Abstract
The Mre11–Rad50–Nbs1 (MRN) heterotrimer plays various and complex roles in DNA damage repair and checkpoint signaling. Its role in activating Ataxia-Telangiectasia Mutated (ATM), the central checkpoint kinase in the metazoan double-strand break response, has been well studied. However, its function in the checkpoint independent of ATM activation, as well as functions that are completely checkpoint independent, are less well understood. In fission yeast, DNA damage checkpoint signaling requires Rad3, the homolog of the ATR (ATM and Rad3-related) kinase, not Tel1, the ATM homolog, allowing us to dissect MRN's ATM-independent S-phase DNA damage checkpoint roles from its role in ATM activation. We find that MRN is involved in Rad3 (ATR)-dependent checkpoint signaling in S phase, but not G2, suggesting that MRN is involved in ATR activation through its role in replication fork metabolism. In addition, we define a role for MRN in the S-phase DNA damage checkpoint-dependent slowing of replication that is independent of its role in checkpoint signaling. Genetic interactions between MRN and Rhp51, the fission yeast Rad51 homolog, lead us to suggest that MRN participates in checkpoint-dependent replication slowing through negative regulation of recombination.
CELLS must respond to DNA damage appropriately to maintain genomic fidelity, prevent mutation, and successfully duplicate and segregate genetic information. Cells employ checkpoints to respond to DNA damage and coordinate cell cycle progression with repair (Hartwell and Weinert 1989; Kastan and Bartek 2004). One of the central responses to DNA damage is the S-phase DNA damage checkpoint, which slows replication in response to DNA damage during S phase (Sancar et al. 2004). In metazoans, this checkpoint acts through either the Ataxia-Telangiectasia Mutated (ATM) or ATM and Rad3-related (ATR) kinase, depending on the type of damage; ATM responds to double-strand breaks and ATR to replication blocking lesions, such as those caused by UV, methyl methane sulfonate (MMS), or camptothecin (McGowan and Russell 2004). In yeast, the checkpoint responds to both types of DNA damage though the ATR homolog—Rad3 in the fission yeast Schizosaccharomyces pombe and Mec1 in budding yeast Saccharomyces cerevisiae (Rhind and Russell 1998).
In fission yeast, the S-phase and G2 DNA damage checkpoints are activated through a conserved protein kinase cascade in which Rad3 phosphorylates and activates one of two downstream effector kinases (Rhind and Russell 2000). Rad3 phosphorylates the S-phase-specific checkpoint kinase Cds1 in response to DNA damage during replication and Chk1 in response to G2 damage (Walworth and Bernards 1996; Lindsay et al. 1998; Martinho et al. 1998; Brondello et al. 1999). The downstream checkpoint targets for prevention of mitosis are well understood (Sancar et al. 2004). However, the downstream players in the S-phase DNA damage checkpoint and mechanisms utilized to slow replication are still unclear.
Checkpoint regulation of cell-cycle progression allows cells the opportunity to repair DNA damage. The Mre11–Rad50–Nbs1 (MRN) recombinational-repair complex serves roles in both checkpoint activation and DNA damage repair. MRN is a heterotrimer composed of proteins with diverse biochemical functions: Mre11 is a single-strand endo- and 3′ exonuclease; Rad50 is an SMC-like protein with DNA binding, ATPase, and adenylate kinase activities; and Nbs1 is the putative regulatory subunit (D'amours and Jackson 2002). Mre11's nuclease activity is involved in processing DSBs for homologous recombinational repair and signaling (Lobachev et al. 2002; Lewis et al. 2004; Jazayeri et al. 2006). Rad50 constitutively associates with Mre11 and may tether DNA molecules in close proximity for efficient repair (de Jager et al. 2001). Nbs1 is required for MRN nuclear localization and DNA damage- and replication-dependent focus formation (Mirzoeva and Petrini 2001). Nbs1 stimulates the DNA binding and nuclease activities displayed by the Mre11–Rad50 heterodimer (Paull and Gellert 1999). MRN is involved in an array of recombinational-repair pathways. However, its role seems to be modulatory or redundant, because although MRN mutations disrupt many repair pathways, they do not eliminate any of them (Symington 2002).
MRN is required for the S-phase DNA damage checkpoint response, which slows replication in response to DNA damage (D'amours and Jackson 2002; Falck et al. 2002; Chahwan et al. 2003). Two pathways contribute to checkpoint-dependent replication slowing in mammals: the first inhibits origin firing via the degradation of Cdc25 in a Chk2-dependent manner, the second is MRN-dependent (Falck et al. 2002). This MRN activity appears to be independent of its role in checkpoint signaling since the Chk2 target Cdc25A is degraded normally in Nijmegen breakage syndrome cells, which are mutant for the Nbs1 subunit (Buscemi et al. 2001; Falck et al. 2002). Although its role in slowing replication is unknown, MRN associates with sites of ongoing replication and forms foci at stalled forks, suggesting that MRN acts to slow replication in a fork-dependent manner (Maser et al. 2001; Mirzoeva and Petrini 2003).
In metazoa, MRN acts both as an upstream activator and downstream target of the checkpoint kinase ATM and ATR. MRN is involved in recognition of double-stand breaks and the activation of ATM, placing it at the very earliest steps in checkpoint signaling (Uziel et al. 2003; Horejsi et al. 2004; Lisby et al. 2004; Lee and Paull 2007). MRN is also a target of ATM phosphorylation in response to ionizing radiation (IR) (Lim et al. 2000) and serves a downstream role in slowing replication independent of its role in checkpoint signaling (Falck et al. 2002). MRN serves independent up- and downstream roles during S-phase, being required for both strong ATR activation and slowing replication in response to UV (Olson et al. 2007). These multiple roles for MRN in metazoan checkpoint response to DNA damage have made it difficult to determine how MRN contributes to the regulation of replication in response to DNA damage.
Schizosaccharomyces pombe offers a convenient system to study the role of the MRN complex in the S-phase DNA damage checkpoint. First, unlike in metazoa, MRN is not essential, allowing the analysis of null alleles. Second, although MRN's role in the activation of ATM is conserved in fission yeast, Tel1, the fission yeast ATM homolog, is not required for checkpoint activation. Therefore, given the involvement of MRN in the S-phase DNA damage checkpoint in both humans and fission yeast, and the fact that Tel1(ATM) is not required for this checkpoint in fission yeast, we can directly investigate the roles of MRN in the checkpoint that are independent of its role in ATM activation.
MATERIALS AND METHODS
Strains used in this study are listed in Table 1; they were constructed using previously published alleles or those described below (Nakamura et al. 2002; Akamatsu et al. 2003; Chahwan et al. 2003; Willis and Rhind 2009). Yeast strains were grown in yeast extract with supplements (YES) at 25° or 30° and manipulated using standard methods (Forsburg and Rhind 2006). Strains containing the cds1 overexpression cassette were grown in Edinburgh minimal medium supplemented with leucine, uracil, adenine, and histidine (EMM LUAH) with or without 15 μm of thiamine.
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| yFS105 | h− leu1-32 ura4-D18 |
| yFS162 | h− leu1-32 ura4-D18 ade6-210 cdc10-m17 |
| yFS199 | h− leu1-32 ura4-D18 cds1∷ura4 |
| yFS206 | h− leu1-32 ura4-D18 chk1∷pBF206 (chk1-9myc2HA6His ura4) |
| yFS249 | h− leu1-32 ura4-D18 nbs1∷kanMX6 |
| yFS260 | h− leu1-32 ura4-D18 cdc10-m17 rad3∷ura4 |
| yFS263 | h− leu1-32 ura4-D18 ade6-m210 his3-D1 cdc10-m17 rad32∷kanMX6 |
| yFS265 | h− leu1-32 ura4-D18 ade6-m210 his3-D1 cdc10-m17 rad50∷kanMX6 |
| yFS270 | h+ leu1-32 ura4-? rad3∷ura4 |
| yFS281 | h− leu1-32 ura4-D18 ade6-m210 his3-D1 cdc10-m17 tel1∷kanMX4 |
| yFS299 | h− ura4-D18 cds1∷ura4 chk1∷pBF206 (chk1-9myc2HA6His ura4) |
| yFS543 | h+ leu1-32 ura4-D18 ade6-210 cdc10-m17 cds1∷ura4 |
| yFS553 | h+ leu1-32 ura4-D18 his3-D1 cdc10-m17 swi5∷his3 |
| yFS556 | h+ leu1-32 ura4-D18 ade6-? his7-366 cdc10-m17 rhp51∷natMX6 |
| yFS639 | h+ leu1-32 ura4-D18 cdc10-m17 nbs1∷kanMX6 rhp51∷natMX6 |
| yFS640 | h− leu1-32 ura4-D18 his3-D1 cdc10-m17 nbs1∷kanMX6 swi5∷his3 |
| yFS641 | h− leu1-32 ura4-D18 ade6-? cdc10-m17 cds1∷ura4 nbs1∷kanMX6 |
| yFS689 | h+ leu1-32 ura4-D18 nbs1∷kanMX6 chk1∷pBF206 (chk1-9myc2HA6His ura4) |
| yFS690 | h− leu1-32 ura4-D18 rad3∷ura4 chk1∷pBF206 (chk1-9myc2HA6His ura4) |
| yFS704 | h− leu1-32 ura4-D18 ade6-? cdc10-m17 rad32∷kanMX4 rad51∷natMX6 |
| yFS705 | h+ leu1-32 ura4-D18 ade6-? cdc10-m17 rad50∷kanMX6 rad51∷natMX6 |
| yFS707 | h− leu1-32 ura4-D18 his3-D1 cdc10-m17 nbs1∷kanMX6 sfr1∷ura4 |
| yFS708 | h+ leu1-32 ura4-D18 his3-D1 cdc10-m17 nbs1∷kanMX6 rad51∷natMX6 sfr1∷ura4 |
| yFS709 | h+ leu1-32 ura4-D18 ade6-M210 cdc10-m17 nbs1∷kanMX6 swi1∷kanMX6 |
| yFS723 | h− leu1-32 ura4-? ade6-? rad3∷ura4 tel1∷kanMX4 |
| yFS724 | h− leu1-32∷pBF162 (nmt1:GST-cds1 leu1) ura4-D18 ade6-M210 cdc10-M17 mik1∷ura4 nmt1:pyp3 (kanMX6) rad3∷ura4 nbs1∷kanMX6 tel1∷kanMX4 |
| yFS730 | h+ leu1-32 ura4-D18 nbs1∷kanMX tel1∷leu2 |
| yFS731 | h+ leu1-32 ura4-D18 ade6-210 cdc10-M17 nbs1∷kanMX tel1∷leu2 |
Cell synchronization:
For S-phase analysis, strains were synchronized, harvested, and prepared for flow cytometry or Cds1 in vitro kinase assay as previously described (Willis and Rhind 2009). Briefly, G1 arrested cells were synchronized by centrifugal elutriation, selecting for a population of cells that replicate quickly upon release. cdc10-M17 cells were arrested for 2 hr at 35° and the smallest cells, those that had most recently arrested in G1, were collected by centrifugal elutriation and released to 25°. Cells were exposed to 200 Gray (Gy) ionizing radiation using a Faxitron RX-650 (10 Gy per minute starting at 5 min postelutriation) or treated with 0.03% methyl methane sulfonate (MMS) (Sigma M4016) or 10 mm hydroxyurea (HU) (Sigma H8627). At 20-min intervals after elutriation, 0.5 OD of cells were collected and fixed in 70% ethanol.
Sample preparation for flow cytometry:
Whole cells display significant background fluorescence when analyzed by flow cytometry, and this background increases as cells elongate in a cell-cycle arrest (Sazer and Sherwood 1990). To reduce this background and improve the signal-to-noise ratio in our experiments, we adapted a protocol to analyze isolated nuclei DNA content (Carlson et al. 1997; Willis and Rhind 2009). Briefly, ethanol-fixed cells were pelleted, washed, and spheroplasted. Spheroplasts were Triton washed, treated with RNase A, and disrupted by sonication to release intact nuclei. Sonicated cells were stained with Sytox Green (Invitrogen S7020) and analyzed on a Becton-Dickinson FACScan cytometer.
S-phase progression analysis:
S-phase progression was measured using CellQuest software (Becton-Dickinson version 3.3). The unreplicated value was determined by measuring DNA content of freshly elutriated G1 cultures and fully replicated values were determined by measuring nuclear DNA content from asynchronous, prearrested cultures and untreated cultures having completed replication. Mean values for S-phase peaks through out time courses were compared with these unreplicated and fully replicated values. Progression through S phase was determined using the equation: % progression = (C − A)/(B − A), where A = 1C, B = 2C, and C = mean histogram value. Error bars represent the standard error of the mean when n > 2 and show the variance of the data when n = 2.
Cds1 in vitro kinase assay:
Cds1 kinase activity was measured as previously described (Lindsay et al. 1998; Kai et al. 2005; Willis and Rhind 2009). Briefly, whole-cell lysates were prepared from 5 OD of cells and total protein normalized between samples and incubated with anti-Cds1 antibodies (a gift from T. Wang) bound to protein-A sepharose beads (GE Health Sciences). Immunoprecipitated Cds1 was washed and incubated with myelin basic protein (Sigma M1891) in kinase reaction buffer with 32P-ATP. Reactions were resolved by SDS–PAGE and analyzed on a phosphorimager. Sample Cds1 activity was normalized to the Cds1 kinase activity displayed by MMS-treated wild-type control cells at 120 min after elutriation. Each experimental set contained a wild-type control allowing for the comparison of strains used in independent experiments. Cds1 kinase activity was also normalized to percentage of G1 (%G1) cells in the freshly elutriated cultures of each strain, as determined by flow cytometry. This normalization is required because G2 cells treated with 0.03% MMS do not complete mitosis and thus do not contribute to measured Cds1 activity (our unpublished results). For comparison of Cds1 activity elicited by different treatments, raw phosphoimager counts were normalized first to the %G1 content of each treated strain and then to the wild-type control treated with MMS for each assay. For the asynchronous kinase assays, asynchronous cultures were grown to 0.5 OD/ml and then treated with 0.015% MMS for 4 hr or left untreated. Ten OD cells were pelleted and prepared as described above. It was assumed that all cells capable of slowing during S phase will be in mid-S phase after 4 hr of MMS exposure, therefore no normalization for cell cycle position was performed.
cds1 overexpression:
cds1 was overexpressed from the strong, thiamine-repressible nmt1 promoter. nmt1:GST-cds1 cells were maintained in EMM LUAH with 15 μm of thiamine to repress cds1 expression. Sixteen hours prior to elutriation, cells were transferred to EMM LUAH without thiamine to induce cds1 expression. To prevent a Cds1-induced G2 arrest, we used a genetic background in which the G2 arrest is overridden by deletion of mik1, a Cdc2 inhibitory tyrosine kinase activated by Cds1, and overexpression of Pyp3, a Cdc2 activating tyrosine phosphatase that can constitutively drive the G2/M transition (Rhind and Russell 2000). The cells also carry the cdc10-M17 allele allowing for G1 synchronization, as described above.
Chk1 Western blotting:
Cells were synchronized in G2 by centrifugal elutriation and immediately exposed to 200 Gy ionizing radiation using a Faxitron RX-650 (10 Gray per minute starting at 5 min postelutriation). A total of 15 OD pellets were collected and frozen in liquid nitrogen. Frozen cells were thawed in 200 μl ice-cold lysis buffer (150 mm NaCl, 50 mm Tris-HCl pH 8.0, 5 mm EDTA pH 8.0, 10% glycerol, 1% NP-40, 50 mm NaF, 5 μg/ml leupeptin, 1 mm PMSF, 1 mm Na3VO4) and broken by vortexing with silica beads for 15 min. Crude cell lysates were cleared by centrifugation (3000 × g for 5 min) and lysate protein concentration determined using the BCA Protein Assay kit (Pierce 23225). A total of 150 μg total protein was separated using SDS–PAGE on 8% 16 × 18 cm gels run 18 hr at 10 mA. Bis-acrylamide:acrylamide ratio of 1:100 was used to separate phosphorylated from unphosphorylated Chk1. Chk1 was detected using mouse anti-HA (Covance MMS-101P) antibody diluted 1:1000 in 5% milk TBST (0.05% Tween-20) overnight at 4°, followed by goat anti-mouse secondary conjugated to HRP 1:3000 in 5% milk TBST for 3 hr at room temperature.
RESULTS
MRN mutants display partial defects in slowing:
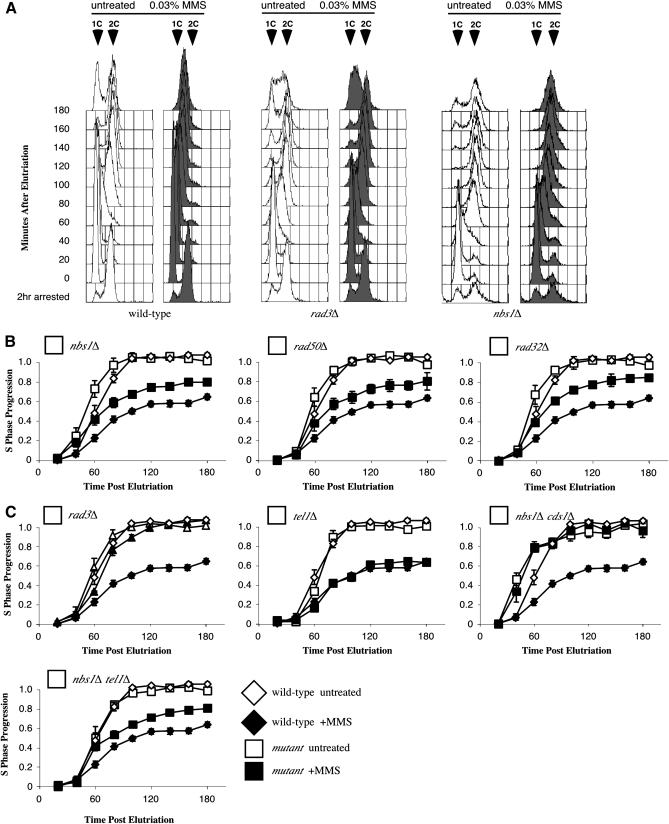
We previously reported that MRN is required for checkpoint-dependent replication slowing in fission yeast (Chahwan et al. 2003). In higher eukaryotes, MRN is only partially required for slowing in response to DNA damage (Falck et al. 2002). To more carefully examine the role of MRN in the checkpoint, we refined our flow cytometry protocol, analyzing isolated nuclei rather than whole cells (Willis and Rhind 2009). Using this more sensitive assay, we found that the MRN mutants retain some ability to slow replication in response to DNA damage. Histogram overlays of the collected data are shown in Figure 1A. Unperturbed wild-type cells displayed a shift from 1C unreplicated nuclear DNA content to 2C replicated content ∼60 min after release from G1. Wild-type cells treated with 0.03% MMS instead display a gradual shift toward 2C over the course of the assay. In contrast, rad3Δ mutants fail to slow replication in response to MMS. MRN mutants display an intermediate phenotype with some reduction in replication in response to DNA damage (Figure 1A). Quantitation of mean peak shift from 1C toward 2C nuclear DNA content better illustrates the difference between wild type and MRN mutants (Figure 1B). Both wild-type and MRN mutant cells replicate in a similar manner in the absence of DNA damage. However, wild-type strains slow replication by ∼40% in the presence of MMS, while nbs1Δ, rad50Δ, and rad32Δ (the fission yeast homolog of MRE11) mutants slow by only ∼25% (Figure 1B, open vs. closed symbols). The slowing displayed in MRN mutants requires the presence of the Cds1 checkpoint kinase; nbs1Δ cds1Δ double mutants display a complete defect in slowing (Figure 1C). Replication slowing in fission is dependent upon the central checkpoint kinase Rad3 (ATR) and not the related kinase Tel1 (ATM); rad3Δ mutants fail to slow while tel1Δ mutants slow well in response to MMS and the nbs1Δ tel1Δ double mutant still retains some ability to slow (Figure 1C).
Figure 1.—
The Rad32–Rad50–Nbs1 complex is required for full replication slowing in response to DNA damage in fission yeast. (A) S-phase flow cytometry histogram overlays comparing wild-type (yFS162), nbs1Δ (yFS267), and rad3Δ (yFS260) strains. G1 cells were synchronized by cdc10-m17 arrest and elutriation and released in the presence or absence of 0.03% MMS. Cells were fixed every 20 min and nuclear DNA content measured by flow cytometry. Wild-type cells treated with 0.03% MMS display replication slowing as shown by a lack of significant peak shifting after release. nbs1Δ strains display intermediate slowing as shown by less reduction in peak shifting toward 2C than the wild-type control, while rad3Δ cells fail to slow, displaying similar profiles in the presence and absence of MMS. (B) All three MRN mutants display partial defects in slowing in response to DNA damage. Progression through S phase is plotted with respect to time by measuring the shifting of the mean of S-phase peaks from unreplicated 1C toward fully replicated 2C values. Wild-type cells (yFS162, n = 3) slow bulk S-phase progression while strains harboring deletion alleles of nbs1Δ (yFS267, n = 7), rad32 (yFS263, n = 2) and rad50 (yFS265, n = 2) display intermediate slowing in response to DNA damage during S phase. (C) Slowing displayed in nbs1Δ mutants is checkpoint dependent. The rad3Δ mutant (yFS260, n = 3) fails to slow displaying a complete checkpoint defect while tel1Δ mutants slow normally (yFS281, n = 4). nbs1Δ cds1Δ double mutants (yFS641, n = 3) also fail to slow while the nbs1Δ tel1Δ (yFS731, n = 3) mutant acts like the nbs1Δ single mutant.
MRN mutants display a partial defect in checkpoint signaling:
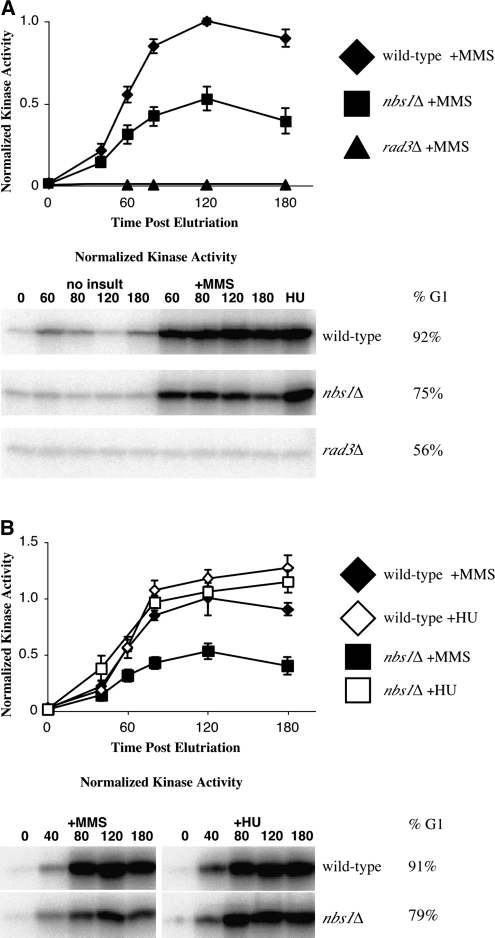
To investigate the cause of the slowing defect in MRN mutants, we focused on the nbs1Δ mutant strain. As previously described, null mutants for nbs1, rad50, and rad32 display identical phenotypes in fission yeast (Chahwan et al. 2003; Ueno et al. 2003). We first investigated the role of Nbs1 in S-phase checkpoint signaling by measuring Cds1 kinase activity using an in vitro assay. nbs1Δ strains display intermediate checkpoint signaling in response to MMS (Figure 2A). Like wild-type, nbs1Δ cells display weak periodic Cds1 activity in unperturbed cells and increased Cds1 activity in response to MMS exposure during replication. However, nbs1Δ strains display only ∼50% of the MMS-induced Cds1 activity of wild-type cells (Figure 2A). In contrast to both wild-type and nbs1Δ mutants, rad3Δ mutants display a total defect in Cds1 activation in response to MMS and HU (Lindsay et al. 1998 and Figure 2). Despite nbs1Δ signaling defects in response to MMS, nbs1Δ mutants retain strong signaling in response to HU (Figure 2B).
Figure 2.—
The Rad32–Rad50–Nbs1 complex is required for full S-phase DNA damage checkpoint activation during replication in response to MMS. (A) nbs1 mutation reduces checkpoint signaling in response to MMS. nbs1Δ (yFS267, n = 8, G1 enrichment range 28–91%) mutants display reduced Cds1 kinase activity relative to wild-type (yFS162, n = 8, G1 enrichment range 54–92%) cells in response to MMS. rad3Δ (yFS260, n = 1) cells display a complete defect in Cds1 activity in response to DNA damage. (B) nbs1Δ (yFS267, n = 6, G1 enrichment range 47–91%) mutants display proficient checkpoint signaling relative to wild-type (yFS162, n = 8, G1 enrichment range 70–94%) cells in response to HU arrest. The MMS data is from A. Signal was normalized within each experiment to the 120-min MMS-treated wild-type culture. Representative raw data and G1 enrichment is shown.
MRN mutant slowing defect is recombinase dependent:
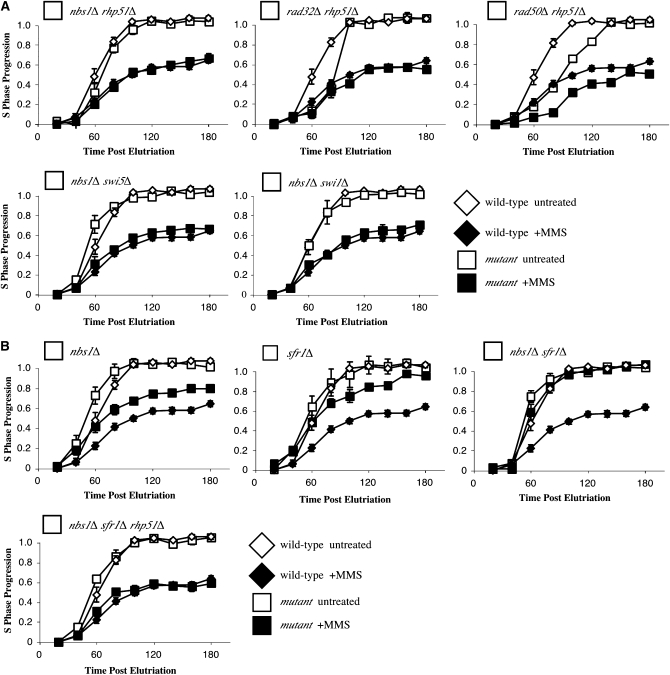
Previously, we have shown regulation of recombination is required for checkpoint-dependent slowing of replication in fission yeast (Willis and Rhind 2009). In particular, loss of Rhp51, the fission yeast homolog of Rad51, suppresses the defect in checkpoint-dependent replication slowing seen in rqh1Δ mutants, a helicase involved in regulation of recombination and processing of stalled replication forks. Since the MRN complex is involved in recombinational repair and thought to act at stalled replication forks, we tested whether the nbs1Δ slowing defect was also recombination dependent. nbs1Δ rhp51Δ and nbs1Δ swi5Δ double mutants both slow replication in response to MMS treatment, showing that loss of the Rhp51 recombinase or the Swi5 mediator, which is required for Rhp51 activity, suppress the MRN mutants' slowing defect (Figure 3A). Slowing defects exhibited by the other two MRN complex mutants are also suppressed by rhp51 removal (Figure 3A). The Swi1 replication fork protection complex is also required for slowing defects exhibited by several strains including rqh1Δ (Willis and Rhind 2009). We found the same true for the nbs1Δ mutant; nbs1Δ swi1Δ double mutants slowed well (Figure 3A).
Figure 3.—
Abrogation of recombination suppresses the nbs1Δ slowing defect. (A) The slowing defect displayed by MRN mutants is recombination dependent. The partial slowing defect seen in MRN mutants is suppressed in the nbs1Δ rhp51Δ (yFS639, n = 3), rad32Δ rhp51Δ (yFS704, n = 2), and rad50Δ rhp51Δ (yFS705, n = 1) double mutants. nbs1Δ defects are also suppressed in the nbs1Δ swi5Δ (yFS640, n = 4) and nbs1Δ swi1Δ (yFS709, n = 2) double mutants. (B) Both nbs1Δ (yFS267, data from 1B) and sfr1Δ (yFS201, n = 3) mutants display a partial defect in slowing while the nbs1Δ sfr1Δ (yFS707, n = 1) mutant displays a complete defect. This defect is suppressed in the nbs1Δ sfr1Δ rhp51Δ (yFS708, n = 2) triple mutant.
Nbs1 and Sfr1 define two independent functions in the checkpoint pathway:
Like nbs1Δ cells, sfr1Δ and rqh1Δ cells display a partial slowing defect in response to MMS that is suppressed by rhp51Δ (Willis and Rhind 2009). These results raise the question of whether MRN cooperates with Rqh1 and Sfr1 or acts independently to slow replication. To distinguish between these possibilities, we tested whether the nbs1Δ and sfr1Δ mutant slowing defects are additive; rqh1Δ cannot be tested because rqh1Δ and nbs1Δ are synthetically lethal. The nbs1Δ sfr1Δ double mutant displayed a complete defect in slowing, suggesting that Nbs1 and Sfr1 function independently to slow replication (Figure 3B). The additive slowing defect was suppressed by removing recombination (Figure 3B). Together, these results suggest that Nbs1 and Sfr1 define two independent functions that allow for replication slowing by negatively regulating Rhp51-depedent recombination.
MRN mutant checkpoint signaling defect is not recombinase dependent:
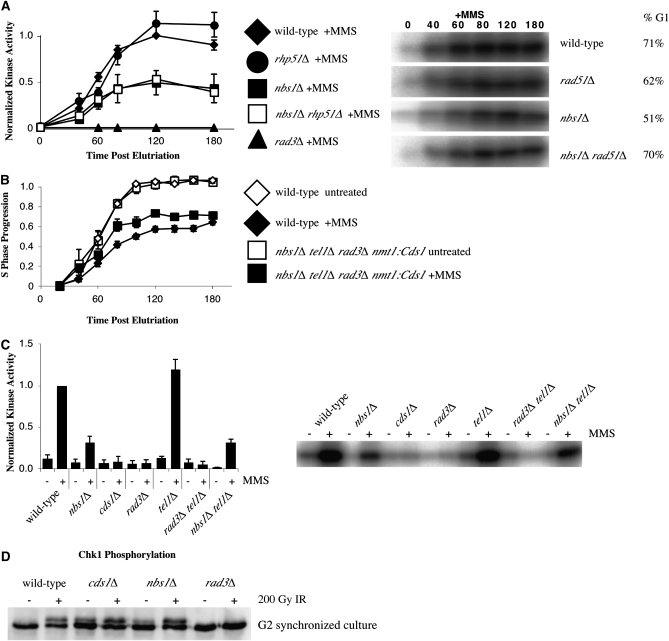
To determine whether removing recombination rescued both the slowing and signaling phenotypes of the nbs1Δ strain, Cds1 activation was compared between the nbs1Δ rhp51Δ double mutant, both single mutants, and wild type. Although both wild-type and rhp51Δ strains display strong Cds1 activity in the presence of MMS, both nbs1Δ and nbs1Δ rhp51Δ mutants display reduced Cds1 activation (Figure 4A). This data shows that Rhp51-dependent recombination is not involved in MRN's signaling function and suggests that the signaling defect exhibited by nbs1Δ cells is not directly responsible for their slowing defect.
Figure 4.—
The nbs1Δ slowing defect is not a direct consequence of its checkpoint signaling defect. (A) Removing recombination does not rescue the checkpoint signaling defect exhibited by nbs1Δ cells. rhp51Δ (yFS556, n = 4, G1 enrichment range 51–83%) and wild-type (yFS162, data from 2A, n = 8, G1 enrichment range 54–92%) cells display similar checkpoint signaling in response to MMS while nbs1Δ (yFS267, data from 2A, n = 8, G1 enrichment range 28–91%) and the nbs1Δ rhp51Δ (yFS639, n = 5, G1 enrichment range 25–70%) double mutant display reduced checkpoint signaling. Representative raw data and G1 enrichment is shown. (B) Overproduction of Cds1 fails to rescue the slowing defect exhibited in the nbs1Δ tel1Δ rad3Δ nmt1:Cds1 mutant (yFS724). (C) Tel1 is not required for Cds1 activation in response to MMS during S phase as measured by asynchronous kinase assay. Wild type (yFS105) displays strong Cds1 activity while cds1Δ (yFS199), rad3Δ (yFS270), and rad3Δ tel1Δ (yFS723) mutants all display a complete defect in Cds1 activation. The nbs1Δ (yFS249) and nbs1Δ tel1Δ (yFS730) mutants display a partial signaling defect and tel1Δ displays strong Cds1 activation. (D) Nbs1 is not involved in phosphorylation of the G2 DNA damage checkpoint kinase Chk1. The nbs1Δ strain (yFS639) displays wild-type levels of phosphorylation of Chk1 in response to 200 Gy ionizing radiation when compared with the rad3Δ (yFS690), cds1Δ (yFS299), and wild-type (yFS06) controls.
Constitutive Cds1 signaling does not suppress the MRN mutant slowing defect:
As an independent test of whether the nbs1Δ signaling defect contributes to its slowing defect, we constitutively activated Cds1 signaling by overproduction of the kinase. Overproduction of Cds1 constitutively activates its G2 arrest function (Brondello et al. 1999). Moreover, the slowing defects exhibited by rad3Δ and mrc1Δ mutants are suppressed by Cds1 overproduction, confirming that these genes act upstream of cds1 (Willis and Rhind 2009). To test whether constitutive activation of Cds1 suppresses the nbs1Δ slowing defect, we overexpressed cds1 from the strong nmt1 promoter in a nbs1Δ tel1Δ rad3Δ strain and assayed whether the strain could slow replication in response to MMS. The rad3Δ and tel1Δ mutations together insure that there is no MMS-induced checkpoint signaling; in addition, the strain is designed to be unable to arrest in G2 in response to constitutive Cds1 activation (see materials and methods). We found that overproducing Cds1 does not suppress the slowing defect displayed in the nbs1Δ tel1Δ rad3Δ nmt1:cds1 strain (Figure 4B). The fact that nbs1Δ still prevents full replication slowing in cells that constitutively overproduce Cds1 suggests that the nbs1Δ slowing defect is independent of Nbs1's role in Cds1 activation and that nbs1 acts downstream of cds1 to regulate replication slowing.
MMS-induced checkpoint signaling is independent of Tel1:
As an independent test of whether Tel1 contributes to Cds1 signaling, and if MRN is involved in activation of Cds1 in a Tel1-dependent manner in response to MMS, we measured Cds1 activity in a number of checkpoint kinase null mutants. Asynchronous kinase assays were performed comparing wild-type, nbs1Δ, cds1Δ, rad3Δ, tel1Δ, rad3Δ tel1Δ, and nbs1Δ tel1Δ mutants (Figure 4C). As shown previously, the nbs1Δ mutant displays a partial defect in signaling and both the cds1Δ and rad3Δ controls display a complete lack of signaling. The tel1Δ mutant displays strong signaling and the nbs1Δ tel1Δ double displays a partial defect in signaling, indicating that Tel1 is not required for activating Cds1 in response to MMS either in the absence or presence of the MRN complex. As shown in Figure 1, tel1Δ mutants are proficient in slowing replication in response to MMS. The fact that nbs1Δ mutants display signaling and slowing defects while the tel1Δ mutant does not, suggests that the nbs1Δ slowing defect is independent of any upstream role in Tel1 activation and that Tel1 does not play a significant role in checkpoint signaling or response to insult during replication in fission yeast.
MRN is not required for checkpoint signaling in G2:
The requirement of Nbs1 for full Rad3-dependent signaling in S phase raised the possibility that MRN may be involved in all Rad3-dependent signaling. We have previously shown that MRN is not required for a DNA-damage-induced G2 cell-cycle delay, but the fact that MRN is involved in DNA repair means that cells lacking MRN actually delay longer than wild-type cells, complicating the analysis (Chahwan et al. 2003). To directly test whether MRN is required for Rad3-dependent checkpoint signaling in G2, we measured the phosphorylation of Chk1, a direct Rad3 target, in response to ionizing radiation (Walworth and Bernards 1996; Martinho et al. 1998). Because endogenous DNA damage accumulates in nbs1Δ cells, asynchronous cultures display constitutive Chk1 phosphorylation (data not shown). We used elutriation to select the smallest, and thus not checkpoint arrested, G2 cells from asynchronous cultures. We treated these cells with 200 Gy ionizing radiation and found that nbs1Δ cells phosphorylated Chk1 to the same extent as wild-type cells (Figure 4D). These results show that MRN is not required for Rad3-dependent checkpoint activation in G2.
DISCUSSION
We show that the MRN complex has both up- and downstream roles in the fission yeast S-phase DNA damage checkpoint. Much like MRN mutants in metazoa, fission yeast MRN mutants display reduced ability to slow replication, as well as reduced checkpoint signaling in response to DNA damage. However, our results are more straightforward to interpret because checkpoint signaling in fission yeast relies on the ATR homolog Rad3, not the ATM homolog Tel1. This situation allows us to investigate the roles of MRN that are independent of its role in ATM activation. Our results place MRN in a well-defined genetic pathway that regulates replication slowing and suggest that MRN acts by negatively regulating recombinational repair of DNA during S phase.
MRN acts upstream of checkpoint signaling in the S-phase DNA damage checkpoint:
We find that MRN is required for Rad3 (ATR) dependent signaling in S phase. In nbs1Δ cells, Rad3-dependent activity of Cds1 reaches only half the level seen in wild-type cells (Figure 2A). However, nbs1Δ mutants do display much stronger signaling in response to HU arrest (Figure 2B). This result shows that MRN's role in checkpoint signaling is not confined to its role in the activation of ATM and is consistent with other reports of MRN-dependent activation of ATR and Mec1 (Nakada et al. 2004; Stiff et al. 2005). However, it also shows that MRN's requirement for Rad3 activation is only partial, suggesting that MRN is not intimately involved in Rad3 activation. Furthermore, although MRN is required for full phosphorylation of Cds1 in response to S-phase DNA damage, MRN is not required for checkpoint signaling in response to replication arrest by HU (Figure 2B) or G2 in response to IR (Chahwan et al. 2003 and Figure 3D). Taken together, these results suggest that MRN's role in activating Rad3 is indirect, perhaps through its role in replication fork metabolism.
MRN acts downstream of or parallel to Cds1 in the S-phase DNA damage checkpoint:
Although nbs1Δ cells show partial defects in both checkpoint signaling and replication slowing, our results suggest that the slowing phenotype is independent of the signaling phenotype. The slowing defect in response to MMS can be suppressed by deletion of rhp51 without suppressing the signaling defect (Figures 3A and 4A). As discussed below, we have established a similar epistatic relationship with other recombinational regulators (Willis and Rhind 2009). In addition, the nbs1Δ partial slowing defect is not suppressed by constitutively activating Cds1 signaling by over production (Figure 4B). Although Cds1 over-production rescues the slowing defects of both rad3Δ and mrc1Δ mutants, nbs1Δ strains continue to fail to slow (Figure 4B and Willis and Rhind 2009). These results show that the nbs1Δ signaling defect is neither necessary nor sufficient for its slowing defect and thus suggest that MRN's checkpoint-dependent role in slowing replication is downstream of or parallel to Cds1 checkpoint signaling. The fact that nbs1Δ blocks replication slowing in cells that produce constitutively active Cds1 leads us to prefer the description of MRN as downstream of cds1. However, this description is in the genetic sense and does not imply that MRN is necessarily downstream of Cds1 in the biochemical sense of being a target of Cds1 phosphorylation.
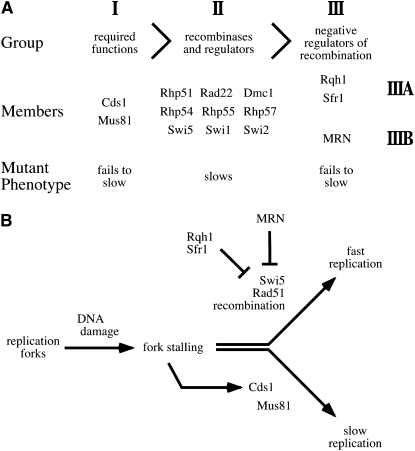
MRN defines a new epistasis group in the S-phase DNA damage checkpoint pathway:
We have recently shown that checkpoint-dependent replication slowing requires the Mus81 endonuclease and the Rqh1 helicase (Willis and Rhind 2009). Furthermore, rhp51Δ suppresses the slowing defect of rqh1Δ but not mus81Δ. This epistatic relationship allowed us to define three epistasis groups in the S-phase DNA damage checkpoint pathway: Group I contains Cds1, Mus81 and its binding partner Eme1, Group II contains Rhp51 and other recombinational regulators; and Group III contains Rqh1 and Sfr1 (Willis and Rhind 2009 and Figure 5A). The suppression of the MRN mutants' slowing defects by rhp51Δ places MRN in group III. However, the additive nature of the nbs1Δ and sfr1Δ slowing phenotypes allows us to subdivide group III into IIIA containing Rqh1 and Sfr1 and IIIB containing MRN. This analysis suggests that MRN acts independently of, but in parallel to, Rqh1 and Sfr1 to negatively regulate Rhp51-dependent recombination.
Figure 5.—
The genetic structure of the S-phase DNA damage pathway and a model for its function. (A) The placement of MRN in the epistasis relationship of the genes involved in checkpoint-dependent slowing of replication (Willis and Rhind 2009). The mus81Δ rhp51Δ double mutant has the same slowing-defective phenotype as mus81Δ, thus mus81Δ is epistatic to rhp51Δ. Likewise, the rqh1Δ rhp51Δ double mutant has the same slowing-proficient phenotype as rhp51Δ, thus rhp51Δ is epistatic to rqh1Δ. The analysis in this article places the MRN mutations in epistasis group III with rqh1Δ and sfr1Δ. However, the additive nature of the sfr1Δ and nbs1Δ phenotypes allow us to subdivide the group into subgroups IIIA and IIIB. (B) A model for the checkpoint-dependent regulation of replication slowing. Cds1 induces Mus81-dependent slowing. However, this slowing requires Rhp51-dependent recombination be held in check by a combination of MRN, Sfr1, and Rqh1.
The role of MRN in the S-phase DNA damage checkpoint:
The epistasis analysis described above suggests that MRN negatively regulates Rhp51-dependent recombination, which, if not inhibited, disrupts Mus81-dependent replication slowing (Figure 5B). This result, along with observation that the nbs1Δ sfr1Δ double mutant completely fails to slow, suggests that the checkpoint in fission yeast is largely dependent on the regulation of recombination. A role in the regulation of recombination is consistent with recombinational phenotypes of MRN mutants, which include altered recombination patterns, reduced recombination between sister-chromatids and increased use of homologs for recombinational repair (Bressan et al. 1999; Paques and Haber 1999; Hartsuiker et al. 2001; Symington 2002; Van et al. 2002; Gonzalez-Barrera et al. 2003). However, the MRN nuclease activity is not required for slowing in response to DNA damage in fission yeast (de Jager et al. 2001; Limbo et al. 2007; Porter-Goff and Rhind 2009). Therefore, we speculate that MRN alters Rhp51-mediated recombination through its DNA-binding activity at forks, possibly by bridging of the two nascent sister chromatids during replication (de Jager et al. 2001; Tittel-Elmer et al. 2009).
In mammals, MRN is thought to work in parallel with a Chk2-Cdc25A pathway that regulates origin firing (Falck et al. 2002; Yazdi et al. 2002). Origin firing is also regulated in fission yeast (Kumar and Huberman 2009). However, Cdc25 regulation is not required in fission yeast to slow replication (Kommajosyula and Rhind 2006). Furthermore, our results show that slowing is abrogated in nbs1Δ sfr1Δ double mutants or mus81Δ single mutants (Figure 3B and Willis and Rhind 2009). These results suggest that slowing is largely dependent on the regulation of recombination and that origin regulation does not contribute significantly to the checkpoint-dependent, MMS-induced bulk slowing of replication seen in fission yeast.
The budding yeast homolog of MRN is not required to slow replication in response to MMS-induced DNA damage (Andrews and Clarke 2005). We suspect that this observation reflects a mechanistic difference in the way the two distantly related yeast deal with such damage. Budding yeast slow bulk replication in response to 0.03% MMS less extensively than fission yeast; S phase is lengthened by between 1 and 2 hr in budding yeast as compared to more than 5 hr for fission yeast (Chang et al. 2002; Andrews and Clarke 2005 and our unpublished data). Previously, the budding yeast rad50Δ mutant was described as displaying a slowing defect (Chang et al. 2002). However, the poor synchrony caused by the sickness of the rad50Δ strain complicates the interpretation of that data. Furthermore, the bulk slowing in budding yeast is dependent on the regulation of origin firing (Tercero and Diffley 2001). We propose that budding yeast relies less on checkpoint-dependent, recombination-based fork slowing and therefore do not display a strong requirement for MRN in the slowing process.
We show the MRN complex plays both upstream and downstream roles in the fission yeast S-phase DNA damage checkpoint. However, epistatic analysis of the nbs1Δ and rhp51Δ mutants suggests that, although MRN mutants display reduced checkpoint signaling, loss of the downstream function negatively regulating recombination is responsible for the slowing defect displayed by MRN mutants. Our results place MRN in a well-defined genetic pathway for the checkpoint-dependent regulation of replication in response to DNA damage that will provide an important framework for future mechanistic investigation of MRN and the S-phase DNA damage checkpoint.
Acknowledgments
We thank P. Russell and H. Iwasaki for kindly providing us strains used in this work, T. Wang for the kind gift of the anti-Cds1 antibody, O. Limbo for help optimizing the Chk1 Westerns, and members of the Rhind lab for helpful discussions and experimental assistance. This work was funded by National Institutes of Health GM069957 (to N.R.).
References
- Akamatsu, Y., D. Dziadkowiec, M. Ikeguchi, H. Shinagawa and H. Iwasaki, 2003. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc. Natl. Acad. Sci. USA 100 15770–15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, C. A., and D. Clarke, 2005. MRX (Mre11/Rad50/Xrs2) mutants reveal dual intra-S-phase checkpoint systems in budding yeast. Cell Cycle 4 1073–1077. [PubMed] [Google Scholar]
- Bressan, D., B. Baxter and J. Petrini, 1999. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 19 7681–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondello, J., M. Boddy, B. Furnari and P. Russell, 1999. Basis for the checkpoint signal specificity that regulates Chk1 and Cds1 protein kinases. Mol. Cell. Biol. 19 4262–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscemi, G., C. Savio, L. Zannini, F. Micciché, D. Masnada et al., 2001. Chk2 activation dependence on Nbs1 after DNA damage. Mol. Cell. Biol. 21 5214–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, C., B. Grallert, R. Bernander, T. Stokke and E. Boye, 1997. Measurement of nuclear DNA content in fission yeast by flow cytometry. Yeast 13 1329–1335. [DOI] [PubMed] [Google Scholar]
- Chahwan, C., T. Nakamura, S. Sivakumar, P. Russell and N. Rhind, 2003. The fission yeast Rad32 (Mre11)-Rad50-Nbs1 complex is required for the S-phase DNA damage checkpoint. Mol. Cell. Biol. 23 6564–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, M., M. Bellaoui, C. Boone and G. Brown, 2002. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl. Acad. Sci. USA 99 16934–16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'amours, D., and S. Jackson, 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell. Biol. 3 317–327. [DOI] [PubMed] [Google Scholar]
- de Jager, M., J. van Noort, J. van Gent, C. Dekker, R. Kanaar et al., 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8 1129–1135. [DOI] [PubMed] [Google Scholar]
- Falck, J., J. Petrini, B. Williams, J. Lukas and J. Bartek, 2002. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat. Genet. 30 290–294. [DOI] [PubMed] [Google Scholar]
- Forsburg, S., and N. Rhind, 2006. Basic methods for fission yeast. Yeast 23 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Barrera, S., F. Cortes-Ledesma, R. Wellinger and A. Aguilera, 2003. Equal sister chromatid exchange is a major mechanism of double-strand break repair in yeast. Mol. Cell 11 1661–1671. [DOI] [PubMed] [Google Scholar]
- Hartsuiker, E., E. Vaessen, A. Carr and J. Kohli, 2001. Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J. 20 6660–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell, L., and T. Weinert, 1989. Checkpoints: controls that ensure the order of cell cycle events. Science 246 629–634. [DOI] [PubMed] [Google Scholar]
- Horejsi, Z., J. Falck, C. J. Bakkenist, M. B. Kastan, J. Lukas et al., 2004. Distinct functional domains of Nbs1 modulate the timing and magnitude of ATM activation after low doses of ionizing radiation. Oncogene 23 3122–3127. [DOI] [PubMed] [Google Scholar]
- Jazayeri, A., J. Falck, C. Lukas, J. Bartek, G. C. Smith et al., 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell. Biol. 8 37–45. [DOI] [PubMed] [Google Scholar]
- Kai, M., M. Boddy, P. Russell and T. Wang, 2005. Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes Dev. 19 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan, M., and J. Bartek, 2004. Cell-cycle checkpoints and cancer. Nature 432 316–323. [DOI] [PubMed] [Google Scholar]
- Kommajosyula, N., and N. Rhind, 2006. Cdc2 tyrosine phosphorylation is not required for the S-phase DNA damage checkpoint in fission yeast. Cell Cycle 5 2495–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., and J. Huberman, 2009. Checkpoint-dependent regulation of origin firing and replication fork movement in response to DNA damage in fission yeast. Mol. Cell. Biol. 29 602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., and T. Paull, 2007. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 26 7741–7748. [DOI] [PubMed] [Google Scholar]
- Lewis, L., F. Storici, S. Van Komen, S. Calero, P. Sung et al., 2004. Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics 166 1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, D. S., S. T. Kim, B. Xu, R. S. Maser, J. Lin et al., 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404 613–617. [DOI] [PubMed] [Google Scholar]
- Limbo, O., C. Chahwan, Y. Yamada, R. A. de Bruin, C. Whittenberg et al., 2007. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell 28 134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, H. D., D. J. Griffiths, R. J. Edwards, P. U. Christensen, J. M. Murray et al., 1998. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 12 382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby, M., J. Barlow, R. Burgess and R. Rothstein, 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118 699–713. [DOI] [PubMed] [Google Scholar]
- Lobachev, K., D. Gordenin and M. Resnick, 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108 183–193. [DOI] [PubMed] [Google Scholar]
- Martinho, R., H. D. Lindsay, G. Flaggs, A. J. DeMaggio, M. F. Hoekstra et al., 1998. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 17 7239–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser, R., O. K. Mirzoeva, J. Wells, H. Olivares, B. R. Williams et al., 2001. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 21 6006–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan, C., and P. Russell, 2004. The DNA damage response: sensing and signaling. Curr. Opin. Cell Biol. 16 629–633. [DOI] [PubMed] [Google Scholar]
- Mirzoeva, O., and J. Petrini, 2001. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzoeva, O., and J. Petrini, 2003. DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol. Cancer Res. 1 207–218. [PubMed] [Google Scholar]
- Nakada, D., Y. Hirano and K. Sugimoto, 2004. Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol. Cell. Biol. 24 10016–10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, T., B. Moser and P. Russell, 2002. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics 161 1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, E., C. Nievera, A. Lee, L. Chen and X. Wu, 2007. The Mre11-Rad50-Nbs1 complex acts both upstream and downstream of ataxia telangiectasia mutated and Rad3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J. Biol. Chem. 282 22939–22952. [DOI] [PubMed] [Google Scholar]
- Paques, F., and J. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull, T., and M. Gellert, 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter-Goff, M. E., and N. Rhind, 2009. The role of MRN in the S-phase DNA damage checkpoint is independent of its Ctp1-dependent roles in double-strand break repair and checkpoint signaling. Mol. Biol. Cell. 20 2096–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind, N., and P. Russell, 1998. Mitotic DNA damage and replication checkpoints in yeast. Curr. Opin. Cell Biol. 10 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind, N., and P. Russell, 2000. Chk1 and Cds1: linchpins of the DNA damage and replication checkpoint pathways. J. Cell Sci. 113 3889–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kacmaz and S. Linn, 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73 39–85. [DOI] [PubMed] [Google Scholar]
- Sazer, S., and S. Sherwood, 1990. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J. Cell Sci. 97 509–516. [DOI] [PubMed] [Google Scholar]
- Stiff, T., C. Reis, G. K. Alderton, L. Woodbine, M. O'Driscoll et al., 2005. Nbs1 is required for ATR-dependent phosphorylation events. EMBO J. 24 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington, L., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero, J., and J. Diffley, 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412 553–557. [DOI] [PubMed] [Google Scholar]
- Tittel-Elmer, M., C. Alabert, P. Pasero and J. Cobb, 2009. The MRX complex stabilizes the replisome independently of the S phase checkpoint during replication stress. EMBO J. 28 1142–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno, M., T. Nakazaki, Y. Akamatsu, K. Watanabe, K. Tomita et al., 2003. Molecular characterization of the Schizosaccharomyces pombe nbs1+ gene involved in DNA repair and telomere maintenance. Mol. Cell. Biol. 23 6553–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel, T., Y. Lerenthal, L. Moyal, Y. Andegeko, L. Mittelman et al., 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 22 5612–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van, D. B. M., P. Lohman and A. Pastink, 2002. DNA double-strand break repair by homologous recombination. Biol. Chem. 383 873–892. [DOI] [PubMed] [Google Scholar]
- Walworth, N., and R. Bernards, 1996. Rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science 271 353–356. [DOI] [PubMed] [Google Scholar]
- Willis, N., and N. Rhind, 2009. Mus81, rhp51(rad51), and rqh1 form an epistatic pathway required for the s-phase DNA damage checkpoint. Mol. Biol. Cell 20 819–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdi, P. T., Y. Wang, S. Zhao, N. Patel, E. Y. Lee et al., 2002. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 16 571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]