Abstract
Developing an effective HIV-1 vaccine will require strategies to enhance antigen presentation to the immune system. In a previous study we demonstrated a marked increase in immunogenicity of the highly glycosylated HIV-1 gp120 protein following enzymatic addition of α-gal epitopes to the carbohydrate chains. In the present study we determined whether gp120αgal can also serve as an effective platform for targeting other HIV-1 proteins to APC and thus increase immunogenicity of both proteins. For this purpose we produced a recombinant fusion protein between gp120 and the HIV-1 matrix p24 protein (gp120/p24). Multiple α-gal epitopes were synthesized enzymatically on the gp120 portion of the fusion protein to generate a gp120αgal/p24 vaccine. Immune responses to gp120αgal/p24 compared to gp120/p24 vaccine lacking α-gal epitopes were evaluated in knockout (KO) mice. These mice lack α-gal epitopes and, therefore, are capable of producing the anti-Gal antibody. T cell responses to p24, as assessed by ELISPOT and by CD8+ T cells intracellular staining assays for IFNγ, was on average 12 and 10-fold higher, respectively, in gp120αgal/p24 immunized mice than in mice immunized with gp120/p24. In addition, cellular and humoral immune responses against gp120 were higher by 10 to 30-fold in mice immunized with gp120αgal/p24 than in gp120/p24 immunized mice. Our data suggest that the α-gal epitopes on the gp120 portion of the fusion protein can significantly augment the immunogenicity of gp120, as well as that of the fused viral protein which lacks α-gal epitopes. This strategy of anti-Gal mediated targeting to APC may be used for production of effective HIV-1 vaccines comprised of various viral proteins fused to gp120.
Introduction
Effective protection against HIV-1 infection may be achieved by prophylactic vaccines that elicit a combined humoral immune response against envelope proteins and a cellular immune response against matrix and core proteins. The humoral immune response comprises primarily of neutralizing anti-gp120 (anti-Env) antibodies that prevent the infection of cells by HIV-1, whereas the cellular immune response includes the activity of cytotoxic T lymphocytes (CTLs) that destroy HIV-1 infected host cells [1]. One of the major factors determining the efficacy of such prophylactic vaccines is the number of memory B and T cells that circulate post-vaccination. These memory cells are rapidly activated in early stages following transmission of the virus, while the number of the infected cells is relatively low. In the absence of a rapid immune response, the infecting HIV-1 replicates and mutates before anti-Env antibodies are produced in sufficiently high titers to prevent viral spread to a large number of cells. Mutations in envelope glycoproteins enable HIV-1 to escape the neutralizing antibodies without losing receptor binding activity [2–11]. In order to elicit effective humoral and cellular immune responses, vaccine antigens must be effectively taken up by antigen presenting cells (APC) which process and present the immunogenic peptides on class I and class II MHC molecules in order to activate the corresponding cytotoxic and helper T cells. In our previous study we described a method that exploits the multiple carbohydrate chains on gp120 of HIV-1 for anti-Gal mediated targeting of a gp120 vaccine to APC [12].
Anti-Gal is the only natural antibody known to be abundantly produced in humans, comprising ~1% of total serum IgG [13]. Anti-Gal interacts specifically with a carbohydrate antigen termed the α-gal epitope (Galα1-3Galβ1-4GlcNAc-R) [14, 15] which is produced by the activity of the glycosylation enzyme α1,3 galactosyltransferase (α1,3GT). Non-primate mammals, prosimians and New World monkeys carry the active α1,3GT gene and naturally produce large amounts of α-gal epitopes on cell surface and secreted glycoproteins [16, 17]. In contrast, humans, apes and Old World monkeys lack α-gal epitopes because they lack active α1,3GT genes [18] and naturally produce anti-Gal in high titers [17]. Natural anti-Gal antibodies can be exploited to effectively target microbial or cancer vaccines carrying α-gal epitopes to APC [19–21]. Such vaccines form immune complexes with anti-Gal at the vaccination sites and are effectively targeted for uptake by APC as a result of the interactions between the Fc portion of the immunocomplexed anti-Gal and Fcγ receptors (FcγR) on APC [22].
We previously showed that the multiple N-linked carbohydrate chains of complex type (i.e. with terminal SA-Galβ1-4GlcNAc-R) on recombinant gp120 can be converted into α-gal epitopes by incubation with neuraminidase (for removal of terminal sialic acid [SA]), recombinant α1,3GT and UDP-galactose (UDP-Gal) as the sugar donor [12]. This enzymatic reaction results in the synthesis of ~30 α-gal epitopes per gp120 molecule. The immunogenicity of gp120 carrying α-gal epitopes (gp120αgal) can be studied in the α1,3GT gene knockout mouse (referred to as KO mice). These mice lack α-gal epitopes and, therefore, are capable of producing anti-Gal (31). In previous studies we reported that KO mice producing anti-Gal and immunized with gp120αgal displayed ~100-fold higher titer of anti-gp120 antibodies than KO mice immunized with gp120 lacking α-gal epitopes [12]. The activities of neutralizing anti-gp120 antibodies were also significantly higher in KO mice immunized with gp120αgal than in those immunized with gp120. We further observed a similar ~100-fold increase of immune responses against influenza virus in KO mice immunized with inactivated influenza virus expressing α-gal epitopes, in comparison with KO mice immunized with the same dose of inactivated virus lacking these carbohydrate epitopes [23].
We hypothesized that gp120 processed to express multiple α-gal epitopes may serve as a platform for delivering other less immunogenic viral proteins to APC, in order to enhance immune responses against HIV-1. Matrix or core proteins of HIV-1 may serve as vaccine antigens since they are highly conserved compared to the viral envelope gp120. Inclusion of other viral proteins such as Nef, Tat and Rev in an HIV-1 vaccine may further increase efficacy since they may elicit broad immune responses against the virus, as observed in long-term non-progressors [21, 24]. The internal proteins of HIV-1 cannot be enzymatically modified to display α-gal epitopes since they lack carbohydrate chains. Anti-Gal mediated targeting of the various HIV-1 internal proteins to APC may be achieved by fusion of the genes encoding them with the gp120 gene and production of the fusion protein in mammalian cell expression systems for the synthesis of carbohydrate chains of the complex type (e.g. CHO or 293 cells). The glycosylated gp120 portion of the protein containing multiple N-linked carbohydrate chains is subsequently subjected to enzymatic synthesis of α-gal epitopes by using neuraminidase and recombinant α1,3GT. Upon vaccination, such fusion protein vaccines would form immune complexes with anti-Gal antibodies in vivo and therefore will be targeted for uptake by APC via Fc/FcγR interactions.
In the present study we evaluated the effect of α-gal epitopes on the immunogenicity of a fusion protein between gp120 and the HIV-1 matrix protein p24. The immunogenicities of both p24 and gp120 in gp120αgal/p24 were compared to that of gp120/p24 in the KO mice producing anti-Gal. Our data suggest that this strategy of fusing gp120αgal to other viral proteins can enhance both humoral and cellular immune responses to candidate HIV-1 vaccines.
MATERIALS AND METHODS
Supplies
Recombinant α1,3GT was produced in the Pichia pastoris expression system as described previously [25]. Neuraminidase from Vibrio cholera was purchased from Sigma (St. Louis MO). Recombinant gp120BAL was received as a gift from NIH AIDS Research and Reference Reagent Program. The monoclonal anti-Gal antibody designated M86 was obtained in tissue culture supernatants of the hybridoma M86 cells, as previously described [26]. Horseradish-peroxidase (HRP) conjugated anti-mouse IgG and anti-mouse IgM antibodies were purchased from Accurate Laboratories (Westbury, NY). The Ribi adjuvant, Trehalose Dicorynomycolate (TDM), was purchased from Corixa (Hamilton, MT).
Mice and immunization procedures
The mice used as the experimental model were α1,3GT knockout (KO) mice on H-2bxd genetic background [27] which are bred and maintained at the animal facility of the University of Massachusetts Medical School. Studies were performed with both males and females which yielded similar results. Production of anti-Gal IgG in the KO mice was achieved by three i.p. immunizations with 50 mg of pig kidney membrane (PKM) homogenate. These injections, administered in one week intervals, induced anti-Gal production in titers similar to those of anti-Gal in humans (titers of 1:200 to 1:2000 as measured by ELISA with α-gal BSA as solid phase antigen) [28, 29]. Following the demonstration of anti-Gal IgG production, the mice received a subcutaneous injection of gp120/p24 or gp120αgal/p24 at 5 μg per injection in Ribi adjuvant. These injections were repeated after 2 weeks, and the immune responses were evaluated two weeks after the second immunization.
Production of gp120/p24 fusion gene
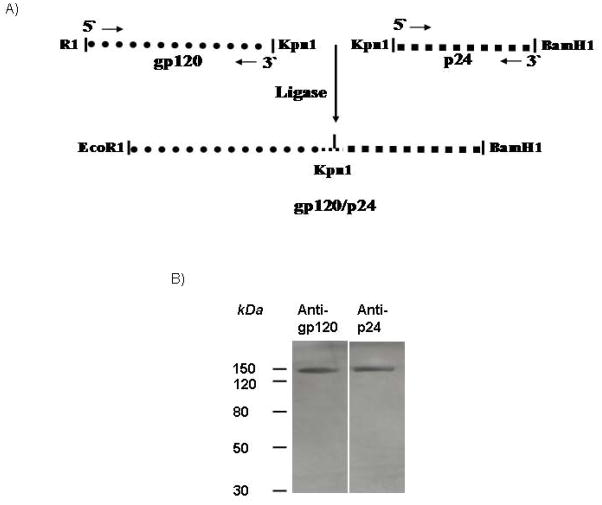
Production of gp120/p24 required the generation of a fusion gene between the coding regions of gp120 and p24. The vector used for this purpose was pCDNA3.1 (−) (Invitrogen). The gp120 JR-FL codon-optimized sequence was reported previously [30]. The p24 DNA gene was cloned from a non-infectious HIV-1 clone pNL4-3.dpol with LTR and partial pol gene deletion [31]. The gp120 region of the HIV-1 env gene was amplified by PCR using the forward primer gp120-jrfl-1 5′-CAGACAGAATTCATGGATGCAATGAAGAGAG-3′ and the reverse primer gp120-jrfl-2 5′-ATCTATGGTACCAGCCACAGCGCGCTTCTCCCTC-3′. The p24 region was amplified by PCR with forward primer NL4-3 p24-1 5′-ATCTATGGTACCGCAATAGTGCAGAACCTCCAGGG-3′ and reverse primer NL4-3 p24-2 5′-TGTGATGGATCCTCATTTATGGCCGGGTCCC-3′. The gp120 primers were used to amplify the codon-optimized JR-FL gene and the p24 primers amplified the NL4-3 p24 gene [30]. The PCR product of gp120 was digested with EcoRI and KpnI, and the p24 gene product was digested by KpnI and BamHI. The digested gp120 and p24 gene fragments were ligated into the vector pCDNA3.1 (−) cut with BamHI and EcoRI. The presence of the gp120-p24 fragment in pCDNA3.1 (−) was confirmed by digesting the vector with EcoRI and BamHI enzymes. The gp120-p24 fragment was further confirmed by sequencing. The p24 gene was fused to the C terminus of gp120 in order to keep the codon-optimized t-PA leader signal proximally upstream of the gp120 gene [32].
Production of gp120/p24 fusion proteins
Gp120-p24 fusion proteins were produced by transfection of 293T cells with pCDNA3.1(−) plasmid expressing the HIV-1 gp120-p24 protein, using Lipofectamine reagent (Invitrogen) as in previous studies. The fusion proteins were purified from the pooled supernatants using ammonium sulfate precipitation and a glycoprotein purification kit according to the manufacturer’s instructions (Qiagen). The purified fusion glycoprotein was stored in aliquots at −20°C until use.
Synthesis of α-gal epitopes on gp120/p24 fusion proteins
Synthesis of α-gal epitopes on gp120/p24 was performed by the use of two enzymes: neuraminidase (extracted from Vibrio cholera) and recombinant α1,3GT [12]. Briefly, gp120/p24 was brought to a concentration of 100 μg/ml in the enzyme buffer containing 0.1 M MES (methyl ethyl morpholino sulfonate) pH 6.0, and 25 mM MnCl2. The terminal SA was removed from the carbohydrate chains on the gp120 portion of gp120/p24 by neuraminidase (1 mU/ml), and α-gal epitopes were synthesized on these carbohydrate chains by α1,3GT (30 μg/ml) and UDP-Gal (1 mM). The control gp120/p24 fusion proteins lacking α-gal epitopes were added in the enzyme solution at 100 mg/ml in the presence of the two enzymes which were previously heat inactivated (10 min at 95°C). The lack of α-gal epitopes on gp120/p24 was confirmed by ELISA, as described below.
ELISA assays
Production of anti-Gal in KO mice immunized with PKM homogenates were determined by ELISA, as previously described [12, 23]. Briefly, ELISA wells were coated with α-gal BSA (10 μg/ml) overnight at 4°C. Plates were washed once with PBS and blocked with 1% BSA in PBS. Serum samples at various dilutions were plated at 50 μl aliquots in the wells for 2 h at 24°C. After washing, HRP-coupled goat anti-mouse IgG was added for 1 h. The colored reactions were developed with ortho-phenylene diamine (OPD) and absorbance measured at 492 nm. KO mice included in the immunization study were those found to display anti-Gal production in titers comparable to those in humans (i.e. 1:200 – 1:2000). Production of anti-gp120 antibodies was assayed by a similar assay, using gp120 (10 μg/ml) as solid-phase antigen. The presence of α-gal epitopes on gp120αgal/p24 was also determined by coating this fusion protein in ELISA plates and detection using monoclonal anti-Gal antibody M86 [26] as a primary antibody followed by HRP-anti-mouse IgM as secondary antibody.
Western blot analysis
The gp120-p24 proteins in supernatants from transiently transfected 293T cells were separated by SDS-PAGE electrophoresis, blotted onto polyvinylidene difluoride membranes (PVDF; Bio-Rad) using 120 amp for 2 h, and blocked overnight at 4°C in blocking buffer (0.2% I-block 0.1% Tween 20 in PBS). Membranes were incubated with rabbit sera containing anti-gp120 or anti-p24 antibodies at 1:20 dilution and incubated for 2 h at room temperature (RT). Subsequently, the membranes were washed and reacted with alkaline phosphatase (ALP)-conjugated goat anti-rabbit IgG (Tropix, Bedford, MA) at a 1:5000 dilution. Signals were detected using the Western-Light Kit (Tropix). Western-light substrate was applied for 5 min to the membranes which were then dried, exposed to X-ray films and developed by a Kodak processor.
Analysis of neutralizing anti-gp120 antibodies
The assay for neutralizing antibodies was previously described [30]. The assay was performed with HIV-1 SF162 pseudotyped virus. Pseudoviruses were generated by the co-transfection of HIV-1 SF162 gp160 envelope with the pSG3ΔEnv backbone. Pseudotyped viruses were added at 200 50% tissue culture infective doses (TCID)/well and incubated with the tested sera or with rabbit sera (positive control) at 37°C for 1 h. TZM-bl cells were then seeded at 10,000 cells/well in a final concentration of 20 μg/ml DEAE-dextran. Plates were incubated at 37°C for 48 h and then developed with Luciferase assay reagent according to the manufacturer’s instructions (Promega). Neutralization was calculated as the percent reduction in luciferase activity in the presence of mouse or rabbit immune sera compared to the luciferase activity induced by the virus in the presence of pre-immune sera as follows: [1 − (luciferase + immune sera)/(luciferase + pre-immune sera)] × 100 [33].
IFNγ ELISPOT assay
ELISPOT assays for IFNγ-secreting cells were performed with a commercial kit (Mabtech, OH, USA), as described previously [12, 23, 34]. Briefly, 96-well ELISPOT plates were coated with 100 μl/well of anti-IFNγ monoclonal antibody AN18 overnight at 4°C. The plates were washed with PBS and blocked with PBS containing 10% fetal calf serum (FCS) for 30 min. at RT. Freshly isolated splenocytes (2×105 cells per well) were plated in triplicates together with gp120 (1JR-FL V3 peptide) or p24189–207 peptides [35]. After overnight incubation at 37°C in 5% CO2, cells were removed by washing with PBS and aliquots of 100 μl of anti-IFNγ-biotin (monoclonal antibody R4-6A2, Mabtech) were added to each well for 2 h at RT. The plates were then washed with PBS and 100 μl of Streptavidin-ALP were added per well and incubated for 1 h at RT. After washing with PBS, 100 μl of chromogenic substrate (NBT-plus, Mabtech) were added to each well for 15 min to allow color development and formation of spots. Color reaction was stopped by addition of water. Wells were then air-dried, and spots were counted with the ELISPOT Automated Reader System (performed by Zellnet, Fort Lee, NJ). Calculated frequencies were based on the average of the triplicates wells. The results are expressed as gp120 or p24 specific IFNγ secreting T cells per 106 splenocytes, i.e. the number of spots after subtraction of the spots number in corresponding control wells.
Intracellular cytokine staining for determining T cell activation
For intracellular cytokine staining (ICS), spleens were harvested from mice, and splenocyte suspensions were adjusted to 2×106 cells per tube for each mouse. Spleen lymphocytes were incubated at 37°C in 5% CO2 with 5 μg of p24 peptide. Recombinant human IL-2 (10 unit/ml) was added to all samples and after 1 h of culture, 10 μg/ml of brefeldin A (Sigma Chemical Co., St. Louis, MO) was added to impair cytokine secretion. After another 5 h of incubation, the cells were washed once in 2% FCS-PBS and kept overnight at 4°C. The following day, these cells were stained for surface markers and intracellular cytokines as described by Bottrel et al. [36]. Briefly, cultured cells were stained for surface markers using fluorescein isothiocyanate-labeled anti-CD3 and PerCP-labeled anti-CD8 monoclonal antibodies (BD Pharmengen, CA) by incubation for 30 min on ice, followed by two washes and fixation using solution A (Fix & Perm kit, Caltag Laboratories). These cells were washed and permeabilized using solution B (Fix & Perm kit, Caltag Laboratories) and stained with PE-labeled anti-IFNγ (BD Pharmingen, CA). After 20 min, cells were washed and resuspended in 2% paraformaldehyde. A minimum of 50,000 splenocyte-gated events were acquired using a lymphocyte gate determined based on size and granularity profiles. Data were analyzed by CELLQuest software (Becton Dickinson, San Jose, CA). Antigen-specific cells were defined as CD3+CD8+ T lymphocytes that co-expressed IFN-γ.
RESULTS
Production of gp120/p24 fusion protein
The gene regions of env coding for gp120 and of gag coding for p24 were fused by ligation of the restriction sites Kpn1 as shown in Fig. 1A. The fused gene was inserted into the plasmid pCDNA3.1 (−) which contains the CMV promoter to enable protein expression in mammalian cells. The fusion gp120/p24 products were produced by transient transfection of 293 cells by Lipofectamine. The fusion proteins secreted from the transfected cells were isolated from the culture medium by ammonium sulfate precipitation. Approximately 0.8 mg of the fusion protein was isolated from a total volume of 450 ml of supernatant from transfected 293 cells. For analysis, the fusion protein was electrophoresed in an SDS- polyacrylamide gel. As shown in Fig. 1B, the size of the obtained band was approximately 150 kDa (the sum of the sizes of gp120 and p24 proteins), and it reacted with both anti-gp120 and ant-p24 antibodies. Unmodified gp120 and p24 proteins were also run in parallel with the fusion protein (data not shown).
Figure 1.
(A) Schematic representation of the fusion gene used for production of gp120/p24. Note: the p24 gene was fused to the C terminus of gp120 in order to keep the codon-optimized t-PA leader signal proximally upstream of the gp120 gene. (B) Detection of the gp120/p24 fusion protein by Western blots. Western blot stained with rabbit anti-gp120 (left lane) or rabbit anti-p24 antibodies (right lane). Note that each antibody detected a band of the expected size of the gp120/p24 protein.
Synthesis of α-gal epitopes on gp120/p24 by recombinant α1,3GT
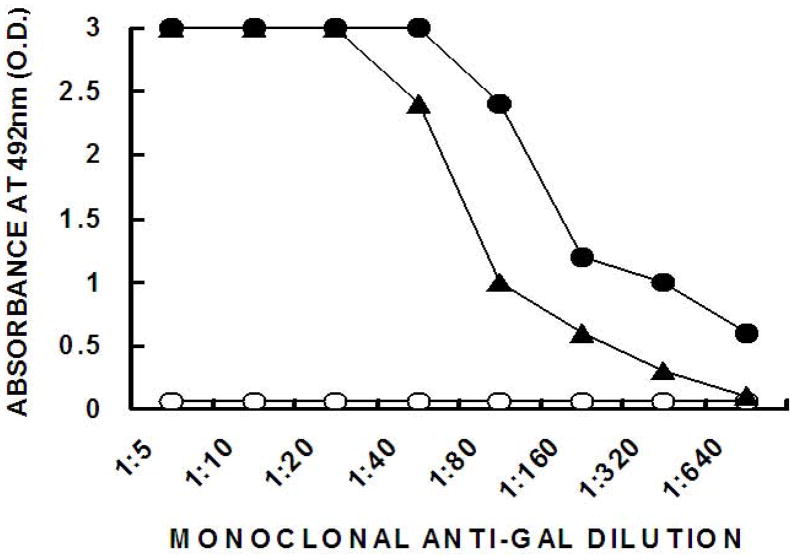
We assumed that since none of the original N-glycosylation sites of gp120 was altered in the fusion protein gp120/p24, the N-linked complex carbohydrate chains on the fusion protein are likely to be similar to those on gp120. The p24 portion of the protein lacks N-glycosylation sites. Many of the N-linked complex carbohydrate chains are capped with SA as SA-Galβ1-4GlcNAC-R. Replacement of SA on the gp120 carbohydrate chains with α-gal epitopes was performed as previously detailed [12] by two enzymes within the same solution: 1. SA was removed from the carbohydrate chains by neuraminidase to expose the penultimate N-acetyllactosamine residues (Galβ1-4GlcNAC-R) on gp120/p24, and 2. The N-acetyllactosamines exposed on the carbohydrate chains functioned as acceptors for recombinant α1,3GT which link to them terminal α1-3galactosyl residues, to form α-gal epitopes. The sugar donor providing galactose to α1,3GT is uridine-diphosphate galactose (UDP-Gal). The de novo expression of α-gal epitopes on gp120αgal/p24 was demonstrated in ELISA wells coated with 1 μg/ml of the fusion proteins and with the monoclonal anti-Gal antibody M86 [26]. As shown in Fig. 2, monoclonal anti-Gal displayed no binding to the gp120/p24 that was incubated with heat-inactivated enzymes, since the original molecule produced in the human 293T cells have no α-gal epitopes. Carbohydrate chains of the complex type of gp120 produced in human cells are capped with SA. In contrast, the monoclonal anti- Gal antibody readily bound to gp120αgal/p24 due to the multiple α-gal epitopes synthesized on this fusion protein. In a previous study (1) we estimated that the number of α-gal epitopes synthesized on each original (i.e. non-fused) gp120αgal molecule to be ~30. It is probable that the number of α-gal epitopes synthesized on gp120αgal/p24 is similar. This is supported by the finding that the binding of monoclonal anti-Gal to BSA with linked 12 synthetic α-gal epitopes is 2 to 3-fold lower than that of gp120αgal/p24 (Fig. 2).
Figure 2. Binding of monoclonal anti-Gal to gp120/p24 and to gp120gal/p24 in ELISA.
Binding of the monoclonal anti-Gal M86 to gp120/p24 (○); gp120αgal/p24 (●) and to α-gal BSA that expresses 10 synthetic α-gal epitopes (■), as measured by ELISA with different amounts of glycoproteins coating the ELISA wells.
Analysis of gp120 specific T cell responses by ELISPOT
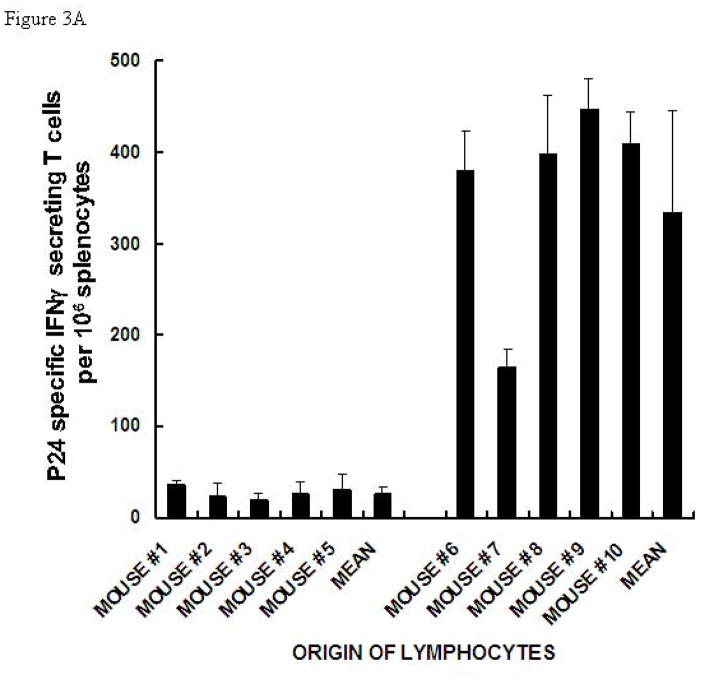
Our previous immunization studies with gp120αgal indicated that the anti-Gal mediated targeting of this vaccine to APC resulted in increased T cell responses, as evaluated by ELISPOT detecting IFNγ secreting T cells (i.e. gp120 specific TH1 and CD8+ T cells) [12]. The main objective in the present study was to determine whether gp120αgal can target the fused p24 to APC, thereby increasing the efficacy of activation of p24 specific T cells. For this purpose anti-Gal producing KO mice were immunized twice with a two week interval as described in the methods. The splenic lymphocytes harvested two weeks after the second immunization were incubated with the p24 immunodominant peptide p24189–207 for 24 h and tested for secretion of INFγ by ELISPOT. As shown in Fig. 3A, in all mice immunized with gp120αgal/p24, the number of T cells secreting IFNγ in response to the p24 peptide was much higher than in mice immunized with gp120/p24. On average, the number of p24 specific T cells in gp120αgal/p24 immunized mice was 12-fold higher than in gp120/p24 immunized mice. The results were compared using the Student’s t-test and found to be statistically significant with t = 5.9 and P-value < 0.001.
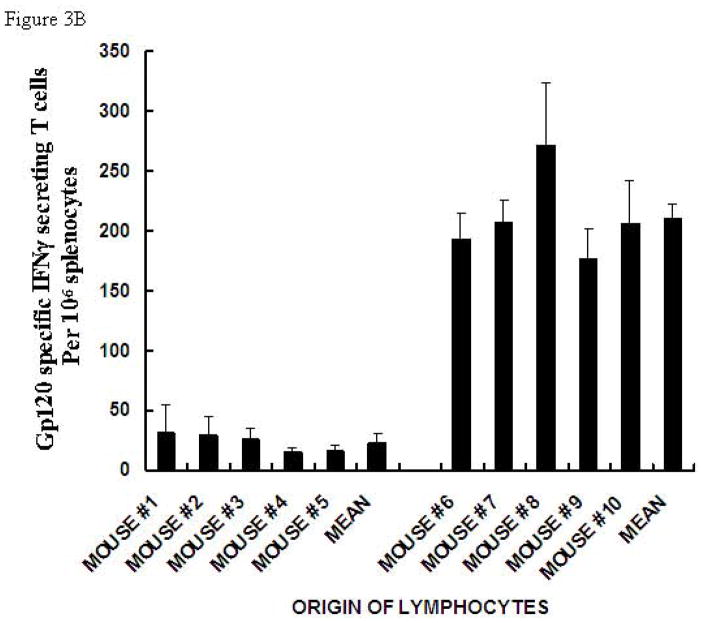
Figure 3. ELISPOT analysis of IFNγ secretion by T cells in KO mice immunized with gp120/p24 or gp120αgal/p24, and in response to p24 peptide.
(A) or gp120 peptide (B). Presentation of ELISPOT data of 6 mice immunized twice with gp120/p24 (mice #1-#6) and 6 mice immunized twice with gp120αgal/p24 (mice #7-#12) as the number of spots per 106 splenocytes. Splenocytes were stimulated with 5 μg of p24 peptide. Data are presented as mean ± standard deviation of triplicate wells.
The fusion of p24 to gp120 did not affect the increase in immunogenicity of the gp120 portion of the molecule when it carried α-gal epitopes. Thus, immunization with gp120αgal/p24 resulted in higher numbers of gp120 specific T cells secreting IFNγ than in mice immunized with gp120/p24 (Fig. 3B). Mice immunized with the fused protein carrying α-gal epitopes displayed ~10 fold higher gp120 specific T cells than those immunized with the fusion protein lacking this epitope (Fig. 3B). The results were compared using the Student’s t-test and found to be statistically significant with t = 11.3 and P-value < 0.01.
We have observed previously a similar increase in T cell responses to gp120 vaccine when the vaccinating protein contained α-gal epitopes (1). The present ELISPOT results imply that the effective anti-Gal mediated targeting of the fusion protein to APC increases the processing and presentation by APC of peptides in both regions of the vaccine, the region with α-gal epitopes and that lacking the anti-Gal binding epitope.
Confirmation of increased T cell responses to p24 by intracellular cytokine staining (ICS)
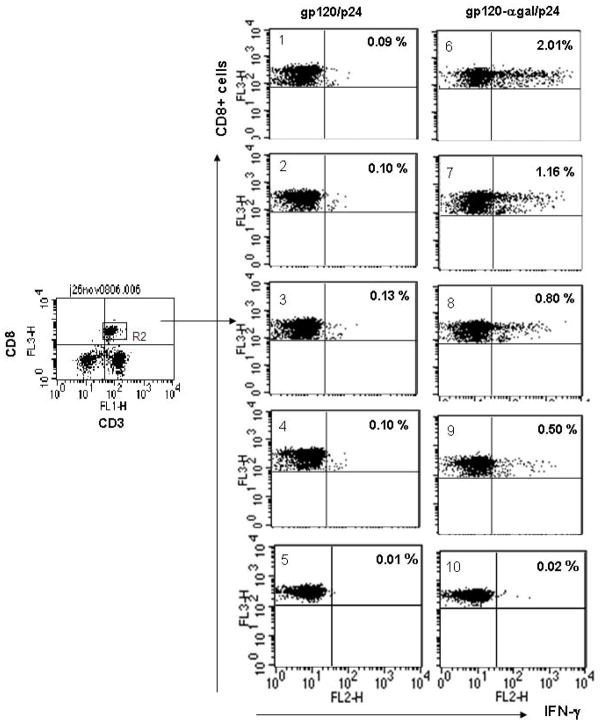
The ELISPOT data described above did not differentiate between TH1 and CD8+ T cell response to p24. In order to determine whether there is an increased response of CD8+ T cells to p24 in mice immunized with gp120αgal/p24, we performed ICS analysis for detection of IFNγ production in activated T cells that are also stained with CD8, and CD3 specific antibodies. The splenic lymphocytes obtained from mice immunized with gp120αgal/p24 or gp120/p24 were pulsed overnight with 5 μg of the immunodominant p24189–207 peptide. Subsequently, CD3+, CD8+ T cells were measured for production of intracellular IFNγ as a result of activation by the p24189–207 peptide. As shown in Fig. 4, four of five mice immunized with gp120αgal/p24 displayed between 0.5–2.0% of activated CD8+ T cells (mice #1- #4), whereas one of the five mice immunized with gp120- gal/p24 displayed no CD8+ activation. In contrast, in mice immunized with gp120/p24 lacking α-gal epitopes (mice #6-#10), only 0.09–0.1% of CD8+ T cells were activated by the p24 peptide. On average, the proportion of p24 specific CD8+ T cells was found in this assay to be 11-fold higher in mice immunized with the gp120αgal/p24 vaccine than in mice immunized with the gp120/p24 vaccine. These results suggest that CD8+ T cells were effectively primed in vivo by the gp120αgal/p24 vaccine and thus, were activated in vitro by the p24 peptides presented on APC. The low p24 specific CD8+T cell activation in gp120/p24 vaccinated mice indicate that the unmodified fusion protein antigen has low immunogenicity, likely due to poor uptake by APC.
Figure 4. Analysis of intracellular staining of IFNγ in CD8+ T cells from KO mice immunized with gp120/p24 or gp120αgal/p24 in response to p24 peptide.
ICS analysis of IFNγ production in CD8+ T cells in response to p24 peptide from mice immunized with gp120/p24 (left panels mice #1 - #5) and mice immunized with gp120αgal/p24 (right panels mice #6 - #10). Lymphocytes were stained for CD3+, CD8+ membrane markers and intracellular IFNγ. Gated CD3+/CD8+ positive events were analyzed for IFNγ production. The % of CD8+ T cells with intracellular IFNγ is indicated in the upper right quadrant for each mouse. Note that 4 of the 5 mice immunized with gp120αgal/p24 (#6-#9) displayed higher proportions of IFNγ positive CD8+ T cells in comparison to mice immunized with gp120/p24.
Increased anti-gp120 antibody responses in mice immunized with gp120αgal/p24
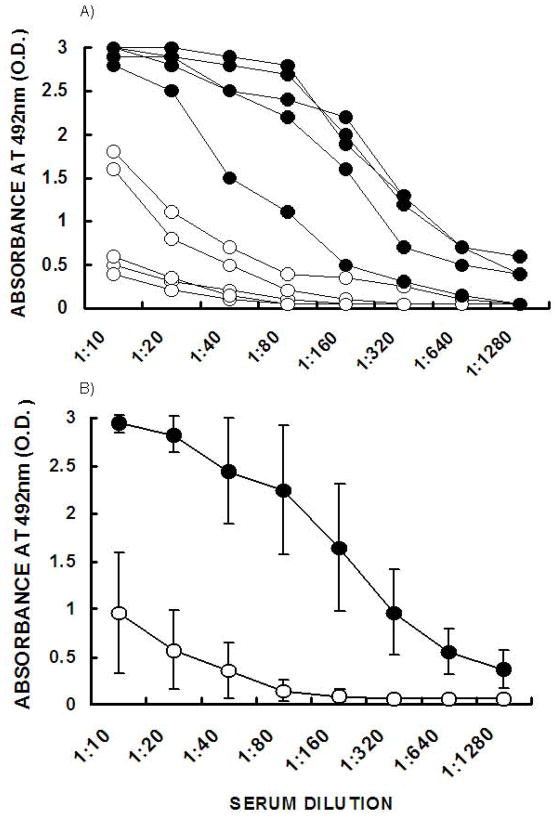
The proposed vaccine is aimed at eliciting a combined cellular immune response against core or matrix virus proteins, such as p24, and both humoral and cellular immune responses against the envelope glycoprotein gp120. Thus, it was of interest to determine whether anti-gp120 antibody activity in gp120αgal/p24 immunized mice was significantly higher than that in gp120/p24 immunized mice. The serum obtained prior to euthanasia of the immunized mice was analyzed by ELISA for the presence of IgG antibodies binding to gp120. Figure 5A describes the anti-gp120 antibody activity in the individual immunized mice. In 4 out of the 5 mice immunized with gp120αgal/p24, the anti-gp120 antibody activity was 15 to 100-fold higher than in the gp120/p24 immunized group. In the fifth gp120αgal/p24 immunized mouse, the antibody activity was only 4 to 8-fold higher than in gp120/p24 immunized mice. The average anti-gp120 antibody response in gp120αgal/p24 immunized mice was 32-fold higher than in gp120/p24 immunized mice (Fig. 5B). The means were compared using the Student’s t-test and found to be statistically significant with P-values < 0.01.
Figure 5. Anti-gp120 response in KO mice immunized with gp120/p24 or gp120αgal/p24.
A. Production of anti-gp120 antibodies in KO mice immunized twice two weeks apart in KO mice either with gp120/p24 (○) or gp120αgal/p24 (●). Note that KO mice immunized with gp120/p24 produced low titers of anti-gp120 antibodies or completely lacked such antibodies, whereas extensive anti-gp120 antibody production was observed in mice immunized with gp120αgal/p24. B. The mean values ± standard deviation calculated from Figure 5A.
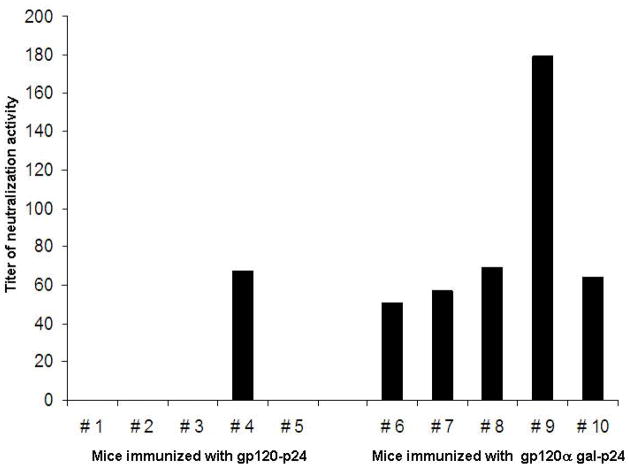
The increased anti-gp120 antibody response as a result of anti-Gal mediated targeting of the vaccine to APC was further demonstrated in an assay analyzing neutralizing antibody activity. As shown in Fig. 6, serum from all mice immunized with gp120αgal/p24 displayed high neutralization activities (mouse #6 to #10). In contrast, serum from mice immunized with gp120–p24 displayed no neutralizing activity with the exception of mouse #4. The response in this one mouse may be associated with variations between individual mice. A similar variation is seen in the group immunized with gp120αgal/p24 where the response in mouse #9 was more than double that in the other mice in this group. The results were compared between the two groups using the Chi square test and found to be statistically significant, Chi square (x2) = 6.67 and P-value < 0.01.
Figure 6. HIV neutralization activity in mice immunized with gp120/p24 or gp120αgal/p24.
HIV neutralization activity in mice immunized with gp120/p24 (mice 1 to 5) or gp120αgal/p24 (mice 6 to 10). Titer is defined as the reciprocal of the serum dilution displaying 50% neutralization
Discussion
The present study demonstrates that gp120αgal can serve as an effective platform for targeting HIV-1 p24 protein to APC and thereby increase its immunogenicity. The gp120 portion of Env has an unusually high number of N- (asparagine -Asn) linked carbohydrate chains relative to its size. The significance of these carbohydrate chains in protecting HIV-1 from immunological assaults was demonstrated by Wei et al [11]. These carbohydrate chains may generate a protective hydration layer that surrounds the virus and affect the access of lymphocytes and antibodies to antigenic peptides on the virus. In addition, the SA capping on these carbohydrate chains can induce electrostatic repulsion with SA on glycoproteins and glycolipids on the APC cell membrane, thereby reducing the uptake of the vaccinating Env by APC. The conversion of the complex carbohydrate chains from SA carrying epitopes into α-gal epitopes can convert the effect of the carbohydrate chains from decreasing to increasing the immunogenicity of the virus or of gp120 by formation of immune complexes (i.e. opsonization) of gp120 vaccines with highly abundant anti-Gal IgG molecules. APC, such as macrophages, skin Langerhans cells and dendritic cells (DC), all express Fcγ receptors (FcγR). Therefore, the Fc/FcγR interactions between immune complexes of anti-Gal and vaccines with FcγR on APC result in effective uptake of the vaccine by these cells [37–39]. Such interactions between immune complexes and the FcγR of APC, are in fact, the most effective mechanism by which APC internalize Ags to stimulate the immune system [37–39].
The ability to express multiple α-gal epitopes on gp120 makes this HIV protein an optimal platform for the targeting proteins of low immunogenicity to APC. In general, the low immunogenicity in various vaccines is in large part due to the lack of markers on the vaccine for optimal uptake by APC. In a recent study[40], we showed that in vivo binding of anti-Gal to liposomes expressing multiple α-gal epitopes increases uptake, processing, presentation and transport by APC to draining lymph nodes of ovalbumin (OVA) encapsulated in these liposomes. The APC further demonstrated effective cross-presentation of immunogenic OVA peptides. This results in a marked increase in immunogenicity of the encapsulated OVA as indicated by increases in CD4+ and CD8+ T cell activations and increased antibody production [40]. In HIV-1 vaccines for eliciting a combined anti-p24 and anti-gp120 immune response, a similar effect can be achieved by fusion of p24 to gp120 and synthesis of α-gal epitopes on the carbohydrate chains of gp120. As shown in the present study, all mice immunized with gp120αgal/p24 fusion protein displayed a much higher number of T cells secreting IFNγ in response to the p24 peptide than mice immunized with gp120/p24 (Fig. 3A). Accordingly, the ICS analysis indicated that the proportion of CD8+ T cells activated in vitro by p24 peptides presented on APC and producing IFNγ was much higher in gp120αgal/p24 immunized mice than in gp120/p24 immunized mice (Fig. 4). These observations strongly suggest that the binding of anti-Gal to α-gal epitopes on the fusion protein vaccine enables the effective internalization of p24 by APC. In the absence of immune complexes with anti-Gal, such uptake is likely to be relatively low and is mediated only by random pinocytosis. The increases in T cell and antibody responses to gp120 within the fusion protein gp120αgal/p24 also implies that the fusion of p24 to gp120 does not diminish the immunogenicity of gp120, which is markedly increased as a result of α-gal epitopes expression. Moreover, the observed increase in anti-gp120 antibody response following immunization with gp120αgal/p24 suggests that CD4+ T cells are also effectively activated by the proposed vaccine, since activation of B cells producing these antibodies is dependent on effective CD4+ T cell help.
The collective experience accumulated in development of HIV vaccines in the recent two decades has resulted in the notion that vaccination only with gp120 may not suffice for conferring resistance to HIV-1 infections in large populations [2–11]. The high mutation rate of HIV-1 gp120 leads to the generation of escape mutants that can evade recognition by CTLs and detrimental effects of neutralizing antibodies. The inclusion of other viral proteins of low mutability in HIV-1 vaccines, such as rev, p17 or p24 may be beneficial for eliciting a protective cellular immune response that destroys HIV-1 infected cells. However, poor uptake of those proteins by APC would render them ineffective vaccine targets. Fusion of these proteins with gp120 and expression of α-gal epitopes on the gp120 portion of the vaccine may result in prophylactic vaccines that elicit a combined humoral immune response against envelope proteins and a cellular immune response against matrix and/or core proteins.
The proposed strategy of fusion proteins comprised of envelope glycoproteins expressing α-gal epitopes and other immunogenic proteins of a given virus, may also be useful in eliciting protective immune responses against other viral infections, as well as other microbial infections. One example may be influenza virus. The envelope hemagglutinin (HA) molecule has ~7 N-linked glycosylation sites. We have shown that expression of α-gal epitopes on HA of an inactivated influenza virus results in a marked increase in immunogenicity and a much higher protection against viral challenge than with a flu vaccine lacking α-gal epitopes [23]. The present study suggests that a vaccine comprised of a fusion protein between HA and other viral proteins of low variation and mutability may result in induction of an immune response that is protective against a wide range of strains which have HA molecules that differ from that used in the vaccine.
The ubiquitous presence of anti-Gal antibodies in large amounts in humans ensures the effective in vivo targeting of any vaccine expressing α-gal epitopes to APC. Therefore, increased immunogenicity of fusion protein vaccines that express α-gal epitopes in HIV-1, influenza and potentially other viruses, will likely be observed in wide populations of vaccinees.
Acknowledgments
This work was supported by the University of Massachusetts Center for AIDS Research (CFAR) (P30 AI042845).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bucy RP, Kilby JM. Perspectives on inducing efficient immune control of HIV-1 replication--a new goal for HIV therapeutics? Aids. 2001;15 (Suppl 2):S36–42. doi: 10.1097/00002030-200102002-00007. [DOI] [PubMed] [Google Scholar]
- 2.Berzofsky JA, Ahlers JD, Janik J, Morris J, Oh S, Terabe M, et al. Progress on new vaccine strategies against chronic viral infections. J Clin Invest. 2004;114(4):450–62. doi: 10.1172/JCI22674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buge SL, Ma HL, Amara RR, Wyatt LS, Earl PL, Villinger F, et al. Gp120-alum boosting of a Gag-Pol-Env DNA/MVA AIDS vaccine: poorer control of a pathogenic viral challenge. AIDS Res Hum Retroviruses. 2003;19(10):891–900. doi: 10.1089/088922203322493067. [DOI] [PubMed] [Google Scholar]
- 4.Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, et al. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5(3):233–6. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 5.Goulder PJ, Watkins DI. HIV and SIV CTL escape: implications for vaccine design. Nat Rev Immunol. 2004;4(8):630–40. doi: 10.1038/nri1417. [DOI] [PubMed] [Google Scholar]
- 6.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420(6916):678–82. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 7.Letvin NL. Progress toward an HIV vaccine. Annu Rev Med. 2005;56:213–23. doi: 10.1146/annurev.med.54.101601.152349. [DOI] [PubMed] [Google Scholar]
- 8.Pantaleo G, Koup RA. Correlates of immune protection in HIV-1 infection: what we know, what we don’t know, what we should know. Nat Med. 2004;10(8):806–10. doi: 10.1038/nm0804-806. [DOI] [PubMed] [Google Scholar]
- 9.Pincus SH, Wehrly K, Tschachler E, Hayes SF, Buller RS, Reitz M. Variants selected by treatment of human immunodeficiency virus-infected cells with an immunotoxin. J Exp Med. 1990;172(3):745–57. doi: 10.1084/jem.172.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomson MM, Perez-Alvarez L, Najera R. Molecular epidemiology of HIV-1 genetic forms and its significance for vaccine development and therapy. Lancet Infect Dis. 2002;2(8):461–71. doi: 10.1016/s1473-3099(02)00343-2. [DOI] [PubMed] [Google Scholar]
- 11.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 12.Abdel-Motal U, Wang S, Lu S, Wigglesworth K, Galili U. Increased immunogenicity of human immunodeficiency virus gp120 engineered to express Galalpha1–3Galbeta1–4GlcNAc-R epitopes. J Virol. 2006;80(14):6943–51. doi: 10.1128/JVI.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galili U, Rachmilewitz EA, Peleg A, Flechner I. A unique natural human IgG antibody with anti-alpha-galactosyl specificity. J Exp Med. 1984;160(5):1519–31. doi: 10.1084/jem.160.5.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galili U. Evolution and pathophysiology of the human natural anti-alpha-galactosyl IgG (anti-Gal) antibody. Springer Semin Immunopathol. 1993;15(2–3):155–71. doi: 10.1007/BF00201098. [DOI] [PubMed] [Google Scholar]
- 15.Galili U, Macher BA, Buehler J, Shohet SB. Human natural anti-alpha-galactosyl IgG. II. The specific recognition of alpha (1----3)-linked galactose residues. J Exp Med. 1985;162(2):573–82. doi: 10.1084/jem.162.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galili U, Clark MR, Shohet SB, Buehler J, Macher BA. Evolutionary relationship between the natural anti-Gal antibody and the Gal alpha 1----3Gal epitope in primates. Proc Natl Acad Sci U S A. 1987;84(5):1369–73. doi: 10.1073/pnas.84.5.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galili U, Shohet SB, Kobrin E, Stults CL, Macher BA. Man, apes, and Old World monkeys differ from other mammals in the expression of alpha-galactosyl epitopes on nucleated cells. J Biol Chem. 1988;263(33):17755–62. [PubMed] [Google Scholar]
- 18.Larsen RD, Rivera-Marrero CA, Ernst LK, Cummings RD, Lowe JB. Frameshift and nonsense mutations in a human genomic sequence homologous to a murine UDP-Gal:beta-D- Gal(1,4)-D-GlcNAc alpha(1,3)-galactosyltransferase cDNA. J Biol Chem. 1990;265(12):7055–61. [PubMed] [Google Scholar]
- 19.Galili U, LaTemple DC. Natural anti-Gal antibody as a universal augmenter of autologous tumor vaccine immunogenicity. Immunol Today. 1997;18(6):281–5. doi: 10.1016/s0167-5699(97)80024-2. [DOI] [PubMed] [Google Scholar]
- 20.LaTemple DC, Abrams JT, Zhang SY, Galili U. Increased immunogenicity of tumor vaccines complexed with anti-Gal: studies in knockout mice for alpha1,3 galactosyltransferase. Cancer Res. 1999;59(14):3417–23. [PubMed] [Google Scholar]
- 21.Manches O, Plumas J, Lui G, Chaperot L, Molens JP, Sotto JJ, et al. Anti-Gal-mediated targeting of human B lymphoma cells to antigen-presenting cells: a potential method for immunotherapy using autologous tumor cells. Haematologica. 2005;90(5):625–34. [PubMed] [Google Scholar]
- 22.Clynes R, Takechi Y, Moroi Y, Houghton A, Ravetch JV. Fc receptors are required in passive and active immunity to melanoma. Proc Natl Acad Sci U S A. 1998;95(2):652–6. doi: 10.1073/pnas.95.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdel-Motal UM, Guay HM, Wigglesworth K, Welsh RM, Galili U. Immunogenicity of influenza virus vaccine is increased by anti-gal-mediated targeting to antigen-presenting cells. J Virol. 2007;81(17):9131–41. doi: 10.1128/JVI.00647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maness NJ, Yant LJ, Chung C, Loffredo JT, Friedrich TC, Piaskowski SM, et al. Comprehensive immunological evaluation reveals surprisingly few differences between elite controller and progressor Mamu-B*17-positive Simian immunodeficiency virus-infected rhesus macaques. J Virol. 2008;82(11):5245–54. doi: 10.1128/JVI.00292-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen ZC, Tanemura M, Galili U. Synthesis of alpha-gal epitopes (Galalpha1–3Galbeta1–4GlcNAc-R) on human tumor cells by recombinant alpha1,3 galactosyltransferase produced in Pichia pastoris. Glycobiology. 2001;11(7):577–86. doi: 10.1093/glycob/11.7.577. [DOI] [PubMed] [Google Scholar]
- 26.Galili U, LaTemple DC, Radic MZ. A sensitive assay for measuring alpha-Gal epitope expression on cells by a monoclonal anti-Gal antibody. Transplantation. 1998;65(8):1129–32. doi: 10.1097/00007890-199804270-00020. [DOI] [PubMed] [Google Scholar]
- 27.Thall AD, Maly P, Lowe JB. Oocyte Gal alpha 1,3Gal epitopes implicated in sperm adhesion to the zona pellucida glycoprotein ZP3 are not required for fertilization in the mouse. J Biol Chem. 1995;270(37):21437–40. doi: 10.1074/jbc.270.37.21437. [DOI] [PubMed] [Google Scholar]
- 28.Henion TR, Gerhard W, Anaraki F, Galili U. Synthesis of alpha-gal epitopes on influenza virus vaccines, by recombinant alpha 1,3galactosyltransferase, enables the formation of immune complexes with the natural anti-Gal antibody. Vaccine. 1997;15(11):1174–82. doi: 10.1016/s0264-410x(96)00300-3. [DOI] [PubMed] [Google Scholar]
- 29.LaTemple DC, Galili U. Adult and neonatal anti-Gal response in knock-out mice for alpha1,3 galactosyltransferase. Xenotransplantation. 1998;5(3):191–6. doi: 10.1111/j.1399-3089.1998.tb00027.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang S, Arthos J, Lawrence JM, Van Ryk D, Mboudjeka I, Shen S, et al. Enhanced immunogenicity of gp120 protein when combined with recombinant DNA priming to generate antibodies that neutralize the JR-FL primary isolate of human immunodeficiency virus type 1. J Virol. 2005;79(12):7933–7. doi: 10.1128/JVI.79.12.7933-7937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu S, Santoro JC, Fuller DH, Haynes JR, Robinson HL. Use of DNAs expressing HIV-1 Env and noninfectious HIV-1 particles to raise antibody responses in mice. Virology. 1995;209(1):147–54. doi: 10.1006/viro.1995.1238. [DOI] [PubMed] [Google Scholar]
- 32.Wang S, Farfan-Arribas DJ, Shen S, Chou TH, Hirsch A, He F, et al. Relative contributions of codon usage, promoter efficiency and leader sequence to the antigen expression and immunogenicity of HIV-1 Env DNA vaccine. Vaccine. 2006;24(21):4531–40. doi: 10.1016/j.vaccine.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 33.Derby NR, Kraft Z, Kan E, Crooks ET, Barnett SW, Srivastava IK, et al. Antibody responses elicited in macaques immunized with human immunodeficiency virus type 1 (HIV-1) SF162-derived gp140 envelope immunogens: comparison with those elicited during homologous simian/human immunodeficiency virus SHIVSF162P4 and heterologous HIV-1 infection. J Virol. 2006;80(17):8745–62. doi: 10.1128/JVI.00956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdel-Motal UM, Gillis J, Manson K, Wyand M, Montefiori D, Stefano-Cole K, et al. Kinetics of expansion of SIV Gag-specific CD8+ T lymphocytes following challenge of vaccinated macaques. Virology. 2005;333(2):226–38. doi: 10.1016/j.virol.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 35.Qiu JT, Song R, Dettenhofer M, Tian C, August T, Felber BK, et al. Evaluation of novel human immunodeficiency virus type 1 Gag DNA vaccines for protein expression in mammalian cells and induction of immune responses. J Virol. 1999;73(11):9145–52. doi: 10.1128/jvi.73.11.9145-9152.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bottrel RL, Dutra WO, Martins FA, Gontijo B, Carvalho E, Barral-Netto M, et al. Flow cytometric determination of cellular sources and frequencies of key cytokine-producing lymphocytes directed against recombinant LACK and soluble Leishmania antigen in human cutaneous leishmaniasis. Infect Immun. 2001;69(5):3232–9. doi: 10.1128/IAI.69.5.3232-3239.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Unkeless JC. Function and heterogeneity of human Fc receptors for immunoglobulin G. J Clin Invest. 1989;83(2):355–61. doi: 10.1172/JCI113891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fanger NA, Wardwell K, Shen L, Tedder TF, Guyre PM. Type I (CD64) and type II (CD32) Fc gamma receptor-mediated phagocytosis by human blood dendritic cells. J Immunol. 1996;157(2):541–8. [PubMed] [Google Scholar]
- 39.Schmitt DA, Hanau D, Bieber T, Dezutter-Dambuyant C, Schmitt D, Fabre M, et al. Human epidermal Langerhans cells express only the 40-kilodalton Fc gamma receptor (FcRII) J Immunol. 1990;144(11):4284–90. [PubMed] [Google Scholar]
- 40.Abdel-Motal UM, Wigglesworth K, Galili U. Mechanism for increased immunogenicity of vaccines that form in vivo immune complexes with the natural anti-Gal antibody. Vaccine. 2009;27(23):3072–82. doi: 10.1016/j.vaccine.2009.03.019. [DOI] [PubMed] [Google Scholar]