Abstract
At steady state, most Rho GTPases are bound in the cytosol to Rho Guanine nucleotide Dissociation Inhibitors (RhoGDI) 1. RhoGDIs have generally been considered to passively hold Rho proteins in an inactive state within the cytoplasm. Here we describe an evolutionarily conserved mechanism by which RhoGDI1 controls the homeostasis of Rho proteins in eukaryotic cells. We found that depletion of RhoGDI1 promotes misfolding and degradation of the cytosolic geranylgeranylated pool of Rho GTPases while unexpectedly activating the remaining membrane-bound fraction. Since RhoGDI1 levels are limiting, and Rho proteins compete for binding to RhoGDI1, overexpression of an exogenous Rho GTPase displaces endogenous Rho proteins bound to RhoGDI1, inducing their degradation and inactivation. These results raise important questions about the conclusions drawn from studies that manipulate Rho protein levels. In many cases the response observed may arise not simply from the overexpression per se, but from additional effects on the levels and activity of other Rho GTPases due to competition for binding to RhoGDI1, and may require a re-evaluation of previously published studies that rely exclusively on these techniques.
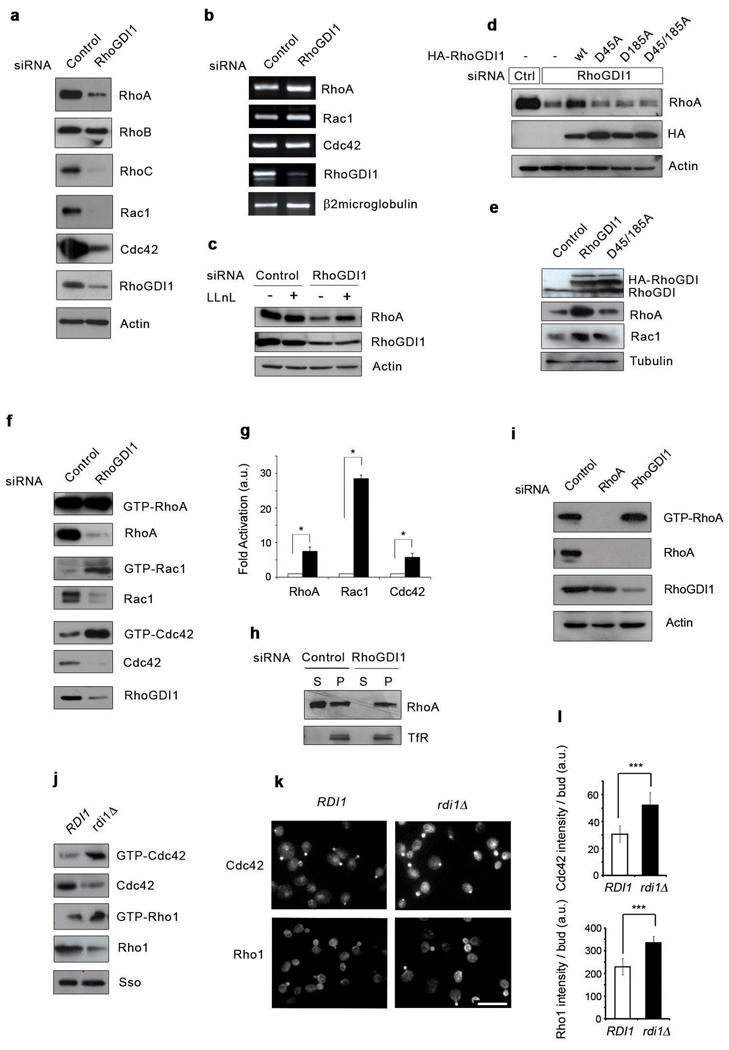
The RhoGDI family has been generally relegated to a secondary role in the regulation of Rho GTPases1. To explore the function of RhoGDI1, we analyzed the effect of depleting RhoGDI1 in mammalian cells. Upon RhoGDI1 depletion by siRNA, the protein levels of the major Rho GTPases, including RhoA, RhoC, Rac1 and Cdc42 were dramatically decreased (Fig.1a). This occurred for multiple cell types, including HeLa, fibroblasts, breast epithelial, melanoma and endothelial cells (Fig.1a, Supplementary Fig.S1a–d). RhoB, which does not bind RhoGDI1, was unaffected by RhoGDI1 silencing (Fig.1a and Supplementary Fig.S2a,b). The mRNA levels of RhoA, Rac1 and Cdc42 remained constant suggesting that post-transcriptional mechanisms account for the decrease in Rho proteins (Fig.1b). Indeed, proteasomal inhibitors such as LLnL, but not calpain or cathepsin inhibitors, partially rescued Rho proteins from degradation suggesting without RhoGDI1, the GTPases are being cleared by the proteasome (Fig.1c and data not shown). Rescue from degradation was achieved by expressing a siRNA-resistant RhoGDI1 (Fig.1d and Supplementary Fig. S2d). A second independent RhoGDI1 siRNA generated similar effects (data not shown). Interaction between the Rho-GTPases and RhoGDI is essential to prevent GTPase degradation since siRNA-resistant mutants that cannot bind Rho GTPases2 failed to rescue the effects of RhoGDI1 depletion (Fig.1d). Overexpression of wildtype RhoGDI1 but not the GTPase binding-deficient mutant increased the total level of RhoA and Rac1 (Fig.1e). These results indicate that RhoGDI1 stabilizes Rho proteins, protecting them from degradation.
Figure 1. RhoGDI1depletion triggers both degradation and activation of Rho proteins in eukaryotic cells.
(a) Lysates from control or RhoGDI1 siRNA transfected HeLa cells were resolved by SDS-PAGE and analyzed by Western blotting. (b) Total RNAs were purified from control or RhoGDI1 siRNA transfected cells. RT-PCR was performed on DNAse I treated RNA using specific primers. RT-PCR products were resolved by agarose gel electrophoresis. (c) Lysates from control or RhoGDI1 siRNA transfected HeLa cells treated with LLnL for 8 hours were resolved by SDS-PAGE and analyzed by Western blotting. (d) HeLa cells were co-transfected with control or RhoGDI1 siRNA and HA-tagged RhoGDI1 RNAi resistant mutants. Cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. All results are representative of at least 3 independent experiments. Note that the degradation of RhoA in RhoGDI1-depleted cells is rescued by expression of RhoGDI1 resistant to the siRNA, but not by RhoGDI1 mutants that do not bind Rho GTPases. (e) Lysates from HeLa cells transfected with HA-tagged wt RhoGDI1 or RhoGDI1 D45/185A were resolved by SDS-PAGE and analyzed by Western blotting. All siRNA experiments were analyzed at 72 h after transfection. For overexpression experiments, cells were transfected with the indicated cDNA 48 h after siRNA transfection and incubated for an additional 24 h. (f) HeLa cells were transfected with control or RhoGDI1 siRNA for 72 h. Active Rho GTPases were pulled-down from cell lysates with GST-RBD or GST-PBD beads. Bound proteins and total cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. (g) Quantitation of the activation of RhoA, Rac1 and Cdc42 in RhoGDI1-depleted cells (black bars) relative to control cells (white bars). * p=0.0108, p=0.0146 and p=0.0229 respectively for RhoA, Rac1 and Cdc42 in a two-tail unpaired student’s t test with error bars representing standard error of the mean (s.e.m.) from three independent experiments. (h) Control or RhoGDI1 siRNA transfected cells were fractionated into cytosol and membrane fractions, resolved by SDS-PAGE and analyzed by Western blotting. TfR stands for transferrin receptor. (i) HeLa cells were infected with a RhoA miR shRNA-expressing adenovirus or transfected with RhoGDI1 siRNA for 72 h. Active RhoA was pulled-down with GST-RBD. Total cell lysates and GST-RBD bound proteins were resolved by SDS-PAGE and analyzed by Western blotting. (j) Active Rho GTPases from wild-type or Rdi1d yeast strains were pulled-down with GTP-RBD or GST-PBD beads. Bound proteins and total cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. The Sso protein is used as loading control. (k) Immunofluorescence staining of Cdc42 and Rho1 in control wild-type (RDI1) and rdi1Δ strains. Scale bar is 8µm. (l) Quantitation of the intensity of polarized bud staining in control wild-type (RDI1) and rdi1Δ strains. Data were analyzed by two-tail student’s t test (p < 0.005) with error bars representing standard deviation from three experiments.
Even though RhoA, Rac1 and Cdc42 levels were drastically reduced after RhoGDI1 depletion, the amount of active Rho proteins was either significantly increased (Rac1 and Cdc42) or unchanged (RhoA) (Fig.1f), which indicates that a much larger fraction of the remaining pool of GTPase is in the active GTP-bound state. While the activity increases by up to 2 fold, the ratio between active and total GTPase increases by up to 10 fold due to the decrease in GTPase protein level (Fig.1g). At steady state, the active GTPase represented 1% of total RhoA while in the absence of RhoGDI1, it peaked at 10% (Fig.1f and g). After RhoGDI1 depletion most RhoA is in the membrane fraction, suggesting that most of the degraded protein derives from the cytosolic pool (Fig.1h) and that the active fraction is associated with membranes. In contrast when an individual GTPase is silenced, both the protein levels and the activity are proportionally reduced (Fig.1i). These results highlight the importance of GDI proteins in regulating both the protein level and activation state of Rho GTPases. RhoGDI depletion in other cell types (WM2664 and HUVECs) caused similar decreases in Rho protein levels (Supplementary Fig. S1a–d), while the activity of the remaining fraction was either elevated or unchanged (Fig. 2d and Fig.S1d). Studying HUVECs, Gorovoy et al. also found that RhoGDI knockdown promoted RhoA activation, although Rho protein expression was not decreased as much as observed here, probably because the knockdown was only allowed to proceed for 30 hours3. More complete RhoGDI1 knockdown and Rho GTPase degradation is achieved at longer time intervals (Supplementary Fig. S1e).
Figure 2. RhoGDI1 depletion impairs cell migration.
(a) Time-course analysis of closure of a wound generated on a confluent monolayer of control or RhoGDI1 siRNA transfected HeLa cells. The graph depicts the wound area with error bars representing standard deviation (n=10). (b) Time-course analysis of closure of a wound generated on a confluent monolayer of control or RhoGDI1 siRNA transfected WM2664 melanoma cells. The graph depicts the wound area with error bars representing standard deviation (n=6). (c) Analysis of the velocity of control or RhoGDI1 siRNA transfected WM2664 melanoma cells. Data were analyzed by two-tailed unpaired student’s t test (p= 1.72×10−14) with error bars representing standard error of the mean (s.e.m.) from respectively n=28 and n=56 cells. The average speed of control and RhoGDI1 knock-down cells was 12.74 µm/min and 5.64 µm/min respectively. (d) WM2664 melanoma cells were transfected with control or RhoGDI1 siRNA for 72h. Active Rac1 and active RhoA were pulled-down from cell lysates with GST-PBD or GST-RBD beads respectively. Bound proteins and total cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. (e) Cell lysates of control or RhoGDI1 siRNA transfected HeLa cells were resolved by SDS-PAGE and analyzed by Western blotting. Notice that Rho protein effectors are not activated despite the strong activation of RhoA and Rac. (f) Images extracted from supplementary information movie 1 depicting one WM2664 melanoma cell transfected with fluorescently labeled RhoGDI1 siRNA (red arrowhead) and two WM2664 cells transfected with unlabelled control siRNA (white and black arrowheads) migrating over time. Panel on right shows an image taken at t0 used to identify the cell transfected with the fluorescently labelled siRNA. Scale bar is 40 µm. (g) Representative XY migration tracks of control (left) or RhoGDI1 (right) siRNA transfected WM2664 melanoma cells. The positions of the cells were recorded every 5 minutes over a period of 2 hours (n=10). (h) HeLa cells transfected with control or RhoGDI1 siRNA for 72 h were fractionated into cytosolic or membrane fractions. Membrane fractions were further separated into plasma membrane (PM) and ER membrane (ER) by centrifugation on an iodixanol density gradient. Each fraction was analyzed by SDS-PAGE and Western blotting. (i) Densitometric analysis of the relative intensity of RhoA, Rac1 and Cdc42 bands in each fraction. Notice that upon RhoGDI1 silencing, the GTPases essentially disappear from the plasma membrane fractions.
To determine whether this aspect of RhoGDI function is evolutionarily conserved, we analyzed the effect of deleting the single RhoGDI gene in S. cerevisiae (RDI1). Strikingly, the loss of Rdi1 protein also resulted in degradation of Cdc42 and Rho1 (RhoA ortholog) while maintaining high levels of activation (Fig.1j). When analyzed by immunofluorescence, both Cdc42 and Rho1 had an increase in protein polarized in the bud, which has previously been correlated with localization of the activated forms of these proteins4, 5 (Fig.1k and l). The increase in polarized bud staining is striking given the overall reduction in Rho1 and Cdc42 protein levels in rdi1Δ strains.
RhoGDI1 knockout mice are viable but display progressive kidney defects leading ultimately to death6. Interestingly, those defects have been attributed to increased Rac1 activation and the kidneys display reduced levels of Rho GTPases7. Renal mesangial cells from RhoGDI knockout mice display altered growth and survival8. However, RhoGDI1 null mouse fibroblasts showed no defects in the reorganization of the actin cytoskeleton upon stimulation with growth factors, or in cell migration measured in a wound healing assay6. Yeast cells lacking rdi1, have a very minor phenotype9. Surprised by the relatively mild phenotype in spite of the marked change in Rho protein levels and activity, we have explored this further. Consistent with previous work, the rate of wound closure was unaffected when RhoGDI1 was knocked down in HeLa cells (Fig.2a). However, the migration velocity of highly motile melanoma cells was significantly decreased upon RhoGDI knockdown, both in a wound healing assay (Fig.2b) and when single cell migration was analyzed (Fig.2c, f, g and Supplementary information movie 1). This decrease in migration speed was unanticipated, given that Rac1 activity was increased (Fig.2d). However, RhoA activity also increased proportionally and may counteract the effects of high Rac1 activity (Fig.2d).
Although RhoGDI1 knockdown affected cell migration, we were surprised by the unexpectedly mild phenotype given the striking level of Rho protein activation following RhoGDI1 depletion. Examining whether RhoGDI2 might compensate for the loss of RhoGDI1 we found that, in cells expressing RhoGDI2 (HeLa and HUVEC), RhoGDI2 levels did not vary after RhoGDI1 KD (Supplementary Fig.S1d and Fig. S3c). In WM2664 and gingival fibroblasts the expression of RhoGDI2 was undetectable and unaffected by RhoGDI1 silencing (Supplementary Fig.S3c). In HeLa cells, silencing GDI-1 and GDI-2 showed no effect on wound closure speed (Supplementary Figure S3a), suggesting the lack of phenotype is not due to a GDI-2 compensation effect.
We hypothesized that upon RhoGDI1 depletion, Rho proteins may fail to properly localize to the plasma membrane (PM) and may remain in the Endoplasmic Reticulum (ER), where the final steps in Rho GTPase maturation occur on the cytoplasmic face of the ER. Here they are postranslationally processed after being prenylated in the cytosol 10. Subcellular fractionation experiments showed that, in control cells RhoA, Rac1 and Cdc42 display a biphasic distribution occurring in both the PM and ER fractions (Fig.2h,i). However, following RhoGDI1 depletion the amount of Rho GTPases found associated with the PM was greatly reduced with the majority of the remaining GTPases cofractionating with the ER (Fig.2h,i). These results reveal that, without RhoGDI1, Rho proteins that are not degraded are not properly delivered to the PM. Instead, they accumulate at the ER suggesting that one function of RhoGDI1 is to shuttle Rho GTPases from the ER to their site of action at the PM. Supporting these results, we did not detect an increase in effector activation following GDI-1 KD, despite the high levels of active GTPases (Fig.2e). These results may explain the mild phenotype resulting from RhoGDI1 depletion in spite of high Rho protein activity.
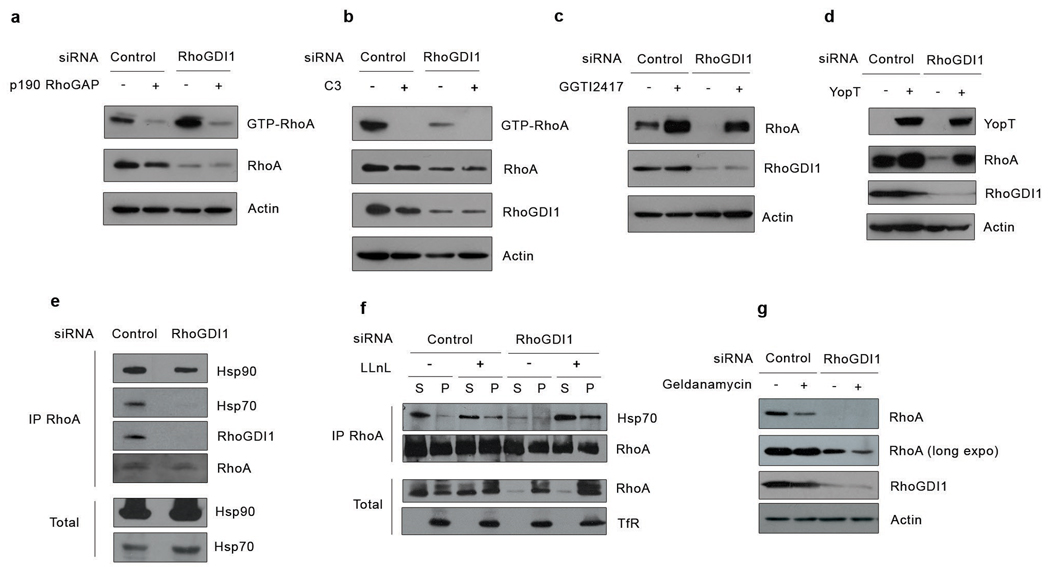
Rho proteins undergo ubiquitylation and degradation by the proteasome upon irreversible activation by the bacterial toxin CNF111. Since RhoGDI1 knockdown promoted a substantial activation of Rho proteins, we explored if degradation occured similarly, following activation. To inhibit activation of RhoA we either overexpressed the RhoA-specific GAP, p190-RhoGAP, or incubated cells with C3-transferase, which inactivates RhoA, and then monitored its degradation following GDI1 silencing. Both treatments reduced RhoA activity to negligible amounts (Fig.3a,b). However, degradation of RhoA after GDI1 depletion still occurred, indicating that this is independent of RhoA activation (Fig.3a,b).
Figure 3. Rho family GTPase degradation following RhoGDI1 depletion does not require activation of the Rho protein, but depends upon their geranylgeranylation and involves the molecular chaperone machinery.
(a) HeLa cells were co-transfected with control or RhoGDI1 siRNA and p190RhoGAP cDNA. Active Rho GTPases were pulled-down from cell lysates with GST-RBD or GST-PBD beads. Bound proteins and total cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. (b) HeLa cells were transfected with control or RhoGDI1 siRNA and treated with cell permeable C3 toxin. Active Rho GTPases were pulled-down from cell lysates with GST-RBD or GST-PBD beads. Bound proteins and total cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. (c) HeLa cells were transfected with control or RhoGDI1 siRNA and treated with the geranylgeranyl transferase inhibitor GGTI 2417. Cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. (d) HeLa cells were co-transfected with control or RhoGDI1 siRNA and a cDNA encoding the bacterial protease YopT. Cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. (e) HeLa cells were transfected with control or RhoGDI siRNA and RhoA was immunoprecipitated from cell lysates. Immunoprecipitated proteins were resolved by SDS-PAGE and analyzed by Western blotting. (f) HeLa cells transfected with control or RhoGDI1 siRNA, with or without the proteasome inhibitor LLnL, were fractionated into cytosolic or membrane fractions and RhoA was immunoprecipitated from cytosolic and membrane fractions. To ensure that similar amounts of RhoA were immunoprecipitated, immunoprecipitations were performed with limiting amount of antibody and saturating amounts of cell lysate. Immunoprecipitated proteins and cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. (g) HeLa cells were transfected with control or RhoGDI1 siRNA and treated with geldanamycin for 12 hours. Cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. All results are representative of at least 3 independent experiments. A short and long exposure of the RhoA blot is shown. All siRNA experiments were analyzed at 72 h after transfection. For overexpression experiments, cells were transfected with the indicated cDNA 48 h after siRNA transfection and incubated for an additional 24 h.
Our results suggest that binding to RhoGDI1 stabilizes and protects Rho GTPases from degradation, and that a GTPase that is not bound to a GDI protein or associated with the membrane is rapidly degraded. Most Rho proteins (RhoA, Rac1 and Cdc42) are posttranslationally modified at the C-terminus with a geranylgeranyl group12. General prenylation inhibitors from the statin family, such as lovastatin, or geranylgeranyl transferase specific inhibitors like GGTI2417 rescued the degradation of GTPases in the absence of GDI (Fig.3c, and data not shown). Similarly, cleavage of the geranylgeranylated C-terminal tail using the bacterial protease YopT13also restored RhoA levels, indicating that newly synthesized Rho proteins are prenylated and fully mature before being degraded and that removing the prenylation is sufficient to stabilize Rho GTPases (Fig.3d). GGTI2417 treatment restores the cytosolic pool of RhoA in the absence of RhoGDI1, suggesting that a Rho GTPase can only exist in the cytosol if it is either bound to RhoGDI1 or not prenylated (Supplementary Fig. S4).
We speculated that, without the RhoGDI1 to protect the prenyl group, this exposed hydrophobic modification might disturb the correct folding of Rho proteins, targeting them to degradation. In cells, proper protein folding is ensured by molecular chaperones14. Proteins that cannot fold properly usually cluster in insoluble aggregates that are targeted for degradation14. We found that under physiological conditions, RhoA associates with the molecular chaperone Hsp70 (Fig.3e). In the absence of RhoGDI1, the levels of Hsp70 increased in the cells indicative of a cellular stress but virtually no Hsp70 was bound to RhoA, suggesting that the remaining membrane-bound RhoA is properly folded and stable (Fig.3e). Indeed, Hsp70 bound mostly to cytosolic RhoA (Fig.3f). When we inhibited the proteasome in RhoGDI1-depleted cells, RhoA and Rac1 accumulated in a triton-insoluble fraction (data not shown). The amount of Hsp70 bound to cytosolic RhoA also increased, suggesting that the pool of free prenylated RhoA was misfolded or partially unfolded and recruited the protein quality control machinery (Fig.3f). These results suggest that prenylation contributes to the misfolding or unfolding of Rho proteins, resulting in their degradation if RhoGDI1 cannot accommodate the lipid moiety and if the molecular chaperones cannot fold them properly. Interestingly, although RhoA co-precipitates with both Hsp70 and RhoGDI1 (Fig.3e), immunoprecipitation of RhoGDI1 indicates that there are two mutually exclusive pools of RhoA associated with either Hsp70 or RhoGDI1 (Supplementary Fig. S2a). Finally, inhibiting molecular chaperones with geldanamycin decreased RhoA levels in the presence or absence of RhoGDI1, arguing that molecular chaperones are required to help stabilize prenylated RhoA (Fig.3g).
In mammalian cells, the level of RhoGDI1 is roughly equivalent to the total levels of RhoA, Rac1 and Cdc42 combined15. Therefore, a competitive balance exists between Rho proteins for binding to RhoGDI1 and any condition that alters the level of one GTPase should disrupt this balance and affect Rho protein homeostasis. Figure 4a shows that silencing RhoA results in decreased levels of RhoA and proportionally increased levels of Rac1 and Cdc42, indicating that reducing one GTPase generates free GDI that can be shared by the others. In contrast, overexpression of myc-tagged RhoA, Rac1 or Cdc42 displaced endogenous RhoA from RhoGDI1 (Fig.4b). Overexpression of a myctagged Cdc42 mutant (R66E) incapable of binding RhoGDI did not affect the level of endogenous RhoA (Fig.4c). The displacement of endogenous Rho proteins resulted in their degradation in a dose-dependent fashion (Fig.4d,g,j). Strikingly, expression of constitutively active (Fig.4e,h,k) or dominant negative Rho proteins (Fig.4f,i,l) also displaced endogenous Rho proteins from RhoGDI1 and caused their degradation. The degradation of RhoA by overexpression of Cdc42 could be prevented by co-expression of RhoGDI1 (Fig.4m). Displacement of Rho proteins from RhoGDI1 by other Rho family members, including RhoA, Rac1 and Cdc42, resulted in their degradation and inactivation (Fig.4n,o,p). This degradation and inactivation did not occur if the overexpressed Rho protein was a mutant (RhoAR68E, Rac1R66E or Cdc42R66E) unable to bind RhoGDI1 (Fig.4n,o,p). The decrease in activity of Rho GTPases displaced from RhoGDI1 is similar to that observed when GTPases are silenced by RNAi (Fig.1i), and contrasts with the activation of Rho GTPases caused by RhoGDI1 depletion.
Figure 4. Competitive interactions with RhoGDI1 regulate the levels and activities of Rho proteins.
(a) HeLa cells were infected with a RhoA miR shRNA adenovirus for 72 h. Cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. Note the increased levels of Rac1 and Cdc42 in cells from which RhoA has been depleted. (b) HeLa cells were transfected with myc-tagged Rho GTPases and endogenous RhoGDI1 was immunoprecipitated. Immunoprecipitated proteins and cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. HeLa cells (c and d) or HEK 293 cells (e, f, g, h, i, j, k and l) were transfected with increasing amount of plasmid encoding for the indicated myc-tagged Rho GTPases. After 24 h, cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. (m) HEK 293 cells were transfected with myc-tagged Cdc42 expressing plasmid with or without HA-tagged RhoGDI1 expressing plasmid for 24 h. Cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. (n, o, and p) HEK 293 cells were transfected with 5µg of myc-tagged Rho GTPase expression plasmid as indicated. After 24 h, active RhoA or active Rac1 were pulled-down from cell lysates with GST-RBD or GST-PBD beads respectively. Bound proteins and total cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. All results shown are representative of at least 3 independent experiments. Note that overexpression of one Rho family member decreases the expression and activity of the other Rho proteins.
One might expect that depletion of RhoGDI1 and competitive displacement of Rho GTPases from RhoGDI1 would have the same effect on Rho protein activity, given that both result in degradation of Rho GTPases. However, in the case of overexpression of an exogenous Rho GTPase, RhoGDI1 is still present and over time will extract and inactivate the remaining endogenous GTPases from the membranes. Competitive displacement of endogenous Rho proteins from RhoGDI1 by overexpressed Rho GTPases will release the endogenous Rho proteins into the cytosol, followed by their degradation without the opportunity for activation. In contrast, during RhoGDI1 depletion, Rho proteins associated with membranes cannot be actively extracted by RhoGDI. These membrane-bound Rho GTPases are likely to become activated by membrane-bound GEFs. Since, RhoGDI1 can inhibit both the activation of the Rho GTPases, as well as GTP hydrolysis16, 17, depletion of RhoGDI1 should relieve this inhibition, thereby promoting activation of the GTPases. In contrast, displacement of Rho proteins from GDI does not alter the cellular content of RhoGDI1, maintaining this inhibitory mechanism on endogenous GTPases. Indeed, we show that after RhoGDI1 depletion, the remaining RhoA that is not targeted for degradation is not bound to GDI (Fig.3e). Furthermore, our results demonstrate that depletion of RhoA by either direct silencing or by overexpressing an exogenous GTPase result in a decrease of both total and active RhoA, whereas RhoGDI silencing decreases RhoA levels but leaves RhoA activity high (Fig.1i and Fig.4o,p).
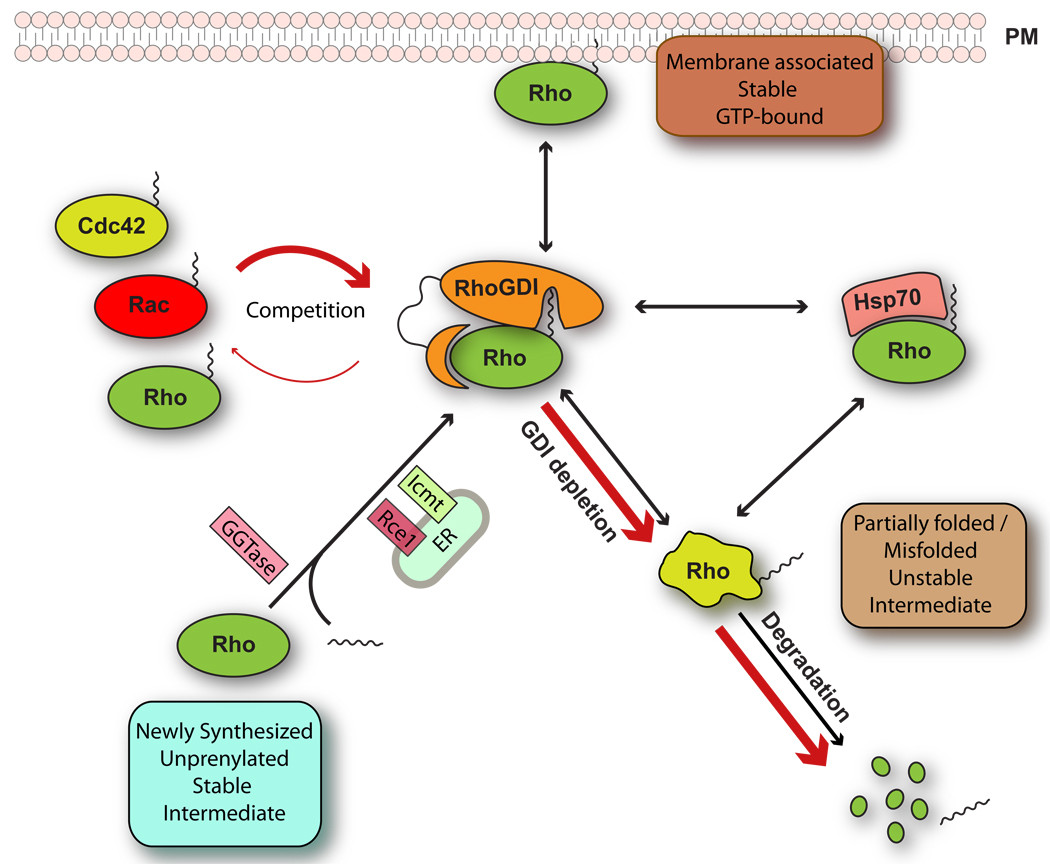
We have shown that RhoGDI1 regulates the stability of Rho GTPases within the cytosol, protecting them from degradation. Our results also reveal an unexpected crosstalk between Rho GTPases through competitive binding to RhoGDI1, illustrated schematically in Figure 5. Rather little is known about changes in expression levels of RhoGDI1 or Rho GTPases during development and disease, although situations have been described in which the expression of a Rho family member occurs, such as the increase of RhoC in metastasis18. In preliminary work, we have found that with different breast cancer cell lines the expression of RhoGDI1 correlates with their invasiveness, as well as with the level and activity of RhoA and Rac1 (Supplementary Fig. S5). However, in situations where the level of RhoGDI stays constant, our results predict that not only overexpressing one GTPase will displace the others from RhoGDI and target them for degradation but it will also affect their activity. One situation where extreme changes occur is in the experimental expression of wildtype or mutant Rho GTPases, which have been the standard tools to study Rho protein functions in different pathways. In many cases, the conclusions drawn from these studies may arise not only from the activity of the overexpressed Rho protein but also from the unanticipated effect of displacing other family members from RhoGDI leading to their degradation and inactivation.
Figure 5. RhoGDI regulates Rho protein homeostasis.
Newly synthesized RhoGTPases are geranylgeranylated and posttranslationally modified in the ER. After geranylgeranylation, Rho proteins associate directly with RhoGDI which sequesters them as a soluble prenylated forming the cytosol and protects them from degradation. Upon depletion of RhoGDI1 or overexpression of Rho proteins, endogenous RhoGTPases are released to the cytosol where they exist as short-lived unstable intermediates that are partially folded or misfolded (red arrows). These can bind to the chaperone complex or be targeted for degradation if they are unable to fold properly. In the absence of GDI, newly synthesized Rho proteins cannot be delivered to the plasma membrane and accumulate in the ER. At steady state, this unstable intermediate is not detected and RhoGTPases are for the most part either associated with cell membranes or bound to GDI, with only a small fraction associated with the chaperone system.
Abbreviations used: ER, endoplasmic reticulum; PM, plasma membrane; GGTase, geranylgeranyl transferase; Rce1, prenyl-protein specific protease; lcmt, isoprenylcysteine carboxyl methyltransferase.
METHODS
Reagents
All chemicals were purchased from Axxora (San Diego, CA, USA) unless mentioned. Cell permeable C3 toxin was from Cytoskeleton (Denver, CO, USA). The geranylgeranyl transferase inhibitor GGTI2417 was a gift from A. Cox (Department of Radiation Oncology, University of North Carolina, Chapel Hill, NC, USA).
Cell culture
HeLa and 293FT cells were grown in low glucose Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum. Gingival Fibroblasts (gift from Dr. Carol Otey, UNC-Chapel Hill) were grown in MEM alpha (Invitrogen) supplemented with 15% FBS (Sigma). WM2664 cells (gift from Dr. Jim Bear, UNC-Chapel Hill) were grown in DMEM (Invitrogen) supplemented with 10% FBS (Sigma). HUVECs were grown in EGM-2 (Clonetics, San Diego, CA, USA) until passage 4.
Yeast Culture and Immunofluorescence
Standard methods for yeast growth and cell permeabilization were as described in Guthrie and Fink 19. Immunofluorescence staining against Cdc42 and Rho1 was as described previously 20. The intensity of fluorescence associated with polarized Cdc42/Rho1 was measured in budded cells using Metamorph software using at least 20 cells of each strain.
Antibodies
The mouse monoclonal anti-RhoA antibody (26C4), the rabbit polyclonal anti-RhoB (119), anti-Rac2 (C11) and anti-RhoGDI1 (A20) antibodies, and the goat polyclonal anti-RhoC antibody (K12) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The mouse monoclonal anti-Rac1 (23A8) and anti-actin (C4) antibodies were from Millipore (Billerica, MA, USA). The mouse monoclonal anti-Cdc42 antibody was from BD Biosciences (San Jose, CA, USA). The mouse monoclonal anti-transferrin receptor, anti-tubulin and anti-PDI antibodies were from Sigma-Aldrich (St Louis, MO, USA). The mouse monoclonal anti-HA antibody (clone 16B12) was from Covance (Princeton, NJ, USA). The mouse monoclonal anit-RhoGDI2 antibody was from BD Pharmingen (BD Bioscience, San Jose, CA, USA). The mouse monoclonal anti-myc antibody (clone 9E10) was a gift from Dr. Thomas Samson (UNC, Chapel Hill, USA). The mouse monoclonal anti-Hsp70 and anti-Hsp90 antibodies were gifts from Dr. Doug Cyr (UNC, Chapel Hill, USA). Affinity purified antibodies against yeast Cdc42 were previously described 20, Monoclonal antibody against yeast Rho1 (UNC3-106.5.1) was isolated in collaboration with the UNC Immunology Core Facility.
DNA constructs, RNA interference, siRNAs and siRNA transfection
The wild type and mutant myc-tagged RhoA, Rac1 and Cdc42 eukaryotic expression constructs were previously described 21, 22. The Cdc42 R66E, RhoGDI1 rescue wt, D45A, D185A and D45/185A mutants were generated by site-directed mutagenesis using the Quickchange II XL Site-Directed Mutagenesis kit (Stratagene). The RhoGDI1 rescue constructs were generated by introducing silent mutations in the sequence targeted by the siRNA duplex. The pCMV-HA-p190 RhoGAP mammalian expression construct was described previously 23. The pGEX-YopT prokaryotic expression construct and the pCMV-HA-RhoGDI1 mammalian expression construct were gifts from S.Ellerbroek (Wartburg College, Waverly, IA, USA). YopT cDNA was subcloned by PCR into the pPTuner IRES2 vector of the ProteoTuner system (Clontech, Mountain View, CA, USA). The pAd-EmGFP-RhoA miR shRNA construct was previously described 24. Short interfering RNAs (siRNAs) were designed using the BIOPREDsi algorithm at www.biopredsi.org 25. SiRNAs were purchased from the UNC Nucleic Acid Core Facility/Sigma-Genosys (Sigma-Aldrich, St Louis, MO, USA). The following siRNAs were used in this study (guide strand only): ARHGDIA#1/RhoGDI1#1 UCAAUCUUGACGCCUUUCCTT, negative control UCACUCGUGCCGCAUUUCCTT. SiRNAs were transfected using a modified calcium phosphate protocol as described previously 26.
RT-PCR
Total RNA was purified from HeLa cells using Trizol (Invitrogen, CA, USA) and was treated with DNAse I (NEB, Ipswich, MA, USA). Reverse transcription was carried out using the iScript cDNA Synthesis kit (Biorad, Hercules, CA, USA) on 1 µg of total RNA. PCR was performed on equal amounts of cDNA using the Taq PCR Master Mix kit (Qiagen, Valencia, CA, USA) and the following sets of oligonucleotides: RhoGDI1 GGAGAGCTTCAAGAAGCAGT and TCAGTCCTTCCAGTCCTTCTT, RhoA, ATGGCTGCCATCCGGAAG and TCACAAGACAAGGCACCCAG, Rac1 ATGCAGGCCATCAAGTGTGT and TTACAACAGCAGGCATTTTCTCT, Cdc42 ATGCAGACAATTAAGTGTGTTGTT and TCATAGCAGCACACACCTGC, and β2 microglobulin CTCGCGCTACTCTCTCTTTCTGG and GCTTACATCTCTCAATCCCACTTAA.
GST-RBD/PBD pull-down
Activation of respectively RhoA or Rac1/Cdc42 was measured in a GST-RBD or GST-PBD pull-down assay as previously described 27, 28. Briefly, cells were lysed in 25 mM Hepes pH 7.3, 150 mM NaCl, 5 mM MgCl2, 0.5% Triton X-100, 4% Glycerol, 10 mM NaF, 5 mM DTT and protease inhibitors for 10 minutes at 4°C. Triton-X-100 insoluble material was removed by centrifugation for 10 minutes at 9500 g and the lysates were incubated with 50 µg of immobilized GST-RBD or GST-PBD for 40 minutes at 4°C, to measure respectively RhoA or Rac1/Cdc42 activity.
Subcellular fractionation
Cells were washed and incubated with ice-cold hypotonic lysis buffer (10 mM Hepes pH 7.3, 1.5 mM MgCl2, 5 mM KCl, 1 mM DTT and protease inhibitors) for 10 minutes. Cells were scraped and homogeneized with 20 strokes of Dounce homogenizer. Homogenates were centrifuged at 700 g for 3 minutes to pellet nuclei and intact cells. The supernatants were spun at 40,000 g for 30 minutes at 4°C and the pellets were gently washed once with hypotonic lysis buffer. Typically, 2 to 5% of the cytosolic fraction and 30 to 40% of the membrane fraction were analyzed by SDS-PAGE and Western blot. Alternatively, membranes were fractionated using an iodixanol (OptiPrep, Axis Shield) discontinuous gradient according to the manufacturer’s instructions. Briefly, the membrane pellets were resuspended thoroughly in homogeneization buffer (0.25 M sucrose, 20 mM Tris/HCl pH 7.4, 25 mM KCl, 5 mM MgCl2) and layered on top of a 5% to 30% discontinuous iodixanol gradient. The samples were the centrifuged for 2 h at 100,000 g. Fractions were then collected and analyzed by western blot.
Immunoprecipitation
Cells were lysed in ice-cold lysis buffer (without DTT) as described in the GST pull-down section. Lysates were centrifuged at 10,000 rpm for 10 minutes. RhoGDI1 was immunoprecipitated with 2 µg of rabbit polyclonal anti-RhoGDI1 antibody for one hour. RhoA was immunoprecipitated with 0.4 µg of mouse monoclonal anti-RhoA antibody (26C4) overnight. Lysates were then incubated with protein A-sepharose beads for 45 minutes and were washed 3 times with lysis buffer.
Single Cell Migration
WM2664 cells were transfected with CTRL siRNA or GDI-1 specific siRNA using siQuest transfection reagent (Mirus). The GDI-1 siRNA was mixed with 1:10 of a fluorescently labeled non-targeting siRNA to identify the transfected cell. In control experiments all cells were transfected with the fluorescent siRNA (not shown). 48h after transfection CTRL and GDI-1 KD cells were trypsinized, mixed 1:1 and plated in a glass bottom 35 mm dish (MatTek) coated with laminin (Sigma). Time-lapse images were acquired using a Nikon BioStation IM live-cell imaging microscope and analyzed using ImageJ (NIH). Images were captured every 5 min for a total of 2 h. Usually 20 different fields were captured in a single experiments (approx. 60 cells).
Wound Healing
HeLa cells transfected with CTRL siRNA or GDI-1 siRNA. After 48 h cells were trypsinized and plated in a 24 well plate. A scratch/wound was made using a pipette tip in each of the wells. The cells were then imaged at the indicated times using a Zeiss Axiovert 200 M microscope equipped with a Hamamatsu ORCA-ERAG digital camera. The wound area was measured at each time point using Metamorph Software (Molecular Devices).
Statistical analysis
Statistical differences between two groups of data were analysed with two-tailed unpaired Student's t-test.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Lisa Sharek for her technical support, Channing Der, Jim Bear and Chloé Féral for comments. This study was supported by National Institutes of Health Grant #GM029860 (to K.B.), #GM054712 (to P.B.), a Department of Defense Breast Cancer Predoctoral Fellowship (#BC051092 to A.D.), a Susan Komen Foundation Postdoctoral Fellowship and a AHA Beginning Grant in Aid (# 5-40078 to R.G.M), and, a Fondation pour la Recherche Medicale Fellowship (E.B), an AHA Postdoctoral Fellowship (#0825333E to E.B.) and an Allocation INSERM InCa/AVENIR (#R08227AS to E.B.).
Footnotes
AUTHOR CONTRIBUTIONS:
E.B. and R.G.M designed, performed experiments and wrote the manuscript. C.G. and A.D. helped with experimental design and procedures. G.R. and P.B. designed and performed the experiments in S. cerevisiae. K.B. directed the project and revised the manuscript. All authors provided detailed comments.
AUTHOR INFORMATION:
Keith Burridge (Keith_Burridge@med.unc.edu).
REFERENCES
- 1.DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Dransart E, Morin A, Cherfils J, Olofsson B. Uncoupling of inhibitory and shuttling functions of rho GDP dissociation inhibitors. J Biol Chem. 2005;280:4674–4683. doi: 10.1074/jbc.M409741200. [DOI] [PubMed] [Google Scholar]
- 3.Gorovoy M, et al. RhoGDI-1 modulation of the activity of monomeric RhoGTPase RhoA regulates endothelial barrier function in mouse lungs. Circ Res. 2007;101:50–58. doi: 10.1161/CIRCRESAHA.106.145847. [DOI] [PubMed] [Google Scholar]
- 4.Abe M, Qadota H, Hirata A, Ohya Y. Lack of GTP-bound Rho1p in secretory vesicles of Saccharomyces cerevisiae. J Cell Biol. 2003;162:85–97. doi: 10.1083/jcb.200301022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tong Z, et al. Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition. J Cell Biol. 2007;179:1375–1384. doi: 10.1083/jcb.200705160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Togawa A, et al. Progressive impairment of kidneys and reproductive organs in mice lacking Rho GDIalpha. Oncogene. 1999;18:5373–5380. doi: 10.1038/sj.onc.1202921. [DOI] [PubMed] [Google Scholar]
- 7.Shibata S, et al. Modification of mineralocorticoid receptor function by Rac1 GTPase: implication in proteinuric kidney disease. Nat Med. 2008;14:1370–1376. doi: 10.1038/nm.1879. [DOI] [PubMed] [Google Scholar]
- 8.Bielek H, Anselmo A, Dermardirossian C. Morphological and proliferative abnormalities in renal mesangial cells lacking RhoGDI. Cell Signal. 2009;21:1974–1983. doi: 10.1016/j.cellsig.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiedje C, Sakwa I, Just U, Hofken T. The Rho GDI Rdi1 regulates Rho GTPases by distinct mechanisms. Mol Biol Cell. 2008;19:2885–2896. doi: 10.1091/mbc.E07-11-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winter-Vann AM, Casey PJ. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat Rev Cancer. 2005;5:405–412. doi: 10.1038/nrc1612. [DOI] [PubMed] [Google Scholar]
- 11.Doye A, et al. CNF1 exploits the ubiquitin-proteasome machinery to restrict Rho GTPase activation for bacterial host cell invasion. Cell. 2002;111:553–564. doi: 10.1016/s0092-8674(02)01132-7. [DOI] [PubMed] [Google Scholar]
- 12.Vega FM, Ridley AJ. Snapshot: Rho family GTPases. Cell. 2007;129:1430. doi: 10.1016/j.cell.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 13.Shao F, et al. Biochemical characterization of the Yersinia YopT protease: Cleavage site and recognition elements in Rho GTPases. 12843403. 2003;100:904–909. doi: 10.1073/pnas.252770599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 15.Michaelson D, et al. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J Cell Biol. 2001;152:111–126. doi: 10.1083/jcb.152.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hart MJ, et al. A GDP dissociation inhibitor that serves as a GTPase inhibitor for the Ras-like protein CDC42Hs. Science. 1992;258:812–815. doi: 10.1126/science.1439791. [DOI] [PubMed] [Google Scholar]
- 17.Ueda T, Kikuchi A, Ohga N, Yamamoto J, Takai Y. Purification and characterization from bovine brain cytosol of a novel regulatory protein inhibiting the dissociation of GDP from and the subsequent binding of GTP to rhoB p20, a ras p21-like GTP-binding protein. J Biol Chem. 1990;265:9373–9380. [PubMed] [Google Scholar]
- 18.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 19.Guthrie C, Fink G, Simon MI, Abelson JN. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–863. [PubMed] [Google Scholar]
- 20.Adamo JE, et al. Yeast Cdc42 functions at a late step in exocytosis, specifically during polarized growth of the emerging bud. J Cell Biol. 2001;155:581–592. doi: 10.1083/jcb.200106065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Mata R, et al. Analysis of activated GAPs and GEFs in cell lysates. Methods Enzymol. 2006;406:425–437. doi: 10.1016/S0076-6879(06)06031-9. [DOI] [PubMed] [Google Scholar]
- 22.Wennerberg K, et al. RhoG signals in parallel with Rac1 and Cdc42. J Biol Chem. 2002;277:47810–47817. doi: 10.1074/jbc.M203816200. [DOI] [PubMed] [Google Scholar]
- 23.Arthur WT, Noren NK, Burridge K. Regulation of Rho family GTPases by cell-cell and cell-matrix adhesion. Biol Res. 2002;35:239–246. doi: 10.4067/s0716-97602002000200016. [DOI] [PubMed] [Google Scholar]
- 24.van Buul JD, et al. RhoG regulates endothelial apical cup assembly downstream from ICAM1 engagement and is involved in leukocyte transendothelial migration. J Cell Biol. 2007;178:1279–1293. doi: 10.1083/jcb.200612053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huesken D, et al. Design of a genome-wide siRNA library using an artificial neural network. Nat Biotechnol. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- 26.Vouret-Craviari V, Boulter E, Grall D, Matthews C, Van Obberghen-Schilling E. ILK is required for the assembly of matrix-forming adhesions and capillary morphogenesis in endothelial cells. J Cell Sci. 2004;117:4559–4569. doi: 10.1242/jcs.01331. [DOI] [PubMed] [Google Scholar]
- 27.Bagrodia S, Taylor SJ, Jordon KA, Van Aelst L, Cerione RA. A novel regulator of p21-activated kinases. J Biol Chem. 1998;273:23633–23636. doi: 10.1074/jbc.273.37.23633. [DOI] [PubMed] [Google Scholar]
- 28.Ren XD, Kiosses WB, Schwartz MA. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. Embo J. 1999;18:578–585. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.