MB-231 cells stimulated with EGF for 10 min (right panel), washed, and immunoblotted with the indicated antibodies. Data shown in this figure are representative results from two independent experiments.
We do not know the nature of the 100-kDa Tyr(P) protein. Due to the lower sensitivity of detection, reprobing the immunoblot with anti-ErbB2 antibody failed to detect the presence of ErbB2 comigrated with the 175-kDa Tyr(P) band in the PTPN9 DA complex although it is very likely that the 175-kDa band is the tyrosyl-phosphorylated ErbB2.
To provide evidence that the PTPN9 DA trapping mutant can coimmunoprecipitate with the phosphorylated ErbB2, 293T cells were transiently cotransfected with plasmid expressing NeuNT, the activated form of rat ErbB2, together with the FLAG-tagged PTPN9 constructs (Fig. 2B). Transfected 293T cells were lysed and incubated with anti-FLAG antibody. Although tyrosyl-phosphorylated ErbB2 was not associated with FLAG-PTPN9 WT, a 175-kDa Tyr(P) protein that comigrated with ErbB2 was associated with FLAG-PTPN9 DA. Immunoblotting with anti-ErbB2 antibodies showed robust association of ErbB2 with the FLAG-PTPN9 DA (Fig. 2B, lane 3) and minimal association of ErbB2 with FLAG-PTPN9 WT. This result indicates that only the PTPN9 DA trapping mutant, not PTPN9 WT, is associated with tyrosyl-phosphorylated ErbB2, strongly suggesting that ErbB2 is a substrate for PTPN9.
To provide evidence that EGFR is a substrate for PTPN9, 293T cells were cotransfected with plasmid-expressing EGFR together with the FLAG-tagged PTPN9 constructs. Transfected cells were stimulated with EGF before being lysed and subjected to immunoprecipitation with anti-FLAG antibody (Fig. 2C). Although the phosphorylated EGFR was not found associated with FLAG-PTPN9 WT, a 170-kDa Tyr(P) protein was associated with FLAG-PTPN9 DA. Immunoblotting with anti-EGFR antibodies revealed that EGFR was comigrated with the 170-kDa Tyr(P) protein in the FLAG-PTPN9 DA immunoprecipitates (Fig. 2C, lane 3), strongly suggesting that EGFR is also a substrate for PTPN9.
It is possible that expression of the PTPN9 DA mutant in cells could affect expression or activity of another protein that may mediate the interaction between the PTPN9 DA mutant and the hyperphosphorylated ErbB2 or EGFR. To exclude this possibility, equal amounts of GST alone, GST-PTPN9 WT, and GST-PTPN9 DA fusion proteins were expressed in bacteria, purified by glutathione-agarose beads (Fig. 2D, left panel), and incubated with cell lysates from 293T cells transiently transfected with NeuNT (Fig. 2D, middle panel) or MDA-MB-231 cells (EGF stimulated) (Fig. 2D, right panel). Consistent with the results from our co-immunoprecipitation experiments (Fig. 2, A-C), GST or GST-PTPN9 WT does not pulldown phosphorylated ErbB2 or EGFR. In contrast, GST-PTPN9 DA pulls down tyrosyl-phosphorylated ErbB2 (Fig. 2D, middle panel) or EGFR (Fig. 2D, right panel), further supporting that PTPN9 DA traps the phosphorylated ErbB2 and EGFR directly. Overall, our data support that ErbB2 and EGFR are direct substrates for PTPN9.
PTPN9 Overexpression Inhibits EGF-induced STAT Activation
Because PTPN9 overexpression specifically dephosphorylates EGFR and ErbB2 (Figs. 1 and 2), we expect that EGF-induced downstream signaling pathways should also be reduced. EGF-induced activation of ERK and Akt in SKBR3 and MDA-MB-231 cells overexpressing PTPN9 was examined by immunoblotting using phosphospecific antibodies against ERK and Akt. To our surprise, EGF-invoked Akt and ERK phosphorylation was not impaired in PTPN9 WT and DA overexpressing cells compared with vector control cells (Fig. 3A). Similarly, in MDA-MB-231 cells, EGF-invoked Akt and ERK phosphorylation was not affected in PTPN9 WT and DA overexpressing cells compared with vector control cells (Fig. 3B). The observation that PTPN9 overexpression has no effect on ERK activation is consistent with the data that PTPN9 WT overexpression has no effect on Shc tyrosyl phosphorylation in SKBR3 and MDA-MB-231 cells (Fig. 1H).
EGF stimulation also leads to activation of STAT3 (23) and STAT5 (24, 25) in breast cancer cells. EGF-induced STAT3 and STAT5 activation was examined in SKBR3 and MDA-MB-231 cells overexpressing PTPN9 by immunoblotting using phosphospecific antibodies against STAT3 and STAT5 (Fig. 3, C and D). In contrast to ERK and Akt activation, EGF-induced STAT3 and STAT5 phosphorylation was dramatically inhibited in PTPN9 WT expressing SKBR3 (Fig. 3C) and MDA-MB-231 cells (Fig. 3D) compared with vector control cells. Interestingly, whereas EGF-induced STAT5 activation was not affected in cells expressing PTPN9 DA compared with vector control, EGF-induced STAT3 activation was significantly reduced in cells expressing PTPN9 DA compared with vector control cells (Fig. 3, C and D). Consistent with the inhibition of STAT3 phosphorylation by PTPN9 overexpression, we found that knockdown of PTPN9 expression by PTPN9 siRNA resulted in an increase of EGF-induced STAT3 phosphorylation in SKBR3 (left panel) and MDA-MB-231 (right panel) cells (Fig. 3E).
Overexpression of PTPN9 WT Inhibits the Oncogenic Growth and Invasion of Breast Cancer Cells
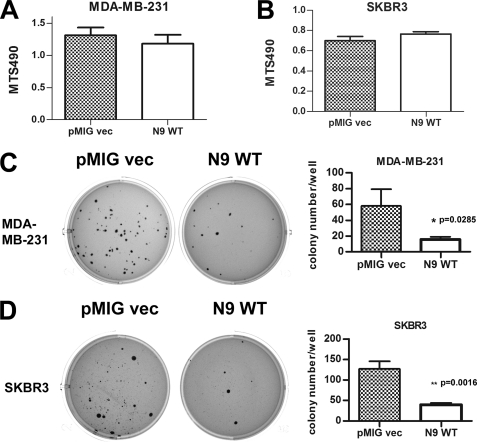
ErbB2 and EGFR play important roles in driving the growth, invasion, and metastasis of breast cancer cells in part through activation of STAT3 and STAT5 (26). We investigated whether expression of PTPN9 affects the growth and invasion of SKBR3 and MDA-MB-231 cells. First, we examined the growth of PTPN9 overexpressing cells in tissue culture plates by MTS, which measures viable cells. On tissue culture plastic plates, for MDA-MB-231 (Fig. 4A) and SKBR3 (Fig. 4B) cells, cell growth in serum containing medium was the same in vector control and PTPN9 overexpressing cells. Similar results were obtained when cells were grown in serum-free medium (data not shown). Next, we examined the effect of PTPN9 WT overexpression on cell growth in soft agar-anchorage independent growth, which is one of the hallmarks of cancer cells. Although there was robust colony formation in the vector control of MDA-MB-231 (Fig. 4C) and SKBR3 (Fig. 4D) cells, the number of colonies were decreased by ∼70% both in PTPN9 WT expressing MDA-MB-231 and SKBR3 cells. These results indicate that increased expression of PTPN9 impairs the oncogenic growth of breast cancer cells.
FIGURE 4.
PTPN9 overexpression inhibits the growth of MDA-MB-231 and SKBR3 cells in soft agar assay. A and B, PTPN9 overexpression does not affect cell growth in tissue culture plastic plates. MDA-MB-231 (A) and SKBR3 (B) cells expressing pMIG vector and PTPN9 WT were grown in tissue culture plates for 3 days in medium containing 10% FBS. Viable cells were assayed using MTS reagent. Shown are representative results from at least three independent experiments. C and D, PTPN9 overexpression inhibits cell growth in soft agar assay. MDA-MB-231 (C) and SKBR3 (D) cells expressing pMIG vector and PTPN9 WT were grown in medium containing 10% FBS and 0.25% agar for 3 weeks. Plates were stained, photographed (C and D, left panels), and visible colonies were counted (C and D, right panels). Representative results in triplicate from two independent experiments are depicted as mean ± S.D.
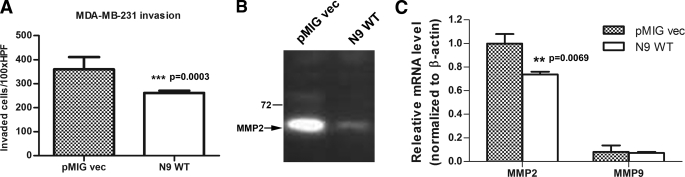
We also examined whether overexpression of PTPN9 affects the invasion of breast cancer cells. MDA-MB-231 cells expressing vector and PTPN9 WT were subjected to the Matrigel invasion assay. Vector control cells showed robust invasion through the Matrigel. In contrast, PTPN9 WT expressing cells displayed a 30% decrease in invasion (Fig. 5A). We cannot assess the effect of PTPN9 expression on the invasion of SKBR3 cells because both the vector control and PTPN9 WT-expressing cells could not invade through the Matrigel in our assay. We also examined the effects of PTPN9 expression on cell migration by performing the wound healing assay. However, we did not find that overexpression of PTPN9 WT affects cell migration in SKBR3 and MDA-MB-231 cells (data not shown).
FIGURE 5.
PTPN9 overexpression inhibits invasion and MMP2 expression in MDA-MB-231 cells. A, MDA-MB-231 cells expressing pMIG vector and PTPN9 WT were subjected to the transwell invasion assay. Shown is the representative result from three independent experiments. B, expression of PTPN9 inhibits the MMP2 activity. Equal number of MDA-MB-231 cells expressing pMIG vector and PTPN9 WT were cultured in medium without FBS for 48 h. Equal volumes of the cultured medium from both cell lines were subjected to gelatin zymography. The arrow indicates the position of the active MMP2. C, expression of PTPN9 inhibits MMP2 mRNA expression. Total RNAs from MDA-MB-231 cells expressing pMIG vector and PTPN9 WT were isolated and reversed transcribed. MMP2 and MMP9 mRNA expression levels were measured by quantitative PCR. The levels of MMP2 and MMP9 mRNA were normalized to β-actin and expressed as relative mRNA compared with MMP2 mRNA in pMIG vector control. Representative results from two independent experiments each run at least in triplicate are shown as mean ± S.D.
Cancer cells invade through its extracellular matrix environment through secretion of matrix degrading proteases. MDA-MB-231 cells are known to express matrix metalloproteinase 2 (MMP2) (27). Thus, we ask whether PTPN9 expression affects MMP2 expression. Gelatin zymography was used to examine the expression of MMP2 and MMP9 proteins from MDA-MB-231 cells expressing vector and PTPN9 WT (Fig. 5B). MMP2 protein was highly expressed in vector control cells. In contrast, MMP2 expression was dramatically reduced in PTPN9-expressing cells. We could not detect any MMP9 protein expression both in vector control and PTPN9 expressing MDA-MB-231 cells. Quantitative PCR was used to assess MMP2 mRNA expression (Fig. 5C). Consistent with zymography data (Fig. 5B), PTPN9 WT overexpression resulted in reduced MMP2 mRNA expression. In addition, MMP9 mRNA was found to be expressed at a very low level in MDA-MB-231 cells. These data suggest that PTPN9 expression inhibits the invasion of MDA-MB-231 cells likely through the suppression of MMP2 gene expression.
DISCUSSION
EGFR and ErbB2 are involved in various types of human cancers including breast cancer. Thus, identification of protein-tyrosine phosphates that can inhibit EGFR and ErbB2 function by direct dephosphorylation should provide new information to design novel cancer therapies. In this article, for the first time, we provide evidence that PTPN9 is a protein-tyrosine phosphatase specific for EGFR and ErbB2 and that negatively regulates EGFR and ErbB2 signaling.
Our study is the first report that PTPN9 inhibits receptor PTK activation by direct dephosphorylation of EGFR and ErbB2. First, knockdown of PTPN9 expression by siRNA results in an increase of tyrosyl phosphorylation of ErbB2 (Fig. 1A) and EGFR (Fig. 1B). Second, overexpression of PTPN9 WT inhibits tyrosyl phosphorylation of EGFR and ErbB2 (Fig. 1, C-F) without affecting EGFR and ErbB2 protein expression. Importantly, PTPN9 WT overexpression does not affect tyrosyl phosphorylation of ErbB3 (Fig. 1G), Shc (Fig. 1H), and activation of Src PTK (data not shown). In contrast, expression of the PTPN9 substrate trapping mutant DA enhances tyrosyl phosphorylation of EGFR and ErbB2 in these same assays (Fig. 1, C and D). Third, only the substrate trapping mutant of PTPN9 DA, not PTPN9 WT, is coimmunoprecipitated with (Fig. 2, A-C) and can pulldown (Fig. 2D) the phosphorylated EGFR or ErbB2. These results strongly suggest that the PTPN9 DA mutant is able to form a stable complex with the phosphorylated EGFR and ErbB2 because the DA phosphatase domain likely binds the phosphotyrosine residues in EGFR or ErbB2 but cannot catalyze the dephosphorylation reaction. Currently, we cannot exclude the possibility that PTPN9 DA coimmunoprecipitates with tyrosyl-phosphorylated EGFR and ErbB2 via another tyrosine-phosphorylated protein such as the p100 Tyr(P) protein although the level of tyrosine phosphorylation on p100 is lower compared with that of ErbB2 in the PTPN9 DA complex (Fig. 2A). Although a previous report shows that PTPN9 inhibits insulin receptor phosphorylation and insulin-induced Akt activation in insulin responsive tissues, it is still not clear whether insulin receptor is a direct substrate of PTPN9 (18).
Our result also reveals PTPN9 as one of the PTPs that can dephosphorylate EGFR and ErbB2, and modulate EGFR and ErbB2 induced signaling and biological responses. EGFR has been reported to be the substrate for tyrosine phosphatase Shp-1 and PTP1B. EGFR is shown to be a potential substrate for Shp-1 only in cotransfection experiments with overexpression of Shp-1 in 293 cells (10). Shp-1 via its SH2 domain binds Tyr1173 in EGFR and dephosphorylates other tyrosine residues in EGFR, and inhibits EGF-induced ERK activation (10). However, our results indicate that overexpression of Shp-1 WT or CS mutant does not affect tyrosyl phosphorylation of EGFR (Fig. 1I) and EGF-induced ERK activation (data not shown) in MDA-MB-231 cells. By fluorescence resonance energy transfer imaging, PTP1B has been shown to dephosphorylate EGFR on the surface of endoplasmic reticulum after endocytosis of the activated EGFR (28). ErbB2 has been reported to be a substrate for PTPN13 in HeLa and 293 cells (11). Besides its effect on STAT3 activation, PTPN13 expression also inhibits EGF-induced ERK and Akt activation (11). Our results show that PTPN9 specifically dephosphorylates EGFR and ErbB2 (Fig. 1), and inhibits EGF-induced STAT activation without affecting ERK and Akt activation (Fig. 3). Our studies using subcellular fractionation and immunofluorescent staining of PTPN9 indicate that overexpressed PTPN9WT and endogenous PTPN9 show similar pattern of subcellular localization, which is highest in the low density microsomes rich in transporting vesicles, medium in the high density microsomes, low in the plasma membrane, and undetectable in cytosol (supplemental Fig. S3). This is consistent with the reported localization of PTPN9 being mainly present in secretory vesicles. However, our data also reveal that a small fraction of PTPN9 is present in plasma membrane. These data suggest that PTPN9 may interact and/or dephosphorylate EGFR and ErbB2 both in the plasma membrane and intracellular membrane compartments, which will be the future focus of our study. These results indicate that PTPN9 has distinct role in regulating the activation status of EGFR and ErbB2 compared with PTP1B, Shp-1, and PTPN13.
Published reports show that EGFR can activate STAT3 and STAT5 in two ways (26). EGFR-activated Src can promote phosphorylation of STAT3 and STAT5. Alternatively, Tyr845 in EGFR can be phosphorylated by Src upon EGF stimulation, which recruits and activates STAT3 and STAT5 (26). A recent study by quantitative proteomics indicates that phosphorylated Tyr998 in EGFR can also recruit STAT5 directly (29). We found that PTPN9 expression did not affect EGF-induced Src activation in SKBR3 and MDA-MB-231 cells (data not shown). However, phosphorylation of Tyr845 and Tyr998 in EGFR was inhibited by PTPN9 expression in MDA-MB-231 cells (Fig. 1D). Therefore, we favor the working model that PTPN9 expression results in impaired phosphorylation on tyrosine residues (Tyr845 and Tyr998) in EGFR that recruit and activate STAT3 and STAT5 in MDA-MB-231 cells. In SKBR3 cells that overexpress ErbB2, EGF stimulation induces heterodimerization of EGFR and ErbB2. Because PTPN9 can also dephosphorylate and inhibits ErbB2 activation, PTPN9 expression could directly dephosphorylate Tyr845 and Tyr998 in EGFR or indirectly by inhibiting ErbB2 activation in SKBR3 cells. In any event, our data do not support that PTPN9 directly dephosphorylates STAT3 and STAT5. Supporting this notion, we found that EGF-induced STAT3 and STAT5 phosphorylation is reduced in cells expressing PTPN9 DA trapping mutant compared with vector control cells (Fig. 3, C and D). If STAT3 and STAT5 were substrates of PTPN9, we would find enhanced STAT3 and STAT5 phosphorylation in PTPN9 DA-expressing cells compared with vector control cells because expression of the substrate trapping mutant of PTPN9 should protect its substrates from dephosphorylation such as in the case of hyperphosphorylated ErbB2 and EGFR in the presence of PTPN9 DA expression (Fig. 1, C and D). These data are consistent with the idea that PTPN9 WT dephosphorylates the phosphorylated tyrosine residues in EGFR that recruit and activate STAT3 and STAT5.
Activation of ERK and phosphatidylinositol 3-kinase/Akt pathways play important roles in mediating EGFR/ErbB2 evoked biological responses such as cell growth, migration, and invasion in breast cancer cells (1). However, recent studies indicate that EGFR can activate STAT3 and STAT5, which turn on gene expression critical for the proliferation and survival of breast cancer cells (26). Because PTPN9 expression specifically inhibits EGF-induced STAT3 and STAT5 activation without affecting ERK and Akt activation in SKBR3 and MDA-MB-231 cells (Fig. 3), it is very likely that impaired STAT3 and STAT5 activation contributes to the reduced cell growth in soft agar when PTPN9 was overexpressed (Fig. 4). In addition, our data also indicate that PTPN9 overexpression reduces invasion and decreases MMP2 gene expression in MDA-MB-231 cells. It was reported that STAT3 activation promotes invasion, metastasis, and MMP2 gene expression in melanoma cells. STAT3 activates MMP2 gene transcription by binding to the high affinity STAT3 binding site in the promoter region of the MMP2 gene (30). Because PTPN9 expression inhibits STAT3 activation (Fig. 3), MMP2 gene expression (Fig. 5C), and invasion (Fig. 5A), it is likely that PTPN9 expression impairs cell invasion through down-regulation of the STAT3-MMP2 pathway in MDA-MB-231 cells. A recent study indicates that subclones of MDA-MB-231 cells, specifically metastasis to the brain and lung in the xenograft model, overexpress the EGFR ligand HB-EGF (31), indicating the importance of EGFR signaling in metastasis of breast cancer cells. Our result suggests that the EGFR-induced STAT3-MMP2 pathway may contribute to brain and lung metastasis of MDA-MB-231 cells, and the level of PTPN9 expression may affect breast cancer metastasis in vivo.
In this study, we find that expression of PTPN9 inhibits the oncogenic growth and invasion of breast cancer cells by specifically dephosphorylating EGFR and ErbB2 and impairing EGFR and ErbB2 signaling. It will be important to examine whether the PTPN9 expression level can predict the disease outcome in ErbB2/Her2 positive and triple negative breast cancers in a future study. Furthermore, it will be also interesting to explore the role of PTPN9 in other human cancers such as lung and brain cancers where EGFR is the critical driver for the malignant progression. Last, therapeutic approaches that can increase PTPN9 expression may be useful in treating breast cancer patients with EGFR and/or ErbB2 overexpression.
Acknowledgment
We thank Dr. A. Sorkin (University of Colorado Denver) for EGFR reagents.
This work was supported, in whole or in part, by National Institutes of Health Grants HL076309 and HL079441 (to Z. J. Z) and the start-up fund from the Department of Pathology, University of Colorado Denver (to H. G).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- PTK
- protein-tyrosine kinase
- PTP
- protein-tyrosine phosphatase
- Tyr(P)
- tyrosyl-phosphorylated
- FBS
- fetal bovine serum
- DMEM
- Dulbecco's modified Eagle's medium
- GPF
- green fluorescent protein
- GST
- glutathione S-transferase
- siRNA
- small interfering RNA
- MMP
- matrix metalloproteinase
- STAT
- signal transducer and activator of transcription
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- ERK
- extracellular signal-regulated kinase
- EGFR
- epidermal growth factor receptor
- CMV
- cytomegalovirus
- WT
- wild type.
REFERENCES
- 1.Hynes N. E., Lane H. A. (2005) Nat. Rev. Cancer 5, 341–354 [DOI] [PubMed] [Google Scholar]
- 2.Campos S. M. (2008) Cancer Invest. 26, 757–768 [DOI] [PubMed] [Google Scholar]
- 3.Hayes D. F., Thor A. D. (2002) Semin. Oncol. 29, 231–245 [DOI] [PubMed] [Google Scholar]
- 4.Bièche I., Onody P., Tozlu S., Driouch K., Vidaud M., Lidereau R. (2003) Int. J. Cancer 106, 758–765 [DOI] [PubMed] [Google Scholar]
- 5.Holbro T., Beerli R. R., Maurer F., Koziczak M., Barbas C. F., 3rd, Hynes N. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8933–8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tonks N. K. (2006) Nat. Rev. Mol. Cell Biol. 7, 833–846 [DOI] [PubMed] [Google Scholar]
- 7.Liu F., Chernoff J. (1997) Biochem. J. 327, 139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haj F. G., Markova B., Klaman L. D., Bohmer F. D., Neel B. G. (2003) J. Biol. Chem. 278, 739–744 [DOI] [PubMed] [Google Scholar]
- 9.Flint A. J., Tiganis T., Barford D., Tonks N. K. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keilhack H., Tenev T., Nyakatura E., Godovac-Zimmermann J., Nielsen L., Seedorf K., Böhmer F. D. (1998) J. Biol. Chem. 273, 24839–24846 [DOI] [PubMed] [Google Scholar]
- 11.Zhu J. H., Chen R., Yi W., Cantin G. T., Fearns C., Yang Y., Yates J. R., 3rd, Lee J. D. (2008) Oncogene 27, 2525–2531 [DOI] [PubMed] [Google Scholar]
- 12.Arias-Romero L. E., Saha S., Villamar-Cruz O., Yip S. C., Ethier S. P., Zhang Z. Y., Chernoff J. (2009) Cancer Res. 69, 4582–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Julien S. G., Dubé N., Read M., Penney J., Paquet M., Han Y., Kennedy B. P., Muller W. J., Tremblay M. L. (2007) Nat. Genet. 39, 338–346 [DOI] [PubMed] [Google Scholar]
- 14.Bentires-Alj M., Neel B. G. (2007) Cancer Res. 67, 2420–2424 [DOI] [PubMed] [Google Scholar]
- 15.Huynh H., Bottini N., Williams S., Cherepanov V., Musumeci L., Saito K., Bruckner S., Vachon E., Wang X., Kruger J., Chow C. W., Pellecchia M., Monosov E., Greer P. A., Trimble W., Downey G. P., Mustelin T. (2004) Nat. Cell. Biol. 6, 831–839 [DOI] [PubMed] [Google Scholar]
- 16.Wang Y., Vachon E., Zhang J., Cherepanov V., Kruger J., Li J., Saito K., Shannon P., Bottini N., Huynh H., Ni H., Yang H., McKerlie C., Quaggin S., Zhao Z. J., Marsden P. A., Mustelin T., Siminovitch K. A., Downey G. P. (2005) J. Exp. Med. 202, 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu M. J., Sui X., Zhao R., Dai C., Krantz S. B., Zhao Z. J. (2003) Blood 102, 4354–4360 [DOI] [PubMed] [Google Scholar]
- 18.Cho C. Y., Koo S. H., Wang Y., Callaway S., Hedrick S., Mak P. A., Orth A. P., Peters E. C., Saez E., Montminy M., Schultz P. G., Chanda S. K. (2006) Cell Metab. 3, 367–378 [DOI] [PubMed] [Google Scholar]
- 19.Dankort D., Jeyabalan N., Jones N., Dumont D. J., Muller W. J. (2001) J. Biol. Chem. 276, 38921–38928 [DOI] [PubMed] [Google Scholar]
- 20.Timms J. F., Carlberg K., Gu H., Chen H., Kamatkar S., Nadler M. J., Rohrschneider L. R., Neel B. G. (1998) Mol. Cell. Biol. 18, 3838–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Battini J. L., Rasko J. E., Miller A. D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1385–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yip S. S., Crew A. J., Gee J. M., Hui R., Blamey R. W., Robertson J. F., Nicholson R. I., Sutherland R. L., Daly R. J. (2000) Int. J. Cancer 88, 363–368 [PubMed] [Google Scholar]
- 23.Sartor C. I., Dziubinski M. L., Yu C. L., Jove R., Ethier S. P. (1997) Cancer Res. 57, 978–987 [PubMed] [Google Scholar]
- 24.Kloth M. T., Laughlin K. K., Biscardi J. S., Boerner J. L., Parsons S. J., Silva C. M. (2003) J. Biol. Chem. 278, 1671–1679 [DOI] [PubMed] [Google Scholar]
- 25.Boerner J. L., Biscardi J. S., Silva C. M., Parsons S. J. (2005) Mol. Carcinog. 44, 262–273 [DOI] [PubMed] [Google Scholar]
- 26.Silva C. M. (2004) Oncogene 23, 8017–8023 [DOI] [PubMed] [Google Scholar]
- 27.Baum O., Hlushchuk R., Forster A., Greiner R., Clézardin P., Zhao Y., Djonov V., Gruber G. (2007) Int. J. Oncol. 30, 325–332 [PubMed] [Google Scholar]
- 28.Haj F. G., Verveer P. J., Squire A., Neel B. G., Bastiaens P. I. (2002) Science 295, 1708–1711 [DOI] [PubMed] [Google Scholar]
- 29.Schulze W. X., Deng L., Mann M. (2005) Mol. Syst. Biol. 1, 2005.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie T. X., Wei D., Liu M., Gao A. C., Ali-Osman F., Sawaya R., Huang S. (2004) Oncogene 23, 3550–3560 [DOI] [PubMed] [Google Scholar]
- 31.Bos P. D., Zhang X. H., Nadal C., Shu W., Gomis R. R., Nguyen D. X., Minn A. J., van de Vijver M. J., Gerald W. L., Foekens J. A., Massagué J. (2009) Nature 459, 1005–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]