Abstract
Over the last half century, myoglobin (Mb) has been an excellent model system to test a number of concepts, theories, and new experimental methods that proved valuable to investigate protein structure, function, evolution, and dynamics. Mb's function, most often considered just an oxygen repository, has considerably diversified over the last 15 years, especially because it was shown to have a role in the biochemistry of quenching and synthesizing nitric oxide in the red muscle, thereby protecting the cell. To tackle protein's structural dynamics by innovative biophysical methods, Mb has been the best prototype; laser flash technology made it possible to obtain molecular movies by time-resolved Laue crystallography (with ps resolution). This approach unveiled the complexity of the energy landscape and the structural basis of the stretched interconversion between conformational substates of a protein.
Keywords: myoglobin, H-atom, molecular biology, protein science
Introduction
The three-dimensional structure of myoglobin (Mb), the hydrogen atom of molecular biology, was solved 50 years ago by Kendrew et al. A preliminary report of the structure appeared in Nature in 19581; a more detailed account was published the following year in the Proceedings of the Royal Society2; and the full paper in Nature on 13 February 1960.3
For half a century, Mb has been a popular model system to test a number of concepts, theories, and new experimental methods dealing with protein structure, function, dynamics, and evolution. Hereby, I summarize the novelties in the physiological role of Mb in the heart and the red muscle over-and-above O2 storage and outline some of the experimental and conceptual advances on protein's structural dynamics that emerged from time-resolved Laue crystallography on Mb, with ps time resolution. This short review, a tribute to Sir John C. Kendrew and coworkers, is being published just 50 years since the Nature paper thanks to the collaboration of the Editor of Protein Science.
Just an Oxygen Repository?
Our understanding of the functional role of Mb expanded considerably over the last 15 years, because it became clear that the protein is involved in controlling the fluxes of nitric oxide (NO) in the heart, under physiological and pathological conditions. Generally, Mb's function in most peoples' mind, and indeed even in Biochemistry text books, is limited to its role as an O2 storage, a minimalist misnomer. This depends largely on the (correct) notion that in diving mammals, whose muscles look black because the concentration of Mb is ∼10-fold greater than in humans, its role as an O2 repository is physiologically important; in man and the other mammals, it was calculated4 to be sufficient to sustain aerobic metabolism for approximately a couple of seconds, that is, for O2 supply in between beats.
Probably, more significant but generally ignored is the role of Mb in facilitating O2 diffusion from capillaries to mitochondria, given its high concentration (100–300 μM) and free translational diffusion in the intracellular milieu.5,6 Interestingly, although a transgenic Mb knockout mice (myo−/−) were reported to behave normally,7 histology of a section of the heart showed that the density of capillaries in the knockouts was increased,8 one of the compensatory responses to reduce the effective diffusion path length. This was a striking unexpected evidence that indeed Mb plays a significant role in facilitating O2 diffusion to mitochondria, thereby keeping the sarcoplasmic O2 pressure essentially constant throughout the cell and Mb approximately half-saturated with O2, independently of the workload.6
More recently, NO moved Mb center stage, once it was discovered in 1994 that cytochrome c oxidase, the terminal enzyme of the respiratory chain, is reversibly inhibited by NO.9,10 Even at low physiological fluxes (<0.1 μM), NO is a potent poison of respiration; the apparent inhibition constant depends on O2 concentration because the two gases compete for the oxidase active site, which may explain11 why the apparent Km for O2 in cells and tissues (5–10 μM) is significantly greater than that measured with mitochondria or the purified enzyme (Km < 1 μM). The mechanism of inhibition of cytochrome c oxidase by NO, which is not simple, has been largely clarified by extensive presteady-state and steady-state experiments.12–14 In 1998, Moncada's group15 discovered that also Complex I is inhibited by NO albeit via a different biochemical reaction, which explains why speed of inactivation and recovery in the latter case are both much slower than for the case of oxidase.
Starting from these premises it seemed obvious to me16 that one important and hitherto overlooked function of Mb in the skeletal and cardiac muscle may be scavenging NO, thereby protecting the energy-producing machinery. The mechanism of quenching is simple and effective [Eq. (1a)], because oxygenated MbO2 reacts rapidly and stoichiometrically with NO, yielding nitrate and ferric Mb+17; the presence of an intracellular Mb+ reductase ensures rereduction to ferrous Mb and thereby a catalytic cycle.
| 1a |
| 1b |
Flögel et al.18 published the first convincing evidence in support of this hypothesis. These authors found that typical cardiac physiological parameters (e.g., coronary blood flow and cardiac output) were more severely affected by NO fluxes in the heart of the knockout mice lacking Mb (myo−/−) compared with the wild type. The fact that the same NO flux has greater adverse effects in the hearts from myo−/− mice may be understood assuming a more substantial inhibition of cytochrome c oxidase by NO in the knockouts.
Over the last few years, yet another function of Mb related to NO metabolism in the heart has been substantiated.19,20 It was shown both in vitro and in the heart21 that under anaerobic conditions deoxy Mb is endowed with nitrite reductase activity [see Eq. (1b)], thus producing NO just when NO synthase is inactive because of the lack of O2. The rate of NO synthesis from  (at ∼10 μM in tissues21) depends on deoxy Mb, because it becomes undetectable in the myo−/− knockouts. Under anaerobiosis, Complex I and Complex IV of the respiratory chain are both inhibited by NO9,10,15; thus, reoxygenation is not associated with the classical ischemia-reperfusion ROS production and tissue damage is reduced. The cardioprotective effect is so evident22,23 that clinical trials to assess the positive effect of nitrite administration on the extent of heart damage are under course.
(at ∼10 μM in tissues21) depends on deoxy Mb, because it becomes undetectable in the myo−/− knockouts. Under anaerobiosis, Complex I and Complex IV of the respiratory chain are both inhibited by NO9,10,15; thus, reoxygenation is not associated with the classical ischemia-reperfusion ROS production and tissue damage is reduced. The cardioprotective effect is so evident22,23 that clinical trials to assess the positive effect of nitrite administration on the extent of heart damage are under course.
A Paradigm of Complexity
Mb has been an ideal model system for experiments on the structural dynamics of proteins and its role in controlling function, because it is sufficiently simple to be investigated in detail but complex enough to be intriguing and nontrivial. An extremely useful property of Mb is the photosensitivity of the ligand adduct, the complex of ferrous Mb with O2, CO, NO, and other ligands being light sensitive24,25; thereby hitting liganded Mb with a short and intense laser pulse, breaks the ligand-iron bond and sets into motion the relaxation of the photoproduct, deoxy Mb. In the late 70s, this approach led to the discovery of the so-called geminate recombination, that is, the intramolecular rebinding to the iron of the photodissociated ligand trapped momentarily within the protein matrix (Fig. 1). Although the structural differences between the two equilibrium states (deoxy and liganded Mb) are not large, kinetics showed their functional importance. The kinetics of Mb has been extensively investigated by transient laser spectroscopy and sophisticated modeling by many investigators; among others, pioneering contributions were reported by Q.H. Gibson, H. Frauenfelder, W.A. Eaton, and their groups.27,28,30–34 Below is a summary of the main features of the relaxation kinetics of the whole protein as revealed by time-resolved Laue crystallography, a global approach.
Figure 1.
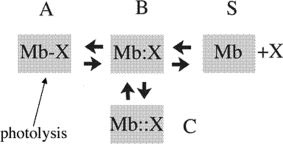
Simplified kinetic scheme depicting events following photolysis of liganded Mb (A). The photolysed ligand (O2, CO, NO, and others) populates the so-called distal pocket (primary docking site, B), and thereafter may: (i) diffuse out into the solvent (S) via the His-gate,26 the most probable path according to Scott et al.27; (ii) rebind to the metal by an intramolecular geminate process B → A; (iii) diffuse into the matrix secondary docking sites (C), following a path traced by the Xe binding cavities (as shown by time-resolved Laue crystallography). Escape out into the solvent is occurring also via other pathways over-and-above the His-gate.28,29
The classical trick to obtain structural information on short-lived states is to freeze intermediates at ultra-low temperatures (e.g., ∼10° K). This approach applied to the photoproduct of MbCO, allowed Schlichting et al.35 to prove that photodissociated CO is indeed located in the so-called distal pocket. Several other papers36–39 exploiting the low-temperature trapping method confirmed and extended this initial finding and substantiated, for example, the migration of CO inside the protein upon increase in the temperature of the crystal.36,37 Nevertheless, it was obvious that direct information to unveil the structural relaxation of the globin was to come from room temperature time-resolved crystallographic data.
This difficult task has been for 2 decades the mission impossible of Keith Moffat and his collaborators, who published in 199640 the first reliable time-resolved Laue diffraction experiment; the quality of those results bears no comparison with current data, but it was nevertheless an important breakthrough showing that the experiment was possible and informative. In the following years, substantial improvements in synchrotron technology, data analysis, and protein biochemistry41,42 made it possible to carry out a high-resolution structural characterization of Mb's dynamics from 100 ps after photolysis up to many ms, covering the huge time domain typical of protein relaxations.
Intrinsic difficulties of photolysis in single crystals are (i) the limited time span available to shoot X-ray pulses before CO bimolecular recombination in the dark, and (ii) the low yield of deoxy Mb photoproduct due to geminate rebinding. Site-directed mutagenesis proved valuable to minimize these shortcomings; for example, the triple mutant called YQR-Mb (L29Y/H64Q/T69R43) was a convenient variant because the mutations in the distal pocket abolish CO geminate recombination and slow down (by 10-fold) bimolecular rebinding, extending the time window accessible to interrogation with short (150 ps) X-ray pulses.
The new generation of Laue diffraction experiments led to a complete characterization of the global structural relaxation of Mb after CO photodissociation (Fig. 2). Wild-type sperm whale Mb and three mutants were investigated in detail.44–48 Data obtained by the different groups were by-and-large consistent and allowed to unveil a few features of general significance for the structural dynamics of proteins at large. In all cases, the structural relaxation of the deoxy Mb photoproduct was found to be nonexponential extending from ps to μs and above, in agreement with spectroscopic data.49 Subtle differences in dynamic behavior between the mutants and the wild type were largely rationalized and correlated with transient optical spectroscopy in solution.
Figure 2.
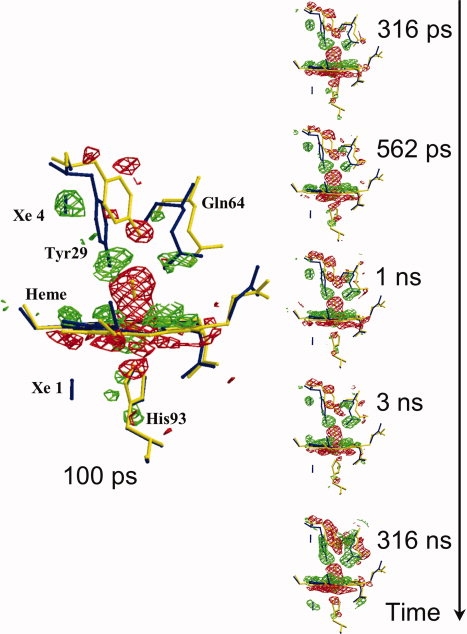
Structural changes in the heme vicinity for the mutant called YQR-Mb,43 from 100 ps to 316 ns after CO photolysis. YQR-Mb is a triple mutant (L29Y/H64Q/T67R) used for this experimental session because of favorable properties (zero CO geminate recombination, slow bimolecular rebinding, and excellent sturdy crystals). (Flight − Fdark), the difference electron density maps, are in red for negative and in green for positive (contoured at 3.0 σ); these are overlaid on models of YQR-MbCO (in yellow) and YQR-Mb (in blue). It may be seen that at 100 ps, the negative density (red) of the Fe2+CO on the distal side of the heme is very prominent; other detectable changes are the initial distortion of the heme and motions of the distal Tyr29 and Gln64. These structural changes continue to grow with time and are fully developed in the frame at 316 ns. In all the frames, a blue bar indicates the position of photolyzed CO migrating initially to the Xe4 pocket (distal, at the top in the 100 ps frame) and later to the Xe1 pocket (proximal, at the bottom, better seen in the frame at 316 ns) (from Bourgeois et al.,44 modified).
The relaxation profile of the heme and of the globin moiety (Fig. 3) extends over a time range from 100 ps (the earliest available frame) to μs and above, until CO rebinding in the dark leads back to the equilibrium state.45 The heme relaxation is heterogeneous, some of the tilt being synchronous with the laser pulse and the rest occurring within several ns; in YQR-Mb heterogeneity was correlated44 to a strain on heme pyrrole C exerted by the E-helix via the CD turn. The larger conformational changes of the globin develop with a lag of ∼50–100 ns, as may be seen by looking at the relaxation of the distal E-helix and several residues of the CD turn [Fig. 3(b,c)]. The stretched relaxation behavior is consistent with the idea of a protein quake (championed by Frauenfelder and coworkers50), whereby photolysis sets into motion faster changes in the immediate environment of the heme, and larger but slower perturbations with rearrangements of the globin involving conformational changes of the helices. This complex relaxation behavior was taken to represent interconversion between sets of conformational substates jumping over multiple barriers, with or without solvent displacement.51
Figure 3.
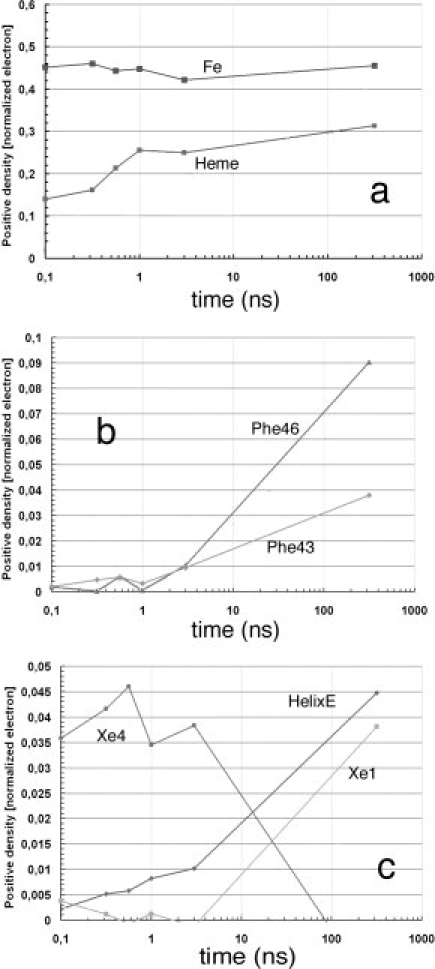
Time dependence of difference electron densities for key structural features, after photolysis of YQR-MbCO. Numerical values represent the integral of the positive electron density beyond 3.0 σ, corrected for variations in photolysis yield and normalized to the negative bound CO feature (arbitrary value of 1). (a) Key features that appear promptly: the density of the Fe popping out of the heme plane without delay remains time independent; while distortion of the heme (see Fig. 2) and motion of Tyr29 follow an heterogeneous time course being already detectable in the first frame (density of a 0.10–0.15 at 0.1 ns) but increasing with time. (b) Amino acid residues involved in the strain of the CD turn clearly lag behind the conformational changes of the heme and of Tyr29 on the distal side (see above). (c) Population of CO in the Xe1 and Xe4 pockets, and conformational changes of the E-helix: CO appears promptly after photolysis in the Xe4 pocket, on the distal side of the heme (with 0.035–0.045 density), and begins to diffuse inside and populate the Xe1 pocket on the proximal side starting from ∼5 ns. The migration of CO is approximately synchronous with larger scale conformational changes of the E-helix, suggesting that long-scale intramolecular diffusion demands more substantial structural changes of the globin. At much longer times (e.g., toward ms) CO rebinds largely via the opening of the His-gate,26 as shown by Scott et al.27 studying many different mutants of sperm whale Mb (from Bourgeois et al.,44 modified).
Associated with these extended conformational changes, migration of the photolyzed ligand inside the protein was observed directly.44–48 CO diffusing from the so-called distal site near the heme to a cavity located just above the distal pocket, the so-called Xe4 (Fig. 2, at 100 ps), and subsequently to another prominent cavity located on the proximal side (so-called Xe1) (Fig. 2, at 316 ns). The electron density of CO while migrating between cavities and its time dependance [Fig. 3(c)] were by-and-large the same for all Mb variants; the recovery of CO was carefully analyzed by Srajer et al.45 and compared to the recombination time course. The crystallographic time-resolved data are in satisfactory agreement with geminate recombination studies on different ligands (CO, NO, and O2) obtained in solution, and particularly with the effects of mutations and Xe binding on the kinetics.27,52
These extended data sets led to some general conclusions. The stretched globin relaxation is a direct and original demonstration of the complexity of the energy landscape of a protein.53 This envisaged a multiplicity of paths as the protein skates along a complex set of conformational coordinates, indicated by the complex kinetics and the effects of mutations, temperature, and viscosity.27,34 A fundamental point yet under debate is whether the stretched structural changes (Fig. 3) are an intrinsic property of each individual molecule relaxing in a smooth extended process or an average behavior of all molecules54; possibly single molecule kinetics may solve this point.
The reliable structural information now available from a few hundred ps allowed a quantitative comparison between experimental data and molecular dynamics simulations. It was rewarding to find out that experiments and simulations55,56 are in good agreement, a result of great value for the future of structural dynamics. Simulations may allow a clearer identification of essential dynamic features that may have escaped examination of the ensemble averaged difference maps; moreover, the necessarily limited number of experimental points along the extended time course can be filled by the simulations. Finally, mutual validation paves the way to wider applications of MD to investigate the stretched conformational relaxation of proteins that are not amenable to laser-triggered Laue diffraction experiments.
The ambition of the structural dynamics approach was to prove in selected cases that direct determination of global conformational changes related to function is feasible, with remarkable time and spatial resolution. The experimental evidence of a stretched relaxation is one of the most convincing proofs of the complexity of the protein energy landscape and the existence of functionally relevant quasi isoenergetic conformational substates. More work is foreseen, such as the acquisition of tertiary relaxation data for the R and T states of hemoglobin, following the path opened by the pioneering studies of Keith Moffat and others exploiting time-resolved Laue crystallography. Based on half a century of honorable service, it seems impossible to ignore the role of Mb as the paradigm of complexity in protein structural dynamics.
Acknowledgments
The author is very grateful to many friends and colleagues who shared his interest in myoglobin; in particular, he is pleased to thank Phil Anfinrud (Bethesda, USA), Dominique Bourgeois (Grenoble), Francesca Cutruzzolà (Rome), and Beatrice Vallone (Rome). It should be noted that the author previously wrote a personal recollection as a tribute to Beatrice and Jonathan Wittenberg, with the same title but different content (in Dioxygen binding and sensing proteins, M. Bolognesi, G. di Prisco, and C. Verde, Eds., ProteinReviews M. Zouhair Atassi, Senior Editor, Vol. 9, pp. 183–189, Springer Verlag, 2008).
References
- 1.Kendrew JC, Bodo G, Dintzis HM, Parrish RG, Wyckoff HW, Phillips DC. A three-dimensional model of the myoglobin molecule obtained by X-ray analysis. Nature. 1958;181:662–666. doi: 10.1038/181662a0. [DOI] [PubMed] [Google Scholar]
- 2.Bodo G, Dintzis HM, Kendrew JC, Wyckoff HW. The crystal structure of myoglobin. V. A low-resolution three-dimensional Fourier synthesis of sperm-whale myoglobin crystals. Proc R Soc Lond A. 1959;253:70–102. [Google Scholar]
- 3.Kendrew JC, Dickerson RE, Strandberg BE, Hart RG, Davies DR, Phillips DC, Shore VC. Structure of myogloba three-dimensional Fourier synthesis at 2 Å resolution. Nature. 1960;185:422–427. doi: 10.1038/185422a0. [DOI] [PubMed] [Google Scholar]
- 4.Millikan GA. Muscle haemoglobin. Physiol Rev. 1939;19:503–523. [Google Scholar]
- 5.Wittenberg BA, Wittenberg JB. Transport of oxygen in muscle. Annu Rev Physiol. 1989;51:857–878. doi: 10.1146/annurev.ph.51.030189.004233. [DOI] [PubMed] [Google Scholar]
- 6.Wittenberg BA, Wittenberg JB. Myoglobin function reassessed. J Exp Biol. 2003;206:2011–2020. doi: 10.1242/jeb.00243. [DOI] [PubMed] [Google Scholar]
- 7.Garry DL, Ordway GA, Lorenz JN, Radford NB, Chin ER, Grange RW, Bassel-Duby R, Williams RS. Mice without myoglobin. Nature. 1998;395:905–908. doi: 10.1038/27681. [DOI] [PubMed] [Google Scholar]
- 8.Gödecke A, Flögel U, Zanger K, Ding Z, Hirchenhain J, Decking UKM, Schrader J. Disruption of myoglobin in mice induces multiple compensatory mechanisms. Proc Natl Acad Sci USA. 1999;96:10495–10500. doi: 10.1073/pnas.96.18.10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown GC, Cooper CE. Nanomolar concentrations of nitric oxide reversibly inhibit synaptosomal respiration by competing with oxygen at cytochrome c oxidase. FEBS Lett. 1994;356:295–298. doi: 10.1016/0014-5793(94)01290-3. [DOI] [PubMed] [Google Scholar]
- 10.Cleeter MWJ, Cooper JM, Darley-Usmar VM, Moncada S, Schapira AHV. Reversible inhibition of cytochrome c oxidase, the terminal enzyme of the mitochondrial respiratory chain, by nitric oxide. FEBS Lett. 1994;345:50–54. doi: 10.1016/0014-5793(94)00424-2. [DOI] [PubMed] [Google Scholar]
- 11.Brown GC. Nitric oxide regulates mitochondrial respiration and cell functions by inhibiting cytochrome oxidase. FEBS Lett. 1995;369:136–139. doi: 10.1016/0014-5793(95)00763-y. [DOI] [PubMed] [Google Scholar]
- 12.Torres J, Darley-Usmar V, Wilson MT. Inhibition of cytochrome c oxidase in turnover by nitric oxide: mechanism and implications for control of respiration. Biochem J. 1995;312:169–173. doi: 10.1042/bj3120169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giuffré A, Sarti P, Buse G, Soulimane T, Brunori M. On the mechanism of inhibition of cytochrome c oxidase by nitric oxide. J Biol Chem. 1996;271:33404–33408. doi: 10.1074/jbc.271.52.33404. [DOI] [PubMed] [Google Scholar]
- 14.Brunori M, Forte E, Arese M, Mastronicola D, Giuffrè A, Sarti P. Nitric oxide and the respiratory enzyme. Biochim Biophys Acta. 2006;1757:1144–1154. doi: 10.1016/j.bbabio.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Clementi E, Brown GC, Feelisch M, Moncada S. Persistent inhibition of cell respiration by nitric oxide: crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc Natl Acad Sci USA. 1998;95:7631–7636. doi: 10.1073/pnas.95.13.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunori M. Nitric oxide, cytochrome-c oxidase and myoglobin. Trends Biochem Sci. 2001;26:21–23. doi: 10.1016/s0968-0004(00)01698-4. [DOI] [PubMed] [Google Scholar]
- 17.Eich RF, Li T, Lemon DD, Doherty DH, Curry SR, Aitken JF, Mathews AJ, Johnson KA, Smith RD, Phillips GN, Jr, Olson JS. Mechanism of NO-induced oxidation of myoglobin and hemoglobin. Biochemistry. 1996;35:6976–6983. doi: 10.1021/bi960442g. [DOI] [PubMed] [Google Scholar]
- 18.Flögel U, Merx MW, Gödecke A, Decking UK, Schrader J. Myogloba scavenger of bioactive NO. Proc Natl Acad Sci USA. 2001;98:735–740. doi: 10.1073/pnas.011460298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundberg JO, Weitzberg E, Gladwin MT. The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat Rev Drug Discov. 2008;7:156–167. doi: 10.1038/nrd2466. [DOI] [PubMed] [Google Scholar]
- 20.Cossins A, Berenbrink M. Physiology: myoglobin's new clothes. Nature. 2008;454:416–417. doi: 10.1038/454416a. [DOI] [PubMed] [Google Scholar]
- 21.Hendgen-Cotta UB, Merx MW, Shiva S, Schmitz J, Becher S, Klare JP, Steinhoff HJ, Gödecke A, Schrader J, Gladwin MT, Kelm M, Rassaf T. Nitrite reductase activity of myoglobin regulates respiration and cellular viability in myocardial ischemia-reperfusion injury. Proc Natl Acad Sci USA. 2008;105:10256–10261. doi: 10.1073/pnas.0801336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez FM, Shiva S, Vincent PS, Ring wood LA, Hsu LY, Hon YY, Aletras AH, Cannon RO, III, Gladwin MT, Arai AE. Nitrite anion provides potent cytoprotective and antiapoptotic effects as adjunctive therapy to reperfusion for acute myocardial infarction. Circulation. 2008;117:2986–2994. doi: 10.1161/CIRCULATIONAHA.107.748814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dezfulian C, Shiva S, Alekseyenko A, Pendyal A, Beiser DG, Munasinghe JP, Anderson SA, Chesley CF, Vanden Hoek TL, Gladwin MT. Nitrite therapy after cardiac arrest reduces reactive oxygen species generation, improves cardiac and neurological function, and enhances survival via reversible inhibition of mitochondrial complex I. Circulation. 2009;120:897–905. doi: 10.1161/CIRCULATIONAHA.109.853267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antonini E, Brunori M. Hemoglobin and myoglobin in their reactions with ligands. Amsterdam, The Netherlands: North Holland Publishing Co; 1971. [Google Scholar]
- 25.Gibson QH. Hemoproteins, ligands, quanta. J Biol Chem. 1989;264:20155–20158. [PubMed] [Google Scholar]
- 26.Perutz MF, Mathews FS. An X-ray study of azide methaemoglobin. J Mol Biol. 1966;21:199–207. doi: 10.1016/0022-2836(66)90088-x. [DOI] [PubMed] [Google Scholar]
- 27.Scott EE, Gibson QH, Olson JS. Mapping the pathways for O2 entry into and exit from myoglobin. J Biol Chem. 2001;276:5177–5188. doi: 10.1074/jbc.M008282200. [DOI] [PubMed] [Google Scholar]
- 28.Varadarajan R, Lambright DG, Boxer SG. Electrostatic interactions in wild-type and mutant recombinant human myoglobins. Biochemistry. 1989;28:3771–3781. doi: 10.1021/bi00435a022. [DOI] [PubMed] [Google Scholar]
- 29.Elber R, Karplus M. Enhanced sampling in molecular dynamics: use of time-dependent Hartree approximation for a simulation of carbon-monoxide diffusion through myoglobin. J Am Chem Soc. 1990;112:9161–9175. [Google Scholar]
- 30.Austin RH, Beeson KW, Eisenstein L, Frauenfelder H, Gunsalus IC. Dynamics of ligand binding to myoglobin. Biochemistry. 1975;14:5355–5373. doi: 10.1021/bi00695a021. [DOI] [PubMed] [Google Scholar]
- 31.Hofrichter J, Sommer JH, Henry ER, Eaton WA. Nanosecond absorption spectroscopy of hemoglobelementary processes in kinetic cooperativity. Proc Natl Acad Sci USA. 1983;80:2235–2239. doi: 10.1073/pnas.80.8.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinbach PJ, Ansari A, Berendzen J, Braunstein D, Chu K, Cowen BR, Ehrenstein D, Frauenfelder H, Johnson JB, Lamb DC, Luck S, Mourant JR, Nienhaus U, Ormos P, Philipp R, Xie A, Young RD. Ligand binding to heme proteins: connection between dynamics and function. Biochemistry. 1991;30:3988–4001. doi: 10.1021/bi00230a026. [DOI] [PubMed] [Google Scholar]
- 33.Jones CM, Ansari A, Henry ER, Christoph GW, Hofrichter J, Eaton WA. Speed of intersubunit communication in proteins. Biochemistry. 1992;31:6692–6702. doi: 10.1021/bi00144a008. [DOI] [PubMed] [Google Scholar]
- 34.Ansari A, Jones CM, Henry ER, Hofrichter J, Eaton WA. Conformational relaxation and ligand binding in myoglobin. Biochemistry. 1994;33:5128–5145. doi: 10.1021/bi00183a017. [DOI] [PubMed] [Google Scholar]
- 35.Schlichting I, Berendzen J, Phillips GN, Jr, Sweet RM. Crystal structure of photolysed carbonmonoxy-myoglobin. Nature. 1994;371:808–812. doi: 10.1038/371808a0. [DOI] [PubMed] [Google Scholar]
- 36.Teng TY, Srajer V, Moffat K. Initial trajectory of carbon monoxide after photodissociation from myoglobin at cryogenic temperatures. Biochemistry. 1997;36:12087–12100. doi: 10.1021/bi971140x. [DOI] [PubMed] [Google Scholar]
- 37.Ostermann A, Waschipky R, Parak FG, Nienhaus GU. Ligand binding and conformational motions in myoglobin. Nature. 2000;404:205–208. doi: 10.1038/35004622. [DOI] [PubMed] [Google Scholar]
- 38.Brunori M, Vallone B, Cutruzzola F, Travaglini-Allocatelli C, Berendzen J, Chu K, Sweet RM, Schlichting I. The role of cavities in protein dynamics: crystal structure of a photolytic intermediate of a mutant myoglobin. Proc Natl Acad Sci USA. 2000;97:2058–2063. doi: 10.1073/pnas.040459697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlichting I, Chu K. Trapping intermediates in the crystal: ligand binding to myoglobin. Curr Opin Struct Biol. 2000;10:744–752. doi: 10.1016/s0959-440x(00)00158-5. [DOI] [PubMed] [Google Scholar]
- 40.Srajer V, Teng T, Ursby T, Pradervand C, Ren Z, Adachi S, Schildkamp W, Bourgeois D, Wulff M, Moffat K. Photolysis of the carbon monoxide complex of myoglobnanosecond time-resolved crystallography. Science. 1996;274:1726–1729. doi: 10.1126/science.274.5293.1726. [DOI] [PubMed] [Google Scholar]
- 41.Rajagopal S, Kostov KS, Moffat K. Analytical trapping: extraction of time-independent structures from time-dependent crystallographic data. J Struct Biol. 2004;147:211–222. doi: 10.1016/j.jsb.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 42.Brunori M, Bourgeois D, Vallone B. Structural dynamics of myoglobin. Methods Enzymol. 2008;437:397–416. doi: 10.1016/S0076-6879(07)37020-1. [DOI] [PubMed] [Google Scholar]
- 43.Brunori M, Cutruzzolà F, Savino C, Travaglini-Allocatelli C, Vallone B, Gibson QH. Structural dynamics of ligand diffusion in the protein matrix: a study on a new myoglobin mutant Y(B10) Q(E7) R(E10) Biophys J. 1999;76:1259–1269. doi: 10.1016/S0006-3495(99)77289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bourgeois D, Vallone B, Arcovito A, Sciara G, Schotte F, Anfinrud PA, Brunori M. Extended subnanosecond structural dynamics of myoglobin revealed by Laue crystallography. Proc Natl Acad Sci USA. 2006;103:4924–4929. doi: 10.1073/pnas.0508880103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srajer V, Ren Z, Teng TY, Schmidt M, Ursby T, Bourgeois D, Pradervand C, Schildkamp W, Wulff M, Moffat K. Protein conformational relaxation and ligand migration in myogloba nanosecond to millisecond molecular movie from time-resolved Laue X-ray diffraction. Biochemistry. 2001;401:3802–3815. doi: 10.1021/bi010715u. [DOI] [PubMed] [Google Scholar]
- 46.Bourgeois D, Vallone B, Schotte F, Arcovito A, Miele AE, Sciara G, Wulff M, Anfinrud P, Brunori M. Complex landscape of protein structural dynamics unveiled by nanosecond Laue crystallography. Proc Natl Acad Sci USA. 2003;100:8704–8709. doi: 10.1073/pnas.1430900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schotte F, Lim M, Jackson TA, Smirnov AV, Soman J, Olson JS, Phillips GN, Jr, Wulff M, Anfinrud PA. Watching a protein as it functions with 150-ps time-resolved X-ray crystallography. Science. 2003;300:1944–1947. doi: 10.1126/science.1078797. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt M, Nienhaus K, Pahl R, Krasselt A, Anderson S, Parak F, Nienhaus GU, Srajer V. Ligand migration pathway and protein dynamics in myogloba time-resolved crystallographic study on L29W MbCO. Proc Natl Acad Sci USA. 2005;102:11704–11709. doi: 10.1073/pnas.0504932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lim M, Jackson TA, Anfinrud PA. Nonexponential protein relaxation: dynamics of conformational change in myoglobin. Proc Natl Acad Sci USA. 1993;90:5801–5804. doi: 10.1073/pnas.90.12.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ansari A, Berendzen J, Bowne SF, Frauenfelder H, Iben IE, Sauke TB, Shyamsunder E, Young RD. Protein states and proteinquakes. Proc Natl Acad Sci USA. 1985;82:5000–5004. doi: 10.1073/pnas.82.15.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frauenfelder H, McMahon BH, Fenimore PW. Myoglobthe hydrogen atom of biology and a paradigm of complexity. Proc Natl Acad Sci USA. 2003;100:8615–8617. doi: 10.1073/pnas.1633688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scott EE, Gibson QH. Ligand migration in sperm whale myoglobin. Biochemistry. 1997;36:11909–11917. doi: 10.1021/bi970719s. [DOI] [PubMed] [Google Scholar]
- 53.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 54.Hagen SJ, Eaton WA. Non-exponential structural relaxations in proteins. J Chem Phys. 1996;104:3395–3398. [Google Scholar]
- 55.Hummer G, Schotte F, Anfinrud PA. Unveiling functional protein motions with picosecond X-ray crystallography and molecular dynamics simulations. Proc Natl Acad Sci USA. 2004;101:15330–15334. doi: 10.1073/pnas.0405295101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bossa C, Amadei A, Daidone I, Anselmi M, Vallone B, Brunori M, Di Nola A. Molecular dynamics simulation of sperm whale myoglobeffects of mutations and trapped CO on the structure and dynamics of cavities. Biophys J. 2005;89:465–474. doi: 10.1529/biophysj.104.055020. [DOI] [PMC free article] [PubMed] [Google Scholar]


