Abstract
Warfarin-dosing algorithms incorporating CYP2C9 and VKORC1 −1639G>A improve dose prediction compared with algorithms based solely on clinical and demographic factors. However, these algorithms better capture dose variability among whites than Asians or blacks. Herein, we evaluate whether other VKORC1 polymorphisms and haplotypes explain additional variation in warfarin dose beyond that explained by VKORC1 −1639G>A among Asians (n = 1103), blacks (n = 670), and whites (n = 3113). Participants were recruited from 11 countries as part of the International Warfarin Pharmacogenetics Consortium effort. Evaluation of the effects of individual VKORC1 single nucleotide polymorphisms (SNPs) and haplotypes on warfarin dose used both univariate and multi variable linear regression. VKORC1 −1639G>A and 1173C>T individually explained the greatest variance in dose in all 3 racial groups. Incorporation of additional VKORC1 SNPs or haplotypes did not further improve dose prediction. VKORC1 explained greater variability in dose among whites than blacks and Asians. Differences in the percentage of variance in dose explained by VKORC1 across race were largely accounted for by the frequency of the −1639A (or 1173T) allele. Thus, clinicians should recognize that, although at a population level, the contribution of VKORC1 toward dose requirements is higher in whites than in nonwhites; genotype predicts similar dose requirements across racial groups.
Introduction
Warfarin, the most commonly prescribed anticoagulant, exhibits large interpatient variability in dose requirements. Patient-specific factors (eg, age, body size, race, concurrent diseases, and medications) explain some of the variability in warfarin dose, but genetic factors influencing warfarin response explain a significantly higher proportion of the variability in dose.1 Candidate-gene association studies2–22 have identified 2 genes responsible for the main proportion of the genetic effect: CYP2C9, which codes for the enzyme cytochrome P450 2C9 that metabolizes S-warfarin,23,24 and VKORC1, which codes for warfarin's target, vitamin K epoxide reductase.25,26 The influence of CYP2C9 and VKORC1 has also been confirmed by genome-wide association studies among whites.27,28 These studies suggest that identification of common variants in other genes exhibiting influence of magnitude similar to that of CYP2C9 and VKORC1 is unlikely in whites. The most influential CYP2C9 polymorphisms are nonsynonymous coding variants resulting in reduced enzyme activity and decreased metabolic capacity.29–31 In contrast, common VKORC1 variants associated with warfarin dose are noncoding polymorphisms, the effects of which are thought to be mediated through differential expression of the VKOR protein.32 These polymorphisms are within a region of strong linkage disequilibrium (LD) among patients of European ancestry; thus, they may all point to the same common causal polymorphism.10,14 However, neither the causative VKORC1 polymorphism nor the molecular mechanism by which it acts has been completely resolved.
Among whites and Asians, VKORC1 polymorphisms have shown a consistently significant influence on warfarin response, accounting for 11% to 32% of the variability in dose.2–17 Among North American blacks, VKORC1 polymorphisms account for 4% to 10% of the variability in dose.15–18 Given that genetic diversity is known to be greater in persons of African descent,33 investigators have hypothesized that other VKORC1 polymorphisms, or combinations of multiple polymorphisms (haplotypes), may better explain the variation in dose in this group.14,34,35 However, the inadequate representation of blacks in individual cohorts and the limited number of VKORC1 polymorphisms assessed have hindered previous efforts in addressing this hypothesis.
The International Warfarin Pharmacogenetics Consortium (IWPC), through collaborative efforts of 21 research groups from 4 continents, pooled data on 5700 chronic warfarin users to develop a dosing algorithm based on clinical and demographic factors and CYP2C9 and VKORC1 genotypes.36 This dataset (now expanded with 1 additional research group and 556 additional participants) provides a unique opportunity to assess the influence of genes across racial groups. Herein, we assess the influence of common polymorphisms in VKORC1 on warfarin dose among Asians, blacks, and whites. We evaluate whether other VKORC1 polymorphisms and VKORC1 haplotypes explain additional variation in warfarin dose beyond that explained by VKORC1 −1639G>A, the one single nucleotide polymorphism (SNP) included in the IWPC dosing algorithm.
Methods
Data collection
Detailed de-identified participant information was provided by IWPC investigators, including information on clinical and demographic factors influencing warfarin dose requirements, such as age, weight, height, and concomitant use of amiodarone or CYP2C9 inducers.36 All studies were approved by the institutional ethics review committees at the participating organizations, and all participants provided informed consent in accordance with the Declaration of Helsinki.
Main outcome measures
The outcome variable for each patient was the stable therapeutic warfarin dose, defined as the dose of warfarin required to maintain an International Normalized Ratio in a target range. The target range was either prespecified as being between 2 and 3, or, if no prespecified range was reported, the measured International Normalized Ratio was considered to be within target if it fell between 1.7 and 3.3.
Genotyping and linkage disequilibrium calculations
Genotype information for CYP2C9*2 (rs1799853), *3 (rs1057910), and from some sites *5 (rs28371686), *6 (rs9332131), and *11 (rs28371685), plus at least 1 of the 7 VKORC1 SNPs −4451C>A (rs17880887), −1639G>A (rs9923231), 497T>G (rs2884737), 1173C>T (rs9934438), 1542G>C (rs8050894), 2255C>T (rs2359612), 3730G>A (rs7294) was available for all participants. Genotyping quality control was performed as detailed previously.36 The overall genotyping accuracy was greater than 97.7% as assessed by genotyping a random 10% sample of patient DNAs at Academia Sinica, Taiwan. Values for linkage disequilibrium between all pairs of VKORC1 SNPs were calculated with Haploview37 and expressed as r2.
Statistical analysis
Differences in allele frequencies and clinical characteristics among racial groups were compared with Fisher exact tests or Kruskal-Wallis tests for categorical and continuous variables, respectively. Deviation from Hardy-Weinberg equilibrium was evaluated separately for each SNP within each racial group using the χ2 test. Missing values of height and weight were imputed with best-subsets regression, which determines the best fit (highest R2) multiple regression model based on the predictor variables used for the imputation. For the height variable, weight, race, and sex were used for the imputation. For the weight variable, height, race, and sex were used.
For all analyses, the outcome variable warfarin dose (in mg per week) was square root–transformed to attain normality. Evaluation of the effects of individual VKORC1 SNPs and haplotypes on warfarin dose used both univariate and multivariable linear regressions. Covariates included in multivariable linear regression were the same as those included in the IWPC pharmacogenetic dosing algorithm: CYP2C9 *2 and *3, age, height, weight, and concurrent use of CYP2C9 enzyme inducers and amiodarone.36 Because the analyses reported here were done in race-stratified subgroups of the expanded IWPC cohort, the race variables included in the IWPC algorithm were rendered uninformative and were not used. For some analyses, the IWPC algorithm was used to generate warfarin dose predictions and residuals for the patients in the expanded IWPC cohort.
To assess the effect of VKORC1 −1639G>A minor allele frequency (MAF) on R2 (proportion of variance in warfarin dose explained), genotypes with given MAFs were simulated for a cohort of 200 patients.1 The mean dose (and SD) for each unique −1639G>A genotype from the entire IWPC cohort, irrespective of race, was used to randomly draw a normally distributed dose given each patient's individually simulated genotype. This was repeated 1000 times per MAF, and for each simulated study R2 was calculated with the use of linear regression. The resulting R2 distribution of the 1000 simulations was summarized with descriptive statistics. This process was repeated for MAFs ranging from 0 to 1.
Haplotype analysis
VKORC1 haplotypes were inferred with the use of SAS Genetics (SAS Institute). Because different contributing sites genotyped different numbers of VKORC1 SNPs in their patients, the IWPC dataset contains a substantial proportion of missing genotype data. Although we attempted to impute missing genotype information with the use of SAS, PHASE,38 MACH,39 and BEAGLE,40 these algorithms produced inconsistent results, and imputation on a simulated dataset with LD and missing data patterns modeled on the IWPC dataset produced unacceptably high error rates for some of the SNPs. We therefore abandoned this approach.
Genotypes for −4451C>A were missing in 82% of participants. Given the high rate of missing data for this SNP, and the fact that it showed little association with warfarin dose in univariate analysis, it was excluded from haplotype inference. Therefore, haplotype inference was restricted to participants with complete genotype data for the remaining 6 VKORC1 SNPs (−1639G>A, 497T>G, 1173C>T, 1542G>C, 2255C>T, 3730G>A). Complete genotype information on all 6 SNPs was available for 1168 chronic warfarin users, thus restricting the association between VKORC1 haplotype and dose to 247 Asians, 365 blacks, and 556 whites. Haplotypes with frequencies less than 1% in all groups were combined into a single category. The informativeness of VKORC1 haplotypes in a multivariable model was compared with that of the −1639G>A SNP alone in a similar multivariable model with the use of the F statistic. To allow valid comparison between the single SNP- and haplotype-based models, both analyses were conducted in the same set of subjects.
Worldwide haplotype survey
To generate a worldwide survey of VKORC1 haplotype frequencies, VKORC1 haplotypes were inferred from genotype data for 6 SNPs (−1639G>A, 497T>G, 1173C>T, 1542G>C, 2255C>T, 3730G>A) for a total of 8751 subjects. This included 1306 subjects from the IWPC cohort (138 with genotype information only); 317 subjects from 6 different Asian countries with genotype information only41; 316 subjects from 5 countries in South America, Africa, and the Middle East with genotype information only who were recruited as part of the PharmacoGenetics for Every Nation Initiative42; and 6812 participants (2108 non-Hispanic blacks, 2631 non-Hispanic whites, and 2073 Mexican Americans) ascertained as part of the Third National Health and Nutrition Examination Survey (NHANES), a population-based, racially representative cross-sectional study in the United States43 (http://www.cdc.gov/nchs/nhanes.htm).
All analyses were performed with SAS Version 9.1 (SAS Institute), Stata v10 (Stata Corporation; www.stata.com) and R (www.r-project.org) at a nondirectional α of 0.05. Because residual analyses (to identify additional SNP effects) and haplotype analysis involve multiple testing, a more stringent α of 0.0025 was implemented to control for type I error.
Results
Cohort characteristics
The IWPC dataset was expanded to include 6256 chronic warfarin users, incorporating an additional 20 Asians, 230 blacks, 302 whites, and 4 participants of unreported race. A total of 4886 patients of known race (1103 Asians, 670 blacks, and 3113 whites) with complete clinical data comprised the cohort for the current analyses. Indian subjects (n = 91) exhibited substantially different VKORC1 allele frequencies compared with other Asians and were therefore excluded from all analysis (see “Evaluating global haplotype distribution to facilitate application of findings”). There were significant differences in sex and age distribution, body size, and concomitant use of potentially interacting medications among racial groups (Table 1). Warfarin dose requirements were significantly higher among blacks and lower among Asians compared with whites (P < .001).
Table 1.
Clinical and demographic characteristics for the analysis cohort of 4886 participants by race
| Asian (n = 1103) | Black (n = 670) | White (n = 3113) | P* | |
|---|---|---|---|---|
| Clinical characteristics† | ||||
| Warfarin dose, mg/wk | 21 (17.5-28.0) | 40 (30.0-52.5) | 31.5 (21.0-42.0) | < .001 |
| INR achieved | 2.2 (2.0-2.5) | 2.4 (2.2-2.7) | 2.5 (2.2-2.6) | < .001 |
| Height, cm | 161.0 (154.8-167.9) | 170.2 (162.6- 179.0) | 172.7 (163.0-178.0) | < .001 |
| Weight, kg | 60.0 (53.2-69.3) | 87.3 (74.6-104.5) | 82.1 (71.2-95.3) | < .001 |
| Demographic characteristics‡ | ||||
| Male | 596 (54) | 295 (44) | 1899 (61) | < .001 |
| Age | .001 | |||
| 10-19 y | 2 (0.3) | 2 (0.3) | 6 (0.2) | |
| 20-29 y | 15 (1.4) | 21 (3.1) | 69 (2.2) | |
| 30-39 y | 49 (4.5) | 50 (7.5) | 107 (3.4) | |
| 40-49 y | 153 (14.1) | 111 (16.6) | 234 (7.5) | |
| 50-59 y | 247 (22.8) | 163 (24.3) | 524 (16.8) | |
| 60-69 y | 328 (30.3) | 154 (23.0) | 709 (22.8) | |
| 70-79 y | 242 (22.4) | 120 (17.9) | 961 (30.9) | |
| 80-89 y | 45 (4.2) | 43 (6.4) | 478 (15.4) | |
| 90 y and older | 1 (0.1) | 6 (0.9) | 25 (0.8) | |
| Concurrent CYP2C9 inducers | 4 (0.3) | 14 (2.1) | 38 (1.2) | .001 |
| Concurrent amiodarone | 14 (1.3) | 33 (5.0) | 198 (6.4) | < .001 |
Information was missing on age for 21 Asians and on stabilized INR for 4 Asians, 4 blacks, and 431 whites.
INR indicates International Normalized Ratio.
Based on Kruskal-Wallis or Fisher exact test.
Values are medians (interquartile ranges).
Values are n (%).
Genotype distributions for CYP2C9 and VKORC1 were in Hardy-Weinberg equilibrium within each racial group. Although genotype frequencies differed significantly across racial groups (P < .001 for all SNPs), both CYP2C9 and VKORC1 showed significant association with dose in all racial groups (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). As previously reported, LD between VKORC1 SNPs was very strong among whites and Asians but weaker among blacks (supplemental Figure 1). However, 2 SNPs (−1639G>A and 1173C>T) showed strong LD in all races (r2 = 0.97 in Asians, r2 = 1.00 in blacks, and r2 = 0.99 in whites).
Evaluating the performance of the IWPC algorithm by race
The performance of the published IWPC pharmacogenetic algorithm,36 which accounts for the influence of CYP2C9 and VKORC1 −1639G>A, as well as clinical and demographic variables, was evaluated in each of the 3 racial groups in the expanded cohort. The variability in warfarin dose explained by the algorithm (as measured by the coefficient of determination, R2) was higher in whites than in Asians and blacks (Table 2). The mean absolute error of prediction was highest in blacks and lowest in Asians, but in each racial group, the mean absolute error represented between 28% and 31% of the average predicted dose.
Table 2.
Average predicted dose and measures of model fit by racial groups with the IWPC algorithm
| Average predicted dose, mg/wk | Median predicted dose, mg/wk | P* | MAE | SE of MAE | IWPC algorithm R2, % | |
|---|---|---|---|---|---|---|
| Asians | 24.01 | 21.97 | 7.41 | 0.20 | 23.94 | |
| Blacks | 40.14 | 39.74 | < .001 | 11.63 | 0.42 | 24.77 |
| Whites | 32.67 | 32.25 | 9.25 | 0.61 | 40.03 |
The International Warfarin Pharmacogenetics Consortium (IWPC) algorithm adjusts for the influence of VKORC1 −1639G>A, CYP2C9 genotype, age, height, weight, CYP2C9 enzyme inducer status, and amiodarone status on warfarin dose. Model parameters are estimated on the square root of warfarin dose, and the resulting R2 value is calculated by squaring the correlation between the retransformed predicted dose and observed dose.
MAE indicates mean absolute error; and SE, standard error.
P value from a Kruskal-Wallis test comparing predicted dose with racial group.
Which VKORC1 SNP explains the highest proportion of variability in warfarin dose?
To determine which of the 7 VKORC1 SNPs (−4451C>A, −1639G>A, 497T>G, 1173C>T, 1542G>C, 2255C>T, 3730G>A) individually explains the highest proportion of variability in warfarin dose, the influence of each SNP on dose was assessed with the use of univariate analysis in all subjects with genotype information for that SNP (supplemental Table 2). Although providing a general indication of SNPs with large and small effects, the resulting R2 values cannot be directly compared between SNPs, because different subsets of subjects were genotyped for each SNP as a result of different subsets of the 7 VKORC1 SNPs having been genotyped at each contributing site. Therefore, to ensure valid comparisons of predictive ability of individual SNPs, the 4 SNPs (−1639G>A, 1173C>T, 1542G>C, 2255C>T) showing the strongest association with dose were further assessed in a subset of 1690 participants with genotype information for all 4 SNPs. The results of univariate and multivariable analyses adjusting for CYP2C9 genotype and clinical factors are shown in Table 3. Within the Asian and white racial groups, all 4 SNPs were essentially equally predictive of dose, a result in accord with the nearly complete linkage disequilibrium between the SNPs in these groups. In blacks, where LD between some of the SNPs is lower (supplemental Figure 1), it is possible to discriminate between the effects of different SNPs. In this group, −1639G>A and 1173C>T explained the greatest variance in warfarin dose and were essentially equally informative. Given that these 2 SNPs are maximally informative across all the racial groups, and given the nearly complete LD between −1639G>A and 1173C>T within each racial group, and the incorporation of the −1639G>A SNP into several algorithms (including the IWPC algorithm) and multiple clinical genotyping platforms, we chose this polymorphism as the comparator for subsequent analyses.
Table 3.
MAFs and variation in warfarin dose explained (R2) by individual VKORC1 SNPs among Asians, blacks, and whites
| SNP | No. of participants* | MAF, % | Univariate R2, % | Multivariable R2, %† |
|---|---|---|---|---|
| −1639G>A | ||||
| Asian | 641 | 91.03 | 18.4 | 26.7 |
| Black | 368 | 10.05 | 4.2 | 26.3 |
| White | 681 | 37.81 | 22.5 | 48.5 |
| 1173C>T | ||||
| Asian | 641 | 91.03 | 18.4 | 26.7 |
| Black | 368 | 10.19 | 4.5 | 26.2 |
| White | 681 | 37.81 | 22.8 | 48.9 |
| 1542G>C | ||||
| Asian | 641 | 91.03 | 18.4 | 26.7 |
| Black | 368 | 26.36 | 0.2 | 23.0 |
| White | 681 | 38.33 | 22.4 | 48.3 |
| 2255C>T | ||||
| Asian | 641 | 91.03 | 18.4 | 26.7 |
| Black | 368 | 21.33 | 2.6 | 24.7 |
| White | 681 | 37.89 | 22.5 | 48.4 |
Model parameters are estimated on the square root of warfarin dose, and the resulting R2 value is calculated by squaring the correlation between the retransformed predicted dose and observed dose.
SNP indicates single nucleotide polymorphism; and MAF, frequency of the minor allele.
Participants with genotype information on all 4 VKORC1 SNPs.
Restricted to 1638 participants (619 Asian, 354 blacks, and 665 whites) with genotype information on all 4 VKORC1 SNPs and complete data on age, height, weight, CYP2C9 enzyme inducer status, and amiodarone status.
Do combinations of VKORC1 SNPs explain a greater proportion of the dose variability?
Although VKORC1 −1639G>A explains the highest percentage of variation in warfarin dose, other SNPs may have independent effects and may explain additional variation not accounted for by −1639G>A. To determine whether inclusion of other VKORC1 SNPs further improved dose prediction beyond that provided by −1639G>A, their additional contributions were assessed against residuals derived from the IWPC algorithm (supplemental Table 3). Among Asians and blacks, incorporation of an additional SNP did not significantly improve dose prediction (all P > .18) beyond that explained by −1639G>A. In whites the addition of 497T>G, 1173C>T, or 1542G>C improved dose prediction by 0.3% to 1%, with the latter 2 being statistically significant (P < .002) after correction for multiple testing. These minor improvements in prediction are unlikely to be clinically relevant.
Do haplotypes explain a greater proportion of the dose variability than the −1639G>A SNP?
We next explored whether inclusion of VKORC1 haplotypes increased the percentage of variability in dose accounted for by VKORC1 (supplemental Table 4). Haplotypes were inferred from multilocus genotypes for 6 VKORC1 SNPs in all subjects with complete genotype data for those SNPs (247 Asians, 365 blacks, and 556 whites). The predictive ability of VKORC1 haplotypes was compared with that of −1639G>A, after adjustment for CYP2C9 genotype and clinical factors (Table 4). VKORC1 haplotypes did not explain additional variability in warfarin dose in Asians (P = .9), blacks (P = .96), or whites (P = .15) over that explained by −1639G>A SNP alone. Irrespective of race, VKORC1 −1639G>A and VKORC1 haplotypes were equally predictive of dose requirements (P = .99). On the basis of these results the most parsimonious model includes a single informative VKORC1 SNP (−1639G>A or 1173C>T) along with CYP2C9 and a limited set of clinical variables.
Table 4.
Comparative influence of VKORC1 haplotype versus single predictive SNP on warfarin maintenance dose
| Predicted mean dose ± SD (mg/wk) | R2full model % | R2VKORC1 %* | |
|---|---|---|---|
| Asians (n = 247) | |||
| −1639G>A (rs9923231) | 21.55 ± 6.56 | 46.09 | 28.58 |
| VKORC1 haplotype | 21.55 ± 6.52 | 46.08 | 28.57 |
| Blacks (n = 365) | |||
| −1639G>A (rs9923231) | 40.79 ± 8.18 | 27.02 | 4.15 |
| VKORC1 haplotype | 40.80 ± 8.13 | 27.41 | 4.54 |
| Whites (n = 556) | |||
| −1639G>A (rs9923231) | 33.23 ± 11.02 | 50.94 | 23.16 |
| VKORC1 haplotype | 33.15 ± 11.06 | 51.53 | 23.75 |
The full, multivariable model is adjusted for CYP2C9 genotype, age, height, weight, CYP2C9 enzyme inducer status, and amiodarone status. Haplotype analysis and the “SNP versus haplotype comparison” were restricted to include participants with complete genotype data on all 6 SNPs.
Semi-partial R2 for VKORC1 SNP and haplotype in the multivariable model.
Understanding racial differences in the influence of VKORC1 −1639G>A on warfarin dose
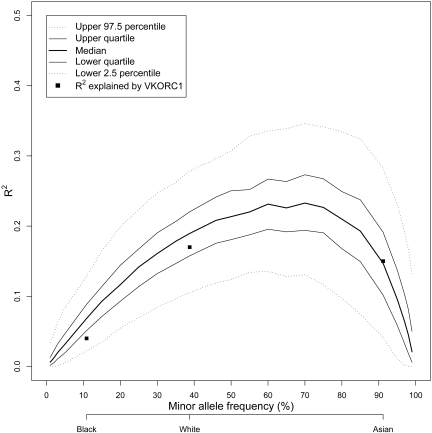
Although possession of the VKORC1 −1639A allele is associated with a similar decrease in the individual warfarin dose requirement irrespective of race (supplemental Figure 2), the variability in dose explained by VKORC1 at a population level varies by race. To explore whether this is due to the differences in MAFs across racial groups, we simulated the effect of −1639A allele frequency on the proportion of variation in warfarin dose explained. Figure 1 shows that as the MAF increases, the percentage of variation in dose explained by −1639A allele increases, with the highest variance explained at MAF of 60% to 70%. These results show that the differences in the percentage of variation explained by VKORC1 across race are driven by the MAF.
Figure 1.
Effect of VKORC1 −1639G>A minor allele frequency in explaining warfarin dose variability at the population level. Results of a simulation study showing the influence of the frequency of the A allele for −1639G>A on the amount of variance in warfarin dose explained (R2). Quartiles and upper and lower 2.5th percentiles of the distribution of R2 values from the simulation are plotted as a function of minor allele frequency. The highest variability in dose explained would occur in a population with an A allele frequency of approximately 60% to 70%. The actual R2 estimates for each racial group in the IWPC data are plotted as squares at the observed MAF for each group.
Evaluating global haplotype distribution to facilitate application of findings
Given the importance of MAF in determining the effect of VKORC1 on warfarin dose at a population level and recognizing global population diversity, we undertook a worldwide survey of VKORC1 haplotype frequencies. Haplotypes from 8751 subjects (including 1168 with warfarin data) representing global population diversity were summarized by the subjects' country of origin (supplemental Table 5). Haplotypes found in at least one country of origin group at a frequency of greater than 2.5% are listed individually as global haplotypes 1 to 7 (GH1-GH7). All other haplotypes are combined into a single category. For the United States, the IWPC subjects are divided into black and white subgroups, and the NHANES subjects are divided into non-Hispanic black, non-Hispanic white, and Mexican American subgroups.
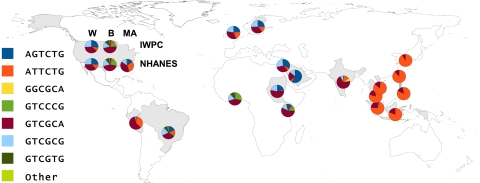
Figure 2 displays haplotype frequency by country of origin. Haplotype distribution within the Asian, black, and white racial groups was consistent across geographically separated populations and differed substantially among the 3 racial groups. Compared with other Asian groups, South Indians showed substantial differences in haplotype distribution, resulting in their exclusion from the analysis of the effect of VKORC1 on warfarin dose.
Figure 2.
Worldwide haplotype distribution. The frequencies of 7 globally distributed haplotypes for 6 SNPs in the VKORC1 gene are represented by pie charts over the country of origin of population samples from which they were derived. The SNPs are listed in the order in which they occur along the VKORC1 gene: −1639G>A, 497T>G, 1173C>T, 1542G>C, 2255C>T, 3730G>A. All haplotypes occurring at frequencies of less than 2.5% in all population samples tested are grouped together in the “Other” category. For the United States, the population samples are broken down into white (W), black (B), and Mexican American (MA) subgroups and by the study under which they were collected: International Warfarin Pharmacogenetics Consortium or National Health and Nutrition Examination Survey III. Detailed data underlying this figure can be found in supplemental Table 5.
Discussion
Warfarin pharmacogenetics has become a case study for personalized medicine. Algorithms incorporating selected SNPs in 2 genes, CYP2C9 and VKORC1, show improved dose prediction compared with algorithms based solely on clinical and demographic factors.8 However, the performance of these algorithms differs between racial groups, with a higher proportion of variability in dose explained in whites than in Asians or blacks.44,45 To explore potential genetic reasons for these differences, we undertook analyses of additional VKORC1 SNPs and haplotypes in a large, multiracial cohort with the aim of determining whether this approach could improve dose prediction in 1 or more of the 3 racial groups. To our knowledge this is the first comprehensive assessment of the influence of 6 common VKORC1 SNPS and haplotypes on warfarin dose among Asians, blacks, and whites with the use of the largest, racially diverse cohort studied to date.
Concordant with prior reports, 2 VKORC1 SNPs (−1639G>A, 1173C>T) were essentially equally informative in capturing the genetic influence of VKORC1 on warfarin dose within each racial group.4,9,14,35 Other common VKORC1 SNPs did not significantly improve model performance in any racial group, either individually or in combination with −1639G>A, nor did haplotypes. The equally informative nature of −1639G>A and 1173C>T across populations enables the use of a common and limited set of polymorphisms that can improve warfarin dose prediction in a racially diverse population.
Although the variability in dose explained by VKORC1 differed by racial group at a population level, at an individual level possession of the minor allele (−1639A or 1173T) was associated with a similar decrease in warfarin dose requirement irrespective of race. This seeming paradox was explained by our simulation study, which showed that the ability of the −1639G>A polymorphism to predict dose at a population level is driven largely by population differences in minor allele frequency. The actual R2 values obtained for the 3 racial groups in our dataset were in line with those expected, based on the simulation. Thus, we caution the reader against deducing that nonwhite patients receive less benefit from genetic-based dosing on the basis of R2 values.
VKORC1 −1639G>A genotype and the 6 SNP haplotype distributions across racial and geographic populations were consistent with prior reports4,9,11,14,35 and with human evolutionary and migratory patterns.46 Blacks (from Africa and the United States) had the lowest frequency of the −1639A allele and exhibited the most diverse haplotype distribution. The East and Southeast Asians were very homogenous, whereas haplotype and −1639A frequencies were substantially different among South Indians. This is consistent with the population substructure in India.47 Within the United States, the genotype and haplotype frequencies among whites and blacks were consistent across the IWPC36 and the NHANES43 cohorts. These similarities increase the generalizability of our findings to the US population at large, because the NHANES cohort was designed as a representative sample of US racial and ethnic subpopulations regardless of health status at the time of ascertainment. Thus, the ascertainment criteria for at least the US components of the IWPC cohort appear not to have introduced substantial selection bias in relation to the US population as a whole.
The NHANES cohort also provided a unique opportunity to understand VKORC1 −1639A frequency and haplotype distribution among Mexican Americans. The changing population demographics in the United States underscore the importance of understanding drug response in this ethnic group. Although we lack a sufficient amount of warfarin data on Hispanics in the IWPC cohort to analyze them as a separate group, our data showing the consistency of genotype effects on dose across racial groups suggest that dose reductions associated with the VKORC1 genotypes are likely to hold true in this ethnic group as well. Moreover, given our results showing the import of the MAF and the high frequency of the −1639A allele in this group (45.3%), the influence of VKORC1 on warfarin dose refinement among Mexican Americans is likely to be substantial.
Of the 2 genes known to have the largest effect at a population level on warfarin dose in white subjects, VKORC1 and CYP2C9, this study has focused primarily on the effects of different SNPs and haplotypes of VKORC1, incorporating CYP2C9 only as a covariate in multiple regression models. For VKORC1, although many studies have shown an effect of individual SNPs or haplotypes on dose,2–22 the causative variant has not previously been identified with certainty, different studies have analyzed different SNPs, generally in relatively small sample cohorts, and the most informative SNP or haplotype for prediction of warfarin dosing has not been clearly delineated, particularly in nonwhite racial groups. In contrast, the effect of CYP2C9 is known to be mediated by nonsynonymous coding SNPs, which result in an enzyme with reduced function.29–31 Any effects of noncoding SNPs in the CYP2C9 gene were shown in 1 study in white subjects to be explained entirely by linkage disequilibrium between these SNPs and the SNPs corresponding to the *2 and *3 alleles, which are known to affect enzyme function.10 Although evaluating additional coding and noncoding SNPs in CYP2C9, particularly in nonwhite cohorts, would be desirable, genotypes for these SNPs were not available in the IWPC dataset. Aggregate allele frequencies for CYP2C9 coding variants that affect function are known to differ between racial groups, ranging in the IWPC dataset from 4% in Asians and 5% in blacks to 20% in whites, and it is to be expected that these differences may account for some of the population-level difference in performance of warfarin-dosing algorithms between racial groups. However, given the smaller range of allele frequencies across racial groups for CYP2C9 compared with that for VKORC1, it is probable that the relative contribution to racial differences in dosing is larger for VKORC1 than for CYP2C9.
Despite the strengths, we recognize limitations of our study. First, because of nonrandom patterns of missing data across study sites and therefore racial groups, we could not include cultural and lifestyle factors, such as dietary intake of vitamin K, smoking, alcohol consumption, and medication adherence. Population differences with respect to these factors could explain some of the differences in warfarin dosing across racial groups. Second, there were also nonrandom patterns of missing genotypes, because different sites genotyped different subsets of the VKORC1 SNPs. Although we attempted to impute missing genotype data, inconsistency in performance of imputation algorithms forced restriction of haplotype analysis to a subset of the cohort with complete genotype data. We acknowledge that this restriction probably decreased the power to detect smaller effects of individual haplotypes. However, this restriction maintained the validity of the haplotype analysis.
Third, we did not assess rare, coding variants in VKORC1 that produce extreme warfarin resistance, the frequency of which may vary between racial groups.22,26,48 In addition, we only assessed VKORC1 SNPs within or in close proximity to the gene; therefore, we cannot rule out the additional influence of regulatory polymorphisms far from the gene. Finally, we did not account for other genetic factors with a minor influence on warfarin dose such as apolipoprotein E,10,49 γ-glutamyl carboxylase,9,50 and cytochrome P450 4F2.20,28 In general, the other genes identified to date as influencing warfarin dose have relatively small population-level effect sizes, and results from independent studies often produce inconsistent results. However, most of these genes have not been evaluated in large population cohorts with broad racial representation, and some of these genes may account for a portion of the population-level difference in performance of warfarin-dosing algorithms between racial groups. For example, one study comparing US whites and African Americans showed a significant effect of APOE genotype only in the African American group.49 CYP4F2, which has the largest effect size in whites after VKORC1 and CYP2C9, accounting for approximately 1% to 2% of variation in warfarin dose,20,28 did not show an association with dose in a Han Chinese cohort.51 Although genome-wide association studies in white participants suggest that other genes with effect sizes as large as those of VKORC1, CYP2C9, and CYP4F2 are unlikely to be found,27,28 genome-wide studies of warfarin dosing have yet to be performed in nonwhite racial groups, and it is possible that additional genes with large effect sizes may be found that will help to explain more of the variation in warfarin dosing in these groups. The IWPC is currently undertaking such a genome-wide association study in blacks.
In summary, the influence of common genetic variation in VKORC1 on variability in warfarin dose was captured by a single polymorphism (either −1639G>A or 1173C>T) across all racial groups. Incorporation of additional VKORC1 SNPs or haplotypes did not improve dose prediction. Therefore, current evidence supports the use of −1639G>A (or 1173C>T) to capture dose variability related to VKORC1. Both VKORC1 and CYP2C9 influenced warfarin dose among individual patients in all 3 racial groups studied. Although the contribution of genetic variation in VKORC1 to the proportion of variability in dose explained at a population level was higher in whites than in blacks and Asians, our analysis shows that these differences were largely explained by differences in allele frequencies across populations. Thus, clinicians should recognize that nonwhite patients probably have similar benefit from genetic-based dosing as do white patients despite lower population R2 values.
Acknowledgment
We thank Jennifer Evertsen (Center for Urban Population Health, Milwaukee, WI) for reviewing the manuscript.
This work was supported by grants from the National Institutes of Health (NIH/NIGMS Pharmacogenetics Research Network: GM61374, GM074492, GM63340, HL65962; NHLBI: HL092173, HL068834, HL066176, HL074724, HL71083; NIGMS: GM081488; NINDS: NS053646, NS45598); HHMI Research Training Fellowship; National Research Program for Genomic Medicine, National Science Council, Taiwan (National Clinical Core, NSC95-3112-B-001-010; National Genotyping Center, NSC95-3112-B-001-011); Shin Korea Health 21 R&D Project, Ministry for Health, Welfare and Family Affairs, Korea (A030001), and The Medical Research Center Program, the Ministry of Science and Technology, Korea (R13-2007-023-00000-0); Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq); Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ); Financiadora de Estudos e Projetos (FINEP); American Association of Colleges of Pharmacy; American Heart Association; University of Illinois at Chicago Hans Vahlteich Research Award; United Kingdom Department of Health; Sweden: the Science Council (Medicine), Heart and Lung Foundation, Society of Medicine, Foundation for Strategic Research, the Söderberg, Thuréus and Selander Foundations, the Clinical Research Support (ALF) at Uppsala University, and the Wellcome Trust.
Our complete dataset of genotypes and clinical variables will be available on publication of this manuscript to registered PharmGKB users at http://www.pharmgkb.org (full dataset accession no. PA165109609).
Appendix
International Warfarin Pharmacogenetics Consortium (listed alphabetically by institution): Ming-Ta M. Lee, Yuan-Tsong Chen, Liang-Suei Lu (Academia Sinica, Taiwan, ROC); Ming-Shien Wen (Chang Gung Memorial Hospital, Chang Gung University, Taiwan, ROC); Yoseph Caraco, Idit Achache, Simha Blotnick, Mordechai Muszkat (Hadassah Medical Organization, Israel); Jae-Gook Shin, Ho-Sook Kim (Inje University, Korea); Guilherme Suarez-Kurtz, Jamila Alessandra Perini (Instituto Nacional de Câncer, Brazil); Edimilson Silva-Assunção (Instituto Nacional de Cardiologia Laranjeiras, Brazil); Jeffrey L. Anderson, Benjamin D. Horne, John F. Carlquist (Intermountain Healthcare, United States); Michael D. Caldwell, Richard L. Berg, James K. Burmester (Marshfield Clinic, United States); Boon Cher Goh, Soo-Chin Lee (National University Hospital, Singapore); Farhad Kamali, Elizabeth Sconce, Ann K. Daly (Newcastle University, United Kingdom); Alison Motsinger-Reif (North Carolina State University, United States); (Nita A. Limdi (University of Alabama, United States); Russ B. Altman, Hersh Sagrieya, Teri E. Klein, Balaji S. Srinivasan (Stanford University, United States); Alan H. B. Wu (University of California, San Francisco, United States); Julie A. Johnson, Taimour Y. Langaee, Hua Feng (University of Florida, United States); Larisa Cavallari, Kathryn Momary (University of Illinois, Chicago, United States); Munir Pirmohamed, Andrea Jorgensen, Cheng Hok Toh, Paula Williamsom (University of Liverpool, United Kingdom); Michael J. Wagner, Howard McLeod, James P. Evans, Karen E. Weck (University of North Carolina, United States); Stephen E. Kimmel, Colleen Brensinger (University of Pennsylvania, United States); Yusuke Nakamura, Taisei Mushiroda (University of Tokyo and RIKEN Center for Genomic Medicine, Japan); David Veenstra, Lisa Meckley, Mark J. Rieder, Allan E. Rettie (University of Washington, United States); David Page, Eric Lantz, Tim Chang (University of Wisconsin-Madison, United States); Mia Wadelius, Niclas Eriksson, Håkan Melhus (Uppsala University, Sweden); C. Michael Stein, Dan M. Roden, Ute Schwartz, Daniel Kurnik, Marylyn Ritchie (Vanderbilt University, United States); Brian F. Gage, Elena Deych, Petra Lenzini, Charles Eby (Washington University in St Louis, United States); Leslie Y. Chen, Panos Deloukas (Wellcome Trust Sanger Institute, United Kingdom).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.A.J., T.E.K., and M.J.W. designed the research; L.C., D.C.C., B.F.G., J.A.J., A.J., S.E.K., M.-T.M.L., N.A.L., M.P., H.S., J.-G.S., G.S.-K., M.W., M.J.W., and A.H.B.W. collected data; L.C., D.C.C., N.E., J.A.J., T.E.K, M.-T.M.L., N.A.L., A.M.-R., H.S., M.W., M.J.W., and A.H.B.W. analyzed and interpreted data; L.C., C.-H.C., D.C.C., N.E., N.L., A.M.-R., and H.S. performed statistical analysis; L.C., D.C.C., N.E., J.A.J., T.E.K, N.A.L., A.M.-R., M.W., and M.J.W. wrote the manuscript; and all authors reviewed the final version of the manuscript.
Conflict-of-interest disclosure: L.C. has received funding from Osmetech Molecular Diagnostics, Pasadena, CA. B.F.G. has received grant support from Osmetech Molecular Diagnostics, Pasadena, CA. J.A.J. is the Vice Chair of the steering committee and a site principal investigator for the NHLBI COAG trial and is on an advisory board for Medco. S.E.K. has received consulting fees from Pfizer, GlaxoSmithKline, Novartis, and Centocor and grant support from the Aetna Foundation, Shire, and Pfizer. M.J.W. has received an honorarium from Eli Lilly. The remaining authors declare no competing financial interests.
Correspondence: Michael J. Wagner, Institute for Pharmacogenomics and Individualized Therapy, University of North Carolina at Chapel Hill, Genetic Medicine Bldg, Rm 1092, 120 Mason Farm Rd, Chapel Hill, NC 27599-7361; e-mail: michael_wagner@unc.edu.
References
- 1.Wadelius M, Pirmohamed M. Pharmacogenetics of warfarin: current status and future challenges. Pharmacogenomics J. 2007;7(2):99–111. doi: 10.1038/sj.tpj.6500417. [DOI] [PubMed] [Google Scholar]
- 2.D'Andrea G, D'Ambrosio RL, Di Perna P, et al. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood. 2005;105(2):645–649. doi: 10.1182/blood-2004-06-2111. [DOI] [PubMed] [Google Scholar]
- 3.Sconce EA, Khan TI, Wynne HA, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106(7):2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 4.Veenstra DL, You JH, Rieder MJ, et al. Association of Vitamin K epoxide reductase complex 1 (VKORC1) variants with warfarin dose in a Hong Kong Chinese patient population. Pharmacogenet Genomics. 2005;15(10):687–691. doi: 10.1097/01.fpc.0000174789.77614.68. [DOI] [PubMed] [Google Scholar]
- 5.Carlquist JF, Horne BD, Muhlestein JB, et al. Genotypes of the cytochrome p450 isoform, CYP2C9, and the vitamin K epoxide reductase complex subunit 1 conjointly determine stable warfarin dose: a prospective study. J Thromb Thrombolysis. 2006;22(3):191–197. doi: 10.1007/s11239-006-9030-7. [DOI] [PubMed] [Google Scholar]
- 6.Obayashi K, Nakamura K, Kawana J, et al. VKORC1 gene variations are the major contributors of variation in warfarin dose in Japanese patients. Clin Pharmacol Ther. 2006;80(2):169–178. doi: 10.1016/j.clpt.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Borgiani P, Ciccacci C, Forte V, Romano S, Federici G, Novelli G. Allelic variants in the CYP2C9 and VKORC1 loci and interindividual variability in the anticoagulant dose effect of warfarin in Italians. Pharmacogenomics. 2007;8(11):1545–1550. doi: 10.2217/14622416.8.11.1545. [DOI] [PubMed] [Google Scholar]
- 8.Gage BF, Eby C, Johnson JA, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84(3):326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wadelius M, Chen LY, Downes K, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5(4):262–270. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- 10.Wadelius M, Chen LY, Eriksson N, et al. Association of warfarin dose with genes involved in its action and metabolism. Hum Genet. 2007;121(1):23–34. doi: 10.1007/s00439-006-0260-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wadelius M, Chen LY, Lindh JD, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009;113(4):784–792. doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Y, Shennan M, Reynolds KK, et al. Estimation of warfarin maintenance dose based on VKORC1 (-1639 G>A) and CYP2C9 genotypes. Clin Chem. 2007;53(7):1199–1205. doi: 10.1373/clinchem.2006.078139. [DOI] [PubMed] [Google Scholar]
- 13.Cho HJ, Sohn KH, Park HM, et al. Factors affecting the interindividual variability of warfarin dose requirement in adult Korean patients. Pharmacogenomics. 2007;8(4):329–337. doi: 10.2217/14622416.8.4.329. [DOI] [PubMed] [Google Scholar]
- 14.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352(22):2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 15.Limdi NA, Arnett DK, Goldstein JA, et al. Influence of CYP2C9 and VKORC1 polymorphisms on warfarin dose, anticoagulation attainment and maintenance among European American and African Americans. Pharmacogenomics. 2008;9(5):511–526. doi: 10.2217/14622416.9.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schelleman H, Chen Z, Kealey C, et al. Warfarin response and vitamin K epoxide reductase complex 1 in African Americans and Caucasians. Clin Pharmacol Ther. 2007;81(5):742–747. doi: 10.1038/sj.clpt.6100144. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi H, Wilkinson GR, Nutescu EA, et al. Different contributions of polymorphisms in VKORC1 and CYP2C9 to intra- and inter-population differences in maintenance dose of warfarin in Japanese, Caucasians and African-Americans. Pharmacogenet Genomics. 2006;16(2):101–110. doi: 10.1097/01.fpc.0000184955.08453.a8. [DOI] [PubMed] [Google Scholar]
- 18.Momary KM, Shapiro NL, Viana MA, Nutescu EA, Helgason CM, Cavallari LH. Factors influencing warfarin dose requirements in African-Americans. Pharmacogenomics. 2007;8(11):1535–1544. doi: 10.2217/14622416.8.11.1535. [DOI] [PubMed] [Google Scholar]
- 19.Aquilante CL, Langaee TY, Lopez LM, et al. Influence of coagulation factor, vitamin K epoxide reductase complex subunit 1, and cytochrome P450 2C9 gene polymorphisms on warfarin dose requirements. Clin Pharmacol Ther. 2006;79(4):291–302. doi: 10.1016/j.clpt.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 20.Caldwell MD, Awad T, Johnson JA, et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111(8):4106–4112. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mushiroda T, Ohnishi Y, Saito S, et al. Association of VKORC1 and CYP2C9 polymorphisms with warfarin dose requirements in Japanese patients. J Hum Genet. 2006;51(3):249–253. doi: 10.1007/s10038-005-0354-5. [DOI] [PubMed] [Google Scholar]
- 22.Scott SA, Edelmann L, Kornreich R, Desnick RJ. Warfarin pharmacogenetics: CYP2C9 and VKORC1 genotypes predict different sensitivity and resistance frequencies in the Ashkenazi and Sephardi Jewish populations. Am J Hum Genet. 2008;82(2):495–500. doi: 10.1016/j.ajhg.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaminsky LS, Zhang ZY. Human P450 metabolism of warfarin. Pharmacol Ther. 1997;73(1):67–74. doi: 10.1016/s0163-7258(96)00140-4. [DOI] [PubMed] [Google Scholar]
- 24.Rettie AE, Korzekwa KR, Kunze KL, et al. Hydroxylation of warfarin by human cDNA-expressed cytochrome P-450: a role for P-4502C9 in the etiology of (S)-warfarin-drug interactions. Chem Res Toxicol. 1992;5(1):54–59. doi: 10.1021/tx00025a009. [DOI] [PubMed] [Google Scholar]
- 25.Li T, Chang CY, Jin DY, Lin PJ, Khvorova A, Stafford DW. Identification of the gene for vitamin K epoxide reductase. Nature. 2004;427(6974):541–544. doi: 10.1038/nature02254. [DOI] [PubMed] [Google Scholar]
- 26.Rost S, Fregin A, Ivaskevicius V, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427(6974):537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 27.Cooper GM, Johnson JA, Langaee TY, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112(4):1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeuchi F, McGinnis R, Bourgeois S, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5(3):e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crespi CL, Miller VP. The R144C change in the CYP2C9*2 allele alters interaction of the cytochrome P450 with NADPH:cytochrome P450 oxidoreductase. Pharmacogenetics. 1997;7(3):203–210. doi: 10.1097/00008571-199706000-00005. [DOI] [PubMed] [Google Scholar]
- 30.Haining RL, Hunter AP, Veronese ME, Trager WF, Rettie AE. Allelic variants of human cytochrome P450 2C9: baculovirus-mediated expression, purification, structural characterization, substrate stereoselectivity, and prochiral selectivity of the wild-type and I359L mutant forms. Arch Biochem Biophys. 1996;333(2):447–458. doi: 10.1006/abbi.1996.0414. [DOI] [PubMed] [Google Scholar]
- 31.Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics. 1994;4(1):39–42. doi: 10.1097/00008571-199402000-00005. [DOI] [PubMed] [Google Scholar]
- 32.Wang D, Chen H, Momary KM, Cavallari LH, Johnson JA, Sadee W. Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 (VKORC1) affects gene expression and warfarin dose requirement. Blood. 2008;112(4):1013–1021. doi: 10.1182/blood-2008-03-144899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The International HapMap Consortium. Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geisen C, Watzka M, Sittinger K, et al. VKORC1 haplotypes and their impact on the inter-individual and inter-ethnical variability of oral anticoagulation. Thromb Haemost. 2005;94(4):773–779. doi: 10.1160/TH05-04-0290. [DOI] [PubMed] [Google Scholar]
- 35.Limdi NA, Beasley TM, Crowley MR, et al. VKORC1 polymorphisms, haplotypes and haplotype groups on warfarin dose among African-Americans and European-Americans. Pharmacogenomics. 2008;9(10):1445–1458. doi: 10.2217/14622416.9.10.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.International Warfarin Pharmacogenetics Consortium. Klein TE, Altman RM, Eriksson N, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360(8):753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 38.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73(6):1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Abecasis G. Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- 40.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81(5):1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee MT, Chen CH, Chuang HP, et al. VKORC1 haplotypes in five East-Asian populations and Indians. Pharmacogenomics. 2009;10(10):1609–1616. doi: 10.2217/pgs.09.80. [DOI] [PubMed] [Google Scholar]
- 42.Marsh S, Van Booven DJ, McLeod HL. Global pharmacogenetics: giving the genome to the masses. Pharmacogenomics. 2006;7(4):625–631. doi: 10.2217/14622416.7.4.625. [DOI] [PubMed] [Google Scholar]
- 43.Chang MH, Lindegren ML, Butler MA, et al. Prevalence in the United States of selected candidate gene variants: Third National Health and Nutrition Examination Survey, 1991-1994. Am J Epidemiol. 2009;169(1):54–66. doi: 10.1093/aje/kwn286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cavallari LH, Limdi NA. Warfarin pharmacogenomics. Curr Opin Mol Ther. 2009;11(3):243–251. [PubMed] [Google Scholar]
- 45.Schelleman H, Chen J, Chen Z, et al. Dosing algorithms to predict warfarin maintenance dose in Caucasians and African Americans. Clin Pharmacol Ther. 2008;84(3):332–339. doi: 10.1038/clpt.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461(7263):489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aklillu E, Leong C, Loebstein R, Halkin H, Gak E. VKORC1 Asp36Tyr warfarin resistance marker is common in Ethiopian individuals. Blood. 2008;111(7):3903–3904. doi: 10.1182/blood-2008-01-135863. [DOI] [PubMed] [Google Scholar]
- 49.Kimmel SE, Christie J, Kealey C, et al. Apolipoprotein E genotype and warfarin dosing among Caucasians and African Americans. Pharmacogenomics J. 2008;8(1):53–60. doi: 10.1038/sj.tpj.6500445. [DOI] [PubMed] [Google Scholar]
- 50.Rieder MJ, Reiner AP, Rettie AE. Gamma-glutamyl carboxylase (GGCX) tagSNPs have limited utility for predicting warfarin maintenance dose. J Thromb Haemost. 2007;5(11):2227–2234. doi: 10.1111/j.1538-7836.2007.02744.x. [DOI] [PubMed] [Google Scholar]
- 51.Lee MT, Chen CH, Chou CH, et al. Genetic determinants of warfarin dosing in the Han-Chinese population. Pharmacogenomics. 2009;10(12):1905–1913. doi: 10.2217/pgs.09.106. [DOI] [PubMed] [Google Scholar]