Abstract
Background
Genome-wide association studies (GWAS) have identified multiple genetic variants associated with susceptibility to prostate cancer (PrCa). In the two-stage Cancer Genetic Markers of Susceptibility (CGEMS) prostate cancer scan, a single-nucleotide polymorphism (SNP) rs10486567 located within intron 2 of JAZF1 gene on chromosome 7p15.2 showed a promising association with PrCa overall (p = 2.14×10−6) with a suggestion of stronger association with aggressive disease (p = 1.2×10−7).
Methods
In the third stage of GWAS, we genotyped 106 JAZF1 SNPs in 10,286 PrCa cases and 9,135 controls of European ancestry.
Results
The strongest association was observed with the initial marker, rs10486567, which now achieves genome-wide significance (p = 7.79×10−11, ORHET 1.19; 95%CI = 1.12 – 1.27 and ORHOM 1.37; 95%CI = 1.20 – 1.56). We did not confirm a previous suggestion of a stronger association of rs10486567 with aggressive disease (p = 1.60×10−4 for aggressive cancer, n=4,597; p = 3.25×10−8 for non-aggressive cancer, n=4,514). Based on a multi-locus model with adjustment for rs10486567, no additional independent signals were observed at chromosome 7p15.2. There was no association between PrCa risk and SNPs in JAZF1 previously associated with height (rs849140, p = 0.587), body stature (rs849141, tagged by rs849136, p = 0.171), risk of type 2 diabetes and systemic lupus erythematosus (rs864745, tagged by rs849142, p = 0.657).
Conclusions
rs10486567 remains the most significant marker for PrCa risk within JAZF1 in individuals of European ancestry.
Impact
Future studies should identify all variants in high LD with rs10486567 and evaluate their functional significance for PrCa.
INTRODUCTION
Prostate cancer (PrCa) is the most common non-cutaneous cancer in the developed world and the second leading cause of cancer death in men (1, 2). The disease is highly treatable when detected early, with an encouraging 5-year survival rate (3). The established risk factors for PrCa are age, ethnicity and family history (3). Heritable factors have been estimated to explain 42% (29–50%) of PrCa risk in individuals of European ancestry (4). PrCa diagnostics based on the blood level of prostate-specific antigen (PSA) can result in 23–45% of overdiagnosis, that is in detection of disease with mild or insignificant clinical manifestations that don’t require treatment (5). Several genome-wide association studies (GWAS) have been performed to determine genetic factors that can identify individuals with increased risk of PrCa. More than 25 genomic regions that harbor genetic risk factors for PrCa have been identified to date (6–10).
The discovery component of the Cancer Genetic Markers of Susceptibility project (CGEMS) has reported a two-stage GWAS for PrCa (6). In the first stage, 1,172 individuals with PrCa and 1,157 controls of European ancestry were genotyped for 527,869 SNPs. In the second stage, 26,958 SNPs selected based on their association in stage 1 (p<0.068), were genotyped in a total of 4,020 cases and 4,028 controls (6). The second stage of CGEMS identified SNPs within the 8q24 region, HNF1B (TCF2) gene, MSMB gene and the 11q13 region to be significantly associated with PrCa (6). An observed association for markers in the C-terminal binding protein 2 (CTBP2) gene, as well as in the juxtaposed with another zinc finger protein 1 (JAZF1), did not reach the threshold of genome-wide significance (p = 1.7×10−7 for CTBP2 and p = 2.14×10−6 for JAZF1). For both genes there was a suggestion that the signal was more strongly associated with the risk of aggressive cancer, defined as Gleason score >7 or disease stage III (p = 2.7×10−8 for CTBP2 and p = 1.2×10−7 for JAZF1) (6).
The goals of the third stage of CGEMS GWAS have expanded to include testing for the presence of independent signals within established candidate regions and conducting a first order mapping of regions identified in CGEMS stage 2. Similar approach has resulted in identification of independent signals within the 8q24 region (10), chromosome 11q13 (11) and HNF1B gene on chromosome 17q12 (12). As a part of stage 3, we genotyped 106 JAZF1 tag SNPs in 10,286 PrCa patients and 9,135 controls of European ancestry. We aimed to confirm the suggestive association observed in stage 2 in a larger data set and to test for presence of stronger or independent association signals within JAZF1. We tested for association of rs10486567 with age of cancer diagnosis, family history and aggressiveness of disease. Additionally, we tested whether the variants of JAZF1 associated with other traits, were also associated with risk of PrCa.
MATERIALS AND METHODS
Subjects
Subjects were prostate cancer patients and controls drawn from 10 studies conducted in the US and Europe: Prostate, Lung, Colon and Ovarian (PLCO) (13); American Cancer Society Cancer Prevention Study II Nutrition Cohort (CPS-II) (6); Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC) (6); CeRePP French Prostate Case-Control Study (CeRePP) (6); Health Professionals Follow-up Study (HPFS) (6); The European Prospective Investigation into Cancer and Nutrition (EPIC)(14); Cohort of Norway (CONOR) (15); The Multiethnic Cohort Study (MEC) (16); Johns Hopkins University (JHU) (17); The Cancer Prostate in Sweden study (CAPS) (18). The counts and characteristics of each study are presented in Supplementary Table 1. The two main PrCa subtypes were defined as non-aggressive (Gleason score < 7 and stage < III) and aggressive (Gleason score ≥ 7 or stage ≥ III) cancer at the time of diagnosis.
SNP selection
The design, SNP selection and analysis of the study were described in details in Supplementary Material, Yeager et al. 2009 (10). In brief, of 7,034 SNPs were selected based on the previous GWAS results (6), and 115 of these SNPs were from JAZF1 gene. The JAZF1 targeted region was selected based on 0.2 cM HapMap recombination map centered on rs10486567 (chr7: 27290918–27826082). A preliminary set of 7 tags was selected using rs10486567 as an obligate-include and tagging the region at a D2 > 0.6 based on genotypes from HapMap CEU (European ancestry) (19). A final set of 127 tags was chosen by using rs10486567 as an obligate-include and by tagging the preliminary list of tags at an r2 > 0.8 in CEU (European), YRI (African) and CHB/JPT (Asian) HapMap samples (Supplementary Table 2A for list of all SNPs). All 7,034 SNPs selected for stage 3, were genotyped using a custom Illumina iSelect™ assay chip in 9,135 controls and 10,286 prostate cancer cases. Of 7,034 selected SNPs 6,313 SNPs passed manufacture and QC and provided > 90% genotype calls. Of 115 JAZF1 SNPs, 9 SNPs failed to provide genotype data or resulted in low genotype call rates (<90%) and were excluded from the analysis. Genotype quality control, assessment of call rates, assessment of unique subjects, analysis of duplicate DNA samples, fitness for Hardy–Weinberg proportion in control DNA and subject exclusions are described in detail in Supplementary Methods from Yeager et al., 2009 (10).
Admixture estimation
Of 7,034 SNPs on the stage 3 iSelect™ assay chip, 1,399 SNPs were selected and used for the detection of population structure as previously described (6). The population stratification analysis was done using the STRUCTURE program by merging the genotypes from all studies with those of the reference HapMap populations. The number of clusters (the “k” parameter) was set to three and the CEU, YRI and JPT+CHB samples were each specified to a different cluster schematically representing populations of European, African and Asian origin, respectively. The origin of the study samples was left unspecified. A total of 372 subjects (1.8%) were estimated to have less than 80% European ancestry and were excluded from analysis. All individuals that had at greater than 80% European ancestry were retained for the study, regardless of their self-reported origin.
Principal components analyses
Principal components analysis (PCA) was performed using the same 1,399 SNPs included for population stratification. These results were based on the remaining subjects after removal of individuals with admixed ancestry as described above. A Wilcoxon rank test was performed to check correlations with the case/control status for the top 5 eigenvectors. PC1 in JHU and top 3 PCs in CAPS studies showed significant differences between cases and controls. These principal components were used as covariates for association studies in JHU and CAPS sample sets.
Statistical software
Tag SNP selection was performed using the GLU software package (20) using the HapMap CEU, JPT+CHB and YRI data. Models were adjusted for study and significant principal components per study. Population stratification analysis was performed with STRUCTURE software (21), Principal Component Analysis was performed with EIGENSTRAT software (22). χ2 tests, Student T-tests, and logistic regression were used to compare the basic characteristics between cases and controls using SAS/STAT® system (SAS Institute Inc.). For single-marker case-control analyses, logistic regression under an additive genetic model was performed for each SNP adjusting for study site, and significant principle components using PLINK (23). A conditional analysis was also performed under additive model to account for the effect of SNP rs10486567 (0, 1 or 2 risk alleles). The association and LD plots were generated with snp.plotter, version 0.2 (24).
RESULTS
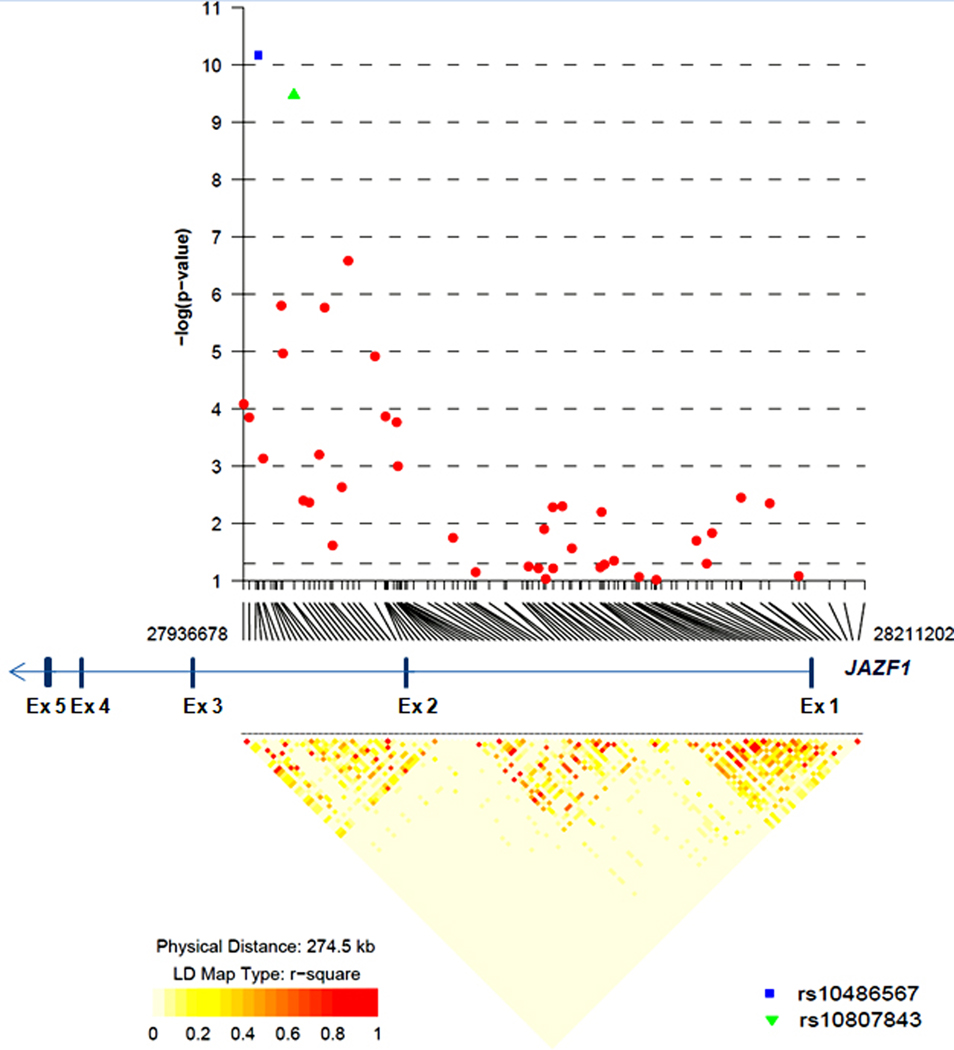
We performed the third stage of CGEMS in 10,286 PrCa patients and 9,135 controls of European ancestry drawn from ten studies conducted in Europe and the United States (10) (also described in Methods and Supplementary Table 1). As part of this effort, we genotyped 106 tag JAZF1 SNPs chosen on the basis of a tiered tagging strategy, the details of which have been published separately (10). First, the target region was bounded by the 0.2 cM HapMap recombination map (chr7: 27290918–27826082). A preliminary set of 7 tags was selected using rs10486567 as an obligate-include and tagging the region at a D2 > 0.6 based on genotypes from HapMap CEU (European ancestry) (19). A final set of 127 tags was chosen by using rs10486567 as an obligate-include and by tagging the preliminary selected 7 tags at an r2 > 0.8 in CEU (European), YRI (African) and CHB/JPT (Asian) HapMap samples (Supplementary Table 2A for list of all SNPs). 21 of these selected SNPs failed design, or showed poor performance. The successfully genotyped 106 SNPs covered a region of 275 Kb, starting from 25 Kb upstream of JAZF1 and well into intron 2 (Figure 1).
Figure 1.
LD structure and association results for 106 SNPs distributed over 275 Kb covering 25 Kb of promoter region through part of intron 2 of JAZF1 gene. LD plot is based on all samples (10,286 prostate cancer patients and 9,135 controls) of European ancestry.
Based on the genotype association test adjusted for study and significant principal components per study to account for subtle differences in population substructure (described in Method section), the strongest association was observed for the originally reported SNP, rs10486567 (6), with the compelling level of association below genome-wide significance (p = 7.79×10−11, ORHET 1.19; 95%CI = 1.12 – 1.27 and ORHOM 1.37; 95%CI = 1.20 – 1.56) (Table 1 and Supplementary Tables 3, and 2A). The second strongest association was observed for SNP rs10807843, also located within intron 2, approximately 16 Kb from rs10486567 (D’ = 1.0, r2 = 0.893 in 9,135 controls from CGEMS) (Table 1). The results for all SNPs are presented in Supplementary Table 2A.
Table 1.
Prostate cancer risk association results for top 3 JAZF1 SNPs in 10,286 patients and 9,135 controls
| SNP | Location | r2 with rs104865 67 |
Risk allele freq. | χ2 statistics | 1p-value | Heterozygous | Homozygous | |||
|---|---|---|---|---|---|---|---|---|---|---|
| (Risk allele) | Controls | Cases | OR | 95% CI | OR | 95% CI | ||||
| rs10486567 (G) | 27,943,088 | -- | 0.759 | 0.788 | 46.55 | 7.79 × 10−11 | 1.19 | 1.12–1.27 | 1.37 | 1.20–1.56 |
| rs10807843 (T) | 27,958,915 | 0.893 | 0.777 | 0.805 | 43.88 | 2.96 × 10−10 | 1.2 | 1.12–1.27 | 1.35 | 1.17–1.56 |
| rs38504 (C) | 27,982,859 | 0.463 | 0.79 | 0.812 | 26.54 | 1.73 × 10−6 | 1.17 | 1.10–1.24 | 1.18 | 1.02–1.36 |
Results for top 3 JAZF1 SNPs of 106 tested.
p-values are for genotype association test (additive model) adjusted for study and significant principal components (see Supplementary Methods).
We also explored several outcomes with respect to rs10486567 and found no association with age of diagnosis (p = 0.365), family history (p = 0.640) or aggressiveness of PrCa (p = 0.324, Table 2). The association for rs10486567 was stronger for non-aggressive PrCa (Gleason score <7 or stage < III), p = 1.60×10−4 for aggressive cancer, n = 4,597 and p = 3.25×10−8 for non-aggressive cancer, n = 4,514 (Table 2 and Supplementary Tables 2B and 2C). Our sample set was enriched for individuals older than 60 years and without family history of PrCa, but the odds ratios (ORs) for association of rs1086567 with PrCa were comparable in each of the subgroups (Table 3).
Table 2.
Association of rs10486567 with clinical traits in prostate cancer patients
| Trait | rs10486567 genotypes (number of cases) |
1p-value | ||
|---|---|---|---|---|
| AA 451 |
AG 3423 |
GG 6338 |
||
| Age at diagnosis |
66.56 ± 7.58 | 66.36 ± 7.50 | 66.23 ± 7.49 | 0.365 |
| Positive family history, % |
17.15 | 17.18 | 17.97 | 0.64 |
| Aggressive disease, % |
52.1 | 51.32 | 49.82 | 0.324 |
p-values are for χ2 test or analysis of variances
Table 3.
Prostate cancer risk for rs10486567 of JAZF1 stratified by potential risk factors
| Number Controls/ |
Risk allele freq. | 1p-value | Heterozygous | Homozygous | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stratified variable | Cases | Controls | Cases | OR | 95%CI | 3p-value | OR | 95%CI | 3p-value | |
| Age < 60 | 1853/1761 | 0.767 | 0.799 | 0.07 | 1.12 | 0.77–1.63 | 0.602 | 1.31 | 0.91–1.89 | 0.549 |
| Age ≥ 60 | 6539/7771 | 0.756 | 0.785 | 2.74 × 10−8 | 1.14 | 0.98–1.34 | 1.37 | 1.17–1.59 | ||
| No family history | 6480/7087 | 0.758 | 0.787 | 5.75 × 10−8 | 1.11 | 0.95–1.31 | 0.335 | 1.34 | 1.14–1.57 | 0.384 |
| With family history | 642/1521 | 0.759 | 0.794 | 0.041 | 1.39 | 0.91–2.14 | 1.62 | 1.07–2.45 | ||
| 2Non-aggressive cancer | 9109/4510 | 0.754 | 0.792 | 3.25 × 10−8 | 1.21 | 1.12–1.31 | 0.528 | 1.41 | 1.19–1.68 | 0.25 |
| 2Aggressive cancer | 9109/4598 | 0.754 | 0.781 | 1.60 × 10−4 | 1.14 | 1.05–1.23 | 1.29 | 1.10–1.53 | ||
p-values are for additive logistic regression model adjusted for study and significant principle component per study
compared to all controls;
Breslow-Day test for homogeneity
To test for presence of additional independent association signals within JAZF1, we performed a genotype association test adjusting for rs10486567. Only a weak association was observed for SNP rs3919460 (p = 0.0053, Supplementary Table 2A). We also examined whether the variants of JAZF1 associated with other traits would affect the genetic susceptibility to PrCa. In our samples, rs849140, previously associated with height (p = 5.3×10−8) (25), was not associated with PrCa (p = 0.587); rs849141, previously reported to be associated with height and body stature (p = 3.26×10−11) (26), was well tagged by rs849136 (r2 = 1.0 with rs849141 in CEU HapMap) but was not associated with PrCa (p = 0.171); the T2D-associated SNP rs864745 (p = 5.0×10−14) (27) was tagged by rs849142 (r2 = 1.0 with rs864745 in CEU HapMap), which was also associated with SLE (p = 1.54×10−10) (28) but showed no association with PrCa (p = 0.657, Table 4).
Table 4.
Association results for SNPs of JAZF1 previously associated with other traits
| *SNP | Location | MAF | χ2 statistics | χ2 | Heterozygous | Homozygous | Adjusted for rs10486567 |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| (Minor allele) | Controls | Cases | p-value | OR | 95% CI | OR | 95% CI | 1p-value | ||
| rs849140 (T) | 28,150,227 | 0.397 | 0.396 | 1.07 | 0.587 | 1.03 | 0.96-1.09 | 0.99 | 0.91-1.08 | 0.829 |
| rs849136 (A) | 28,141,482 | 0.281 | 0.287 | 3.53 | 0.17 | 1.06 | 0.99-1.13 | 1.03 | 0.92-1.15 | 0.199 |
| rs849142 (G) | 28,152,416 | 0.496 | 0.498 | 0.84 | 0.657 | 1.03 | 0.96-1.11 | 1.01 | 0.93-1.10 | 0.899 |
rs849140 is associated with height; rs849136 is in r2=1.0 with rs849141 associated with body stature;
rs849142 is in r2=1.0 with rs864745 associated with type 2 diabetes and systemic lupus erythematosus (SLE).
p-values are from logistic additive genetic model adjusted for study and significant principal component per study;
rs10486567 was included as a covariate with 0, 1 and 2 allele dosage coding.
DISCUSSION
Our study clearly confirms common SNPs in JAZF1 are associated with risk for PrCa overall and establishes this candidate gene for PrCa susceptibility in individuals of European ancestry. We show that rs10486567, previously reported as a promising association (p = 2.14×10−6) (6), is now conclusively associated with risk of PrCa (p = 7.79×10−11). The association was for risk of PrCa overall and was not specific for cases with aggressive cancer, as was previously suggested (6), or cases with family history or early/late age of disease diagnosis. Association analysis for 106 JAZF1 SNPs with adjustment for rs10486567 failed to reveal any independent signal, indicating that rs10486567 is a marker representing a single common allele associated with PrCa risk within JAZF1.
Located in intron 2 of JAZF1, rs10486567 is not predicted to affect mRNA expression, splicing, or transcription factor or miRNA-binding sites. Further deep resequencing efforts in PrCa patients together with information provided by the 1000 Genomes project (29) will help to catalog all common and rare variants in strong LD with rs10486567 in order to determine the optimal markers for investigation of their possible functional effects for PrCa.
The risk allele G of rs10486567 is the major allele in Europeans (0.73) and Africans (0.68), while it is rare in East Asians (0.16) based on allele frequencies in HapMap (19). Of note, PrCa is less frequent in individuals of Asian ancestry (30). Despite differences in allele frequencies, an association between rs10486567 and PrCa has been noted in African-Americans, Latinos, Japanese Americans and Native Hawaiians, but due to the small sample sizes in non-Caucasian populations the results often did not reach conclusive statistical significance (31). In the first stage of GWAS for PrCa conducted by PRACTICAL consortium that included 1,854 cases and 1,894 controls of European ancestry, rs10486567 did not meet the criteria (p < 0.05 or p-trend < 0.01) to be followed up in larger sample sets (8). It is important to note that the first stage of the GWAS mentioned above is not sufficiently powered to detect most variants with modest effect size, nominally in the rage of 1.1 to 1.25, as has been observed for rs10486567. This SNP has also been associated with PrCa in an independent set of 1,725 cases and 35,392 controls of European ancestry with OR 1.13 and p = 4.4×10−3 (32).
Currently, there is no biological explanation for the functional implications for JAZF1 in prostate carcinogenesis. Intra-chromosomal fusions between JAZF1 and SUZ12 (33) or JAZF1 and PHF (34) have been identified in endometrial cancer but there are no reports on these or other fusions of JAZF1 in PrCa. It will be important to determine the molecular functions of JAZF1 that can be important for several traits with which genetic variants within JAZF1 have recently been associated. JAZF1 is a large gene of approximately 350 kb. Rs10486567, associated with risk of PrCa, is located in intron 2. Several SNPs located close to each other in another LD block within intron 1 and ~210 Kb centromeric from rs10486567, have been associated with height (25), body stature (26) and increased risk of type 2 diabetes (T2D) (27) and systemic lupus erythematosus (SLE) (28). We agnostically tested all these SNPs in our set of samples but observed no association with PrCa. An inverse correlation between the risk of PrCa and T2D has been observed in epidemiological studies (35–37), a finding consistent with the results for genetic variants within the HNF1B (TCF2) gene (6, 38, 39). Although the inverse correlation between PrCa and T2D has also been suggested for variants in JAZF1 (40), our results did not show an effect of the T2D risk variant on susceptibility to PrCa.
In conclusion, our study has established rs10486567 within JAZF1 on chromosome 7p15.2 as a bona-fide marker for association with susceptibility to PrCa in individuals of European ancestry. Here, we intentionally studied only tag SNPs from the region to test for presence of independent association signals. Future studies should be conducted to identify all common and uncommon variants by deep sequence analysis that are in strong LD with rs10486567 in order to nominate the optimal variants for evaluation of functional significance for susceptibility to PrCa.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to all individuals participated in this study. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The study was also funded by grants to the NCI Breast & Prostate Cancer Cohort Consortium, UO1-CA98233, UO1-CA98710, UO1-CA98216, and UO1-CA98758. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
REFERENCES
- 1.Ferlay J, Autier P, Boniol M, Heanue M, Colombet M, Boyle P. Estimates of the cancer incidence and mortality in Europe in 2006. Ann Oncol. 2007;18:581–592. doi: 10.1093/annonc/mdl498. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Crawford ED. Epidemiology of prostate cancer. Urology. 2003;62:3–12. doi: 10.1016/j.urology.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 4.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 5.Shteynshlyuger A, Andriole GL. Prostate Cancer: To Screen or Not to Screen? Urol Clin North Am. 37:1–9. doi: 10.1016/j.ucl.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 7.Witte JS. Prostate cancer genomics: towards a new understanding. Nat Rev Genet. 2009;10:77–82. doi: 10.1038/nrg2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eeles RA, Kote-Jarai Z, Al Olama AA, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collin SM, Metcalfe C, Zuccolo L, et al. Association of folate-pathway gene polymorphisms with the risk of prostate cancer: a population-based nested case-control study, systematic review, and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2009;18:2528–2539. doi: 10.1158/1055-9965.EPI-09-0223. [DOI] [PubMed] [Google Scholar]
- 10.Yeager M, Chatterjee N, Ciampa J, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet. 2009 doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng SL, Stevens VL, Wiklund F, et al. Two independent prostate cancer risk-associated Loci at 11q13. Cancer Epidemiol Biomarkers Prev. 2009;18:1815–1820. doi: 10.1158/1055-9965.EPI-08-0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J, Zheng SL, Wiklund F, et al. Evidence for two independent prostate cancer risk-associated loci in the HNF1B gene at 17q12. Nat Genet. 2008;40:1153–1155. doi: 10.1038/ng.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 14.Riboli E, Hunt KJ, Slimani N, et al. European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public Health Nutr. 2002;5:1113–1124. doi: 10.1079/PHN2002394. [DOI] [PubMed] [Google Scholar]
- 15.Naess O, Sogaard AJ, Arnesen E, et al. Cohort profile: cohort of Norway (CONOR) Int J Epidemiol. 2008;37:481–485. doi: 10.1093/ije/dym217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolonel LN, Henderson BE, Hankin JH, et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol. 2000;151:346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng SL, Sun J, Cheng Y, et al. Association between two unlinked loci at 8q24 and prostate cancer risk among European Americans. J Natl Cancer Inst. 2007;99:1525–1533. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]
- 18.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 19.Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. http://code.google.com/p/glu-genetics/
- 21. http://pritch.bsd.uchicago.edu/structure.html.
- 22. http://genepath.med.harvard.edu/~reich/EIGENSTRAT.htm.
- 23.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. http://cbdb.nimh.nih.gov/~kristin/snp.plotter.html.
- 25.Johansson A, Marroni F, Hayward C, et al. Common variants in the JAZF1 gene associated with height identified by linkage and genome-wide association analysis. Hum Mol Genet. 2009;18:373–380. doi: 10.1093/hmg/ddn350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soranzo N, Rivadeneira F, Chinappen-Horsley U, et al. Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5:e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gateva V, Sandling JK, Hom G, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009 doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. http://www.1000genomes.org/page.php.
- 30.Robbins AS, Koppie TM, Gomez SL, Parikh-Patel A, Mills PK. Differences in prognostic factors and survival among white and Asian men with prostate cancer, California, 1995–2004. Cancer. 2007;110:1255–1263. doi: 10.1002/cncr.22872. [DOI] [PubMed] [Google Scholar]
- 31.Waters KM, Le Marchand L, Kolonel LN, et al. Generalizability of associations from prostate cancer genome-wide association studies in multiple populations. Cancer Epidemiol Biomarkers Prev. 2009;18:1285–1289. doi: 10.1158/1055-9965.EPI-08-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koontz JI, Soreng AL, Nucci M, et al. Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proc Natl Acad Sci U S A. 2001;98:6348–6353. doi: 10.1073/pnas.101132598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Micci F, Panagopoulos I, Bjerkehagen B, Heim S. Consistent rearrangement of chromosomal band 6p21 with generation of fusion genes JAZF1/PHF1 and EPC1/PHF1 in endometrial stromal sarcoma. Cancer Res. 2006;66:107–112. doi: 10.1158/0008-5472.CAN-05-2485. [DOI] [PubMed] [Google Scholar]
- 35.Kasper JS, Giovannucci E. A meta-analysis of diabetes mellitus and the risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:2056–2062. doi: 10.1158/1055-9965.EPI-06-0410. [DOI] [PubMed] [Google Scholar]
- 36.Kasper JS, Liu Y, Giovannucci E. Diabetes mellitus and risk of prostate cancer in the health professionals follow-up study. Int J Cancer. 2009;124:1398–1403. doi: 10.1002/ijc.24044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waters KM, Henderson BE, Stram DO, Wan P, Kolonel LN, Haiman CA. Association of diabetes with prostate cancer risk in the multiethnic cohort. Am J Epidemiol. 2009;169:937–945. doi: 10.1093/aje/kwp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winckler W, Weedon MN, Graham RR, et al. Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes. 2007;56:685–693. doi: 10.2337/db06-0202. [DOI] [PubMed] [Google Scholar]
- 39.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 40.Frayling TM, Colhoun H, Florez JC. A genetic link between type 2 diabetes and prostate cancer. Diabetologia. 2008;51:1757–1760. doi: 10.1007/s00125-008-1114-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.