Abstract
Modification of proteins by small ubiquitin-like modifier (SUMO) is emerging as an important control of transcription and RNA processing. The human factor TCERG1 (also known as CA150) participates in transcriptional elongation and alternative splicing of pre-mRNAs. Here, we report that SUMO family proteins modify TCERG1. Furthermore, TCERG1 binds to the E2 SUMO-conjugating enzyme Ubc9. Two lysines (Lys-503 and Lys-608) of TCERG1 are the major sumoylation sites. Sumoylation does not affect localization of TCERG1 to the splicing factor-rich nuclear speckles or the alternative splicing function of TCERG1. However, mutation of the SUMO acceptor lysine residues enhanced TCERG1 transcriptional activity, indicating that SUMO modification negatively regulates TCERG1 transcriptional activity. These results reveal a regulatory role for sumoylation in controlling the activity of a transcription factor that modulates RNA polymerase II elongation and mRNA alternative processing, which are discriminated differently by this post-translational modification.
Keywords: Protein/Post-translational Modification, RNA/Messenger RNA/mRNA, RNA/Polymerase, RNA/Splicing, Transcription/Coactivators, Transcription/Elongation Factors, Transcription/Regulation, Transcription/RNA Polymerase II
Introduction
Splicing and transcriptional elongation are physically and functionally interconnected processes (1, 2). Although both processes can occur autonomously, their coupling and coordination may be important for regulation of gene expression. Coupling of these two processes may influence splicing and alternative splicing regulation. Indeed, promoter composition, transcriptional elongation efficiency, chromatin environment, and recruitment of specific coregulators to the transcriptional complex have been shown to affect alternative splicing decisions in a number of experimental systems (3–6). Yet the molecular mechanisms at work are not understood. The unique carboxyl-terminal domain (CTD)4 of the large subunit of RNAPII seems to play a central role in the coupling of splicing, as well as other RNA processing functions, to transcription (7, 8). To provide a framework, we consider two models as follows: the recruiting and kinetic models, which are not mutually exclusive. In the “recruiting model,” the CTD functions as a “landing pad” for specific subsets of RNA processing factors in a manner dependent on its phosphorylation pattern and therefore on the functional state of the transcription complex (9). A number of independent research lines have suggested that RNA splicing factors can interact with RNAPII molecules that are hyperphosphorylated on their CTD (10–14), although these associations might be highly dynamic and transient in vivo. The integrity of the RNAPII CTD has also been shown to influence the recruitment of splicing factors to active transcription sites in the nucleus (15). The “kinetic model” (1) proposes that the rate of elongation of the nascent transcript affects specific alternative splicing decisions by modulating the probability of simultaneous presentation of competing splicing sites. Thus, modulation of transcription elongation efficiency at specific, alternatively spliced regions of genes might constitute a mechanism to regulate splicing decisions (5). Several nuclear factors have been shown to play dual roles both in transcription and splicing regulation and/or to exhibit physical interaction with components of both machineries, thus revealing themselves as potential “coupling mediators” or “cross-talk” factors; these include among others PSF, p54nrb/NONO, TAT-SF1, Prp40, FBP11, and TCERG1. PSF and p54nrb/NONO influence both transcription and splicing and are components of high molecular weight complexes that include processive RNAPII as well as a broad subset of splicing factors (16, 17). Tat-SF1 is an elongation regulator factor that interacts with small nuclear ribonucleoprotein complexes (18). Yeast Prp40 and mammal FBP11 are related proteins that contain tandem repeats of WW and FF domains, a feature that might define a subset of transcription and splicing-related factors; consistent with this hypothesis, they both interact with the CTD of RNAPII and U1 small nuclear ribonucleoprotein (13, 19–21).
TCERG1 (previously designated CA150) was first described as a transcriptional elongation regulator found in human immunodeficiency virus, type 1, Tat-responsive HeLa nuclear extract fractions (22). Transient overexpression of TCERG1 reduces expression from the human immunodeficiency virus, type 1, and α-4 integrin promoters by inhibiting elongation efficiency. This repression is promoter-specific, and it is also dependent on a specific functional TATA-box element (23). The primary sequence of TCERG1 contains three WW domains in the amino-terminal half and six FF repeat motifs in the carboxyl-terminal half. TCERG1 interacts directly with the phosphorylated CTD of RNAPII via its FF repeats (24) and binds to several elongation-related factors in HeLa nuclear extracts (25). Based on those data, TCERG1 appears to function in the elongation stage of transcription. However, accumulating evidence indicates a potential role of TCERG1 in splicing and hence in the coupling between transcription and splicing. TCERG1 copurifies with in vitro assembled spliceosomes and was identified in spliceosomal subcomplexes (26–28). In addition, we and others have found multiple interactions with components of the splicing machinery (25, 29–31). The subnuclear distribution of TCERG1 resembles that of an RNA metabolism-related factor with enrichment in the peripheral regions of the splicing factor-rich nuclear speckles (25). Importantly, TCERG1 can affect alternative pre-mRNA splicing of β-globin, β-tropomyosin, and CD44 splicing reporters (30, 32, 33) and in putative cellular targets identified upon TCERG1 knockdown by microarray analysis (33).
TCERG1 may be regulated at multiple levels. TCERG1 forms multiple protein complexes, subpopulations of which may differ in their functional properties and biochemical associations. Compartmentalization in the nuclear subdomains may control TCERG1 function. Post-translational modifications may also influence TCERG1 function; for instance, a recent report demonstrated that TCERG1 interacts with the spinal muscular atrophy protein SMN when methylated by CARM1. This modification modulated the functional interaction of TCERG1 and CARM1 to affect alternative splicing of a CD44 exon 4 (32). Phosphorylation of specific motifs on TCERG1 sequence has also been reported, although its functional significance remains unknown (34).
A number of small ubiquitin-like modifier paralogs have been described in higher eukaryotes as follows: SUMO-1, SUMO-2, and SUMO-3 (SUMO-2 and SUMO-3 are ∼96% identical and we refer to them as SUMO-2/3). They seem to modify different, partially overlapping subsets of cellular factors (35), and a recent study points to the compensatory utilization of SUMO-2 and/or SUMO-3 for sumoylation of SUMO-1 targets in vivo (36). In a cyclic process highly related to ubiquitination, SUMO modifiers are activated by an E1 activity (Uba-AOS heterocomplex), directed to the target substrate by E2 activity (Ubc9), and covalently attached to the lysine residue that is usually embedded in a minimal motif ΨKX(D/E) in the substrate primary sequence, where Ψ indicates a large hydrophobic amino acid and X indicates any residue. This last step often requires an E3 group of substrate-specific ligases, at least in vivo (37). Unlike monoubiquitination, SUMO modification does not normally target proteins for proteolytic degradation. Instead, sumoylation modulates a wide range of properties of the protein substrates, including subnuclear localization, protein stability, and functional interactions. Sumoylation has been shown to constitute a pivotal mechanism of transcriptional regulation, and its transcriptome-wide effect has been proposed to be transcription-inhibitory (38). The molecular basis of SUMO-driven transcriptional modulation is not well understood. Sumoylation of components of the transcription machinery promotes recruitment of chromatin remodeling complexes, such as histone deacetylases (39). Sumoylation can also modify the affinity of factors for target DNA sequences (40). RNA processing factors are also SUMO targets, but the consequences have not been elucidated. For example, SUMO is implicated in the regulation of assembly of the 3′ end processing machinery (41), thus revealing the potential importance of these modifiers as general mRNA metabolism regulators.
In this study, we investigated the role of sumoylation in modulating the function of TCERG1 in transcription and mRNA processing. We identified TCERG1 as a target for sumoylation in vivo. Sumoylation occurs on at least two sites in the amino terminus of TCERG1. Mutation of the lysines abolishes sumoylation both in vivo and in vitro. Arginine substitution of those lysines did not interfere with TCERG1 localization to the splicing factor-rich nuclear speckles nor was the activity of TCERG1 in mRNA processing affected. Instead, mutation of the sumoylation sites increased the transcriptional activity of TCERG1 in cell reporter assays. Thus, our data strongly support a role for SUMO modification in regulating specific aspects of TCERG1 function.
EXPERIMENTAL PROCEDURES
Plasmids
The eukaryotic expression plasmids pEFBOST7-TCERG1(1–1098) and -(1–662) and bacterial expression vectors pGEX2TK-TCERG1(234–662) and -(631–1098) have been described previously (23, 24). The plasmid pEFBOST7-TCERG1(616–1098) was created by cloning a PCR product with BglII ends into the BamHI site of the parental vector pEFBOST7 (42). The pEFBOST7-TCERG1 full-length single mutants K503R and K608R and the double mutant K503R,K608R were obtained by using the QuikChange XL II-directed mutagenesis kit (Stratagene), and mutations were verified by sequencing. The enhanced cyan fluorescent protein (ECFP) fragment was obtained by PCR from pECFP-C1 plasmid (Invitrogen) and inserted in an EcoRI-digested pEFBOST7 fragment (4677 bp) yielding an intermediate vector pEF-ECFP. pEFBOS/ECFP/T7-TCERG1, pEFBOS/ECFP/T7-TCERG1/K503R, pEFBOS/ECFP/T7-TCERG1/K608R, and pEFBOS/ECFP/T7-TCERG1/K503R,K608R were obtained using EcoRI fragments obtained from the corresponding pEFBOST7 parental vectors by standard cloning procedures. pGAL4-TCERG1 wild-type and mutants (K503R, K608R, and K503R,K608R) were constructed by inserting appropriate sequences into SacI/EcoRI-digested pSG424 vector (43). For expression of recombinant proteins, pGEX2TK-TCERG1(234–662) and pGEX2TK-TCERG1(631–1098) have been described previously (24). The TCERG1 deletion expression vectors pGEX2TK-TCERG1(134–641), wild-type, and SUMO mutants (K503R, K608R, and K503R,K608R), were cloned by inserting TCERG1 PCR fragments into the BamHI/EcoRI sites of pGEX2TK vector (Amersham Biosciences). Expression vectors pcDNA3-HA·SUMO1 and SUMO-2/3 were kindly provided by Dr. R. T. Hay (University of Dundee). For HA·hUbc9 and SUMO-2 recombinant expression and native purification, BamHI/EcoRI fragments were obtained by PCR or enzymatic digestion from pSUbc9V5 (R. T. Hay) or pcDNA3- HA·SUMO-2/3 expression vectors, respectively, and inserted in a BamHI/EcoRI-digested pGEX2TK vector. EDI splicing reporter minigenes pSVEDAmFN (fibronectin promoter) and pSVEDATot (α-globin promoter) were the kind gift from A. R. Kornblihtt (Universidad de Buenos Aires, Argentina) and F. E. Baralle (International Centre for Genetic Engineering and Biotechnology, Trieste, Italy), respectively, and have been described previously (44, 45). The reporter construct pSVEDA-TK contains the herpes simplex virus (HSV) thymidine kinase (TK) promoter and was constructed by inserting a HindIII/BglII-digested pRL-TK (Promega) fragment into a ScaI/BssHII-digested and blunted pSVEDATot plasmid. pSL3b-based Tau exon 10 splicing reporter has been described previously (46, 47) and was kindly provided by Tom Misteli (National Institutes of Health, Bethesda). p3X-κB-L and pRL-TK plasmids were kindly provided by M.A. Garcia-Blanco (Duke University, Durham, NC).
Antibodies
Antibodies against TCERG1 have been described previously (25). Immunoglobulins were purified from antiserum on protein A-Sepharose columns (GE Healthcare) by following standard procedures and used at dilutions of 1:10,000. To affinity-purify the antibodies, antiserum was incubated with nitrocellulose containing immobilized TCERG1 protein as described by Lin et al. (30). Affinity-purified anti-GST antibodies were obtained by standard affinity chromatography through glutathione-Sepharose columns covalently attached to purified GST (48). The anti-SUMO1 monoclonal antibody (Santa Cruz Biotechnology) was provided by M. Lafarga (Universidad de Cantabria) and used at dilutions of 1:100 in Western blot analysis. The anti-T7 (Bethyl) or anti-HA 12CA5 (Roche Applied Science) antibodies were used at dilutions of 1:30,000 and 1:4,000 to detect T7 or HA epitope-tagged proteins, respectively. Antibody against cyclin T1 (Santa Cruz Biotechnology) was used at a dilution of 1:500. For immunofluorescence studies, we used anti-T7 and anti-SC35 (Sigma) antibodies at dilutions of 1:1,000 and 1:2,000, respectively.
Cell Culture, Transfection, and Reporter Gene Assays
HEK293T cells were grown in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (Invitrogen), l-glutamine at 4 mm (Invitrogen), and penicillin/streptomycin to 100 units and 100 μg per ml, respectively (Invitrogen). For luciferase assays, transfections were performed in 35-mm diameter plates (Nunc). Each plate was seeded with ∼1 × 105 cells 20 h prior to transfection. Cells were grown to ∼60–70% confluence and transfected with the appropriate amounts of the indicated constructs by using calcium phosphate and/or Polyfect reagent (Qiagen Inc.) according to the manufacturer's protocols. Transfections were carried out with at least two different preparations of each plasmid DNA purified using kits from Qiagen. Approximately 48 h after transfection, cells were harvested and lysed in 0.2 ml of T7 buffer (20 mm HEPES, pH 7.9, 150 mm NaCl, 5 mm EDTA, 1% Nonidet P-40, 1 mm dithiothreitol (DTT), 1 mm phenylmethylsulfonyl fluoride (PMSF)) for 30 min at 4 °C. The luciferase activities (firefly luciferase for the reporter and Renilla luciferase for the internal control) were measured by using the Dual-Luciferase assay system (Promega) and a standard manual luminometer (Berthold). All experiments were performed in triplicate and at least three independent experiments were considered to calculate means ± S.D. For RT-PCR analysis, total RNA was isolated from HEK293T cells by using the TRIzol reagent (Invitrogen) and then reverse-transcribed by using Moloney murine leukemia virus reverse transcriptase (Invitrogen). The resulting RT reaction product was analyzed for EDI and tau splicing products by radioactive PCR as described previously (44, 49).
Quantification of EDI transcripts (Fig. 4I) was carried out by real time RT-PCR using the MasterMix SYBR Green reagent (Bio-Rad), the iCycler thermal cycler station (Bio-Rad), and protocols described previously (50, 51). Statistical analysis of data was performed using the Prism 4.0 software package (GraphPad).
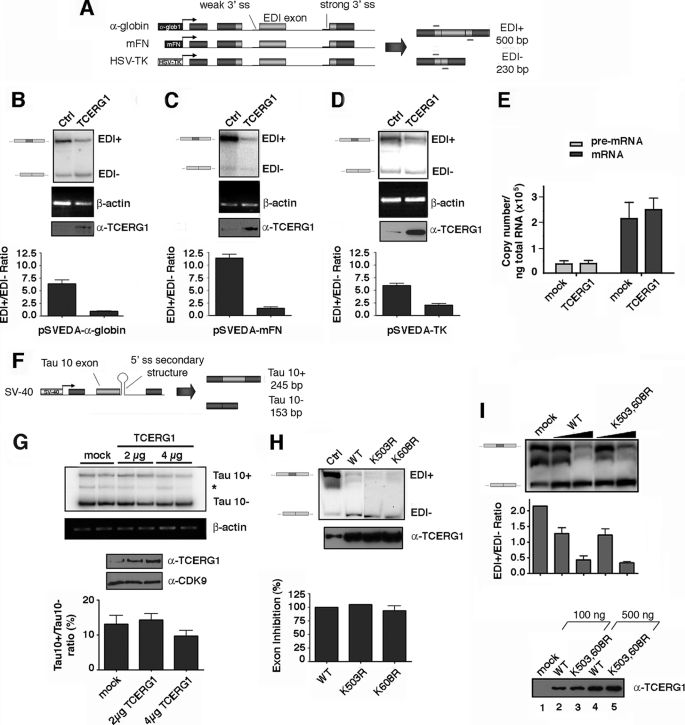
FIGURE 4.
Effect of wild-type TCERG1 and sumoylation mutants on EDI alternative splicing. A, schemes of the minigenes carrying the different promoters. Exons are represented by boxes and introns by lines. Alternative EDI isoforms generated by inclusion (EDI+, 500 bp) or skipping (EDI−, 230 bp) of EDI exon are indicated. Also indicated are the position of the primers used to amplify the mRNA splicing variants. α-globin, human α-globin; mFN, human fibronectin; HSV-TK, herpes simplex virus-thymidine kinase; ss, single strand. B–D, effect of TCERG1 overexpression on alternative splicing elicited by minigenes carrying the α-globin (A), fibronectin (B), and HSV-TK (C) promoters. RNA splicing variants were detected by radioactive RT-PCR, and the products of amplification were separated by polyacrylamide gels. β-Actin was amplified as a control. A sample of cell lysate was immunoblotted with anti-TCERG1 antibody to demonstrate the expression levels. Ratios between radioactivity in EDI+ bands and EDI− bands from two independent experiments are shown below the panels. Ctl, control. E, quantitative analysis of the amount of nascent transcript and total mRNA species derived from the α-globin EDI minigene upon cotransfecting with wild-type TCERG1 by real time PCR. Quantification of the experimental data from four independent experiments is shown in graphic form. F, schematic representation of the tau exon 10 minigene system. Alternative spliced mRNAs containing tau exon 10 (Tau 10+, 245 bp) and mRNAs with tau exon 10 skipped (Tau 10−, 153 bp) are shown in the figure. G, effect of TCERG1 overexpression in the regulation of tau exon 10. RT-PCR analysis of the cotransfections of tau 10 minigene with TCERG1. β-Actin was amplified as a control. A sample of cell lysate was immunoblotted with anti-TCERG1 antibody to demonstrate the expression levels. Ratios between radioactivity in tau 10+ bands and tau− bands from two independent experiments are shown below the panel. H, cotransfection assays were carried out with wild type and sumoylation mutants of TCERG1 and the α-globin EDI minigene. Patterns of EDI minigene splicing were analyzed as in previous panels. I, curve dose-response for wild-type (WT) TCERG1 and double mutant variant (K503R,K608R (K503,608R)) on EDI alternative splicing. The α-globin EDI minigene and the indicated TCERG1 constructs were cotransfected at 100 (lanes 2 and 4) and 500 (lanes 3 and 5) ng. RNA splicing variants were detected by radioactive RT-PCR, and the products of amplification were separated by polyacrylamide gels (top panel). Experimental data from two independent experiments were quantified and are shown in graphic form (middle panel). A sample of cell lysates were immunoblotted with anti-TCERG1 antibody to demonstrate the expression levels (bottom panel).
For RNA interference transfections (supplemental Fig. 2), 300 ng of the human fibronectin EDI splicing reporter minigene were cotransfected with one of the following small interfering RNA duplexes: small interfering enhanced green fluorescent protein 5′CUA-CAA-CAG-CCA-CAA-CGU-C 3′ and small interfering TCERG1 5′GGA-GUU-GCA-CAA-GAU-AGU-U3′ (33) at a final concentration of 100 nm, and with 250 ng of TCERG1 wild type, the K503R,K608R double mutant, or empty vector as a control using Lipofectamine 2000 (Invitrogen). Cells were harvested 48 h post-transfection and processed for Western blotting and RT-PCR.
Immunofluorescence
Immunofluorescence studies were performed as described previously (25) using a spectral laser confocal microscope Leica SP5. Images were analyzed and digitally processed for presentation using LAS AF software.
In Vivo Sumoylation Assays
Transfected cells were harvested in phosphate-buffered saline and lysed in 0.2 ml of ice-cold RIPA-375 buffer (50 mm Tris-HCl, pH 8, 375 mm NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 5 mm EDTA, 1 mm DTT, 1 mm PMSF, and protease inhibitor mixture (Complete, Roche Applied Science)) for 20 min. Chromatin was sheared by repeated passage of the extracts through 23-gauge needles, and debris was cleared by centrifugation at 14,000 rpm for 5 min at 4 °C. Ten percent of the lysate was saved as input samples (WCE), and the remaining extract was diluted to a 1-ml final volume with ice-cold RIPA-375. Thirty microliters of anti-T7 tag monoclonal antibody that was covalently coupled to cross-linked agarose beads (Novagen) was added to the diluted extract and then incubated with end-over-end rotation for 6 h at 4 °C. After five washes with 1.5 ml of RIPA-375 and once with 1.5 ml of T7 buffer, proteins bound to the antibody resin were boiled in 30 μl of 2× SDS-PAGE sample buffer. Fifteen microliters of each input and pellet sample were subjected to 7.5% SDS-PAGE, transferred to a polyvinylidene difluoride (Bio-Rad) or nitrocellulose (Amersham Biosciences) membrane, and then incubated with the specific antibody. After being washed, the membrane was incubated with a peroxidase-conjugated secondary antibody, and bound antibodies were detected by enhanced chemiluminescence (PerkinElmer Life Sciences).
To assess steady-state sumoylation of endogenous TCERG1, ∼3 × 105 HEK293T transfected cells were lysed in buffer T7 with or without 25 mm N-ethylmaleimide (NEM) and 1% SDS. Lysates were subjected to Western blotting analysis as described above. Immunoprecipitation of endogenous TCERG1 was conducted as follows: ∼5 × 106 HEK293T cells were lysed in SDS-HS buffer (1% SDS, 0.5% Triton X-100, 50 mm Tris-HCl, pH 8, 5 mm EDTA, 1 mm DTT, 500 mm NaCl, 0.5 mm DTT, 25 mm NEM, 1 mm PMSF, and protease inhibitor mixture (Complete, Roche Applied Science)), and chromatin was sheared by repeated passage of the extracts through a 23-gauge needle. The cell extracts were diluted 10 times in non-SDS-HS buffer (0.5% Triton X-100, 50 mm Tris-HCl, pH 8, 5 mm EDTA, 500 mm NaCl, 0.5 mm DTT, 25 mm NEM, 1 mm PMSF, and protease inhibitor mixture (Complete, Roche Applied Science)), and split into 2 equal volumes. Fifty microliters of anti-TCERG1 IgG antibodies (100 μg) that was covalently coupled (25) to cross-linked protein A-Sepharose beads (4FastFlow, Amersham Biosciences) was added to the diluted samples and incubated with end-over-end rotation for 4 h at 4 °C. Purified nonspecific rabbit IgGs were used as a control. After extensive washing with RIPA-375 buffer, immunoprecipitates were boiled in 50 μl of 2× SDS-PAGE sample buffer and analyzed by Western blotting.
Protein Purification
Proteins were expressed in BL21 (D3) cells and purified on glutathione-agarose beads (Sigma) following standard procedures. The release of the fusion protein and the cleavage to yield free proteins were carried out as described by Smith and Johnson (52). hUbc9 protein was dialyzed against phosphate-buffered saline containing glycerol at 20%; SUMO-2 was dialyzed against IVS buffer (20 mm HEPES, pH 7.5, 5 mm Mg, 100 mm NaCl, 0.5 mm DTT, 0.5 mm PMSF); GST-TCERG1 proteins were dialyzed against a buffer described in Lin et al. (30).
In Vitro Interaction and Sumoylation Assays
In-solution interaction assay was performed essentially as described previously (53, 54) with minor modifications. Briefly, 5 μg of HA-hUbc9 were incubated with 20 μg of the appropriate GST fusion protein pre-bound to 20 μl of glutathione-agarose, in a 500-μl final volume of binding buffer (40 mm HEPES, pH 7.9, 150 mm KCl, 5 mm MgCl2, 0.5 mm EDTA, 10% glycerol, 0.1% Nonidet P-40, 1 mm DTT, and 1 mm PMSF) with end-over-end rotation for 4 h at 4 °C. Pellets were washed extensively with binding buffer, boiled in 2× SDS-PAGE buffer, and then separated on SDS-polyacrylamide gels and analyzed by Coomassie staining and Western blotting.
SUMO modification of TCERG1 was performed in vitro using a commercial sumoylation kit (LAE Biotech International). The reaction mixtures were performed in 20 μl of the suggested buffer (20 mm HEPES, pH 7.5, 5 mm magnesium, 100 mm NaCl ± ATP) with 500 ng of the GST-TCERG1 recombinant proteins, incubated at 37 °C for 90 min, and terminated by addition of SDS-PAGE sample buffer. The samples were then subjected to SDS-PAGE and Western blotting with specific antibodies.
RESULTS
TCERG1 Is Modified by SUMO-1 and SUMO-2/3 in Vivo
TCERG1 potentially couples transcription and mRNA processing; however, the molecular mechanisms by which TCERG1 modulates transcriptional elongation and splicing remain to be elucidated. We postulated that the activities of TCERG1 may be regulated by post-translational modification. We began our analysis by examining whether TCERG1 can be modified by sumoylation in vivo. The steady-state level of most sumoylated proteins is very low, presumably caused by strictly regulated modification and/or rapid cycles of modification and de-modification. This often causes a detection problem of sumoylation in vivo. To overcome this obstacle, we have used protocols that involve enrichment of sumoylated proteins under strong denaturing conditions. HeLa whole-cell lysates were obtained in the absence or presence of the cysteine protease inhibitor N-ethylmaleimide (NEM) or 1% SDS buffer, which are usually used to preserve the sumoylation of cellular proteins and then subjected to immunoblotting with anti-TCERG1-specific antibodies. Along with the 150-kDa TCERG1, the antibodies detected a predominant slow migrating band with apparent mass of 200 kDa. This band is dependent on extracts prepared with NEM and 1% SDS buffer (Fig. 1A, lanes 2 and 4). Upon treatment with 1% SDS, a second retarded form of TCERG1, migrating at ∼180 kDa, was detected. This 180-kDa form was unresponsive to NEM (Fig. 1A, lanes 3 and 4). Both the mobility and dependence of these modified TCERG1 species were consistent with potential sumoylation.
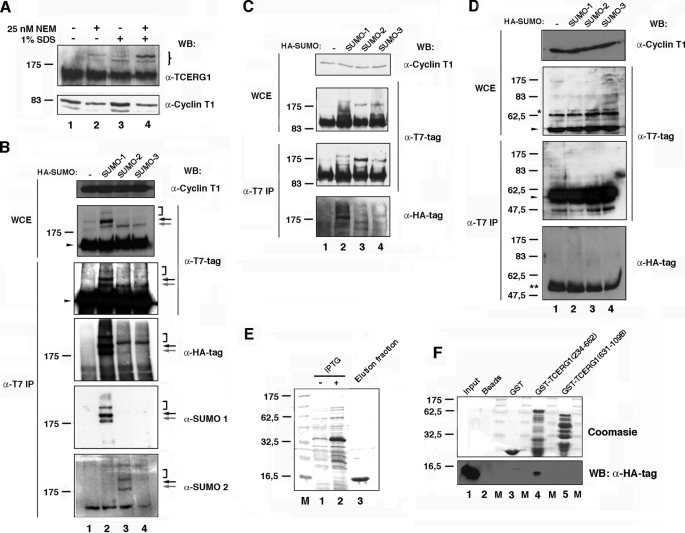
FIGURE 1.
TCERG1 is a sumoylation substrate in vivo and interacts directly with human Ubc9. A, Western blotting (WB) analyses of endogenous TCERG1 from HeLa cells with or without 25 nm NEM and/or 1% SDS. Specific antibodies against TCERG1 and cyclin T1 (as a loading control) were used to localize the proteins. The brace indicates the positions of the two predominant slow migrating bands in the samples treated with 25 nm NEM and 1% SDS buffer. Molecular masses in kDa are indicated to the left. B, HeLa cells were cotransfected with a plasmid encoding T7-tagged TCERG1 along with empty vector (lane 1) or vectors encoding HA-SUMO-1 (lane 2), HA-SUMO-2 (lane 3), or HA-SUMO-3 (lane 4). WCE were directly analyzed by Western blotting or subjected to immunoprecipitation (IP) with T7-specific antibodies followed by SDS-PAGE and Western blotting with anti-T7, anti-HA, anti-SUMO-1, anti-SUMO-2, or anti-cyclin T1 antibodies. Specific higher molecular weight forms of TCERG1 are visible above the position of the 175-kDa marker and are indicated by arrows and bracket (see description in text). The unmodified form of TCERG1 is indicated by a closed arrowhead. C and D, same experiment described for B was repeated but using plasmids encoding T7-tagged amino- (C) and carboxyl-terminal (D) regions of TCERG1. *, nonspecific band present in WCE and reactive to anti-T7 antibodies. **, immunoglobulin fraction eluted from the matrix. E, purification of HA-tagged Ubc9. Lanes 1 and 2, crude E. coli lysates with (+) or without (−) isopropyl 1-thio-β-d-galactopyranoside (IPTG); lane 3, elution fraction after thrombin cleavage (HA-hUbc9). F, amino-terminal region of TCERG1 interacts directly with human Ubc9. HA-tagged Ubc9 was incubated with equal amounts of GST or GST-TCERG1(234–662) and GST-TCERG1(631–1098) bound to glutathione-Sepharose beads. Bound proteins (lanes 2–5) were eluted with SDS-PAGE loading buffer and resolved in a 15% SDS-polyacrylamide gel along a sample of the input (lane 1). The upper part of the gel was stained with Coomassie Blue R-250 to visualize the eluted GST fusion proteins. and the lower part was transferred to a membrane and incubated with an anti-HA-specific antibody to detect Ubc9. Molecular masses (M) in kDa are indicated to the left.
We also performed immunoprecipitation experiments under stringent conditions to preserve SUMO conjugates using affinity-purified anti-TCERG1 antibodies. Anti-SUMO-1 monoclonal antibody detected two specific bands in the anti-TCERG1 immunoprecipitates with apparent molecular weights similar to those of the previously identified putative SUMO-TCERG1 species (supplemental Fig. 1A). Those results further support that TCERG1 is a bona fide target for in vivo SUMO modification.
As mentioned above, the level of sumoylation was very low in most of the target proteins (usually below 5%). To estimate the proportion of modified TCERG1, we carried out semiquantitative experiments in the linear range of the standard curve. We found that TCERG1 sumoylation accounts for a small percentage of total protein, approximately <5%, which is in agreement with most published data (supplemental Fig. 1B and data not shown).
Next, we asked if a T7-tagged human TCERG1 cDNA could also be modified and whether SUMO coexpression elicited TCERG1 sumoylation. First, we transfected HEK293T cells with a plasmid encoding T7-tagged TCERG1 and prepared whole-cell extracts. Upon Western blotting with anti-T7 monoclonal antibody, we observed the expected 150-kDa species of TCERG1. We also observed at least two slower migrating forms with apparent molecular mass of ∼180 and 200 kDa, consistent with the preceding observations with anti-TCERG1 antibody (Fig. 1B, lane 1, WCE panel; gray and black arrows, respectively). To determine whether the modified forms of TCERG1 could be enriched by sumoylation, we independently cotransfected each of the three SUMO isoforms with the T7-tagged TCERG1 into HEK293T cells and probed extracts from these cells (Fig. 1B). Coexpression of SUMO-1 with T7-tagged TCERG1 resulted in an increase of the 180- and 200-kDa species along with a third slower migrating form of apparent molecular size of 300 kDa (Fig. 1B, WCE panel; lane 2, gray and black arrows, and bracket, respectively). Coexpression of SUMO-2 or SUMO-3 with TCERG1 resulted in the appearance of a single slow migrating band of ∼180 kDa immunoreactive to anti-T7 antibody (Fig. 1B, WCE panel; lanes 3 and 4, gray arrow). Higher molecular weight species, which likely represent the poly-SUMO-2/3 chains, were also detectable in most of the experiments (Fig. 1B, bracket).
To confirm that these higher molecular weight species were indeed SUMO-modified TCERG1 proteins, T7-tagged TCERG1 and HA-tagged SUMO-1 and SUMO-2/3 were again coexpressed. TCERG1 was immunoprecipitated from cells with anti-T7 antibody under highly stringent conditions and then blotted with anti-T7 or anti-HA antibody. Similar higher molecular weight species were detected by those antibodies. Coexpression of SUMO-1 with T7-tagged TCERG1 resulted in an increase of the 180- and 200-kDa species (Fig. 1B, α-T7 IP and α-HA-tag panels; lane 2, gray and black arrows, respectively). Coexpression of SUMO-2 or SUMO-3 with TCERG1 resulted in the appearance of the single slow migrating band of ∼180 kDa (Fig. 1B, α-T7 IP and α-HA-tag panels; lanes 3 and 4, gray arrow). Higher molecular weight species were also detectable in most those experiments (Fig. 1B, α-T7 IP and α-HA-tag panels; lanes 2–4, bracket). Finally, we analyzed the TCERG1 immunoprecipitates with specific SUMO-1 and SUMO-2 antibodies and found immunoreactivity that was overlapping with the signal obtained with anti-TCERG1 and anti-HA antibodies (Fig. 1B, α-SUMO-1 and α-SUMO-2 panels). Those data further support the results of the previous experiment. Therefore, both endogenous TCERG1 and transiently expressed TCERG1 can be covalently modified by SUMO paralogs in cells.
To map the sumoylation sites within TCERG1, two T7 epitope-tagged fragments encompassing the amino- and carboxyl-terminal halves of human TCERG1 (containing amino acid residues 1–662 and 616–1098, respectively) were assayed in a similar manner as described above. Both fragments retain the putative nuclear localization signal found in the middle of the protein and localize throughout the nucleus. Like wild type, the TCERG1 carboxyl-terminal fragment localizes in the splicing factor-rich nuclear speckles (Ref. 25 and data not shown). HEK293T cells were transfected with either the amino- and carboxyl-terminal TCERG1 fragments. SUMO-1, SUMO-2, or SUMO-3 was coexpressed with TCERG1 fragments. Only the amino-terminal half of TCERG1 rendered a pattern of bands consistent with that of the full-length protein, either in whole-cell extracts or in pellets of immunoprecipitations with anti-T7 antibody (Fig. 1C), whereas no higher molecular weight species were observed with the carboxyl-terminal fragment of TCERG1 (Fig. 1D). Thus, the major sumoylation sites reside within the first 662 amino acids of TCERG1.
Because most SUMO substrates interact with the E2 enzyme Ubc9 and this interaction is considered a strong indicator of sumoylation (54), we next determined whether TCERG1 interacts with Ubc9 using the GST pulldown assay. We first purified HA-tagged human Ubc9 recombinant protein from bacteria using native conditions (Fig. 1E). The amino- and carboxyl-terminal fragments of TCERG1 were purified from bacteria as GST fusions by binding to glutathione-agarose beads (Fig. 1F, top panel). The bead-bound TCERG1 fragments were then used as baits in pulldown assays with the purified HA·hUbc9. As a negative control, we used GST and beads alone. After extensive washing, the bead-bound proteins were eluted by boiling the samples with SDS-PAGE loading buffer and analyzed by SDS-PAGE and Western blotting to detect HA-tagged hUbc9. The results of the in vitro binding assay revealed a direct interaction between hUbc9 and the amino-terminal 662 amino acids of TCERG1 (Fig. 1F, bottom panel), thus supporting the conclusion that this region of TCERG1 contains required sequences for proper interaction with the sumoylation machinery and conjugation with SUMO.
Lys-503 and Lys-608 Are the Major Acceptor Sites of SUMO Modification in TCERG1
Sumoylation usually takes place on a lysine embedded in the core consensus motif ΨKX(E/D), where Ψ represents a hydrophobic residue and X represents any amino acid residue (37). Recent studies have defined an extended consensus motif for sumoylation, termed the NDSM (negatively charged amino acid-dependent sumoylation motif), which includes clusters of acidic residues located downstream from the core SUMO modification motif ΨKX(E/D) (54). This extended motif can be used to correctly predict targets for sumoylation. Two lysines within the amino-terminal region of TCERG1, Lys-503 and Lys-608, match this sumoylation motif (Fig. 2A). These lysines and surrounding sequences are highly conserved among TCERG1 homologs in metazoa, as shown in the amino acid alignment in Fig. 2A.
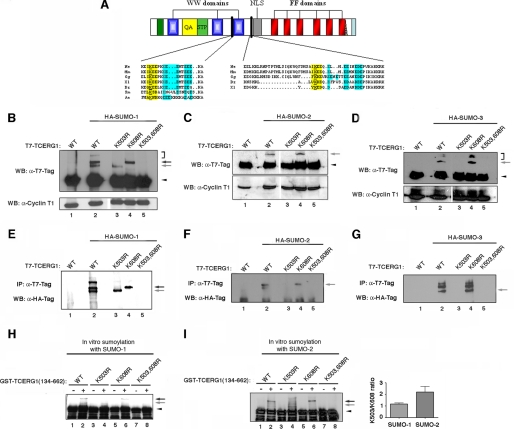
FIGURE 2.
Lys-503 and Lys-608 are the major sites of sumoylation of TCERG1 and are conserved among species. A shows a ClustalW amino acid alignment of TCERG1 homologs that include the putative SUMO acceptor motifs. Conserved acceptor lysines within the core consensus motif (in yellow) and NDSM consensus motif (in blue) features are highlighted. Hs, Homo sapiens; Mm, Mus musculus; Gg, Gallus gallus; Xl, Xenopus laevis; Dr, Danio rerio; Sm, Schistosoma mansoni; Ae, Aedes aegypti; NLS, nuclear localization signal. B–D, HEK293T cells were cotransfected with expression constructs of HA-SUMO-1 (A), HA-SUMO-2 (B), or HA-SUMO-3 (D) and T7-tagged wild-type (WT) TCERG1 or the indicated mutants, and lysates subjected to immunoblotting with anti-T7 and anti-cyclin T1 antibodies. The sumoylated forms of TCERG1 are indicated by arrows (see description in text). The unmodified form of TCERG1 is indicated by a closed arrowhead. WB, Western blot. E–G, HEK293T cells were cotransfected with expression constructs of HA-SUMO-1 (E), HA-SUMO-2 (F), or HA-SUMO-3 (G) and T7-tagged WT TCERG1 or the indicated mutants, and lysates were subjected to immunoprecipitation (IP) with anti-T7 antibodies and Western blotting analysis with anti-HA antibodies. The sumoylated forms of TCERG1 are indicated by arrows. H, in vitro expressed GST-TCERG1(134–662) fusion protein, either wild type or the indicated mutants, was incubated with SUMO-1 reaction components, and the reaction products were analyzed by SDS-PAGE and immunoblotting with specific antibodies against GST. The bands corresponding to the SUMO-conjugated forms of TCERG1(134–662) are indicated by arrows. The unmodified form of recombinant TCERG1 is indicated by a close arrowhead. I, the same experiment described in H was carried out with SUMO-2. Quantification of the experimental data obtained with the double mutant variant from two independent experiments were quantified and are shown in graphic form at the right of the panel. K503,608R is K503R,K608R.
To determine whether these lysines were modified by sumoylation, we generated T7-tagged TCERG1 constructs containing lysine to arginine mutations at either position Lys-503, Lys-608, or both. We then carried out in vivo sumoylation assays by coexpressing the wild-type and mutated TCERG1 proteins with or without SUMO-1, SUMO-2, or SUMO-3 in transient transfection experiments. Both unmodified and sumoylated TCERG1 were detected by immunoblotting of these cell extracts with T7-specific antibodies (Fig. 2, B–D, α-T7-Tag panels). Coexpression of SUMO-1 with T7-tagged TCERG1 resulted in an increase of the 180- and 200-kDa species (Fig. 2B, lane 2, arrows). Mutation of lysine 503 abrogated the 200-kDa band associated with SUMO-1 conjugation (Fig. 2B, lane 3). Mutation of lysine 608 abrogated the formation of a shifted band corresponding to the faster migrating species (180 kDa) modified specifically by SUMO-1 (Fig. 2B, lane 4). The K503R and K608R mutants abrogated the formation of the third shifted band of apparent molecular size of 300 kDa corresponding to the minor SUMO-1-modified form (Fig. 2B, lanes 3 and 4, bracket). The 300-kDa form may represent a double SUMO-1 modification of TCERG1 at different lysine residues. Mutations at lysines 503 and 608 caused changes in the sumoylation pattern, and no species associated with SUMO-1 conjugation were observed with the double mutant (Fig. 2B, lane 5), suggesting that both residues are the major sumoylation acceptor sites for SUMO-1 in vivo. The data also show that the carboxyl-terminal region of TCERG1 is poorly, if at all, modified by SUMO in vivo.
Coexpression of SUMO-2 or SUMO-3 with TCERG1 resulted in the appearance of the single slow migrating band of ∼180 kDa immunoreactive to anti-T7 antibody (Fig. 2, C and D, lane 2, gray arrow). The poly-SUMO-2/3 chains were also detectable (Fig. 2D, bracket, and data not shown). Mutation at lysine 503 and the double mutant abrogated sumoylation by SUMO-2 and SUMO-3 (Fig. 2, C and D, lanes 3 and 5). The sumoylated forms of the proteins produced by SUMO-2/3 in transfected cells were unchanged upon mutating lysine 608 (Fig. 2, C and D, lane 4), supporting that modification by SUMO-2/3 in vivo occurs preferentially at position 503. Lysines residues can also be sites for ubiquitination; however, our transfection data indicate that the identified sumoylation sites do not affect protein stability and are therefore unlikely to be critical for ubiquitination.
To further corroborate those results and to determine which slow mobility species bear SUMO-1 or SUMO-2/3, the HA immunoreactivity of TCERG1 immunoprecipitates was also analyzed. The same specific bands described above were also detected in TCERG1 immunoprecipitates by using an anti-HA monoclonal antibody (Fig. 2, E–G), which further suggest that the lower band of ∼180 kDa likely corresponds to sumoylation at position 608 and the upper band of ∼200 kDa to sumoylation at position 503.
Our findings indicate that Lys-503 and Lys-608 are the major SUMO acceptor sites within TCERG1 in vivo. To further confirm that Lys-503 and Lys-608 are SUMO acceptor sites, we carried out in vitro sumoylation assays. As a substrate, we used a GST-tagged TCERG1 fragment (134–662) that contained the major sumoylation sites. Wild-type TCERG1 or K503R, K608R, and K503R,K608R mutant proteins were affinity-purified from bacteria and incubated in the presence of recombinant SUMO-1, E1, and E2 (Ubc9) enzymes. Sumoylation is ATP-dependent, and therefore we performed the assay in the absence and presence of ATP. Higher molecular weight species of the amino-terminal domain of TCERG1 containing residues 134–662 were observed only when all components were added in the reaction in the presence of ATP (Fig. 2H, lanes 1 and 2), providing further evidence that the amino-terminal region of TCERG1 is a substrate for sumoylation by SUMO-1. Furthermore, the K503R and K608R mutants abrogated the formation of shifted bands corresponding to a single SUMO-1 molecule conjugated to a specific lysine residue (Fig. 2H, lanes 4 and 6), resembling closely the in vivo pattern shown for SUMO-1. The double mutant could not be sumoylated in vitro (Fig. 2H, lane 8), indicating that those lysines are also preferentially targeted for sumoylation in vitro.
We also carried out in vitro sumoylation experiments with SUMO-2-purified protein. Higher molecular weight species were again observed only when all components were added in the reaction in the presence of ATP (Fig. 2I, lanes 1 and 2). Mutation at position 503 abrogated the formation of the major sumoylated band (Fig. 2I, lanes 3 and 4), whereas mutation at position 608 eliminated a minor slow migrating band (Fig. 2I, lanes 5 and 6), thus confirming that those lysines are preferential SUMO acceptor sites. Simultaneous mutation of both lysines to arginine again abolished sumoylation (Fig. 2I, lanes 7 and 8). Here, we were able to detect modification of the lysine 608 residue by SUMO-2, but still significant differences in the relative conjugation efficiency for each site between SUMO-1 and SUMO-2 were observed, supporting that the modification by SUMO-2 occurs preferentially at Lys-503 (Fig. 2I, bar graph).
Nuclear Localization of TCERG1 Is Independent from Modification at Lys-503 and Lys-608 by Sumoylation
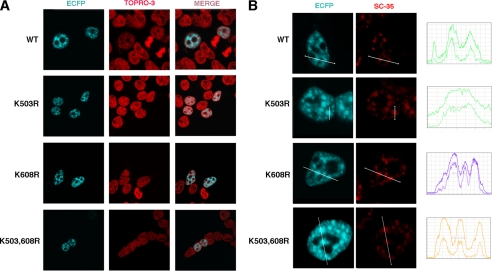
Because sumoylation is known to influence the cellular localization of target proteins, we wished to investigate whether loss of sumoylation at Lys-503 or/and Lys-608 modulates TCERG1 distribution. TCERG1 is distributed in dot-like structures throughout the nucleoplasm and colocalizes with the splicing factor-rich nuclear speckles (25). We expressed wild-type full-length TCERG1 and the lysine-to-arginine mutants tagged with the ECFP at the amino terminus and examined their nuclear localization in HEK293T cells using immunofluorescence microscopy. As shown in Fig. 3, all the TCERG1 constructs tested exhibited a similar nucleoplasm distribution with an increased signal in organized granule-like sites and outside the highly compacted chromatin territories detected with the fluorescent dye TOPRO-3 (Fig. 3A). This pattern is similar to that of endogenous TCERG1 (25). TCERG1 also is enriched in speckles (25); therefore, we investigated the effect of SUMO conjugation at Lys-503 and Lys-608 on the TCERG1 localization to nuclear speckles. Abrogation of SUMO conjugation at those lysines had no effect on the distribution of TCERG1 to nuclear speckles, which were visualized by costaining with antibody against SC35, a splicing factor that is commonly used to define nuclear speckles (Fig. 3B). We analyzed the spatial relationship between wild-type and mutant TCERG1 variants relative to SC-35 by quantitatively scanning specific nuclear regions containing speckles (Fig. 3B, line scans ECFP-TCERG1 and SC-35 in blue and red, respectively). We conclude that SUMO modification at amino acid residues Lys-503 and Lys-608 is dispensable for the natural nuclear distribution of TCERG1.
FIGURE 3.
Nuclear localization of TCERG1 is independent from modification at Lys-503 and Lys-608 by sumoylation. A, immunofluorescence analysis of HEK293T cells transfected with ECFP-TCERG1-WT or the indicated mutant constructs (ECFP, blue). TOPRO-3 labeling was used to stain chromatin (red). Individual and merge images are shown. B, colocalization of the indicated TCERG1 constructs with the essential splicing factor SC35 that commonly serves to define nuclear speckles. Dual labeling of cells with either ECFP-TCERG-WT or SUMO mutants (ECFP, blue) and with the anti-SC35 antibody (SC-35, red) was performed. Line scans showing local intensity distributions of TCERG1 and of SC35 are shown to the right of the panels. Bars indicate the position of the line scans. K503,608R is K503R,K608R.
Sumoylation Decreases TCERG1 Transcriptional Activity but Does Not Affect Its Alternative Splicing Function
Many proteins involved at different steps of regulation of gene expression are modified by sumoylation. TCERG1 modulates the activity of RNAPII complexes and appears to play a role in both transcription elongation and pre-mRNA splicing. We sought to determine the functional significance of TCERG1 sumoylation in the context of those processes.
Recent data have shown that TCERG1 modulates fibronectin EDI exon inclusion (33). The fibronectin EDI minigene displays a weak 3′ splice site followed by a stronger splice site in the downstream intron. As a result, the EDI exon can be alternatively spliced in a regulated fashion and controlled, at least in part, by RNAPII elongation (4, 45, 50). We wished to confirm those results and used this assay to determine whether SUMO modification regulates the effect of TCERG1 on the alternative processing of the fibronectin EDI exon. Given that changes in promoter structure strongly affect EDI splice site selection (44), we performed the experiments using minigenes carrying different promoters (Fig. 4A). HEK293T cells were transiently transfected with the minigenes containing the α-globin, fibronectin, or HSV-thymidine kinase promoter (HSV-TK), and alternative splicing of EDI was assessed by RT-PCR. Inclusion of EDI exon was lower when transcription was driven by the α-globin promoter compared with the fibronectin promoter (compare Fig. 4, B and C), as demonstrated previously (44), thus confirming that the regulation of this system is linked to transcriptional control. A similar result was obtained when transcription was driven by the HSV-TK promoter (Fig. 4D). When TCERG1 cDNA was overproduced by cotransfection, EDI exon inclusion was significantly diminished from all three reporter minigenes (Fig. 4, B–D), even in the case of a promoter previously reported to be resistant to TCERG1 influence such as HSV-TK (23). In our experiments, TCERG1 did not affect the EDI− band (Fig. 4, B–D). These results suggest a direct effect of TCERG1 on splicing decisions in a manner independent of transcription.
In those experiments, it is clear that for all three promoters, the decrease in EDI+ transcript is not fully compensated by an increase in the EDI− transcript. To determine whether TCERG1 inhibits the overall transcription, we have used a previously described approach that quantitatively analyzes the amount of nascent transcript and total mRNA species derived from the EDI minigene by real time PCR (50, 51). No significant alteration in the level of nascent transcript or total mRNA was observed (Fig. 4E). The results further support that TCERG1 influences alternative splicing and does not globally inhibit mRNA synthesis.
The effect of TCERG1 on the alternative splicing of the well characterized tau minigene system was also determined. Human tau exon 10 is regulated by a stem-loop structure that is formed in the pre-mRNA at the downstream 5′ splice site. Normally, exon 10 is included or excluded from the mRNA with roughly equal probability during pre-mRNA splicing. Mutations clustered around the 5′ splice site of exon 10 result in predominant inclusion of exon 10, resulting in a severe neurological disorder (46, 55). In contrast to the EDI situation, no specific change in the alternative splicing of exon 10 was observed upon TCERG1 overexpression (Fig. 4, F and G). The very minor reduction of Tau 10 observed at 4 μg of TCERG1 (Fig. 4G) is not statistically significant. Those results are in agreement with previous observations that link TCERG1 to recognition of specific 3′ splice sites of pre-mRNAs (30, 56).
Having established that TCERG1 alters the alternative splicing of EDI exon, we investigated the potential functional consequences of the conjugation of SUMO to TCERG1. Cells were cotransfected with the α-globin promoter construct and plasmids expressing the wild-type TCERG1 and the K503R, K608R, and K503R,K608R mutant versions. The splicing pattern elicited by the EDI minigene was similarly affected by the expression of the mutant variants that fail to be modified by SUMO (Fig. 4, H and I) even when the levels of TCERG1 used are moderate (Fig. 4I). To exclude a contribution of the endogenous pool of sumoylated TCERG1, which might mask differences between overexpressed wild-type and mutant TCERG1, we examined the effects of overexpressing wild-type and sumoylation-deficient TCERG1 in cells that have been previously depleted of endogenous TCERG1 by RNA interference. Both wild-type and mutant TCERG1 decreased EDI inclusion the same magnitude. Therefore, SUMO modification does not modulate the ability of TCERG1 to regulate alternative splicing of EDI exon, which rules out a putative effect of endogenous TCERG1 sumoylation (supplemental Fig. 2).
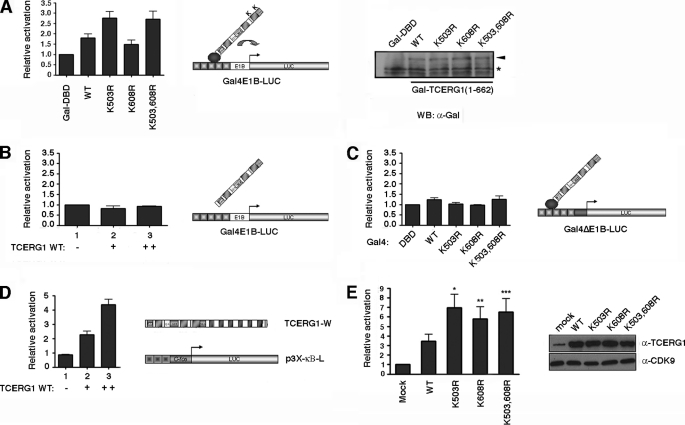
To investigate whether SUMO modification affects the transcriptional activity of TCERG1, we determined the effects of the single and double mutants (K503R, K608R, and K503R,K608R) compared with the wild-type protein. For this purpose, we employed a one-hybrid assay to measure TCERG1 transcriptional activity. This experiment uses a reporter construct that contains five copies of the GAL4 consensus DNA-binding site and a minimal adenovirus E1b promoter that drives the expression of luciferase (GAL4E1B-LUC in Fig. 5A). HEK293T cells were cotransfected with the reporter plasmid and the wild-type amino-terminal domain of TCERG1(134–662) or the mutant variants fused to the GAL4 DNA-binding domain. The wild-type protein activated the expression of luciferase modestly (2-fold) but significantly (Fig. 5A). Interestingly, mutation of the lysine residue at position 503 into an arginine resulted in an increase on the transactivation activity of the protein compared with wild type (Fig. 5A). Mutating the lysine at position 608 had no effect on the transcriptional activity of the protein compared with wild type, and the double mutant form had an activity similar to that of the K503R single mutant (Fig. 5A). Thus, the lysine residue 503 that serves as SUMO-1 and SUMO-2/3 acceptor sites appears to negatively regulate TCERG1 transcriptional activity. Expression of the various TCERG1 proteins was nearly identical, suggesting that the enhancement of TCERG1-K503R transactivation capacity was not due to different protein expression levels (Fig. 5A). Furthermore, the activities exhibited by the different constructs seem to rely on a specific mechanism because the observed modulation of reporter gene activity was dependent upon the GAL4 DNA-binding domain in the effector constructs and the presence of a functional TATA-box sequence in the reporter construct (Fig. 5, B and C). Renilla luciferase was used as an internal control for transfection. The observed effects are consistent with the fact that effects of SUMO on transcriptional regulation are generally much more evident at promoters bearing multiple binding sites (57).
FIGURE 5.
SUMO modification sites negatively regulate the TCERG1-mediated transcriptional activation. A, HEK293T cells were cotransfected with a Gal4-luciferase reporter plasmid (Gal4E1B-LUC), a control plasmid expressing the Gal4 DNA binding domain (Gal4-DBD, 1st lane), the wild-type amino-terminal domain of TCERG1(134–662) (lane 2), or the SUMO mutant variants (K503R, 3rd lane; K608R, 4th lane; K503R,K608R (K503,608R), lane 5) fused to the Gal4 DNA-binding domain. The luciferase activity is shown relative to the Gal4-DBD that was set at 1. The data shown are from four independent experiments performed in triplicate (p (wild type (WT) versus Gal4-DBD) = 0.02; p (wild type versus K503R) = 0.0058; p (wild type versus K608R) = 0.11; p (wild type versus K503R,K608R) = 0.01; paired Student's t test, set at p < 0.05). A sample of cell lysate was immunoblotted with anti-Gal4 antibody to demonstrate the expression levels. B, HEK293T cells were cotransfected with a Gal4-luciferase reporter plasmid (Gal4E1B-LUC) and a control plasmid (lane 1) or increasing amounts of TCERG1(134–662) without the Gal4 DNA-binding domain (lanes 2 and 3). The luciferase activity is shown relative to the control that was set at 1. The data shown are from four independent experiments performed in triplicate. C, HEK293T cells were cotransfected with a Gal4-luciferase reporter plasmid lacking a functional TATA-box (Gal4ΔE1B-LUC), a control plasmid expressing the Gal4 DNA binding domain (Gal4-DBD, 1st lane), the wild-type amino-terminal domain of TCERG1(134–662) (2nd lane) or the SUMO mutant variants (K503R, 3rd lane; K608R, 4th lane; K503,608R, 5th lane) fused to the Gal4 DNA-binding domain. The luciferase activity is shown relative to the Gal4-DBD that was set at 1. The data shown are from four independent experiments performed in triplicate. D, HEK293T cells were cotransfected with the p3X-kb-L reporter plasmid together with increasing concentrations of wild-type TCERG1 expression plasmid. The luciferase activity was calculated relative to the control that was cotransfected with the reporter and empty plasmids. The total DNA amount for transfection was kept the same in each sample by normalizing with empty vector. E, HEK293T cells were cotransfected with the p3X-kb-L reporter plasmid together with empty vector (1st lane), wild-type TCERG1 (2nd lane), and SUMO variants (K503R, 3rd lane; K608R, 4th lane; K503,608R, 5th lane) DNA constructs. The luciferase activity was calculated relative to the control. The data shown are from four independent experiments performed in triplicate (*, p < 0.01; **, p < 0.02; ***, p < 0.04). Cell lysates were analyzed by immunoblotting with the indicated antibodies to detect the TCERG1 and CDK9 proteins.
To lend additional support to the observation that SUMO modification represses transcriptional activation by TCERG1, we examined the transcriptional activity of the full-length wild-type TCERG1 and the sumoylation-impaired mutants in an independent system that does not imply recruitment through GAL4-DBD. Toward this end, we took advantage of a system to study TCERG1-activated NF-κB-dependent transcription, originally developed by the Garcia-Blanco laboratory (Duke University), that uses a luciferase-reporter construct consisting of a minimal murine c-fos promoter and three upstream copies of the major histocompatibility complex class I NF-κB-binding site termed p3X-κB-L (Fig. 5D), previously described by Mitchell and Sugden (58). Transient overexpression of wild-type TCERG1 in HEK293T cells stimulated the expression from p3X-κB-L reporter plasmid in a dose-dependent manner (Fig. 5D). We next measured the transcriptional response to the mutant versions of TCERG1. Both the K503R and K608R mutants exhibited greater transactivation activity than wild-type TCERG1 (Fig. 5E). This enhancement was not the result of differential TCERG1 expression, as confirmed by Western blotting (Fig. 5E). The induction level of p3X-κB-L reporter plasmid by the double mutant (K503R,K608R) was similar to that of the single mutants (Fig. 5E). Together, these findings support our previous data and suggest that the SUMO modification sites negatively modulate TCERG1 transcriptional activity.
DISCUSSION
In this study, we report sumoylation as a novel regulator mechanism of TCERG1 function. We demonstrate that TCERG1 is a substrate for SUMO-1 and SUMO-2/3 conjugation. We observed that sumoylation occurs within the amino-terminal region of TCERG1 (Fig. 1). Furthermore, the amino-terminal region interacts directly with the SUMO E2-conjugating enzyme Ubc9 (Fig. 1F). No additional active sumoylation sites were detected within the carboxyl-terminal moiety of TCERG1 in vivo; however, we cannot rule out other sites. In fact, several strong putative SUMO acceptor sites are predicted in the carboxyl-terminal region of the protein (data not shown). These might reflect on other SUMO conjugation pathways regulated in a cell- or environment-specific manner. Consistent with our results, TCERG1 has two motifs that conform to the extended consensus sequence recognized by Ubc9 and are located at the amino-terminal region. Here, we provide in vivo and in vitro evidence that human TCERG1 is modified by SUMO-1 and SUMO-2/3 on lysine residues 503 and 608 (Fig. 2) following specific patterns and displaying distinct paralog specificities, which may reflect an additional functional specialization of each acceptor site. In relation to this, we have detected a shift from SUMO-1 to SUMO-2/3 conjugation upon heat shock treatment (data not shown). Interestingly, a very recent report describing the heat shock-induced global changes in the sumoylation state of the proteome identified TCERG1 as a target for SUMO-2 modification (59). To note, proteins involved in mRNA transcription and RNA-binding proteins were significantly over-represented in the SUMO-2-modified proteome (59). In this context, it will be of interest to investigate TCERG1 effects on splicing upon cellular stress and whether it depends on TCERG1 sumoylation.
The algorithms that we used to locate putative SUMO acceptor residues predicted other potential sumoylation sites in the amino-terminal region of TCERG1. Among those, a lysine at position 478 significantly scores with the sumoylation consensus sequence. We carried out in vivo and in vitro sumoylation experiments with TCERG1 constructs containing lysine to arginine mutation at position 478, and we found no evidence of SUMO modification on this particular amino acid residue (data not shown).
Having identified sumoylation sites in TCERG1, we investigated its effect on the protein. One documented function of sumoylation is to control targeting of proteins to different cell compartments. We described previously that TCERG1 accumulates in the splicing factor-rich nuclear speckles and that the FF-domains, which are located at the carboxyl-terminal region of the protein, are required for this localization (25). We investigated whether sumoylation of TCERG1 effects its spatial distribution in the nuclei. Our results show that mutant forms of TCERG1 that are not sumoylated localize to the nucleus similarly to the wild-type protein (Fig. 3). Thus, it is unlikely that sumoylation of TCERG1 is required for the spatial distribution of this protein in the cell.
TCERG1 modulates alternative pre-mRNA splicing (Fig. 4) (30, 33) and transcription (Fig. 5) (23), and a role for this protein in coordinating transcription elongation and pre-mRNA processing has been proposed (25, 56). We show here that TCERG1 specifically influences fibronectin EDI splicing independently of the promoter used and without altering the overall levels of full-length transcripts (Fig. 4), which might argue against a coupling mechanism to alter splicing decisions at the EDI exon. Very recently, studies have also reported observations that demonstrate that these two functions can occur independently of each other for regulators that function as transcriptional coactivators that can also regulate alternative splicing (60, 61). SUMO addition could influence these processes. Recently, several heterogeneous nuclear ribonucleoproteins were found in proteomic studies to be modified by SUMO (62) suggesting a role of sumoylation in regulating mRNA metabolism and perhaps splicing regulation. In support of this hypothesis, the list of spliceosome components that are substrates for SUMO conjugation has been recently extended (35). Moreover, sumoylation alters ADAR1 editing activity and the assembly and activity of the pre-mRNA 3′-processing complex (41, 63) emphasizing the importance of this post-translational modification in RNA-processing events. However, no differences were found between wild-type TCERG1 and the mutant forms that are not modified by SUMO in the alternative processing of the fibronectin EDI exon (Fig. 4). Thus, we conclude that post-translational modification of TCERG1 at lysines 503 and 608 by SUMO may not be essential for TCERG1 in mediating these specific alternative splicing decisions, although we cannot rule out potential effects on other mRNAs. In our experiments, TCERG1 did not affect the EDI− band (Fig. 4, B–E). Although this finding remains to be fully investigated, this observation might point toward a role of TCERG1 in the modulation of the spliceosome assembly after exon definition, a poorly understood mechanism for alternative splicing regulation that has been observed recently (64) and that could explain a previously reported intron-retention event with one of the endogenous targets of TCERG1 (33).
We have also taken several approaches to define the consequences of TCERG1 sumoylation for transcription. TCERG1 can activate transcription in cell assays by a mechanism not fully understood but that may include elements that regulate transcriptional control such as P-TEFb (43, 65, 66). In fact, TCERG1 interacts with protein complexes that are essential for establishing a processive transcription (22, 25). Mutating the SUMO acceptor sites enhanced the transactivation capacity of TCERG1. In other examples, sumoylation negatively impacts transcription factor activity (67, 68) by multiple mechanisms (38). At least two repressive mechanisms are consistent with low steady-state levels of transcription factor sumoylation (69). First, sumoylation of the transcription factor can lead to the recruitment of repressive factors with chromatin remodeling activity. If this occurs, a repressive chromatin state will remain even after sumoylation of the transcription factor is lost. Alternatively, sumoylation of a transcription factor can be necessary to initiate the recruitment of inhibitory factors to the promoter. After a stable repressor complex has formed, this complex will remain stable even after SUMO is removed from the initial transcription factor. In both cases, the transcription factor only needs to be sumoylated for a short period of time, during which gene silencing is initiated. Although the precise consequence of sumoylation on TCERG1 function remains to be determined, we speculate that this modification might alter or disrupt protein interactions. Sumoylation may alter TCERG1 association with transcriptional complexes, including RNA polymerase and elongation factors. In fact, structural elements adjacent to the SUMO acceptor motifs participate in interactions with elongation factors and play a role in the transcriptional activity of TCERG1 (25, 29). It is tempting to speculate that SUMO modification might serve as a molecular switch to rearrange transcription processing complexes. Future work should document that SUMO modification affects the transcript level of an endogenous gene whose expression is directly affected by TCERG1 and clarify the precise role of TCERG1 sumoylation in the coupling between transcription and alternative splicing, the regulation of those processes, and the molecular mechanisms that are involved.
There are indications that TCERG1 is involved in the pathogenesis of Huntington disease (70, 71). Given the recent findings implicating sumoylation as an important regulator of aggregation or toxicity of polyglutamine expanded proteins (72, 73), it is tempting to speculate that TCERG1 sumoylation may influence its role in this disease.
Other proteins related to TCERG1 by virtue of their domain architecture (specifically the WW- and FF-domain-containing proteins FBP11, HYP, and FBP21) also possess predicted sumoylation target sites. These putative sumoylation sites match the expanded consensus motif for sumoylation NDSM and are structurally located in a similar position as Lys-503 and Lys-608 in TCERG1 (data not shown). Whether those proteins, which like TCERG1 are involved in transcription and mRNA processing, are regulated by sumoylation remains to be determined.
Acknowledgments
We thank F. E. Baralle, M. A. Garcia-Blanco, R. T. Hay, A. R. Kornblihtt, M. Lafarga, and T. Misteli for providing reagents; B. Del Blanco for helpful advice with the RT-PCR quantitative analysis; I. Montanuy for useful insights and discussion; and A. Goldstrohm and M. A. Garcia-Blanco for critical reading of the manuscript.
This work was supported in part by Spanish Ministry of Science and Innovation Grant BFU2008-01599, Fundación para la Investigación y la Prevención del SIDA en España Grant 36768 (to C. S.), Spanish Ministry of Science and Innovation Grant BFU2008-01651, and Fundación de Investigación Médica Mútua Madrileña (to C. H.-M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- CTD
- carboxyl-terminal domain
- RNAPII
- RNA polymerase II
- SUMO
- small ubiquitin-like modifier
- EDI
- fibronectin extra domain I
- DTT
- dithiothreitol
- PMSF
- phenylmethylsulfonyl fluoride
- HA
- hemagglutinin
- NEM
- N-ethylmaleimide
- WCE
- whole-cell lysate
- HSV
- herpes simplex virus
- TK
- thymidine kinase
- RT
- reverse transcription
- ECFP
- enhanced cyan fluorescent protein
- GST
- glutathione S-transferase.
REFERENCES
- 1.Kornblihtt A. R., de la Mata M., Fededa J. P., Munoz M. J., Nogues G. (2004) RNA 10, 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Proudfoot N. J., Furger A., Dye M. J. (2002) Cell 108, 501–512 [DOI] [PubMed] [Google Scholar]
- 3.Auboeuf D., Hönig A., Berget S. M., O'Malley B. W. (2002) Science 298, 416–419 [DOI] [PubMed] [Google Scholar]
- 4.Nogues G., Kadener S., Cramer P., Bentley D., Kornblihtt A. R. (2002) J. Biol. Chem. 277, 43110–43114 [DOI] [PubMed] [Google Scholar]
- 5.Kornblihtt A. R. (2005) Curr. Opin. Cell Biol. 17, 262–268 [DOI] [PubMed] [Google Scholar]
- 6.Batsché E., Yaniv M., Muchardt C. (2006) Nat. Struct. Mol. Biol. 13, 22–29 [DOI] [PubMed] [Google Scholar]
- 7.Orphanides G., Reinberg D. (2002) Cell 108, 439–451 [DOI] [PubMed] [Google Scholar]
- 8.Hirose Y., Manley J. L. (2000) Genes Dev. 14, 1415–1429 [PubMed] [Google Scholar]
- 9.Phatnani H. P., Greenleaf A. L. (2006) Genes Dev. 20, 2922–2936 [DOI] [PubMed] [Google Scholar]
- 10.Bourquin J. P., Stagljar I., Meier P., Moosmann P., Silke J., Baechi T., Georgiev O., Schaffner W. (1997) Nucleic Acids Res. 25, 2055–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carty S. M., Greenleaf A. L. (2002) Mol. Cell. Proteomics 1, 598–610 [DOI] [PubMed] [Google Scholar]
- 12.Kim E., Du L., Bregman D. B., Warren S. L. (1997) J. Cell Biol. 136, 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris D. P., Greenleaf A. L. (2000) J. Biol. Chem. 275, 39935–39943 [DOI] [PubMed] [Google Scholar]
- 14.Yuryev A., Patturajan M., Litingtung Y., Joshi R. V., Gentile C., Gebara M., Corden J. L. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6975–6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misteli T., Spector D. L. (1999) Mol. Cell 3, 697–705 [DOI] [PubMed] [Google Scholar]
- 16.Emili A., Shales M., McCracken S., Xie W., Tucker P. W., Kobayashi R., Blencowe B. J., Ingles C. J. (2002) RNA 8, 1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kameoka S., Duque P., Konarska M. M. (2004) EMBO J. 23, 1782–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fong Y. W., Zhou Q. (2001) Nature 414, 929–933 [DOI] [PubMed] [Google Scholar]
- 19.Kao H. Y., Siliciano P. G. (1996) Mol. Cell. Biol. 16, 960–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bedford M. T., Reed R., Leder P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10602–10607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen M., Friedler A., Schon O., Bycroft M. (2002) J. Mol. Biol. 323, 411–416 [DOI] [PubMed] [Google Scholar]
- 22.Suñé C., Hayashi T., Liu Y., Lane W. S., Young R. A., Garcia-Blanco M. A. (1997) Mol. Cell. Biol. 17, 6029–6039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suñé C., Garcia-Blanco M. A. (1999) Mol. Cell. Biol. 19, 4719–4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carty S. M., Goldstrohm A. C., Suñé C., Garcia-Blanco M. A., Greenleaf A. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9015–9020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sánchez-Alvarez M., Goldstrohm A. C., Garcia-Blanco M. A., Suñé C. (2006) Mol. Cell. Biol. 26, 4998–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neubauer G., King A., Rappsilber J., Calvio C., Watson M., Ajuh P., Sleeman J., Lamond A., Mann M. (1998) Nat. Genet. 20, 46–50 [DOI] [PubMed] [Google Scholar]
- 27.Deckert J., Hartmuth K., Boehringer D., Behzadnia N., Will C. L., Kastner B., Stark H., Urlaub H., Lührmann R. (2006) Mol. Cell. Biol. 26, 5528–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makarov E. M., Makarova O. V., Urlaub H., Gentzel M., Will C. L., Wilm M., Lührmann R. (2002) Science 298, 2205–2208 [DOI] [PubMed] [Google Scholar]
- 29.Goldstrohm A. C., Albrecht T. R., Suñé C., Bedford M. T., Garcia-Blanco M. A. (2001) Mol. Cell. Biol. 21, 7617–7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin K. T., Lu R. M., Tarn W. Y. (2004) Mol. Cell. Biol. 24, 9176–9185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith M. J., Kulkarni S., Pawson T. (2004) Mol. Cell. Biol. 24, 9274–9285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng D., Côté J., Shaaban S., Bedford M. T. (2007) Mol. Cell 25, 71–83 [DOI] [PubMed] [Google Scholar]
- 33.Pearson J. L., Robinson T. J., Muñoz M. J., Kornblihtt A. R., Garcia-Blanco M. A. (2008) J. Biol. Chem. 283, 7949–7961 [DOI] [PubMed] [Google Scholar]
- 34.Shimada M., Saito M., Katakai T., Shimizu A., Ichimura T., Omata S., Horigome T. (1999) J. Biochem. 126, 1033–1042 [DOI] [PubMed] [Google Scholar]
- 35.Vertegaal A. C., Andersen J. S., Ogg S. C., Hay R. T., Mann M., Lamond A. I. (2006) Mol. Cell. Proteomics 5, 2298–2310 [DOI] [PubMed] [Google Scholar]
- 36.Evdokimov E., Sharma P., Lockett S. J., Lualdi M., Kuehn M. R. (2008) J. Cell Sci. 121, 4106–4113 [DOI] [PubMed] [Google Scholar]
- 37.Hay R. T. (2005) Mol. Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 38.Gill G. (2005) Curr. Opin. Genet. Dev. 15, 536–541 [DOI] [PubMed] [Google Scholar]
- 39.Yang S. H., Sharrocks A. D. (2004) Mol. Cell 13, 611–617 [DOI] [PubMed] [Google Scholar]
- 40.Boyer-Guittaut M., Birsoy K., Potel C., Elliott G., Jaffray E., Desterro J. M., Hay R. T., Oelgeschläger T. (2005) J. Biol. Chem. 280, 9937–9945 [DOI] [PubMed] [Google Scholar]
- 41.Vethantham V., Rao N., Manley J. L. (2007) Mol. Cell. Biol. 27, 8848–8858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizushima S., Nagata S. (1990) Nucleic Acids Res. 18, 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montanuy I., Torremocha R., Hernández-Munain C., Suñé C. (2008) J. Biol. Chem. 283, 7368–7378 [DOI] [PubMed] [Google Scholar]
- 44.Cramer P., Pesce C. G., Baralle F. E., Kornblihtt A. R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11456–11460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kadener S., Cramer P., Nogués G., Cazalla D., de la Mata M., Fededa J. P., Werbajh S. E., Srebrow A., Kornblihtt A. R. (2001) EMBO J. 20, 5759–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grover A., Houlden H., Baker M., Adamson J., Lewis J., Prihar G., Pickering-Brown S., Duff K., Hutton M. (1999) J. Biol. Chem. 274, 15134–15143 [DOI] [PubMed] [Google Scholar]
- 47.Mabon S. A., Misteli T. (2005) PLoS. Biol. 3, e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harlow E., Lane D. (1999) Using Antibodies: A Laboratory Manual, pp. 521–523, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 49.Kalbfuss B., Mabon S. A., Misteli T. (2001) J. Biol. Chem. 276, 42986–42993 [DOI] [PubMed] [Google Scholar]
- 50.de la Mata M., Alonso C. R., Kadener S., Fededa J. P., Blaustein M., Pelisch F., Cramer P., Bentley D., Kornblihtt A. R. (2003) Mol. Cell 12, 525–532 [DOI] [PubMed] [Google Scholar]
- 51.de la Mata M., Kornblihtt A. R. (2006) Nat. Struct. Mol. Biol. 13, 973–980 [DOI] [PubMed] [Google Scholar]
- 52.Smith D. B., Johnson K. S. (1988) Gene 67, 31–40 [DOI] [PubMed] [Google Scholar]
- 53.Yan J., Yang X. P., Kim Y. S., Joo J. H., Jetten A. M. (2007) Biochem. Biophys. Res. Commun. 362, 132–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang S. H., Galanis A., Witty J., Sharrocks A. D. (2006) EMBO J. 25, 5083–5093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hutton M., Lendon C. L., Rizzu P., Baker M., Froelich S., Houlden H., Pickering-Brown S., Chakraverty S., Isaacs A., Grover A., Hackett J., Adamson J., Lincoln S., Dickson D., Davies P., Petersen R. C., Stevens M., de Graaff E., Wauters E., van Baren J., Hillebrand M., Joosse M., Kwon J. M., Nowotny P., Che L. K., Norton J., Morris J. C., Reed L. A., Trojanowski J., Basun H., Lannfelt L., Neystat M., Fahn S., Dark F., Tannenberg T., Dodd P. R., Hayward N., Kwok J. B., Schofield P. R., Andreadis A., Snowden J., Craufurd D., Neary D., Owen F., Oostra B. A., Hardy J., Goate A., van Swieten J., Mann D., Lynch T., Heutink P. (1998) Nature 393, 702–705 [DOI] [PubMed] [Google Scholar]
- 56.Goldstrohm A. C., Greenleaf A. L., Garcia-Blanco M. A. (2001) Gene 277, 31–47 [DOI] [PubMed] [Google Scholar]
- 57.Holmstrom S., Van Antwerp M. E., Iñiguez-Lluhi J. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15758–15763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitchell T., Sugden B. (1995) J. Virol. 69, 2968–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Golebiowski F., Matic I., Tatham M. H., Cole C., Yin Y., Nakamura A., Cox J., Barton G. J., Mann M., Hay R. T. (2009) Sci. Signal. 2, ra24. [DOI] [PubMed] [Google Scholar]
- 60.Harrison M. J., Tang Y. H., Dowhan D. H. (2010) Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amelio A. L., Caputi M., Conkright M. D. (2009) EMBO J. 28, 2733–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li T., Evdokimov E., Shen R. F., Chao C. C., Tekle E., Wang T., Stadtman E. R., Yang D. C., Chock P. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8551–8556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Desterro J. M., Keegan L. P., Jaffray E., Hay R. T., O'Connell M. A., Carmo-Fonseca M. (2005) Mol. Biol. Cell 16, 5115–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.House A. E., Lynch K. W. (2006) Nat. Struct. Mol. Biol. 13, 937–944 [DOI] [PubMed] [Google Scholar]
- 65.Barboric M., Nissen R. M., Kanazawa S., Jabrane-Ferrat N., Peterlin B. M. (2001) Mol. Cell 8, 327–337 [DOI] [PubMed] [Google Scholar]
- 66.Nojima M., Huang Y., Tyagi M., Kao H. Y., Fujinaga K. (2008) J. Mol. Biol. 382, 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Girdwood D., Bumpass D., Vaughan O. A., Thain A., Anderson L. A., Snowden A. W., Garcia-Wilson E., Perkins N. D., Hay R. T. (2003) Mol. Cell 11, 1043–1054 [DOI] [PubMed] [Google Scholar]
- 68.Fernández-Lloris R., Osses N., Jaffray E., Shen L. N., Vaughan O. A., Girwood D., Bartrons R., Rosa J. L., Hay R. T., Ventura F. (2006) FEBS Lett. 580, 1215–1221 [DOI] [PubMed] [Google Scholar]
- 69.Geiss-Friedlander R., Melchior F. (2007) Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 70.Arango M., Holbert S., Zala D., Brouillet E., Pearson J., Régulier E., Thakur A. K., Aebischer P., Wetzel R., Déglon N., Néri C. (2006) J. Neurosci. 26, 4649–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andresen J. M., Gayán J., Cherny S. S., Brocklebank D., Alkorta-Aranburu G., Addis E. A., Cardon L. R., Housman D. E., Wexler N. S. (2007) J. Med. Genet. 44, 44–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mukherjee S., Thomas M., Dadgar N., Lieberman A. P., Iñiguez-Lluhí J. A. (2009) J. Biol. Chem. 284, 21296–21306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Subramaniam S., Sixt K. M., Barrow R., Snyder S. H. (2009) Science 324, 1327–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]