Abstract
The rate of tumor recurrence post resection suggests that there are underlying molecular changes in nearby histologically normal tissue that go undetected by conventional diagnostic methods that utilize contrast agents and histochemistry. MALDI MS is a molecular technology that has the specificity and sensitivity to monitor and identify molecular species indicative of these processes. The current study utilizes this technology to assess molecular distributions within a tumor and adjacent normal tissue in clear cell renal cell carcinoma biopsies. Results indicate that the histologically normal tissue adjacent to the tumor expresses many of the molecular characteristics of the tumor. Proteins of the mitochondrial electron transport system are examples of such distributions. This work demonstrates the utility of MALDI MS for the analysis of tissue biopsies in the elucidation of molecular processes in the tumor microenvironment.
Keywords: MALDI, tumor margins, renal carcinoma, proteomics
Introduction
One of the foremost concerns in clinical oncology and pathology is ensuring complete tumor removal 1. Currently, histopathological assessment of the resected tumor is the primary post-operative, and in some cases intraoperative, method for assessing surgical margin status 2. Despite these assessments, local tumor recurrence remains a problem in many cancers such as lung 3, breast 4, soft tissue 5, 6, head and neck 2, 7, and brain 8. Depending on the tissue type, recurrence may occur a few months to a few years after removal of the primary tumor, suggesting there are underlying molecular processes present in the remaining tissue that go undetected using current histopathological techniques. Since molecular alterations precede phenotypic changes, the environment outside of the tumor margin may appear normal upon histological assessment, which renders conventional histological methods unsatisfactory in being the only determinant of successful resection. The discovery of new molecular markers for such processes has the potential to provide an additional, complimentary approach to identifying an aberrant tissue environment 2.
Previous studies of tumor margins have utilized antibody-directed approaches for several cancer types; however, these targeted approaches limit the information obtainable and may be insufficient for determining the extent of the abnormal cellular environments beyond the histological tumor border 2, 7, 9. Antibody-based approaches for assessing tumor margins often suffer from unknown cross-reactivities while nuclear staining procedures are limited to visualization of cellular morphology 10. Proteomic studies can facilitate the understanding of the changes in the environments of the tumor and adjacent normal tissue and provide an estimate of the distance beyond the histologically-defined margin that these changes occur.
Proteomic tools, such as gel electrophoresis and mass spectrometry (MS), have been successfully applied to facilitate the discovery of potential disease biomarkers 11–14. In particular, direct tissue analysis by profiling and imaging MALDI (matrix-assisted laser desorption ionization) MS provides spatial information in addition to molecular specificity, high sensitivity, and a relatively large dynamic range for the detection of drugs and their metabolites 15 as well as intact proteins and peptides 16, 17. Studies utilizing this technology have resulted in the identification of prognostic and diagnostic markers for patients with brain tumors 18, 19 and non-small-cell lung cancer 20 biomarkers indicative of glomerulosclerosis 21, the prediction of tumor response to molecular therapies 22, further understanding of Parkinson’s disease progression 14 and the effect of TGF-beta in mammary tumor development 23. Recently, the potential applicability of this technology to the analysis of tumor margins was illustrated with a soft tissue sarcoma biopsy 24. The current research utilizes this technology for assessment of the tumor microenvironment in clear cell renal cell carcinoma resection specimens.
Each year there are approximately 36,160 new cases and 12,660 deaths in the United States attributed to cancer of the kidney or renal pelvis 25. Of these, approximately 75–80% 26 are renal cell carcinoma (RCC), which includes a group of unique histological subtypes: clear cell (conventional) RCC, papillary (chromophil) RCC, chromophobe cell RCC, collecting duct RCC, medullary RCC, and other unclassified tumors. The majority (80%) of RCC cases are classified as clear cell RCC (ccRCC), called such because of their high content of lipid and cholesterol, creating a yellow appearance grossly and clear cytoplasm histologically 27, 28. Clear cell RCC is the most malignant of renal tumors, having the worst prognosis of all subtypes 26.
For cancers such as ccRCC, it is critical to study the molecular characteristics of both the tumor and surrounding tumor margin. Such studies would not only facilitate a better understanding of tumor invasion, but also provide potential markers to aid in histological assessments that help ensure complete tumor removal. The radical nephrectomy procedure for ccRCC has become the standard treatment; however for patients with RCC in a solitary functioning kidney or those with RCC arising bilaterally, alternative approaches are needed. Partial nephrectomy procedures are desired for these patients and patients with small tumors as well as for individuals in whom the normal contralateral kidney could be under potential future threat from hypertension, diabetes mellitus, or hereditary cancer syndromes such as von Hippel-Lindau 29. Unlike radical nephrectomies, the oncologic effectiveness of partial nephrectomies has been questioned with concern over tumor involvement of the surgical margin. Standard guidelines are lacking on how large of a surgical margin is needed to ensure complete tumor removal while maximizing the amount of spared kidney 30.
We report studies using profiling/imaging MALDI MS of snap frozen ccRCC resection specimen tissue to examine the characteristics of the tumor microenvironment and tumor margin. Utilizing a high-throughput MS platform, we were able to characterize protein signatures that implicate abnormal tissue in some cases far beyond the histological margin. The subsequent identification of one group of such proteins suggests a major role of the mitochondrial electron transport system in tumor involvement of tissue beyond the ccRCC histological border.
Materials and Methods
Sample Processing
Human kidney samples from patients undergoing nephrectomies were obtained from the Vanderbilt University Ingram Cancer Center-Human Tissue Acquisition and Pathology Resource and the National Institutes of Health Cooperative Human Tissue Network. Each sample (n of 34) contained ccRCC and attached adjacent normal renal tissue. Samples were snap frozen in liquid nitrogen 30 minutes or less after removal from the patient and stored at −80°C until use. For MALDI profile analysis, three 12 um sections were cut and thaw mounted with random positioning onto a gold-coated MALDI target plate followed by fixation by submersion in graded alcohols (70% for 20 s, 70% for 20 s, and 95% for 10 s)31. After drying, MALDI plates were stored in a desiccator until matrix deposition. Sections adjacent to those used for MALDI analysis were cut and mounted onto microscope slides and stained with hematoxylin and eosin according to standard protocols. Stained serial sections were reviewed by a board-certified pathologist (M.E.S.) and regions of tissue (e.g. tumor, normal, and histological border) were marked.
Matrix Deposition and MALDI MS Analysis
A robotic matrix spotter (LabCyte, Sunnyvale, CA) was used to deposit matrix droplets of ~120 pL volumes. A detailed description of the device and its operational conditions are described elsewhere 32. For each sample, the matrix was spotted in an array (~400 μm × 400 μm spot-to-spot) spanning the tumor and attached adjacent normal. Sinapinic acid matrix solution was prepared at 25 mg/ml in 50% acetonitrile/0.1% TFA (v/v). Matrix was deposited on each sample in cycles of 13 drops dispensed at 10 Hz at each designated coordinate in the array. A total of 6 cycles, or passes, was determined to be optimal for matrix coverage on renal tissue. After deposition, the final matrix spots had a diameter of approximately 250 μm.
Spectra were acquired using an Ultraflex II MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, Billerica, MA) equipped with a SmartBeam™ laser. The instrument was operated under automated, linear-mode acquisition parameters with a method optimized for acquisition at a mass-to-charge ratio of 2,000 to 30,000 using a 200 Hz laser. A total of 400 laser shots were collected for each matrix spot in increments of 50 shots beginning at the center of each spot and randomly rastering at different positions within the spot. Automated profile acquisition required the creation of custom plate geometry files that involved the determination of the relative coordinate of each matrix spot from an optical image of the prepared MALDI plate. These x-y coordinates of each spot were then written to file in the native target geometry format of the mass spectrometer control software. Three spots from the outer corners of the array were selected for final plate alignment using a standard instrument protocol.
Spectral Processing and Data Interpretation
Prior to statistical analyses, mass spectra were baseline-corrected, normalized and aligned using ProTS Data software (Biodesix Inc., Steamboat Springs, CO). Multiple spectra from each individual patient were averaged for each of the four regions of the biopsy: tumor, normal, margin-tumor and margin-normal. Peaks lists were extracted from the average spectra using ProTS Data, peaks were binned and peak amplitudes were calculated.
Statistical Analysis: Identification of differentially expressed features
Comparisons of tumor versus normal and margin-normal versus normal were carried out using the Significance Analysis of Microarrays (SAM) and permutation t-test for paired data 33, 34. Features identified as significant by both methods with a false discovery rate (FDR) less than 0.01 were considered as potentially significant. For the differentially expressed features identified in the two comparisons of tumor versus normal and margin-normal versus normal, a support vector machine (SVM) classifier was used to assess the class prediction ability of each individual feature. The prediction accuracy was estimated using a leave-one-out-cross-validation (LOOCV) algorithm (Supporting Information: Tables 1 and 2) 35. The class prediction accuracy of the SVM classifier using multiple input features was also evaluated, and the optimal set of differentially expressed features that attained the maximum prediction accuracy was identified. In order to visualize dissimilarities among tissue samples from different histological regions, the first two multidimensional scaling (MDS) coordinates of the optimal set of features used in SVM classifier were plotted against each other for each tissue sample. Tissue samples from different histological regions were plotted in different colors (Supporting Information: Figures 1A and 1B, respectively).
Statistical Validation of Feature Identification Procedure
A bootstrap procedure was used to validate the procedure of feature identification 35. The original data were randomly sampled with replacement to simulate a virtual sample of size n, where n is equal to the size of the original data set. For each of these bootstrap samples, the same statistical analysis procedure was followed to identify the significant feature. Features identified as significant by both methods with a false discovery rate (FDR) less than 0.01 were considered potentially significant. This process was repeated for 100 bootstrap replications in order to observe how the lists of significant features behave over each repetition. A highly consistent list of significant features from the 100 bootstrap replications implies good reproducibility of the result from the original data and highly consistent performance of the statistical analysis procedure.
Protein Identification
Tissue was homogenized with tissue protein extraction reagent buffer (Pierce Biotechnology, Rockford, IL) according to the manufacturer’s protocol. Proteins from homogenized tissue were digested and loaded onto immobilized pH gradient gel (IPG) strips for isoelectric focusing of peptides. IPG strips were cut into 15 pieces followed by peptide extraction and desalting with C18 resin using a stepwise elution gradient of 20%, 40%, 60% and 80% acetonitrile. The ability to match an identified peptide with its experimental isoelectric point enables elimination of false positive identifications36, 37. In a second method, proteins (~300 μg) from homogenized tissue were separated by RP-HPLC on a C8 column (Grace Vydac, Hesperia, CA) with the following stepwise gradient: 5%–25% B for 5 min, 25–60% B for 55 min, and 60–95% B for 10 min (mobile phase A: 0.1% TFA in H2O, mobile phase B: 0.1% TFA in ACN). Fractions were collected in 1 min intervals. To identify fractions containing the proteins of interest, fractions were vacuumed to dryness, re-suspended in 20–30 μl of ~40% ACN and spotted, with matrix (25 mg/ml sinapinic acid, 50/50/0.1 of H2O/acetonitrile/trifluoroacetic acid (% v/v/v)) on a MALDI plate using an LC-MALDI robotic spotter (Symbiot, Applied Biosystems, Framingham, MA). Each fraction was profiled using MALDI-TOF MS and each m/z value of interest was confined to one well or two consecutive wells. The fraction of interest was further interrogated by one of two methods: an in-solution digest (trypsin, Promega Corporation, Madison, WI) or separation on a 1D polyacrylamide gel (10–20% Tricine, Invitrogen Corporation, Carlsbad, CA) followed by in-gel trypsin digestion. Peptides were then analyzed by LC-MS/MS on a Deca XP Plus ion trap and/or an LTQ linear ion trap mass spectrometer (Thermo Scientific, Waltham, MA), equipped with a nanospray source containing a RP-C18 column (70 μm i.d., mobile phase A: 0.1% formic acid, mobile phase B: 0.1% formic acid and 2% acetonitrile). Sequest38 was used for database (UniProtKB/Swiss-Prot, human-nonredundant) searching and ProteinProphet39, 40 software was used to determine the probability that a protein had been correctly identified based on the available peptide sequence 41. The observed and experimental masses matched within 1 Da.
Feature Mapping
Mass spectra were collected in a way that allows mapping of feature expression (m/z signal) from tumor into adjacent normal tissue. Because tumor margins are not linear, the analysis of these features requires that the distance of each matrix spot to the margin be measured accurately. Thus, a feature can be mapped according to its distance from the histologically-defined tumor margin. The optical image of a hematoxylin and eosin stained section was designed to define the tumor border on the optical image of the spotted tissue. A custom program was designed to calculate the distance of the matrix spot to the closest point along the defined histological margin and a spreadsheet was created containing the distance of each matrix spot from the histological margin and its corresponding mass spectrometry file. To best represent the data, the feature amplitude (log2 scale) from each matrix spot was plotted against its distance from the histological margin. A locally-weighted-regression scatter-plot smoothing (LOWESS) curve was laid over the scatter plot to show the trend of change of feature amplitude across the histological margin. The first derivative of the LOWESS trend line was plotted to show the position where the maximum rate of change of feature amplitude occurs. In an independent analysis (Biodesix, Inc.) the feature amplitude (absolute intensity) from each matrix spot was plotted against its distance from the histological margin. Manual interpretation was used to determine the distance of cellular compromise and correlate with results from the first method.
Results
Molecular Analysis of Tumor Margins by MALDI MS
MALDI MS analysis was performed on frozen tumor tissue from 75 patients with ccRCC and matched normal kidney tissues. Of the tissue samples from the 75 patients, 34 samples consisted of tumor and attached adjacent normal tissue, and 41 samples included separated but patient matched tumor and normal kidney. Overall, approximately 32,000 individual mass spectra were acquired, and approximately 200–300 individual molecular features were observed between the m/z range of 2,000 and 25,000 from each spot (~200 μm in diameter) on the tissue. The intra-patient reproducibility of mass spectral measurements was assessed for both normal and tumor tissue. The average concordance correlation coefficient for the multiple mass spectral measurements from within the same patient was 0.6 (± 2) for tumor tissue and 0.7 (± 1) for normal tissue. These assessments indicated an acceptable level of intra-sample reproducibility of these measurements.
Mass spectral profiles were compared among four different regions of tissue: 34 samples with tumor and attached adjacent normal and 41 samples with separate but patient-matched tumor and normal tissue. Figure 1 highlights the regions of interest for the analysis. These four regions consist of tumor, margin-tumor, margin-normal, and normal. Generally, normal is defined to be approximately 6 mm or greater from the histological margin in samples that contained tumor and adjacent normal. Regions were grouped independently of their respective tumor grade 42 or stage diagnosis 26. Prior to statistical analyses, the mass spectrometric data was processed, averaged by region for each patient, and grouped into four categories. Two statistical methods, the Significance Analysis of Microarrays (SAM) and permutation t-test, were used to identify differential features between different regions. The comparisons included tumor versus normal from all 75 samples (Figure 2A; Supporting Information: Table 1) and margin-normal versus normal from 34 samples (Figure 2B; Supporting Information: Table 2). Features identified by both statistical methods were considered as differentially expressed between two regions.
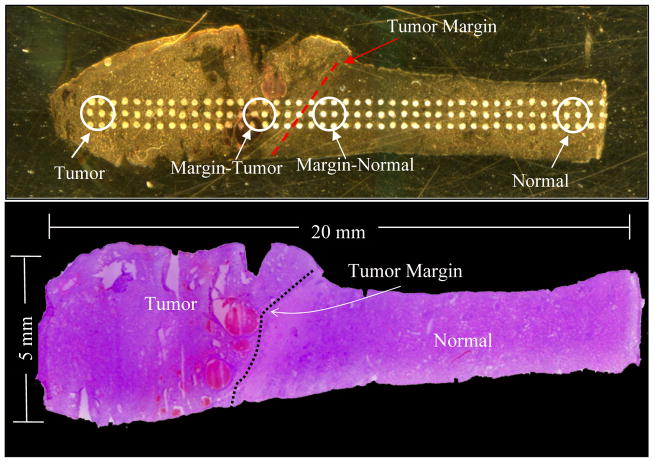
Figure 1.
Regions of interest for statistical analysis of tumor margin profiles. (Top) Optical image of tumor and attached adjacent normal section on a MALDI plate with regions of interest marked; (Bottom) Optical image of a corresponding H&E stained section marked by a pathologist.
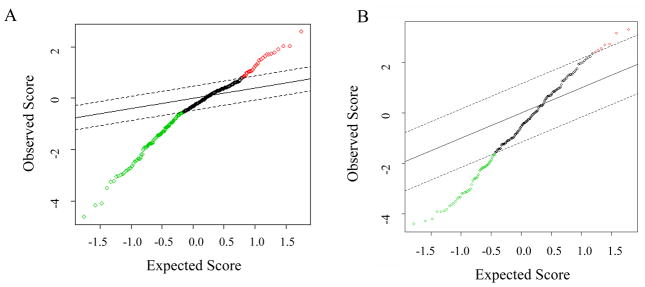
Figure 2.
SAM statistics. (A) SAM statistics plot of results of tumor versus normal tissue (n = 75). Red circles indicate significant features that are over-expressed in tumor. The green circles indicate features significantly under-expressed in the tumor. (B) SAM statistics plot of results of margin normal versus normal tissue (n = 34). Green circles indicate features significantly under-expressed in margin normal tissue. Red circles indicate features significantly over-expressed in the margin normal tissue. Dotted lines represent the significance threshold (Δ) corresponding to FDR <0.01. Features have been arranged by their degree of difference in expression. Points to the right (top) or left (bottom) of the first point outside of the dotted line are called significant.
Statistical analysis of the data demonstrated that there are feature patterns resembling tumor in the margin-normal microenvironment outside of the histological tumor border. Approximately 120 differentiating features between tumor and normal tissue regions were found. A subset of those differential features showing the best classification accuracy is highlighted in bold in Table 1. The estimated prediction accuracy of a support vector machines (SVM) classifier based on these six features was 95%. Comparison of the normal region and the margin-normal region was done to determine if there were significant differences and if any of these features were also significant between the tumor and normal tissue regions. Results of this analysis revealed ~50 features that are under-expressed in the margin-normal as compared to the normal, most (~95%) of which were also under-expressed in the tumor as compared to the normal using the SVM classifier. Nine features, represented in bold in Table 2, were able to predict the samples based as normal and margin-normal with an accuracy of 75%. A plot of the first two multidimensional scaling (MDS) coordinates was used to show the dissimilarities among the tissue samples in terms of the multiple features used in the SVM classifier (Supporting Information: Figures 1A & 1B respectively). The MDS plot of margin-normal versus normal regions illustrates some similarity between the two regions, as would be expected. There is, however, a subset of features that are dissimilar between the two regions.
Table 1.
Summary of proteins identified.
| MW | Protein Name | UniProtKB/Swiss-Prot Accession# | % Coverage | Peptide Residue #’s | Method | Comment |
|---|---|---|---|---|---|---|
| 11072 | S100AA/Calpactin I | P60903 | 18 | 47–53*, 37–46* | In-solution digest; LC-MS/MS | Up in Tumor |
| 10090 | S100-A6/Calcyclin | P06703 | 24 | 41–47, 48–55, 56–62* | In-solution digest; LC-MS/MS | Down in Tumor |
| 8577 | COX6C | P09669 | 31 | 18–38 | In-solution digest; LC-MS/MS | Down in Tumor and MN |
| 9368 | NADH-ubiquinone oxidoreductase MLRQ subunit | O00483 | 53 | 36–47, 48–55, 56–63, 64–72, 77–81 | In-solution digest; LC- MS/MS | Down in Tumor and MN |
| 11651 | S100-A11/Calgizzarin | P31949 | 16 | 37–52* | IPG LC-MS/MS | Up in Tumor |
| 10738 | Ubiquinol-cytochrome c reductase complex 11 kDa protein | P07919 | 21 | 61–78 | IPG LC-MS/MS | Down in Tumor and MN |
| 12272 | Cytochrome C | P99999 | 57 | 28–38*, 39–52, 40–52, 56–73, 80–86, 92–99 | 1D gel; LC-MS/MS | Down in Tumor and MN |
| 10611 | COX5B | P10606 | 24 | 50–56, 57–68, 58–68, 75–86* | 1D gel; LC-MS/MS | Down in Tumor and MN |
| 6720 | COX7A2 | P14406 | 28 | 34–46*, 47–56* | 1D gel; LC-MS/MS | Down in Tumor and MN |
| 5354 | COX7C | P15954 | 14 | 26–34* | 1D gel; LC-MS/MS | Down in Tumor and MN |
| 4890 | COX82 | P10176 | 13 | 26–34* | 1D gel; LC-MS/MS | Down in Tumor and MN |
| 12345 | MIF | P14174 | Other | Up in Tumor | ||
| 4933 | Thymosin β-10 | P63313 | Other | Up in Tumor |
An asterisk indicates a unique peptide.
Letters A and B in the “Methods” column indicates the protein identification method used as described in Figure 5. The data for proteins identified by “other” methods was previously acquired by top down MS/MS analysis on a Bruker Daltonics MALDI fourier transform mass spectrometer and subsequent database (MASCOT) searching.
Assessing Patterns of Molecular Change in Tumor Margins
Another objective of this work was to determine how the features of interest change in and around the tumor margins and how their expression patterns relate to tumor aggressiveness. It was expected that some features would show a correlation to the histological border, while others would show changes occurring beyond the histological border, namely the features whose expression patterns in the margin normal resembled their counterparts in the tumor. A schematic of possible trends is presented in Figure 3 and is further discussed below.
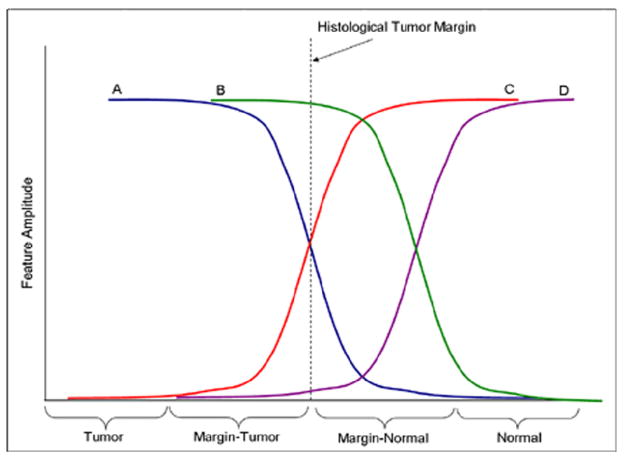
Figure 3.
Expected and observed molecular patterns. This figure illustrates the observed patterns of the molecular features traversing the histological tumor margin. Lines A and C represent features that begin changing at the histological margin; while lines B and D represent features that change after the histological margin.
Mass spectra were acquired from tissue containing tumor and attached normal tissue, which allowed for the mapping of proteins in tumor and adjacent normal tissue. However, since the histological tumor border is usually not a straight line, the analysis of expression patterns required that the distance of each matrix spot to the margin be measured accurately. To obtain the distance, the optical image of a hematoxylin and eosin stained section was used to define the tumor border. A custom program was then used to calculate the distance of the matrix spot to the closest point along the defined histological margin. The information was transferred to a spreadsheet which correlated the distance of each matrix spot with the corresponding mass spectral data.
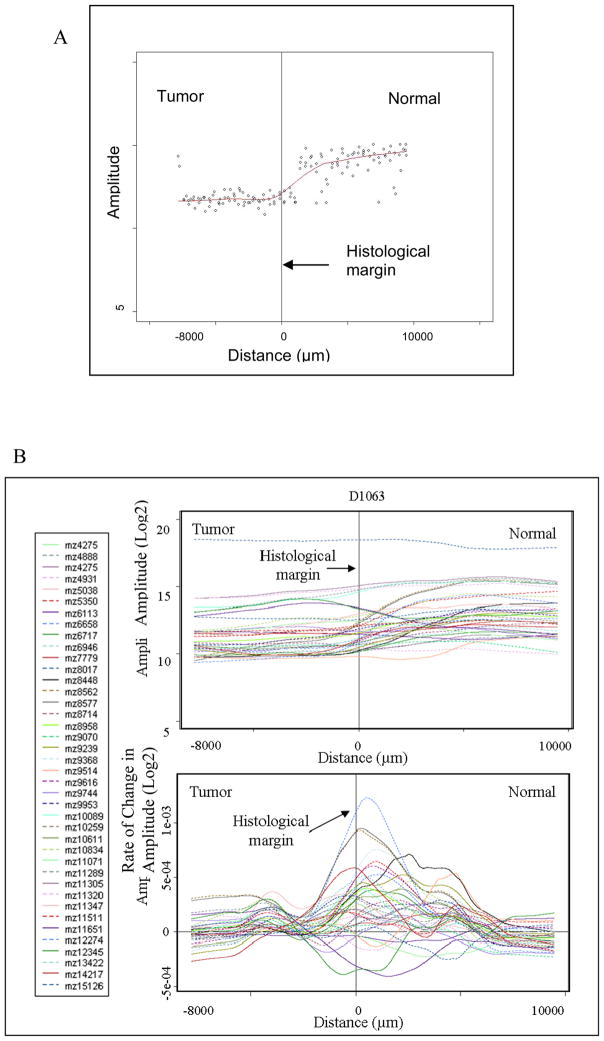
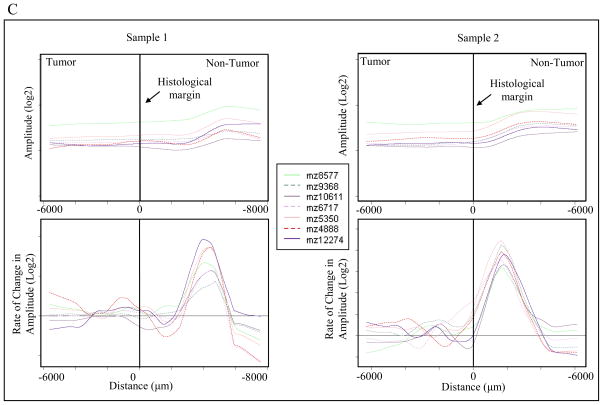
For interpretation of molecular trends, 40 significant features were plotted for 25 patients. For graphical representation, samples with fewer than 4 mm of tumor or less than 7 mm of normal were excluded from the assessment. These features represented m/z values that were determined as under-expressed and over-expressed in the ccRCC tumors as well as aberrant molecular features present outside of the histological margin. To represent the data, the feature amplitude from each matrix spot was plotted against its distance to the histological margin. A locally-weighted-regression scatter-plot smoothing (LOWESS) curve was constructed to show the change in feature amplitude across the histological margin. The first derivative of the LOWESS trend line was plotted to determine the position where the maximum rate of change of feature amplitude occurs [Figure 4]. Together, the LOWESS and first derivative lines suggest that while are numerous features that change near the histological margin, there are several features that are under-expressed in the tumor and the margin-normal as compared to the normal [see line labeled “D” in Figure 3]. The focus of this project was to determine the characteristics in margin normal tissue that resemble the tumor. After reviewing the LOWESS trends and statistics, the features chosen to assess these patterns were m/z values 4888, 5351, 6717, 8577, 9368, 10611, 12274 [Figure 4C]. Identification of these proteins (features) is presented below.
Figure 4.
Demonstration of margin plot analysis. (A) LOWESS fit line of scatter plot data. (B) Top: LOWESS line of 40 selected significant features; Bottom: First Derivative of all LOWESS lines (x-axis error approximately +/−400 μm). (C) LOWESS and corresponding 1st derivative plots of 7 selected features for two representative samples. The notation “mz” represents m/z, or mass-to-charge ratio.
Another goal was the correlation of the molecular changes in the tumor-normal tissue with tumor aggressiveness. As with most tumors, there are no molecular markers for tumor aggressiveness in renal cell carcinoma, thus the attempt was made to determine if these patterns correlate with the current standards of clinical diagnoses, which are Fuhrman grade and TNM stage. Multiple characteristics of the plot and of the patient information were considered in order to find the best trend in the data. Most samples expressed protein patterns exemplified by line “D” in Figure 3, where features are under-expressed in the tumor and the margin-normal as compared to the non-tumor tissue. The characteristics monitored were the point at which the maximum rate of change occurred and the point at which the feature amplitude began to plateau. A summary of observations is listed in Table 3 of the Supporting Information.
There was not a clear correlation between the pathological diagnosis and the distance of compromised tissue beyond the histological tumor border. For the m/z features noted, expression patterns in most samples followed the aforementioned pattern illustrated in Figure 3. There was a small group of samples in which margin trends could not be assessed. This group includes four high stage tumors and three low stage tumors as well as three high grade tumors and four low grade tumors.
Identifying Differentially Expressed Biological Features and Validating Protein Signatures
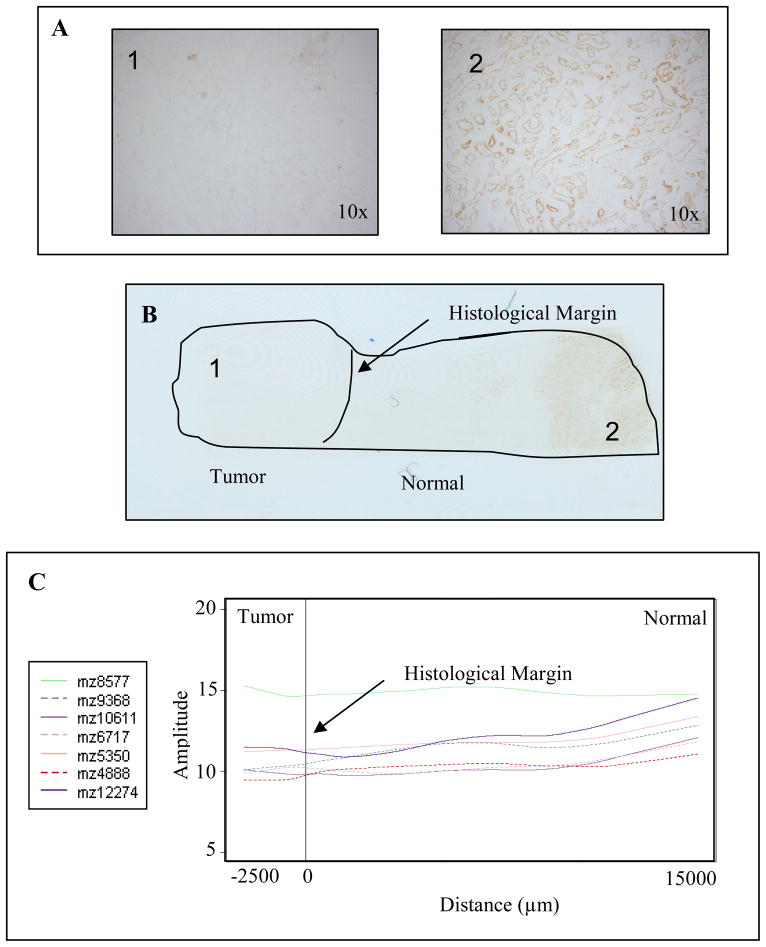
In the protein identification process, emphasis was placed on those signals suggestive of abnormal environments outside of the histological margin, though others were also identified. From this study, 12 proteins were identified [Table 1] and those over-expressed in tumor include calpactin I and calgizzarin. Proteins under-expressed in tumor include calcyclin, cytochrome c, NADH-ubiquinone oxidoreductase MLRQ subunit, a ubiquinol-cytochrome c reductase complex protein, and several cytochrome c oxidase polypeptides (VIC, VB, VIIA2, VIIC, VIII2). The proteins that indicated a compromised environment outside of the histological margin were cytochrome c, the cytochrome c oxidase polypeptides, and the NADH-ubiquinone oxidoreductase MLRQ subunit. To validate the observed molecular signatures, the distribution of cytochrome c oxidase activity was also assessed by immunohistochemistry. As seen in Figure 5, the staining pattern follows the same trend observed in the LOWESS graph.
Figure 5.
Cytochrome oxidase activity assay confirms localization of features. (A) Microscope images of tumor (1) and normal (2) regions of the tissue. (B) Scanned image of stained section shows localization. (C) LOWESS trend of features involved in mitochondrial electron transport.
Discussion
Biological Perspectives
This study utilized MALDI MS to characterize the molecular microenvironment adjacent to histological tumor margins and determine if there are indications of aberrant molecular expression beyond these margins. Molecular features in the histologically normal-appearing tissue adjacent to the histological tumor border that have an increased or decreased expression relative to the same proteins in the tumor may represent regions of aberrant cellular function indicative of an abnormal microenvironment.
Results of this study showed that there were many proteins over-expressed and under-expressed in ccRCC tumors as compared to adjacent normal; however, those of particular interest are the proteins that showed abnormal patterns outside of the histologically defined tumor margin. For example, proteins involved in the mitochondrial electron transport system were consistently under-expressed in the tumor as well as in the histologically normal tissue adjacent to the tumor. The decreased expression of these proteins has been previously observed in tumors of clear cell renal cell carcinoma 43, 44 but has not been reported in the adjacent normal tissue.
Mitochondrial deficiency in cancer cells was recognized almost eighty years ago by Otto Warburg 45. The Warburg effect is described as the dependency of tumors on glycolysis rather than oxidative phosphorylation for ATP, even in the presence of oxygen 46. Although the production of ATP through glycolysis is less efficient than its production through mitochondrial oxidative phosphorylation, cancer cells appear to preferentially utilize glycolysis.
Although the deficiency of the mitochondrial electron transport system and related events in clear cell renal cell carcinoma have been described 47, the mechanisms by which they may facilitate tumor spread or invasion remain unclear. Interestingly, the results of this work suggest that this phenomenon persists into the adjacent normal tissue, although it is not known whether the mitochondrial deficiency is a primary or secondary event. Several mechanisms, including oxygen sensing processes 48–52 and pH-mediated invasion 53, 54, may give rise to the observations reported in this study.
Factors governing tumor mediated molecular changes beyond the histological margin into ‘normal’ tissue may correlate with tumor aggressiveness. Currently, there is no marker to define ccRCC tumor aggressiveness. In this study, tumor stage and grade were both utilized as an assessment of aggression in order to determine if the extent of compromised tissue (i.e. the distance from the histological margin at which molecular changes were significant) could be correlated with aggressiveness. Our results showed that such distance measurements could not be correlated with tumor stage or grade, indicating either that this measurement is independent of tumor aggressiveness or the currently used methods cannot effectively make this distinction. The current clinical inspection of the tumor is based on anatomical location of the tumor and its cellular morphology, which may not best represent the molecular state and aggressiveness of the tumor and that there are molecular events occurring in the tumor and its adjacent tissue that cannot be visualized with current histology procedures. Alternatively, the extent of compromised tissue beyond the histological border may correlate with patient outcome; however, patient follow-up information was not available for this study.
Results from this study demonstrate that many intriguing yet complex events occur at tumor borders that contribute to the aberrant molecular changes occurring outside of the histological margin. One hypothesis is the aberrant cells outside of the tumor margin are infiltrative tumor cells that go undetected by conventional histology. Alternatively, these cells could be pre-cancerous cells that have already undergone malignant transformation at the molecular level yet show no phenotypic characteristics of tumor cells. Another possibility is the aberrant cellular characteristics of these seemingly normal cells result from cross-talk or secretory interactions between the tumor and surrounding tissue. This signaling pathway could cause cells outside of the histological margin to express some molecular features characteristic of the tumor.
Technology Perspective
The applicability of MALDI MS to the rapid evaluation of tumor margins has been proposed 55 and the supportive work presented here is the first, in-depth molecular assessment of tumor margin microenvironments by this technology. Previous studies have resulted in the identification of disease-state and patient prognostic-specific protein patterns. Collectively, these studies suggest that proteomic information will become increasingly important in assessing disease progression, prognosis, and response to therapy.
With the ability to rapidly analyze thin tissue sections and provide for the visualization of hundreds of proteins simultaneously, it is anticipated that profiling and imaging MALDI MS technology will become an invaluable tool in diagnostic pathology and diagnostic labs to enhance the quality of information provided to clinicians 55, 56. Conventional approaches rely on histomorphology and immunohistochemistry procedures that represent a challenge in the interoperative setting. Histochemistry, such as hematoxylin and eosin staining, only allows for morphological characterization. Immunohistochemistry procedures, however, are difficult to standardize, require procedures taking hours or overnight and in most cases must be performed separately for each antibody. For many proteins, antibodies are not readily available and there is no standard, rigorous procedure for assessing their cross-reactivity. Microarray analysis of antibody cross-reactivity produces uncertain results because of numerous cross-reactivities with both monoclonal and polyclonal antibodies, as well as substantial binding to non-specific proteins 10. MALDI MS technology allows for the visualization of many hundreds of individual proteins in the molecular weight range from 2000 to 100,000. Its application to direct tissue analysis allows direct correlation of individual protein signals with their distributions within specific regions of tissue. It is also well suited for discovery studies in that it does not require prior knowledge of protein composition and does not require molecular-specific reagents. MALDI MS technology offers a molecularly specific process for analyzing tumor margins to complement the current clinical diagnostic tools to ensure successful tumor extirpation.
Conclusions
Elucidation of the events that drive tumor recurrence will not only provide additional insights into the molecular mechanisms of tumor invasion, but also facilitate the discovery of molecular markers to aid in current histological procedures that better define molecular tumor margins in an effort to reduce local recurrence. Identifying biological changes and understanding the molecular events in and around tumor margins is vital to understanding therapeutic resistance and designing new therapeutic agents.
Supplementary Material
Acknowledgments
The authors would like to thank Heinrich Roder, Ph.D. and Julia Grigorieva, Ph.D. from Biodesix, Inc. for their input as well as their assistance with data processing and statistical analysis. We would also like to thank Scott Sobecki for custom software, Lisa Zimmerman, Ph.D. and Julie Coleman for the IPG separations, Malin Andersson for assistance with histochemistry, and M. Reid Groseclose and Hans R. Aerni for their respective contributions to the protein identifications. Funding was provided by NIH/NIGMS 5R01 GM 58008, NIH/NCI 5R33 CA86243, and DOD W81XWH-05-01-0179, and the Training Program in Environmental Toxicology T32 ES07028-29.
Footnotes
Supporting Information Available
This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Looser KG, Shah JP, Strong EW. The significance of “positive” margins in surgically resected epidermoid carcinomas. Head Neck Surg. 1978;1(2):107–11. doi: 10.1002/hed.2890010203. [DOI] [PubMed] [Google Scholar]
- 2.Nathan CAO, Amirghahri N, Rice C, Abreo F, Shi R, Stucker S. Molecular Analysis of Surgical Margins in Head and Neck Squamous Cell Carcinoma Patients. The Laryngoscope. 2002;112:2129–2140. doi: 10.1097/00005537-200212000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Masasyesva BG, Tong BC, Brock MV, Pilkington T, Goldenberg D, Sidransky D, Harden S, Westra WH, Califano J. Molecular margin analysis predicts local recurrence after sublobar resection of lung cancer. Int J Cancer. 2005;113:1022–1025. doi: 10.1002/ijc.20683. [DOI] [PubMed] [Google Scholar]
- 4.Balch GC, Mithani SK, Simpson JF, Kelley MC. Accuracy of intraoperative gross examination of surgical margin status in women undergoing partial mastectomy for breast malignancy. Am Surg. 2005;71(1):22–7. discussion 27–8. [PubMed] [Google Scholar]
- 5.Choong PF. Surgical margins for soft tissue sarcoma: size does matter. ANZ J Surg. 2006;76(3):97. doi: 10.1111/j.1445-2197.2006.03686.x. [DOI] [PubMed] [Google Scholar]
- 6.Gronchi A, Casali PG, Mariani L, Miceli R, Fiore M, Lo Vullo S, Bertulli R, Collini P, Lozza L, Olmi P, Rosai J. Status of surgical margins and prognosis in adult soft tissue sarcomas of the extremities: a series of patients treated at a single institution. J Clin Oncol. 2005;23(1):96–104. doi: 10.1200/JCO.2005.04.160. [DOI] [PubMed] [Google Scholar]
- 7.van Houten VM, Leemans CR, Kummer JA, Dijkstra J, Kuik DJ, van den Brekel MW, Snow GB, Brakenhoff RH. Molecular diagnosis of surgical margins and local recurrence in head and neck cancer patients: a prospective study. Clin Cancer Res. 2004;10(11):3614–20. doi: 10.1158/1078-0432.CCR-03-0631. [DOI] [PubMed] [Google Scholar]
- 8.Toms SA, Lin WC, Weil RJ, Johnson MD, Jansen ED, Mahadevan-Jansen A. Intraoperative optical spectroscopy identifies infiltrating glioma margins with high sensitivity. Neurosurgery. 2005;57(4 Suppl):382–91. doi: 10.1227/01.neu.000176855.39826.2d. discussion 382–91. [DOI] [PubMed] [Google Scholar]
- 9.Brennan JA, Mao L, Hruban RH, Boyle JO, Eby YJ, Koch WM, Goodman SN, Sidransky D. Molecular assessment of histopathological staging in squamous-cell carcinoma of the head and neck. N Engl J Med. 1995;332(7):429–35. doi: 10.1056/NEJM199502163320704. [DOI] [PubMed] [Google Scholar]
- 10.Predki PF, Mattoon D, Bangham R, Schweitzer B, Michaud G. Protein microarrays: a new tool for profiling antibody cross-reactivity. Hum Antibodies. 2005;14(1–2):7–15. [PubMed] [Google Scholar]
- 11.Friedman DB, Hill S, Keller JW, Merchant NB, Levy SE, Coffey RJ, Caprioli RM. Proteome analysis of human colon cancer by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics. 2004;4(3):793–811. doi: 10.1002/pmic.200300635. [DOI] [PubMed] [Google Scholar]
- 12.Rahman SM, Shyr Y, Yildiz PB, Gonzalez AL, Li H, Zhang X, Chaurand P, Yanagisawa K, Slovis BS, Miller RF, Ninan M, Miller YE, Franklin WA, Caprioli RM, Carbone DP, Massion PP. Proteomic patterns of preinvasive bronchial lesions. Am J Respir Crit Care Med. 2005;172(12):1556–62. doi: 10.1164/rccm.200502-274OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seliger B, Lichtenfels R, Kellner R. Detection of renal cell carcinoma-associated markers via proteome- and other ‘ome’-based analyses. Brief Funct Genomic Proteomic. 2003;2(3):194–212. doi: 10.1093/bfgp/2.3.194. [DOI] [PubMed] [Google Scholar]
- 14.Pierson J, Norris JL, Aerni HR, Svenningsson P, Caprioli RM, Andren PE. Molecular profiling of experimental Parkinson’s disease: direct analysis of peptides and proteins on brain tissue sections by MALDI mass spectrometry. J Proteome Res. 2004;3(2):289–95. doi: 10.1021/pr0499747. [DOI] [PubMed] [Google Scholar]
- 15.Reyzer ML, Hsieh Y, Ng K, Korfmacher WA, Caprioli RM. Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom. 2003;38(10):1081–92. doi: 10.1002/jms.525. [DOI] [PubMed] [Google Scholar]
- 16.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69(23):4751–60. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 17.Chaurand P, Stoeckli M, Caprioli RM. Direct profiling of proteins in biological tissue sections by MALDI mass spectrometry. Anal Chem. 1999;71(23):5263–70. doi: 10.1021/ac990781q. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz SA, Weil RJ, Johnson MD, Toms SA, Caprioli RM. Protein profiling in brain tumors using mass spectrometry: feasibility of a new technique for the analysis of protein expression. Clin Cancer Res. 2004;10(3):981–7. doi: 10.1158/1078-0432.ccr-0927-3. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz SA, Weil RJ, Thompson RC, Shyr Y, Moore JH, Toms SA, Johnson MD, Caprioli RM. Proteomic-based prognosis of brain tumor patients using direct-tissue matrix-assisted laser desorption ionization mass spectrometry. Cancer Res. 2005;65(17):7674–81. doi: 10.1158/0008-5472.CAN-04-3016. [DOI] [PubMed] [Google Scholar]
- 20.Yanagisawa K, Shyr Y, Xu BJ, Massion PP, Larsen PH, White BC, Roberts JR, Edgerton M, Gonzalez A, Nadaf S, Moore JH, Caprioli RM, Carbone DP. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet. 2003;362(9382):433–9. doi: 10.1016/S0140-6736(03)14068-8. [DOI] [PubMed] [Google Scholar]
- 21.Xu BJ, Shyr Y, Liang X, Ma LJ, Donnert EM, Roberts JD, Zhang X, Kon V, Brown NJ, Caprioli RM, Fogo AB. Proteomic patterns and prediction of glomerulosclerosis and its mechanisms. J Am Soc Nephrol. 2005;16(10):2967–75. doi: 10.1681/ASN.2005030262. [DOI] [PubMed] [Google Scholar]
- 22.Reyzer ML, Caldwell RL, Dugger TC, Forbes JT, Ritter CA, Guix M, Arteaga CL, Caprioli RM. Early changes in protein expression detected by mass spectrometry predict tumor response to molecular therapeutics. Cancer Res. 2004;64(24):9093–100. doi: 10.1158/0008-5472.CAN-04-2231. [DOI] [PubMed] [Google Scholar]
- 23.Xie L, Xu BJ, Gorska AE, Shyr Y, Schwartz SA, Cheng N, Levy S, Bierie B, Caprioli RM, Moses HL. Genomic and proteomic analysis of mammary tumors arising in transgenic mice. J Proteome Res. 2005;4(6):2088–98. doi: 10.1021/pr050214l. [DOI] [PubMed] [Google Scholar]
- 24.Caldwell RL, Gonzalez A, Oppenheimer SR, Schwartz HS, Caprioli RM. Assessment of the Tumor Protein Microenvironment Using Imaging Mass Spectrometry. Cancer Genomics and Proteomics. 2006;3(5):279. [Google Scholar]
- 25.Jemal A, Murray T, Ward E, Samuels A, Tiwari R, Ghafoor A, Feuer E, Thun M. Cancer Statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 26.Grignon DJ, Che M. Clear cell renal cell carcinoma. Clin Lab Med. 2005;25(2):305–16. doi: 10.1016/j.cll.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Kosari F, Parker AS, Kube DM, Lohse CM, Leibovich BC, Blute ML, Cheville JC, Vasmatzis G. Clear cell renal cell carcinoma: gene expression analyses identify a potential signature for tumor aggressiveness. Clin Cancer Res. 2005;11(14):5128–39. doi: 10.1158/1078-0432.CCR-05-0073. [DOI] [PubMed] [Google Scholar]
- 28.Murphy W, Grignon DJ, Perlman E. Tumors of the kidney, bladder, and related urinary structures. American Registry of Pathology; Washington, D.C: 2004. [Google Scholar]
- 29.Sengupta S, Zincke H. Lessons learned in the surgical management of renal cell carcinoma. Urology. 2005;66(5 Suppl):36–42. doi: 10.1016/j.urology.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen TT, Parkinson JP, Kuehn DM, Winfield HN. Technique for ensuring negative surgical margins during laparoscopic partial nephrectomy. J Endourol. 2005;19(3):410–5. doi: 10.1089/end.2005.19.410. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz SA, Reyzer ML, Caprioli RM. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J Mass Spectrom. 2003;38(7):699–708. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- 32.Aerni HR, Cornett DS, Caprioli RM. Automated acoustic matrix deposition for MALDI sample preparation. Anal Chem. 2006;78(3):827–34. doi: 10.1021/ac051534r. [DOI] [PubMed] [Google Scholar]
- 33.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrell FE., Jr . Regression Modeling Strategies. Springer; New York, NY: 2001. [Google Scholar]
- 36.Cargile BJ, Bundy JL, Freeman TW, Stephenson JL., Jr Gel based isoelectric focusing of peptides and the utility of isoelectric point in protein identification. J Proteome Res. 2004;3(1):112–9. doi: 10.1021/pr0340431. [DOI] [PubMed] [Google Scholar]
- 37.Cargile BJ, Talley DL, Stephenson JL., Jr Immobilized pH gradients as a first dimension in shotgun proteomics and analysis of the accuracy of pI predictability of peptides. Electrophoresis. 2004;25(6):936–45. doi: 10.1002/elps.200305722. [DOI] [PubMed] [Google Scholar]
- 38.Enj JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 39.Keller A, Purvine S, Nesvizhskii AI, Stolyar S, Goodlett DR, Kolker E. Experimental protein mixture for validating tandem mass spectral analysis. Omics. 2002;6(2):207–12. doi: 10.1089/153623102760092805. [DOI] [PubMed] [Google Scholar]
- 40.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 41.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 42.Fuhrman SA, Lasky LC, Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol. 1982;6(7):655–63. doi: 10.1097/00000478-198210000-00007. [DOI] [PubMed] [Google Scholar]
- 43.Hervouet E, Demont J, Pecina P, Vojtiskova A, Houstek J, Simonnet H, Godinot C. A new role for the von Hippel-Lindau tumor suppressor protein: stimulation of mitochondrial oxidative phosphorylation complex biogenesis. Carcinogenesis. 2005;26(3):531–9. doi: 10.1093/carcin/bgi001. [DOI] [PubMed] [Google Scholar]
- 44.Simonnet H, Alazard N, Pfeiffer K, Gallou C, Beroud C, Demont J, Bouvier R, Schagger H, Godinot C. Low mitochondrial respiratory chain content correlates with tumor aggressiveness in renal cell carcinoma. Carcinogenesis. 2002;23(5):759–68. doi: 10.1093/carcin/23.5.759. [DOI] [PubMed] [Google Scholar]
- 45.Warburg O. London: Constable Press; 1930. [Google Scholar]
- 46.Herrmann PC, Herrmann EC. Oxygen metabolism and a potential role for cytochrome c oxidase in the Warburg effect. J Bioenerg Biomembr. 2007 doi: 10.1007/s10863-007-9084-z. [DOI] [PubMed] [Google Scholar]
- 47.Godinot C, de Laplanche E, Hervouet E, Simonnet H. Actuality of Warburg’s views in our understanding of renal cancer metabolism. J Bioenerg Biomembr. 2007 doi: 10.1007/s10863-007-9088-8. [DOI] [PubMed] [Google Scholar]
- 48.Acker T, Fandrey J, Acker H. The good, the bad and the ugly in oxygen-sensing: ROS, cytochromes and prolyl-hydroxylases. Cardiovasc Res. 2006;71(2):195–207. doi: 10.1016/j.cardiores.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 49.Giaccia AJ, Simon MC, Johnson R. The biology of hypoxia: the role of oxygen sensing in development, normal function, and disease. Genes Dev. 2004;18(18):2183–94. doi: 10.1101/gad.1243304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guzy RD, Schumacker PT. Oxygen sensing by mitochondria at complex III: the paradox of increased reactive oxygen species during hypoxia. Exp Physiol. 2006;91(5):807–19. doi: 10.1113/expphysiol.2006.033506. [DOI] [PubMed] [Google Scholar]
- 51.Haase VH. The VHL/HIF oxygen-sensing pathway and its relevance to kidney disease. Kidney Int. 2006;69(8):1302–7. doi: 10.1038/sj.ki.5000221. [DOI] [PubMed] [Google Scholar]
- 52.Hervouet E, Simonnet H, Godinot C. Mitochondria and reactive oxygen species in renal cancer. Biochimie. 2007;89(9):1080–8. doi: 10.1016/j.biochi.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 53.Gatenby RA, Gawlinski ET. The glycolytic phenotype in carcinogenesis and tumor invasion: insights through mathematical models. Cancer Res. 2003;63(14):3847–54. [PubMed] [Google Scholar]
- 54.Raghunand N, Gatenby RA, Gillies RJ. Microenvironmental and cellular consequences of altered blood flow in tumours. Br J Radiol. 2003;76(Spec No 1):S11–22. doi: 10.1259/bjr/12913493. [DOI] [PubMed] [Google Scholar]
- 55.Chaurand P, Sanders ME, Jensen RA, Caprioli RM. Proteomics in diagnostic pathology: profiling and imaging proteins directly in tissue sections. Am J Pathol. 2004;165(4):1057–68. doi: 10.1016/S0002-9440(10)63367-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson MD, Floyd JL, Caprioli RM. Proteomics in diagnostic neuropathology. J Neuropathol Exp Neurol. 2006;65(9):837–45. doi: 10.1097/01.jnen.0000235116.67558.24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.