Abstract
RNA editing by adenosine deamination fuels the generation of RNA and protein diversity in eukaryotes, particularly in higher organisms. This includes the recoding of translated exons, widespread editing of retrotransposon-derived repeat elements and sequence modification of miRNA transcripts. Such changes can bring about specific amino acid substitutions, alternative splicing and changes in gene expression levels. Although the overall prevalence of A-to-I editing and its specific functional impact on many of the affected genes are not yet known, the importance of balancing RNA modification levels across time and space is becoming increasingly evident. In particular, transcriptome instabilities in form of too much or too little RNA editing activity, or misguided editing manifest in several human disease phenotypes which disrupt that balance.
Transcript and protein diversity through RNA editing
RNA editing is broadly defined as post-transcriptional alteration of RNA sequences through the insertion, deletion, or modification of nucleotides but not including RNA processing events such as splicing, polyadenylation or degradation of RNA molecules 1. Of the various types of RNA editing (Box 1), adenosine-to-inosine (A-to-I) base modification is the most widespread in higher eukaryotes (for a comprehensive review see 2). Furthermore, both the complexity of the molecular machinery that mediates A-to-I editing and the number of editing targets seem to increase from lower to higher organisms 2–4.
Box 1. Types of RNA editing.
The term “RNA editing” was initially introduced over 20 years ago, after the discovery of mitochondrial mRNA modification in kinetoplastide Protozoa. The different types of editing distinguished today differ substantially in their molecular mechanisms, machineries and species distributions (for review see 1).
Insertion and deletion: Affects most mitochondrial transcripts in kinetoplastids and involves the addition and deletion of non-genomically encoded uridine residues in pre-mRNA transcripts. The required information for site selection and editing extent is provided by short guide RNAs (gRNAs) which are complementary to the fully edited mRNA, and is further mediated by multiprotein complexes. Another type of insertional editing is observed in mitochondria of the slime mold Physarum polycephalum. The majority of the editing events observed in this organelle involve co-transcriptional insertion of cytosines 1.
Substitution: Occurs in both pre-mRNAs and tRNAs. Apart from A-to-I modifications, cytosine deamination is a form of RNA editing also found in mammalian nuclear genes although only a few physiological targets of the C-to-U RNA editing machinery are known. A well characterized C-to-U editing target is human apolipoprotein B (APOB100), which is essential for the removal of low-density lipoproteins (LDL). Tissue-specific APOB100 deamination introduces an in-frame stop codon, generating a truncated protein (ApoB48) with altered physiological functions 1. Intriguingly, the enzymatic component of the C-to-U editing activity is the cytidine deaminase APOBEC1 (APOB mRNA-editing enzyme catalytic polypeptide 1), which is related to the APOBEC 2/3 family of DNA-specific modification enzymes active in retroviral restriction 84. C-to-U base conversion in RNA is more common in plant mitochondria 1.
tRNA editing: Adenosine deamination in tRNAs is found across organisms from prokaryotes to mammals and includes the generation of the wobble base in the tRNA anticodon. A-to-I modification is accomplished by ADATs (Adenosine Deaminase Acting on tRNAs), a family of deaminases that share sequence similarity with the catalytic deaminase domain of ADARs but lack double stranded RNA-binding domains (for review see 95).
Other types of substitution: G-to-A, U-to-C or other conversions are occasionally reported 2. Those types of changes would often require the cleavage and re-ligation of the RNA molecules and to date, neither the molecular mechanism(s) nor the involved enzymes are known.
The diversity generated by A-to-I editing affects gene expression at several levels and targets different types of transcripts. Here we review the emerging insights on molecular diversity generated through RNA editing and the implications of tipping the complex balance of editing patterns in experimental models and in human disease.
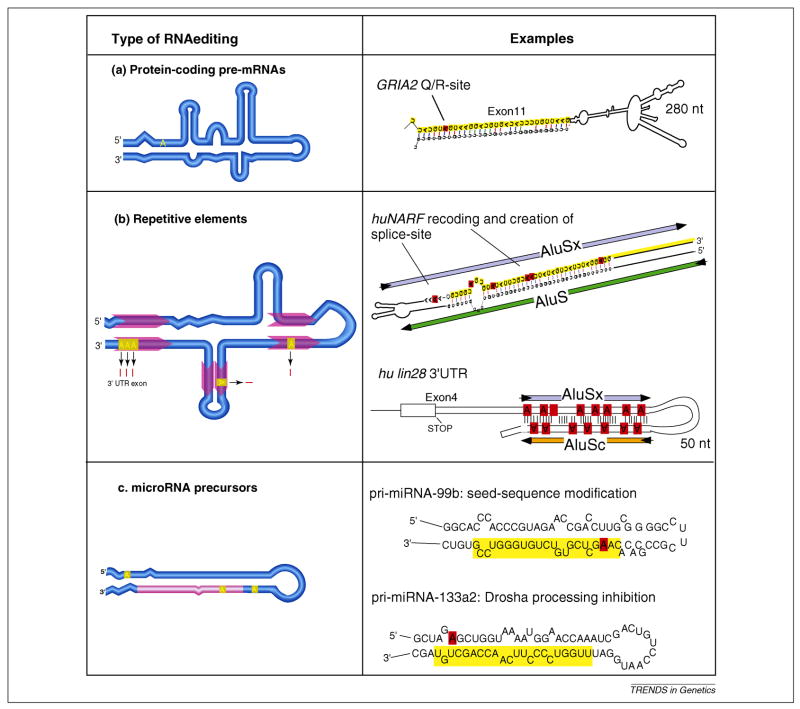
The three major sequence classes undergoing A-to-I editing are protein-coding exons in pre-mRNAs, repetitive sequence elements in untranslated exons and introns and microRNA (miRNA) precursor transcripts (Figure 1). A key distinguishing feature among the three kinds of targets is that the type of RNA secondary structure formed influences how the RNA editing machinery interacts and modifies them. Whereas RNA folds involving repetitive sequence elements are characterized by extended, almost perfectly base-paired duplex structures that undergo heavy and multiple site editing, the secondary structures that lead to miRNA editing consist of short RNA hairpins with small bulges and loops -- a hallmark feature of miRNA precursors. RNA editing events in pre-mRNAs that do not involve repetitive elements are mediated by composite secondary structures with multiple small base-pairing segments separated by bulges and loops. These types of structures often give rise to highly site-selective and high efficiency base modification by adenosine deaminases acting on RNA (ADARs).
Figure 1.
The three major types of A-to-I RNA editing targets and their fates. Panels on the left show schematic of RNA secondary structures highlighting translated exon sequence (dark blue box), untranslated exon sequence (light blue boxes) location of repetitive sequence elements (red arrows), non-coding and intronic RNA sequence (light blue lines) and location of mature miRNA sequence (light red line). (a) Pre-mRNA editing of protein-coding genes with composite RNA secondary structure leads to highly site-selective recoding if it affects a non-synonymous codon site. For example, the glutamate receptor subunit GRIA2 exon 11 Q/R site 2 forms an experimentally validated secondary RNA structure between exon 11 (marked in yellow) and intron 11. (b) Pairs of repetitive elements, such as primate Alus located in coding or non-coding exons or introns can generate RNA secondary structures targeted by the RNA editing machinery. For example, editing of the intramolecular RNA fold between two Alu elements in human nuclear prelamin A recognition factor (NARF), causes recoding within the Alu-exon (marked in yellow) and leads to the creation of the 3′-splice consensus site upstream of the Alu-exon, thereby regulating alternative splicing of this exon. In the case of human lin28, extensive RNA editing within its non-coding, 3′-untranslated region mediated by a pair of Alu-elements, leads to the nuclear retention of the mRNA. (c) The characteristic secondary structure of pre-miRNAs is a frequent target of ADARs. For example, pri-miRNA-99b editing alters a nucleotide within the seed of the mature miRNA (marked in yellow and edited position highlighted in red) and therefore has the potential to alter the target interaction profile of this miRNA 26, whereas the modification of an adenosine outside of the mature miRNA region in pri-miRNA-133a2 causes a change in the processing rate by the RNAse Drosha 26.
Target substrates, functions and fates
Editing within pre-mRNAs can generate or destroy splice sites, regulate alternative splicing events and influence the dynamics of constitutive splice sites 2. Of particular interest are instances in which A-to-I editing within protein-coding exons results in a non-synonymous codon change (reviewed in 5). Usually, the protein sequence of a gene product can be faithfully deduced from the nucleotide sequence of the translated exons. However, this is not the case if the gene is subject to A-to-I RNA editing, because a fraction of the primary transcripts undergo a recoding event. Since inosine (Box 2) is interpreted as a guanosine by the translational machinery, RNA editing may change the meaning of codons. As a result, a fraction of the protein output will carry a single amino acid substitution compared to the non-edited version. Until recently, only a small number of proteins with amino acid substitutions caused by editing were known, most of them identified by chance. Recent studies, facilitated by bioinformatics and deep sequencing approaches, support the notion that hundreds of genes undergo recoding editing resulting in amino acid substitutions 6, 7. However, it appears that many of the recoding events identified more recently display low level modification rates and that despite considerable high-throughput sequencing, relatively few novel sites become validated 6, 7. Thus, because RNA editing is fractional and might be restricted in time and/or space, the comprehensive mapping of all recoding editing sites within the human transcriptome will require the combination of bioinformatics-based editing site prediction with deep sequencing and/or targeted specimen analysis.
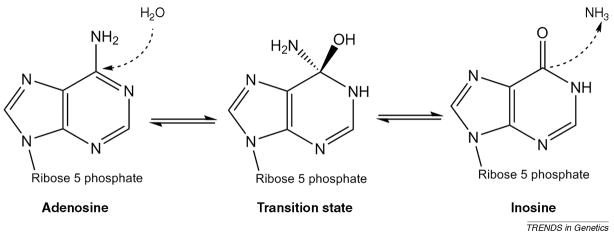
Box 2. A-to-I editing: chemical mechanism and machinery.
A-to-I RNA editing by ADARs proceeds via a hydrolytic deamination mechanism without the requirement for RNA backbone breaks (see proposed mechanism in Figure I). Only adenosines within the context of RNA molecules are targeted by ADARs. Inosine largely behaves like a guanosine in RNA folding and is also interpreted as G by the translation machinery. Editing occurs within sections of RNA that are completely or partially double stranded and does not require any essential co-factors 2, 3.
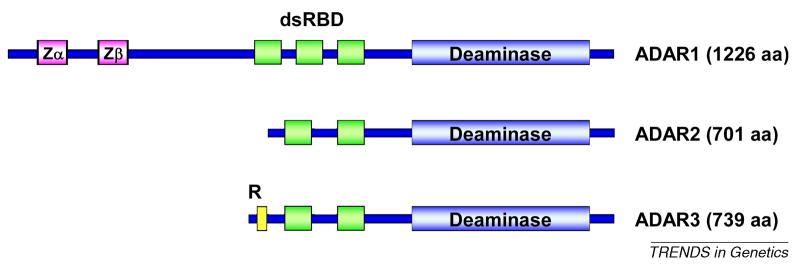
ADAR proteins have been characterized from different organisms, including worms, insects and vertebrates 2, 3. They share a common domain structure with 2 or 3 double-stranded RNA binding domains (dsRBD) and a C-terminal catalytic deaminase domain. The C-terminal deaminase domain, which is highly conserved between ADARs, coordinates a Zn2+-ion in its catalytic center and the functional deaminase fold requires the incorporation of an inositol hexaphosphate IP6 molecule 96.
Three ADARs (ADAR1, ADAR2 and ADAR3) have been identified in humans (Figure II). ADAR1 protein is the largest of the three family members and is expressed in two major splice variants, ADAR1-p150 and ADAR1-p110. ADAR1-p150 contains an extended N-terminus including two Z-DNA/RNA binding motif (Zα and Zβ) 2, 3. ADAR2 and ADAR3 share high sequence similarity (50% protein sequence identity), but to date no catalytic activity has been documented for ADAR3. The ADAR3 R-domain, a 13–15 amino acid, arginine-rich sequence motif, mediates ssRNA binding 2, 3 and serves as a nuclear localization signal 31. ADAR2 can also be expressed to include a N-terminal R-domain 97, which can exhibit a different cellular localization from the major ADAR2 splice form. ADARs undergo both homo- and heterodimerization and are likely catalytically active as dimers 37.
Figure I.
Mechanism of adenosine deamination
Figure II.
ADAR domain structure
For most cases of recoding editing characterized in mammals, subtle to dramatic changes in protein function result from single amino acid differences. Intriguingly, many of the modified codons specify highly conserved residues that might otherwise be excluded from variation through genomic mutations by purifying selection 4, 8. A-to-I RNA editing might therefore play an evolutionary role allowing for the exploration of sequence space at a small, tolerable rate 4, 8.
The highest level of transcript diversity caused by editing is generated within transposon-derived repeat sequences. Especially in primate genomes, the prevalence and genetic properties of Alu-type repeat elements make pairs of Alu elements within primary transcripts the most prominent editing targets. Tens of thousands of individual editing sites in thousands of mRNAs with Alu-elements have been mapped within the human transcriptome 9–12 and deep sequencing analysis further indicates that many more editing events exist in Alu elements already known to harbor editing sites 13. The properties of the intrinsically promiscuous RNA editing machinery paired with the characteristic Alu-pair RNA secondary structure induces highly efficient multiple-site editing in Alu elements. In fact, predictions based on the existing data posit that more than 98% of all pre-mRNAs are subject to Alu-mediated RNA editing 9.
What could be the functional impact of the abundant editing of repetitive sequences, especially primate-specific Alu elements? Most Alu repeats (as well as other types of repeat elements) are located in introns and non-translated exons; in these cases, editing will not directly influence protein function. Still, editing changes within those sequences have the potential to indirectly alter protein expression or function. For example, sometimes editing can induce alternative pre-mRNA splicing through the creation of a cryptic splice donor (AT to IT) or acceptor (AA to AI) site 9, 14, modulate alternative splicing efficiency through modification of splicing enhancer or inhibitor sequences, or eliminate a consensus splice acceptor site (AG to IG). Alternatively, editing can modulate other types of functional RNA elements, such as miRNA binding sites in mRNAs 15. However, most of the time, the outcome of Alu-editing is simply an RNA with multiple inosines present within one or more regions of the pre-mRNA or within the untranslated regions of a spliced mRNA. It seems that there is not a single mechanism, but rather several outcomes, for the fate of such an Alu-edited RNA. On the one hand, experimental evidence shows that Alu-edited RNAs often become sequestered in the nucleus by a protein complex with specific affinity for inosine in RNA molecules 16, 17. On the other hand, these and other Alu-edited transcripts sometimes get exported and associate with polysomes despite being edited 18, 19. So far it is unknown what might regulate such distinct behaviors. In one specific example of a heavily edited 3′-UTR (untranslated region) of the mouse cationic amino acid transporter (Cat2) gene, nuclear retained transcripts become mobilized for export and translation following cellular stress through cleavage of the inosine-containing 3′-UTR from the rest of the mRNA 20. Furthermore, a more general switch activating the retention of inosine-containing RNAs in the nucleus of human cells is provided by the induction of the non-coding RNA nuclear paraspeckle assembly transcript 1 (NEAT1) 19. Human embryonic stem cells do not express NEAT1 and export heavily edited RNAs. By contrast, differentiation induces NEAT1 expression leading to the formation of nuclear structures called paraspeckles, which not only co-localize with the proteins known to bind inosine-containing RNAs, but also prevent the export of heavily edited Alu-containing transcripts 19. Despite these intriguing examples for the regulation of gene expression involving edited Alu-repeats, it will be necessary to elucidate the molecular mechanisms that lead to the nuclear binding, storage, degradation or release of these RNAs to understand the bigger picture of why and when a particular transcript that undergoes Alu-mediated editing enters a specific pathway.
The site-selective modification of miRNA precursor molecules represents another frequent event of A-to-I RNA editing. miRNAs are small, regulatory RNA molecules with diverse roles in development, differentiation and cell cycle regulation 21. Each of the small RNAs is excised from longer, hairpin-structured precursors through the sequential action of the RNAses Drosha and Dicer. Following the initial reports of miRNA sequence editing 22, 23, additional miRNA precursors were subsequently shown to undergo editing and current estimates posit that ~16% of all human miRNA genes are subject to A-to-I modification. The editing of nucleotides in the vicinity of Dicer or Drosha processing sites can prevent further maturation and expression of the miRNA 2, 24, 25. Intriguingly, if A-to-I editing modifies a nucleotide within the miRNA seed sequence that is critical for target recognition, then the edited mature miRNA may exhibit a distinct target profile from the non-edited variant. This is the case for human miR-376 and possibly for four other miRNAs 26, 27. Yet, the predominant outcome of pre-miRNA editing is the modulation of miRNA biogenesis through inhibition of Drosha- or Dicer-mediated cleavage 26. miRNA function could also be influenced through the editing of miRNA binding sites on their target sequences 28. Although this aspect has not been fully explored, human miR-513 and miR-769-3p/−450b-3p provide examples in which A-to-I editing in the target mRNA generates a consensus target sequence 15.
Maintaining the balance: Regulation of RNA editing
Editing of recoding targets is under tight control, and the deregulation of RNA editing in space and/or time is correlated with various human disease phenotypes. The specific molecular mechanisms that govern intracellular RNA editing levels are largely unknown. For example, although ADAR1 and ADAR2 expression is, in principle, ubiquitous, the presence of ADAR mRNA (or even proteins) often does not correlate with the observed intracellular RNA editing activity (reviewed in 2, 3, 29). However, recent insights regarding the developmental as well as cell-type specific modulation of RNA editing in conjunction with ADAR expression and localization studies reveal multiple and complex patterns of regulation on the transcriptional, post-transcriptional, translational, and post-translational levels. For example, the ADAR proteins are expressed in several alternative splice forms that differ with respect to their intracellular localization, enzymatic activity and/or target specificity 2, 3, 29.
Editing of pre-mRNAs often is restricted to the nucleus, in particular for editing events that affect intronic sequences or that are mediated through RNA folds involving intronic regions (such as many of the known recoding cases of editing). Most ADAR proteins localize to the nucleus, with the exception of the ADAR1 p150 variant, which is shuttled between the nucleus and cytoplasm, and might perform specific editing or other functions in the cytosol. The p150 isoform of ADAR1 is expressed from an interferon-induced promoter and carries a unique N-terminal DNA-binding domain 30. In contrast, the nuclear ADAR1 p110 variant as well as the editing enzyme ADAR2 is expressed constitutively. Nuclear RNA editing activity may be regulated through controlled nuclear import of ADAR proteins. This notion is supported by the observed differential interaction of the nuclear import machinery with individual ADARs 31. Furthermore, dsRNA binding co-regulates transportin-1 mediated nuclear import of ADAR1 through the competition of dsRNA and transportin-1 to the ADAR1 dsRNA binding domains (dsRBDs) 32. Within the nucleus, ADARs are shuttled between the nucleoli and nucleoplasma – another potential mechanism for regulating nuclear editing activity 33, 34. Intriguingly, ADAR2 editing activity is further balanced through a feed-back mechanism wherein increased functional ADAR2 expression leads to self-editing of ADAR2 pre-mRNA, which results in the production of inactive, truncated ADAR2 protein 35. Similarly, the single ADAR gene in Drosophila melanogaster is subject to self-editing; however, in this case the modification results in an amino acid substitution that substantially represses RNA editing activity 36.
Although ADAR1 and ADAR2 appear to be fully functional without the requirement of essential co-factors, homodimer (and potentially heterodimer) formation can modulate target specificity and activity 37. On that level even ADAR3 may modulate RNA editing activity through heterodimerization with ADAR1 or ADAR2 37. The ADAR3 protein shares high sequence similarity with ADAR2, but exhibits no detectable deamination activity 2, 3. Furthermore, the recent identification of additional ADAR interaction partners presents further opportunities for cell-type specific regulation of editing activity 31, 38, 39. Although post-translational modification of ADARs has been suggested as a regulatory mechanism, only ADAR1 sumoylation, which represses editing activity, has been documented to date 40.
The regulation of RNA editing extent and specificity also occurs on the level of individual target transcripts and involves competition between and co-regulation of pre-mRNA splicing and editing. In particular, if editing sites are positioned in close proximity to splice consensus sites, the strength of the splicing signal influences RNA editing extent nearby. Similarly, a strong RNA fold mediating editing might promote efficient splicing of only the edited transcript molecules 41–43. It is often difficult to predict the level of interdependence between editing and splicing, as other interactions of RNA-binding proteins (for example splicing enhancers or silencers) with the RNA target can impact its ability to be edited or spliced. ADARs have also been found to physically associate with the RNA polymerase II c-terminus. This co-localization further argues for a close coupling between transcription and editing 44.
For at least one specific case of recoding A-to-I RNA editing, the co-regulation of the target through small nucleolar (sno) RNA binding and modification modulates RNA editing activity. The snoRNA h/mbii-52, a component of the Prader-Willi syndrome (PWS) imprinting cluster, not only regulates alternative splicing of the serotonin receptor 2C through specific interaction with its pre-mRNA 45, but also inhibits the site-selective editing of this RNA and leads to the methylation of the adenosine that is also targeted for editing by ADAR2 46, 47.
Tipping the balance: Insights from genetics
In recent years, various animal models with hyper-, hypo-, or misediting have substantiated the general importance of editing for normal physiology and also revealed some intriguing connections to human disease phenotypes. In flies, which carry a single ADAR gene (dADAR), the genetic inactivation of A-to-I editing activity yields a strong neurological phenotype with locomoter deficiencies, seizures, premature neuro-degeneration, and altered reproductive behavior 48. The ability to both reproduce the phenotype through neuron-specific knock-down of dADAR in adult flies and partially rescue the knock-out phenotype in adults using ADAR transgenes suggests that the recoding of mostly neuronal targets in fully developed individuals is the primary function of dAdar 49. In rodents, the genetic inactivation of Adar1 or Adar2 also leads to severe phenotypes. Indeed, the mouse Adar1 knock-out is embryonic lethal around developmental day E12.5 50, 51. Although the molecular mechanism for this outcome is unknown, a failure of the hematopoetic system and widespread apoptosis is observed in Adar1−/− embryos. Intriguingly, adult-specific Adar1 inactivation demonstrates that ADAR1 is essential for the maintenance, but not for the establishment, of hematopoetic stem cells (HSC) and that the increased rate of cell death upon Adar1 loss is due to a runaway interferon response within these stem cells 52, 53. Of note, ADAR1 p150 is highly expressed in wild-type HSCs and might constitute a negative regulator for interferon induction. Currently, it is unknown if this dependency involves a specific RNA modification event, or stems from an editing-independent function of ADAR1 p150.
ADAR2 is essential for normal murine brain function as homozygous knock-out mice develop epileptic seizures shortly after birth and die within a few weeks of age 41. This phenotype can be attributed entirely to the consequences of the editing deficiency within a single neuron-specific gene, the glutamate receptor subunit GRIA2, which in normal neurons is edited to nearly 100% specifically by ADAR2. Genetic pre-editing of GRIA2 transcripts through genomic mutation completely rescues the phenotype of Adar2−/− mice. This is remarkable as many other RNAs are edited to lower levels in Adar2 deficient mice; however, the ensuing functional changes do not seem to interfere with lifespan or normal physiology. In both Adar1 and Adar2 knock-out mice, the loss of editing activity of one ADAR is partially compensated through the overlapping activity of the other. Moreover, the linkage of editing to other RNA processing events can lead to a partial rescue of editing deficiency being present on the pre-mRNA level. For example, whereas GRIA2 pre-mRNA is edited to only 10% in Adar2−/− knockout mice, the processed mRNA shows editing of 40% because edited primary transcripts are preferentially spliced 41.
Fewer insights are available regarding the consequences of overproducing ADARs in vivo. In Drosophila, the expression of a dADAR mutant that escapes downregulation through self-editing is lethal and displays a hyperediting phenotype 54. Mammalian ADAR2 is also subject to self-editing that leads to a decrease in functional ADAR2 protein 35; however, mutant mice that lack the ability to edit Adar2 pre-mRNA show hyperediting, but do not display a discernable behavioral or neurological phenotype 55. By contrast, the widespread overexpression of a rat Adar2 transgene in mice results in an obese phenotype 56. The molecular mechanism for this outcome is unknown, however, this phenotype might not only be due to the increased production of ADAR2, but also a result of the constitutive misexpression of the editing enzyme in cells that do not produce ADAR2 in wild-type mice 56.
Dyschromatosis Symmetrica Hereditaria (DSH1) is an autosomal dominant trait that has been linked to mutations in human ADAR1 within several Chinese and Japanese families 57. Characterized by hyperpigmentation of the hands and feet, many of the mapped mutations suggest a monoallelic inactivation of the functional deaminase. The dominant phenotype could therefore be related to a gain-of-function of the truncated or otherwise mutant protein, for example due to its altered RNA binding properties 57, 58. In addition, recent studies in several centenarian populations link polymorphisms in either ADAR1 or ADAR2 to human longevity 59.
Connections to cancer
Owing to the diverse impact of RNA editing on gene expression and function, it is possible that its misregulation might play a role in tumorigenesis either by inactivating a tumor suppressor or by activating genes that promote tumor development or progression. This notion is supported by observations that link RNA editing alterations with cancer phenotypes (reviewed in 60). In addition to the general decrease in RNA editing activity detected in several cancer types 61, a specific deficiency in A-to-I editing of glutamate receptor channels is evident in human brain cancers 62, 63. In particular, GRIA2 Q/R site editing, the molecular determinant for Ca2+-permeability of AMPA-type glutamate receptors, shows a reduction in modification rates that seems to correlate with tumor stage and has been linked directly with malignant cell behavior, such as migration and invasion 64. To date, it is unknown if the deregulation in GRIA2 editing is a causal event for tumor development, or represents a marker for tumor classification and progression.
The identification of several cancer-specific editing events within known or potential oncogenes 60, 65, 66 supports the idea that this epigenetic mechanism could contribute, directly or indirectly, to cancer growth. However, a direct link between these editing events and cancerous growth remain to be shown. Recently, a high-throughput analysis of genome and transcriptome evolution of a lobular breast cancer specimen interestingly identified a few novel cases of human A-to-I recoding editing 7. However, even though ADAR1 expression was upregulated within the tumor tissue 7, the detected editing events were not restricted to the cancerous cells. The possibility remains that ADAR1 hyperactivity or deregulation of ADAR2 editing due to ADAR1 ADAR2 heterodimerization might cause aberrant editing.
The frequent A-to-I editing of miRNA transcripts also might contribute to tumorigenesis and cancer progression as RNA editing alters expression levels or the target spectrum of miRNAs that in turn regulate signal transduction pathways involved in cell cycle and growth regulation 67. For example, both miR-376 and miR-142 undergo editing and their deregulation is implicated in molecular signatures of pancreatic cancer and leukemia, respectively 60, 68, 69.
Although several cancer phenotypes are associated with hypoediting 61–63 (Figure 2), there is no apparent causal relationship between decreased RNA editing levels and the initiation of cancerous growth as judged by currently available animal models of RNA editing deficiency 41, 48, 52, 70, 71.
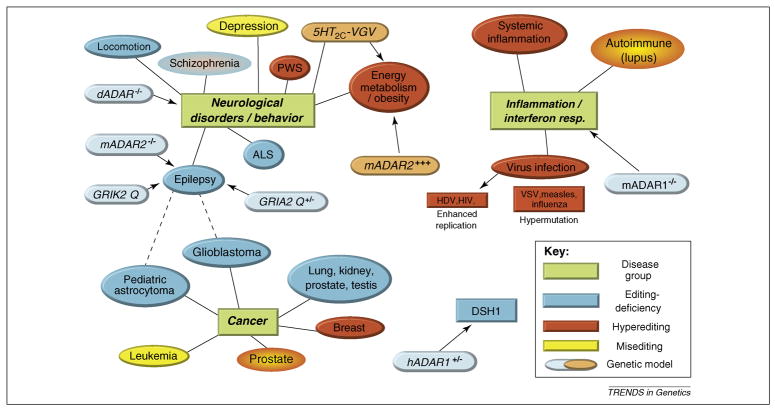
Figure 2. Disruption of the RNA editing balance.
Overview of RNA editing phenotypes in various genetic animal models and correlated observations regarding editing in human diseases. Direct causal relationships are indicated by arrows, correlations are shown as lines, and possible cross-connections as dotted lines. Partial or complete inactivation of editing has been linked to several neurological and neuropsychiatric disorders. Green shaded areas: Main disease groups. Blue shaded areas: General or gene-specific editing deficiency (genetic models with hypoediting are in lightblue boxes with dashed line). Red shaded areas: increased editing activity (genetic models with hyperediting are in light blue with dashed lines). Yellow shaded areas: changes in editing pattern or misediting without general increase or decrease in editing activity. In Prader-Willi syndrome (PWS) loss of imprinted sno RNA mbii-52 leads to increased editing of 5-HT2C receptor transcripts 46. DSH1: Dyschromatosis Symmetrica Hereditaria is linked to haploinsufficiency of ADAR1 57. Related references: Schizophrenia 80; locomotion 48, 98; depression 77, 79, 81; ALS 99; epilepsy 41, 70, 71; glioblastoma 63; pediatric astrocytoma 60, 62; leukemia 100; prostate cancer 66; breast cancer 7; lung, kidney, prostate, testicular cancer 61; systemic inflammation 92; autoimmune (lupus) 90, 91; virus infection 85–88; dADARr−/−48; mADAR2 −/− 41; GRIK2 Q 71; GRIA2 Q+/− 70; 5HT2C−VGV 74; mADAR2+++ 75 ; hADAR1+/− 57; mADAR1−/− 52, 53.
Neurological Disorders and Behavior
Neuronal tissues show high RNA editing activity and many recoding A-to-I editing events affect brain specific genes. Thus, highly complex systems and their complex physiology and behavior might strongly rely on epigenetic sources of variation, such as A-to-I editing 4, 8, 72. In fact, these types of mechanisms could enable and/or accelerate the evolution of highly complex organisms 4, 73. Thus, defects or deregulation in RNA editing might cause or accompany disturbances in higher order function more frequently than they disturb any basic physiological processes. In that respect it is noteworthy that behavioral differences between mouse strains are correlated with distinct RNA editing profiles and that several animal models of editing deregulation display behavioral abnormalities 48, 74–76 (see also Figure 2). Editing of the 5-HT2C serotonin receptor, which has established roles in emotion, locomotion, appetite, metabolic rate control, depression, schizophrenia and drug-addiction, provides an illustrative example 77, 78. The multiple site editing of this receptor subunit regulates the responsiveness of the receptor to serotonin: upon serotonin binding, more strongly edited molecules display decreased coupling efficiency to the downstream G-protein 77. In human patients with depression, changes in the 5-HT2C editing patterns are indeed apparent and intriguingly, treatment of mice with a serotonin uptake inhibitor is accompanied by converse alterations in editing 78, 79. From the analysis of patient specimens, misediting is also observed in some cases of schizophrenia 80. Mice that misexpress solely the fully edited version of the serotonin receptor (5-HT2C-VSV) display increased metabolism, hyperphagia, and growth retardation 74. Although the fully edited serotonin receptor dampens its G-protein coupling efficiency, in this mouse model 5-HT2C neurotransmission is over-sensitive, due to strongly increased functional expression of the receptors 74. Straightforward genetic mutations are clearly not sufficient to fully elucidate the physiological role(s) of serotonin receptor editing. The possibility that RNA editing patterns might display dynamic changes in response to external signals such as stress or medication 81 makes the analysis and interpretation of in vivo models even more complicated. However, at the same time, if this exciting aspect proved to apply to RNA editing in general, new layers in cell-type and time-selective regulation of gene expression through RNA editing could emerge.
ADARs on the radar
Several recent reports suggest that some aspects of ADAR function might be independent of their adenosine deaminase activity. For example, catalytically inactive ADAR2 can suppress the processing of human pri-mir-376a2 without causing editing changes 58, probably based on its selective RNA binding properties that interfere with the association of miRNA processing factors. Similarly, ADAR1 p150 counteracts siRNA function in mouse Adar1−/− MEF cells 82 and in a Drosophila system 58, also in an editing independent fashion. In summary, the range of ADAR RNA targets could be much larger than the number of edited messages, and moreover, the catalytically inactive ADAR3 might exert independent functions that arise from its RNA binding properties.
The functional roles of the ADAR1 p150 isoform are not well understood. It shares properties with antiviral factors: both are interferon induced 83 and largely localized in the cytosol; moreover, ADAR1 p150 editing activity could target viral RNAs thereby inhibiting their replication in a similar manner as C-to-U DNA modifying proteins restrict retroviruses 84. However, recent studies document that ADAR1 can act as a proviral factor during HIV 85, 86, vesicular stomatitis virus 87, and measles 88 infections through both editing-dependent and -independent mechanisms. In several cellular systems, including during measles virus infection, ADAR1 overexpression counteracts PKR kinase activity and inhibits apoptosis 88. In light of these findings, the escalating interferon response and cellular death observed in Adar1 ablated hematopoetic stem cells 52 further supports a role for ADAR1 in downregulating inflammatory response pathways. In that sense, interferon-induced ADAR1 expression does not occur to battle an infection, but instead serves to keep the antiviral response in check. As a result, virus replication is enhanced in cells which express ADAR1. In addition, some viruses might utilize cytoplasmic ADAR1 p150 to further stimulate viral infection or replication through direct editing of their transcripts 85, 88, 89.
Another connection between the interferon-mediated induction of ADAR1 p150 and inhibition of apoptosis might lie in the observation of high ADAR1 levels in T-cells and B-cells of lupus erythematosis patients, a severe, systemic autoimmune disease with signs of aberrant RNA editing events 90, 91. This hyperediting phenotype is also observed in other inflammatory processes, such as endotoxin-induced systemic inflammation 92 and upon cellular treatment with tumor-necrosis-factor α or interferon γ 92. As such, the re-equilibration of ADAR1 activity within immune cells could be an effective strategy for treatment of autoimmune disorders.
Concluding remarks and perspectives
Clearly, A-to-I RNA editing can directly or indirectly affect the expression or function of many genes. Alteration of amino acid codons, splice patterns, stability or localization of protein-coding transcripts, modulation of regulatory RNA biogenesis and function, as well as crosstalk of RNA editing with RNA processing and silencing pathways provides a rich resource for the generation of molecular diversity and for gene regulation. These findings also illustrate that we are only beginning to understand how RNA editing is integrated into the biological networks of gene expression, regulatory pathways and genome evolution.
Recent efforts to identify RNA editing events in the human transcriptome using deep sequencing approaches indicate that many editing sites remain to be discovered. However, most recoding sites might be modified only to levels of less than a few percent 6, suggesting that many recoding events might not be of immediate biological relevance, but could represent a form of noise or be part of a broader evolutionary role of editing 4. Ultimately, the generation of in vivo models of gene-specific editing deficiency or hyperediting should shed light on the physiological significance of particular editing events within the organismal context as exemplified by the neuronal glutamate and serotonin receptor targets 70, 71, 74, 93. However, such a reductionist approach will probably not be appropriate to unravel other aspects of RNA editing biology. For some editing targets, such as repetitive sequence elements, a direct genetic strategy will neither be technically feasible nor expected to yield insights that apply to the whole group of targets. Keeping in mind the complex environment in which RNA editing occurs (highly dynamic RNA folding equilibrium of substrates, divers expression of machinery and targets) and its role in providing additional levels of molecular complexity, it is possible to think of RNA editing as an indicator of complexity states; for example, reflecting higher order brain functions. In diseases where the normal complex states of activity become perturbed, we can therefore expect to observe also a disturbance in RNA editing activity or patterns. We speculate that monitoring the global activity of RNA editing in vivo represents a useful early biomarker to detect disturbances in complex systems (such as the brain) even before clinical symptoms become apparent. In that way, learning about editing patterns and dynamics could enhance the understanding of complex biological systems even before all the molecular targets and consequences of RNA editing are elucidated.
Acknowledgments
We apologize to all whose data could not be cited due to space constraints. Research in the laboratory of S. M. is supported by the National Institutes of Health (grant number NS057739).
Glossary
- Alu-repeat elements
large family of retrotransposon-derived sequence elements, each about 300 nucleotides long, that have entered the primate genomes more than 60 Mya and since expanded in number (reviewed in 94). The human genome harbors about 1.4 million Alu sequences constituting about 10% of the total genome content and leading to an average frequency of about one dozen Alus per gene. Any two Alu sequences are at least 70–80% identical in sequence, which leads to high base complementarity between pairs of Alus that are oppositely oriented within the same RNA molecule. Some Alus are still active in retrotransposition today causing about 1 reinsertion event in human every 100–200 births 94
- Editing site identification
in order to determine if a RNA is subject to A-to-I editing in vivo, the gDNA and cDNA from the gene in question is analyzed from the same specimen in order to exclude any genomic variations from the epigenetic modification. Through gene-specific amplification and sequencing of gDNA and cDNA covering the same region, a mixed signal for A and G is obtained only in the cDNA read and the editing level can be estimated directly from the relative signals for A and G in the sequence electropherogram
- Inosine
the product of adenosine deamination. The properties of inosine closely resemble those of guanosine both during RNA folding and translation of inosine-containing codons. Therefore, any A-to-I change in a protein-coding sequence is equivalent to making an A-to-G mutation. A-to-I editing is the only mechanism known to generate inosine within RNA molecules
- Recoding editing
the alteration through A-to-I editing of non-synonymous codon positions in protein-coding genes, which results in protein variants harboring a single amino acid substitution
- RNA editing frequency
the fraction of edited RNA molecules ranges from a few to almost 100% of the gene’s transcripts. Thus, edited and unedited variants are usually co-expressed within the same cell providing for transcriptome variation without the all-or-nothing effect of DNA mutations in the genome
- Specificity of editing
intrinsically, the A-to-I RNA editing machinery is promiscuous in that it will modify without site-selectivity many of the adenosines that are located within an extended, perfectly double-stranded RNA. The high site-specificity of physiological recoding targets (GluRs, 5-HT2C, Gabra-3 etc.) lies within the intricate three-dimensional RNA fold, which includes base-paired regions, as well as bulges and loops. Although the exact mode of interaction of the editing enzymes with their targets is not known, these partially base-paired RNA structures are believed to guide the machinery to edit a single nucleotide with high efficiency
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- 2.Nishikura K. Functions and Regulation of RNA Editing by ADAR Deaminases. Annual Reviews in Biochemistry. 2010;79:1–29. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gommans WM, et al. RNA Editing: A Driving Force for Adaptive Evolution? BioEssays. 2009;31:1137–1145. doi: 10.1002/bies.200900045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gommans WM, et al. Diversifying Exon Code through A-to-I RNA Editing. In: Smith H, editor. DNA RNA Editing. Wiley & Sons, Inc; 2008. pp. 3–30. [Google Scholar]
- 6.Li JB, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 7.Shah SP, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 8.Reenan RA. Molecular determinants and guided evolution of species-specific RNA editing. Nature. 2005;434:409–413. doi: 10.1038/nature03364. [DOI] [PubMed] [Google Scholar]
- 9.Athanasiadis A, et al. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blow M, et al. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim DD, et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levanon EY, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 13.Barak M, et al. Evidence for large diversity in the human transcriptome created by Alu RNA editing. Nucleic Acids Res. 2009;37:6905–6915. doi: 10.1093/nar/gkp729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lev-Maor G, et al. RNA-editing-mediated exon evolution. Genome Biol. 2007;8:R29. doi: 10.1186/gb-2007-8-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borchert GM, et al. Adenosine deamination in human transcripts generates novel microRNA binding sites. Hum Mol Genet. 2009;18:4801–4807. doi: 10.1093/hmg/ddp443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, et al. Vigilins bind to promiscuously A-to-I-edited RNAs and are involved in the formation of heterochromatin. Curr Biol. 2005;15:384–391. doi: 10.1016/j.cub.2005.01.046. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 18.Hundley HA, et al. C. elegans and H. sapiens mRNAs with edited 3′ UTRs are present on polysomes. RNA. 2008;14:2050–2060. doi: 10.1261/rna.1165008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 22.Luciano DJ, et al. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 24.Kawahara Y, et al. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang W, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawahara Y, et al. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawahara Y, et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang H, Landweber LF. Hypothesis: RNA editing of microRNA target sites in humans? RNA. 2007;13:463–467. doi: 10.1261/rna.296407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maydanovych O, Beal PA. Breaking the central dogma by RNA editing. Chem Rev. 2006;106:3397–3411. doi: 10.1021/cr050314a. [DOI] [PubMed] [Google Scholar]
- 30.Schwartz T, et al. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 31.Maas S, Gommans WM. Identification of a selective nuclear import signal in adenosine deaminases acting on RNA. Nucleic Acids Res. 2009;37:5822–5829. doi: 10.1093/nar/gkp599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fritz J, et al. RNA-regulated interaction of transportin-1 and exportin-5 with the double-stranded RNA-binding domain regulates nucleocytoplasmic shuttling of ADAR1. Mol Cell Biol. 2009;29:1487–1497. doi: 10.1128/MCB.01519-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desterro JM, et al. Dynamic association of RNA-editing enzymes with the nucleolus. J Cell Sci. 2003;116:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- 34.Sansam CL, et al. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc Natl Acad Sci U S A. 2003;100:14018–14023. doi: 10.1073/pnas.2336131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rueter SM, et al. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 36.Palladino MJ, et al. dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. Rna. 2000;6:1004–1018. doi: 10.1017/s1355838200000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valente L, Nishikura K. RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J Biol Chem. 2007;282:16054–16061. doi: 10.1074/jbc.M611392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohta H, et al. ADBP-1 regulates an ADAR RNA-editing enzyme to antagonize RNA-interference-mediated gene silencing in Caenorhabditis elegans. Genetics. 2008;180:785–796. doi: 10.1534/genetics.108.093310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agranat L, et al. The editing enzyme ADAR1 and the mRNA surveillance protein hUpf1 interact in the cell nucleus. Proc Natl Acad Sci U S A. 2008;105:5028–5033. doi: 10.1073/pnas.0710576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desterro JM, et al. SUMO-1 modification alters ADAR1 editing activity. Mol Biol Cell. 2005;16:5115–5126. doi: 10.1091/mbc.E05-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higuchi M, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 42.Reenan RA, et al. The mle(napts) RNA helicase mutation in drosophila results in a splicing catastrophe of the para Na+ channel transcript in a region of RNA editing. Neuron. 2000;25:139–149. doi: 10.1016/s0896-6273(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 43.Ryman K, et al. The C-terminal domain of RNA Pol II helps ensure that editing precedes splicing of the GluR-B transcript. RNA. 2007;13:1071–1078. doi: 10.1261/rna.404407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raitskin O, et al. RNA editing activity is associated with splicing factors in lnRNP particles: The nuclear pre-mRNA processing machinery. Proc Natl Acad Sci U S A. 2001;98:6571–6576. doi: 10.1073/pnas.111153798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 46.Doe CM, et al. Loss of the imprinted snoRNA mbii-52 leads to increased 5htr2c pre-RNA editing and altered 5HT2CR-mediated behaviour. Hum Mol Genet. 2009;18:2140–2148. doi: 10.1093/hmg/ddp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vitali P, et al. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J Cell Biol. 2005;169:745–753. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palladino MJ, et al. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 49.Jepson JE, Reenan RA. Adenosine-to-inosine genetic recoding is required in the adult stage nervous system for coordinated behavior in Drosophila. J Biol Chem. 2009;284:31391–31400. doi: 10.1074/jbc.M109.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hartner JC, et al. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 51.Wang Q, et al. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 52.Hartner JC, et al. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.XuFeng R, et al. ADAR1 is required for hematopoietic progenitor cell survival via RNA editing. Proc Natl Acad Sci U S A. 2009;106:17763–17768. doi: 10.1073/pnas.0903324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keegan LP, et al. Tuning of RNA editing by ADAR is required in Drosophila. EMBO J. 2005;24:2183–2193. doi: 10.1038/sj.emboj.7600691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng Y, et al. Altered RNA editing in mice lacking ADAR2 autoregulation. Mol Cell Biol. 2006;26:480–488. doi: 10.1128/MCB.26.2.480-488.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh M, et al. Hyperphagia-mediated obesity in transgenic mice misexpressing the RNA-editing enzyme ADAR2. J Biol Chem. 2007;282:22448–22459. doi: 10.1074/jbc.M700265200. [DOI] [PubMed] [Google Scholar]
- 57.Tojo K, et al. Dystonia, mental deterioration, and dyschromatosis symmetrica hereditaria in a family with ADAR1 mutation. Mov Disord. 2006;21:1510–1513. doi: 10.1002/mds.21011. [DOI] [PubMed] [Google Scholar]
- 58.Heale BS, et al. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009;28:3145–3156. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sebastiani P, et al. RNA editing genes associated with extreme old age in humans and with lifespan in C. elegans. PLoS ONE. 2009;4:e8210. doi: 10.1371/journal.pone.0008210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gallo A, Galardi S. A-to-I RNA editing and cancer: From pathology to basic science. RNA Biol. 2008:5. doi: 10.4161/rna.5.3.6739. [DOI] [PubMed] [Google Scholar]
- 61.Paz N, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cenci C, et al. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J Biol Chem. 2008;283:7251–7260. doi: 10.1074/jbc.M708316200. [DOI] [PubMed] [Google Scholar]
- 63.Maas S, et al. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci U S A. 2001;98:14687–14692. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishiuchi S, et al. Blockage of Ca(2+)-permeable AMPA receptors suppresses migration and induces apoptosis in human glioblastoma cells. Nat Med. 2002;8:971–978. doi: 10.1038/nm746. [DOI] [PubMed] [Google Scholar]
- 65.Maas S, et al. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martinez HD, et al. RNA editing of androgen receptor gene transcripts in prostate cancer cells. J Biol Chem. 2008;283:29938–29949. doi: 10.1074/jbc.M800534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sotiropoulou G, et al. Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA. 2009;15:1443–1461. doi: 10.1261/rna.1534709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen CZ, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 69.Lee EJ, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brusa R, et al. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science. 1995;270:1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- 71.Vissel B, et al. The role of RNA editing of kainate receptors in synaptic plasticity and seizures. Neuron. 2001;29:217–227. doi: 10.1016/s0896-6273(01)00192-1. [DOI] [PubMed] [Google Scholar]
- 72.Mattick JS, Mehler MF. RNA editing, DNA recoding and the evolution of human cognition. Trends Neurosci. 2008;31:227–233. doi: 10.1016/j.tins.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 73.Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 74.Kawahara Y, et al. Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. J Neurosci. 2008;28:12834–12844. doi: 10.1523/JNEUROSCI.3896-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Singh M, et al. Affect-related behaviors in mice misexpressing the RNA editing enzyme ADAR2. Physiol Behav. 2009;97:446–454. doi: 10.1016/j.physbeh.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gardiner K, Du Y. A-to-I editing of the 5HT2C receptor and behaviour. Brief Funct Genomic Proteomic. 2006;5:37–42. doi: 10.1093/bfgp/ell006. [DOI] [PubMed] [Google Scholar]
- 77.Berg KA, et al. Fine-tuning serotonin2c receptor function in the brain: molecular and functional implications. Neuropharmacology. 2008;55:969–976. doi: 10.1016/j.neuropharm.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmauss C. Regulation of serotonin 2C receptor pre-mRNA editing by serotonin. Int Rev Neurobiol. 2005;63:83–100. doi: 10.1016/S0074-7742(05)63004-8. [DOI] [PubMed] [Google Scholar]
- 79.Gurevich I, et al. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- 80.Sodhi MS, et al. RNA editing of the 5-HT(2C) receptor is reduced in schizophrenia. Mol Psychiatry. 2001;6:373–379. doi: 10.1038/sj.mp.4000920. [DOI] [PubMed] [Google Scholar]
- 81.Englander MT, et al. How stress and fluoxetine modulate serotonin 2C receptor pre-mRNA editing. J Neurosci. 2005;25:648–651. doi: 10.1523/JNEUROSCI.3895-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang W, et al. ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. J Biol Chem. 2005;280:3946–3953. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kawakubo K, Samuel CE. Human RNA-specific adenosine deaminase (ADAR1) gene specifies transcripts that initiate from a constitutively active alternative promoter. Gene. 2000;258:165–172. doi: 10.1016/s0378-1119(00)00368-1. [DOI] [PubMed] [Google Scholar]
- 84.Rosenberg BR, Papavasiliou FN. Beyond SHM and CSR: AID and related cytidine deaminases in the host response to viral infection. Adv Immunol. 2007;94:215–244. doi: 10.1016/S0065-2776(06)94007-3. [DOI] [PubMed] [Google Scholar]
- 85.Doria M, et al. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res. 2009;37:5848–5858. doi: 10.1093/nar/gkp604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Phuphuakrat A, et al. Double-stranded RNA adenosine deaminases enhance expression of human immunodeficiency virus type 1 proteins. J Virol. 2008;82:10864–10872. doi: 10.1128/JVI.00238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nie Y, et al. Double-stranded RNA deaminase ADAR1 increases host susceptibility to virus infection. J Virol. 2007;81:917–923. doi: 10.1128/JVI.01527-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Toth AM, et al. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J Biol Chem. 2009;284:29350–29356. doi: 10.1074/jbc.M109.045146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Casey JL. RNA editing in hepatitis delta virus. Curr Top Microbiol Immunol. 2006;307:67–89. doi: 10.1007/3-540-29802-9_4. [DOI] [PubMed] [Google Scholar]
- 90.Laxminarayana D, et al. Transcript mutations of the alpha regulatory subunit of protein kinase A and up-regulation of the RNA-editing gene transcript in lupus T lymphocytes. Lancet. 2002;360:842–849. doi: 10.1016/s0140-6736(02)09966-x. [DOI] [PubMed] [Google Scholar]
- 91.Orlowski RJ, et al. Altered editing in cyclic nucleotide phosphodiesterase 8A1 gene transcripts of systemic lupus erythematosus T lymphocytes. Immunology. 2008 doi: 10.1111/j.1365-2567.2008.02850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang JH, et al. Widespread inosine-containing mRNA in lymphocytes regulated by ADAR1 in response to inflammation. Immunology. 2003;109:15–23. doi: 10.1046/j.1365-2567.2003.01598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Aronoff R, et al. Neuronal toxicity in Caenorhabditis elegans from an editing site mutant in glutamate receptor channels. J Neurosci. 2004;24:8135–8140. doi: 10.1523/JNEUROSCI.2587-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 95.Gerber AP, Keller W. RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem Sci. 2001;26:376–384. doi: 10.1016/s0968-0004(01)01827-8. [DOI] [PubMed] [Google Scholar]
- 96.Macbeth MR, et al. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maas S, Gommans WM. Novel exon of mammalian ADAR2 extends open reading frame. PLoS ONE. 2009;4:e4225. doi: 10.1371/journal.pone.0004225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Doe CM, et al. Loss of the imprinted snoRNA mbii-52 leads to increased 5htr2c pre-RNA editing and altered 5HT2CR mediated behaviour. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kawahara Y, et al. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 100.Beghini A, et al. RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum Mol Genet. 2000;9:2297–2304. doi: 10.1093/oxfordjournals.hmg.a018921. [DOI] [PubMed] [Google Scholar]