Abstract
How transcription factors interpret the cis-regulatory logic encoded within enhancers to mediate quantitative changes in spatiotemporally restricted expression patterns during animal development is not well understood. Pax6 is a dosage-sensitive gene essential for eye development. Here, we identify the Prep1 (pKnox1) transcription factor as a critical dose-dependent upstream regulator of Pax6 expression during lens formation. We show that Prep1 activates the Pax6 lens enhancer by binding to two phylogenetically conserved lower-affinity DNA-binding sites. Finally, we describe a mechanism whereby Pax6 levels are determined by transcriptional synergy of Prep1 bound to the two sites, while timing of enhancer activation is determined by binding site affinity.
Keywords: Transcription factor, protein-binding microarray, DNA-binding site affinity, eye development, mathematical modeling, gene regulation
The precise spatiotemporal patterns of metazoan gene expression are controlled by cis-regulatory modules (CRMs), which are regions of DNA, typically a few hundred base pairs long, that comprise binding sites for transcriptional regulators (Yuh et al. 1998; Arnosti and Kulkarni 2005; Davidson and Levine 2008). While current genomic approaches have traditionally focused on identifying the highest-affinity transcription factor (TF)-binding sites, new genome-wide location analyses have demonstrated widespread TF binding in the absence of high-affinity sites, thus forcing a re-evaluation of the scope and regulatory significance of lower-affinity sites (Hollenhorst et al. 2007; Gordan et al. 2009).
The affinity of a binding site provides a mechanism to respond to different concentrations of TFs. In development, the ability of CRMs that contain sites of differing affinities to interpret TF gradients has been studied in Drosophilia melanogaster and other organisms for spatial gradients (Driever et al. 1989; Struhl et al. 1989; Jiang and Levine 1993; Scardigli et al. 2003; Segal et al. 2008), and in Caenorhabditis elegans and sea urchin for temporal gradients (Lai et al. 1988; Gaudet and Mango 2002). For example, in C. elegans, altering PHA-4 DNA-binding sites to higher- or lower-affinity sequences altered the onset of pharyngeal gene expression, with higher-affinity sites directing expression earlier in development (Gaudet and Mango 2002). The ability to tune the timing of expression by modulating the affinity of the DNA-binding site(s) for just one TF is conceptually appealing. However, it remains unclear whether such a simple mechanism could operate in the context of a complex vertebrate developmental enhancer.
Pax6 is a key regulator of metazoan eye development, and in mammals is required in a dose-dependent fashion for lens induction and specification (Hill et al. 1991; Grindley et al. 1995; Ashery-Padan et al. 2000; van Raamsdonk and Tilghman 2000; Davis-Silberman et al. 2005). Furthermore, the precise cis regulation of Pax6 is obligatory, as alteration in either the timing or level of Pax6 expression has profound effects on eye development (Schedl et al. 1996; van Raamsdonk and Tilghman 2000; Duncan et al. 2004). Several groups have molecularly dissected regulatory elements contained within a region of Pax6 that contains the P0 promoter and 3.9 kb of upstream sequence (P0-3.9), and identified CRMs that control expression in the lens (denoted as the Pax6 ectodermal enhancer or EE) and endocrine pancreas (Williams et al. 1998; Kammandel et al. 1999; Zhang et al. 2002, 2006). Within the EE, there is a highly conserved minimal 104-base-pair (bp) region sufficient to direct lens-specific expression (Williams et al. 1998), and several TFs—including Sox2, Oct1 (Pou2f1), Meis1/2, and Pax6 itself—have been shown to bind this region (Zhang et al. 2002; Aota et al. 2003; Donner et al. 2007). Here we decode a cis-regulatory logic whereby DNA-binding site affinities of a novel EE regulator, Prep1, are used to control the timing of Pax6 activation in the developing lens.
Results and Discussion
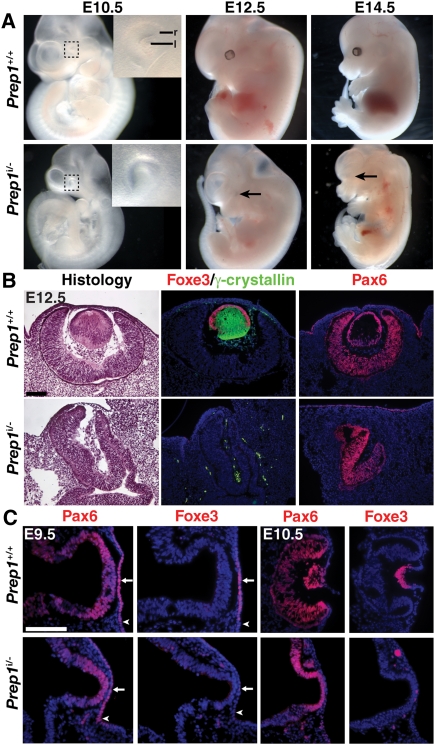
Prep1 (also known as Pknox1) is a homeobox TF in the TALE superfamily that was identified previously along with other closely related Meis TFs as a regulator of the Pax6 pancreatic enhancer (Zhang et al. 2006). Prep1 homozygous-null mouse embryos (Prep1−/−) do not survive to midgestation, while Prep1 hypomorphs, where RNA expression is reduced to ∼2% of wild-type levels due to a retroviral insertion in first intron of Prep1 (Prep1i/i), show highly variable phenotypes (Ferretti et al. 2006). We found that Prep1 trans-heterozygotes (Prep1i/−) generally survive until embryonic days 12.5–14.5 (E12.5–E14.5), and exhibit distinct and highly penetrant phenotypes (Fig. 1A; Supplemental Table 1). Strikingly, Prep1i/− mutants lack an external eye, with an arrest in eye development by E10.5 (Fig. 1A), the same time point at which eye development arrests in Pax6 mutant mice.
Figure 1.
Genetic requirement for Prep1 in lens induction. (A) Whole-mount control or Prep1i/− mutants at E10.5, E12.5, and E14.5. The inset shows high magnification of eye region (boxed). Arrows show the absence of eyes. (B) Sections through E12.5 control or Prep1i/− mutant eyes stained with hematoxylin and eosin to show histology, or stained with antibodies to Foxe3, γ-crystallin, or Pax6 as indicated. (C) Sections through E9.5 or E10.5 control or Prep1i/− mutant embryos stained with antibodies for Pax6 or Foxe3. Arrows indicate presumptive lens ectoderm, and arrowheads indicate nonlens head ectoderm. Bars: B,C, 100 μm. (l) Lens; (r) retina.
Histological analysis at E12.5 revealed an absence of lens tissue, and marker analyses revealed that neither lens epithelial cells (Foxe3+), nor lens fiber cells (γ-crystallin+), nor lens precursor cells (Pax6+) formed (Fig. 1B). Foxe3 is a dose-dependent Pax6 target gene in the presumptive lens epithelium (Fig. 1C) and a definitive marker of lens specification (Blixt et al. 2007). Prep1i/− mutant mice do not initiate Foxe3 expression (Fig. 1C), and while Pax6 is initially expressed in the presumptive lens epithelium of Prep1i/− mutants, its expression is subsequently lost specifically in the lens preplacode, which fails to thicken or invaginate at E10.5 to form the lens placode (Fig. 1C). Conversely, Prep1 is coexpressed with Pax6 in the presumptive lens epithelium, and does not depend on Pax6 for its expression (Supplemental Figs. 1, 2). These data identify Prep1 as a novel regulator of lens induction, and also as a putative regulator of Pax6 expression.
Pax6 regulation during lens development is complex, with multiple enhancers controlling the earliest phase of expression in lens preplacodal epithelium, and the Pax6 EE having a more significant role later during lens induction (Williams et al. 1998; Dimanlig et al. 2001). We evaluated whether Pax6 regulatory elements might be regulated directly by Prep1 by crossing a P0-3.9 Pax6 reporter transgene (Fig. 2C; Rowan et al. 2008) into the Prep1i/− mutant background. Strikingly, the activity of this GFP reporter in the eye was reduced dramatically in E10.0 Prep1i/− mutants, while pancreatic enhancer activity was unaltered (Fig. 2A). The expression of other TFs that reside upstream of the Pax6 EE was retained in Prep1i/− mutants, ruling out indirect regulation via these factors (Supplemental Fig. 3).
Figure 2.
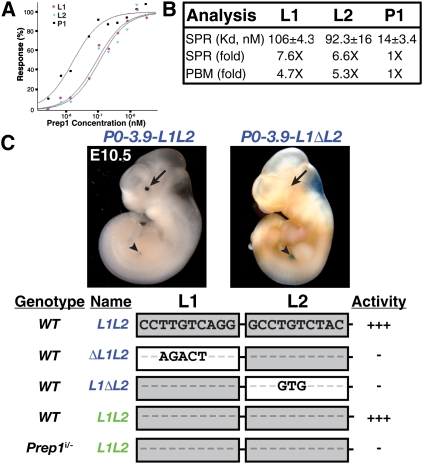
Prep1 is required for Pax6 EE activation and binds multiple highly conserved sites upstream of Pax6. (A) Whole-mount GFP visualization of E10 control or Prep1i/− mutant eyes expressing the P0-3.9-GFPCre transgene, or section through the pancreas of the same embryo stained with antibodies for Pdx1 and GFP. Dotted line marks the optic vesicle. (B) Logo of the Prep1 DNA-binding site motif (Berger et al. 2008), with a box showing the CTGTCA core sequence. (C) Schematic of the Pax6 P0-3.9 genomic region used in the transgenic mouse line. P0 box represents the location of the Pax6 P0 promoter. Shown in the University of California at Santa Cruz Genome Browser view are PBM scores (enrichment score, ≥0.37) from a sliding 8-bp window. Hits in the minimal essential region of the EE (cyan) or Pancreas enhancer (magenta) are labeled L1, L2, and P1; sequences are shown boxed below. Red nucleotides indicate divergence from the boxed CTGTCA core sequence.
We sought to determine whether the P0-3.9 region contained potential Prep1-binding sites. To search the P0-3.9 region for potential Prep1-binding sites, we used protein-binding microarray (PBM) data on Prep1 (Berger et al. 2008), which provide comprehensive and quantitative in vitro assessment of binding to all possible 8-bp DNA sequences. Along with a previously identified binding site in the Pax6 P0-3.9 region that is required for Pax6 pancreatic enhancer activity (denoted P1) (Zhang et al. 2006), we identified several lower-scoring putative binding sites throughout the region, including a pair of sites in the EE (denoted L1, L2) that are separated from each other by 34 bp (Fig. 2B,C). The high-scoring P1 sequence was a perfect match to the Prep1 DNA-binding site motif (Berger et al. 2008), while L1 and L2 each had a single phylogenetically conserved mismatch (L1: TTGTCA; L2: CTGTCT) from the optimal core 6-bp sequence 5′-CTGTCA-3′ (Fig. 2B,C). The single base mismatches in the L1 and L2 sites are consistent with the PBM data, indicating that L1 and L2 are lower-affinity Prep1-binding sites.
Motivated by the PBM data, we used surface plasmon resonance (SPR) to quantitatively measure the binding affinity of Prep1 to genomic sequences encompassing the L1, L2, and P1 sites (Majka and Speck 2007). P1 is a high-affinity Prep1-binding site, with a dissociation constant (Kd) of 14 ± 3.4 nM (mean ± SD), whereas L1 and L2 are of approximately sixfold to sevenfold lower affinity, with Kd values of 106 ± 4.3 and 92 ± 16 nM, respectively (Fig. 3A,B). These relative affinity differences agree well with estimates derived independently from the PBM data (Fig. 3B). Searching for Prep1-binding sites in multiple aligned genomes, we found two lower-scoring sites, spaced 34 bp apart, aligning to L1 and L2 (Fig. 2C; Supplemental Fig. 4) throughout the vertebrate lineage, suggesting conservation of affinity and function.
Figure 3.
L1 and L2 are essential lower-affinity Prep1-binding sites. (A) SPR dose response curves for GST-Prep1 on L1, L2, or P1. (B) Kd values of Prep1 for L1-, L2-, and P1-binding sites and fold change in affinity relative to the P1 site (numbers >1 indicate decreased affinity) as determined by SPR and PBM data. (C) Whole-mount view of β-galactosidase-stained E10.5 embryos expressing wild-type P0-3.9-L1L2 or P0-3.9-L1ΔL2 transgenes. The arrow points to the lens, while the arrowhead points to the pancreas. A summary is shown for Pax6 reporters lacking either L1 or L2 sites (21) or genetically deficient for Prep1 as assayed at E10.5. Only nucleotides that diverge from the wild-type EE sequence are shown. Lightly shaded L1 or L2 boxes indicate lower-affinity sites; white boxes indicate ablated binding sites. Binding site ablations for L1 and L2 are genetically indicated with Δ for deletion, although they are specific point mutations that have been tested by PBM and biochemical analyses to have no detectable Prep1 binding. Activity indicates relative β-galactosidase (name in blue) or GFP (name in green) reporter activity in the lens.
The L1 site was previously mutated and shown to be essential for EE activity (Zhang et al. 2002); hence, we sought to test the functional significance of the L2 site for EE regulation. Pax6 P0-3.9 reporters containing an ablation of the L2 site resulted in loss of reporter activity in the developing lens (Fig. 3C). Additional reporter analyses indicated that L1 and L2 function nonredundantly as critical Pax6 regulators (Fig. 3C). These results suggested three possible models: (1) Prep1 binds cooperatively to the two sites, but when either site is mutated, it is insufficiently bound to activate detectable levels of expression. (2) Prep1 binds independently to each site, with DNA-bound Prep1 proteins acting synergistically to activate detectable levels of transcripts. (3) Individual DNA-bound Prep1 proteins cannot activate expression, and binding to both sites is required to form a larger, multicomponent complex required for Pax6 expression.
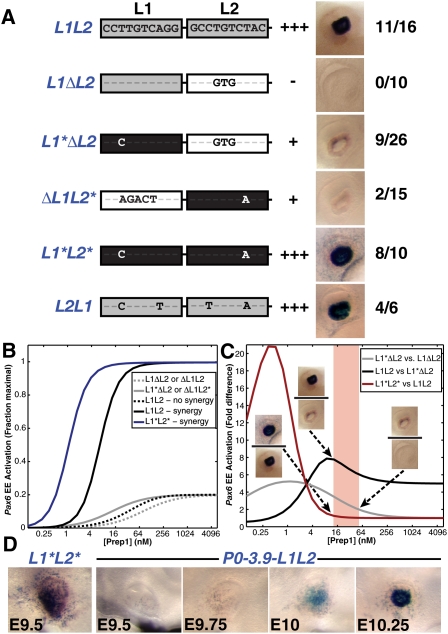
We used SPR to measure the binding of full-length Prep1 to a DNA probe containing the portion of the EE spanning the L1 and L2 sites in the event that regions outside the DNA-binding domain may mediate cooperative protein interactions. We did not detect any cooperative binding (Supplemental Fig. 5), arguing against model 1. Model 2 proposes synergistically acting lower-affinity sites. We reasoned that our mutant reporter constructs (ΔL1L2 and L1ΔL2) might be rescued by substituting the remaining, native lower-affinity site in each of the constructs with a high-affinity site resembling the P1 site (ΔL1L2* and L1*ΔL2). Strikingly, both constructs rescued EE reporter activity, but only to a modest extent and only in a minority of transgenic embryos, while expression from an L1L2 to L2L1 swap or from the L1*L2* double-mutant construct was not significantly different from that of L1L2 (Fig. 4A). These results demonstrate several important features of the EE element: (1) Prep1 occupancy at either site under native conditions is below saturation. (2) Prep1 bound to either L1 or L2 sites can provide the necessary requirements for activation (thereby ruling out model 3). (3) Prep1 molecules bound to L1 and L2 act synergistically and without a preferred order relative to each other to achieve wild-type EE expression levels.
Figure 4.
L1 and L2 interact synergistically to activate the EE. (A) Summary and examples of Pax6 reporter constructs with mutations in L1 or L2 sites that create high-affinity binding sites (asterisks and black boxes). The ratio indicates the number of transgenic embryos with lens β-galactosidase staining as a fraction of all transgenic embryos. Activity indicates relative β-galactosidase reporter activity in the lens. (B) Modeling of EE activation versus Prep1 concentration (log2 scale) for L1L2 modeled with synergy (black solid line) or without synergy (black dashed line), single-site mutations (ΔL1L2, L1ΔL2, gray dashed line), or high-affinity site mutations (ΔL1L2*, L1*ΔL2, gray solid line; L1*L2*, blue solid line). (C) Ratio of activation levels for reporter constructs modeled in B to facilitate the comparison of model predictions and the ratio of relative reporter levels. Single high-affinity site mutations (L1*ΔL2) versus wild-type (L1L2) reporter modeled with synergy (black line) or single-site ablation (L1ΔL2, gray line); double high-affinity site mutation (L1*L2*) versus wild-type (L1L2) reporter modeled with synergy (red line). We conservatively estimate the ratio differences between L1*ΔL2 and L1ΔL2 to be at least twofold, between L1L2 and L1*ΔL2 to be at least fourfold, and between L1*L2* and L1L2 to be at most 1.3-fold. These estimates are based on β-galactosidase staining intensities and histochemical development times. The shaded area defines the predicted range of physiological concentration for Prep1. (D) β-Galactosidase staining of an eye from an E9.5 L1*L2* transgenic embryo was more intense than from wild-type (P0-3.9-L1L2) embryos of increasingly older ages up to E10.25.
We employed a biophysical model (Bintu et al. 2005) of the Pax6 EE to investigate whether a model based on transcriptional synergy between Prep1 bound independently to the L1 and L2 sites could explain the behavior of our reporter constructs and provide further testable predictions. Expression was modeled as equilibrium binding of Prep1, with DNA-bound Prep1 recruiting a required cofactor (e.g., TFIID) or RNA polymerase II. The model was parameterized directly by using our SPR-determined binding affinities, and indirectly by requiring consistency with relative expression levels from our reporter constructs. In the absence of Prep1-binding cooperativity, we find that synergistic activation is required by Prep1 bound at L1 and L2 in order to achieve agreement with the experimental reporter expression data (Fig. 4B,C). The levels of Prep1 protein that are consistent with the relative reporter expression data lie in a narrow range (Fig. 4C, shaded region) that is consistent with published nuclear TF concentrations (Gregor et al. 2007; Giorgetti et al. 2010). For this range of Prep1 concentrations, the model predicts that the EE should be highly sensitive to Prep1 levels. This prediction is consistent with the dramatically reduced EE activity observed in Prep1i/− mutant mice (Fig. 2A).
In light of the model, we re-examined why both Prep1-binding sites are highly conserved as lower-affinity sites, given that substitution of both sites for high-affinity versions (L1*L2*) did not alter reporter activity at E10.5. We found that, at concentrations of Prep1 below those modeled for E10.5, the predicted difference in expression between the L1*L2* and L1L2 constructs is enhanced (Supplemental Figs. 6, 7). Prep1 mRNA and protein levels are known to increase during early mouse development (Ferretti et al. 1999), as does EE activity (Williams et al. 1998). We predicted, therefore, that the L1*L2* construct should be expressed at higher levels than L1L2 at stages preceding E10.5, when Prep1 levels are lower (see Supplemental Figs. 1, 2). We tested this prediction, and indeed found that L1*L2* directed inappropriately high reporter activity at E9.5 (Fig. 4D). Our interpretation, therefore, is that Prep1-binding site affinity provides a mechanism for controlling the timing of Pax6 expression in lens development. Additionally, we detected specific regions of expanded reporter activity in L1*L2* transgenic embryos (Fig. 4; Supplemental Fig. 8), suggesting additional selective pressure against high-affinity Prep1 sites in the EE. These results provide a mechanistic link between Prep1-binding site affinity and the timing of Pax6 expression during eye development.
Deciphering the cis-regulatory logic of enhancers governing the expression of developmental genes requires the description of coordinate action of multiple DNA-bound TFs. To what extent the role of individual sites can be dissected within the context of larger, complex vertebrate enhancers remains an open question. Here we describe a mammalian enhancer using binding site affinity of a single TF, Prep1, to regulate the temporal control of gene expression. Specifically, mutating the lower-affinity, native L1 and L2 Prep1 sites to higher-affinity sites resulted in high-level EE activity at an earlier developmental time point (Fig. 4D).
Coordination of the timing and level of Pax6 expression is of particular importance because of the exquisite sensitivity of eye development to even subtle changes in Pax6 levels; both Pax6 heterozygotes (mouse) and PAX6 hypomorphs (human) exhibit developmental phenotypes (Hill et al. 1991; Glaser et al. 1994; Schedl et al. 1996; van Raamsdonk and Tilghman 2000; Sansom et al. 2009). Lens development, particularly when the EE becomes active (E8.75), is highly sensitive to Pax6 concentration, and a threshold model for Pax6 function has been proposed (van Raamsdonk and Tilghman 2000). The lower affinities of L1 and L2 may have been evolutionarily selected to be most responsive to the developmental concentrations of Prep1 present when increasing levels of Pax6 expression are required to coordinate lens morphogenesis. Other enhancers have been shown to contribute to Pax6 lens expression both earlier and later than the EE (e.g., SIMO element) (Kleinjan et al. 2001), and may function coordinately with the EE to regulate Pax6. It is possible that Prep1 may regulate Pax6 expression via some of these other enhancers and possibly the P0 promoter (see Fig. 2C) in addition to the EE to account for the severe lens phenotype we observed in Prep1i/− mice.
The quantitative measurement and modeling of binding site affinities has led to the development and testing of biophysical models of transcription (Wang et al. 1999; Bintu et al. 2005; Kim and O'Shea 2008; Segal et al. 2008; Gertz et al. 2009; Kim et al. 2009). Here we demonstrate that a biophysical model, parameterized with biochemical and in vivo reporter analyses, can describe the activity of the Pax6 EE as a function of Prep1 levels and binding site affinities. Additionally, our model provides a mechanism for decoupling the timing and levels of gene expression, whereby the affinities of the L1 and L2 sites for Prep1 dictate when Pax6 is expressed, while the level of activation is modulated by the synergistic activity of Prep1 molecules noncooperatively bound to the two sites. The strict maintenance of the presence, affinity, and spacing of the L1 and L2 sites throughout the vertebrate lineage suggests evolutionary conservation of this regulatory mechanism.
Vast numbers of lower-affinity TF-binding sites are present in vertebrate genomes, but the functions of such sites remain unclear. Here we identified a functional role for lower-affinity TF-binding sites in establishing the temporal control of gene expression in the developing mouse. We propose that conservation of TF-binding sites as lower-affinity sites is a signature of CRMs that interpret temporal and/or spatial gradients of their upstream activators. Genomic searches for regulatory regions containing such conserved lower-affinity TF-binding sites (Jaeger et al. 2010) may identify other delicately calibrated enhancers controlling key developmental genes.
Materials and methods
Full descriptions of the Materials and Methods are available in the Supplemental Material.
Mouse genetics
P0-3.9-GFPCre mice were described previously, and were maintained as heterozygotes on an FVB background (Rowan et al. 2008). Pax6Sey-Neu mice were maintained as heterozygotes on a C3H background. Prep1i (gift from Dr. Francesco Blasi) and Prep1− (gift from Dr. Neal Copeland) mice were maintained as heterozygotes on a C57BL/6 background (Ferretti et al. 2006). Transgenic constructs were based on the pLNGLKS reporter plasmid containing the 526-bp Pax6 EE directing lacZ expression (Zhang et al. 2002), except for the P0-3.9-L1ΔL2-lacZ reporter, which was based on the P0-3.9-lacZ reporter line (Zhang et al. 2006). Site-directed mutations were constructed using the QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's directions.
Tissue analyses
For antibody staining, primary antibodies used were mouse anti-Pax6 (1:10; Development Studies Hybridoma Bank, The University of Iowa), rabbit anti-Pax6 (1:1000; Covance Research Products), rabbit anti-Foxe3 (1:1000; gift from Dr. Peter Carlsson), goat anti-γ-crystallin (1:1000; Santa Cruz Biotechnologies), goat anti-Pdx1 (1:5000; gift from Dr. Chris Wright), guinea pig anti-Prep1 (1:200) (Zhang et al. 2006), guinea pig anti-Six3 (1:500; Abcam), rabbit anti-Sox2 (1:1000; Chemicon), and goat anti-Meis1/2 (1:250; Santa Cruz Biotechnologies). Secondary antibodies were generated in donkey versus the appropriate species, and were directly conjugated with Cy3 (Jackson Immunologicals) or Alexa Fluor 488 (Molecular Probes).
Protein expression and SPR
GST-Prep1 homeodomain and full-length ORFs (Berger et al. 2008) were expressed in Escherichia coli. Tagged protein was purified using GSTrap FF affinity columns (GE Healthcare) on an AKTA prime plus FPLC (GE Healthcare). Concentration of purified protein was determined by Coomassie Bradford assay. SPR was performed on a Biacore 3000. Biotinylated oligos were immobilized onto a Sensor Chip SA (Biacore). Serial concentrations of protein samples were diluted in running buffer and applied to the Sensor Chip at 25 μL/min, using the KINJECT option, 250-μL sample (1000-sec dissociation phase). Binding rate constants and equilibrium Kd values were determined using Scrubber2 software (BioLogic Software).
Computational analysis and modeling
The P0-3.9 region was searched for Prep1 DNA-binding site sequences using custom Perl scripts (available on request). Eight-base-pair sequences were scored according to their PBM enrichment score, a statistical measure that ranges from −0.5 to +0.5 and indicates the binding preference of the protein, assayed in a universal PBM experiment, for a particular 8-mer as compared with all other 8-mers (Berger et al. 2008). Prep1 PBM data are available via the UniPROBE database (Newburger and Bulyk 2009).
An equilibrium thermodynamic model, described recently by Bintu et al. (2005), was used to model transcriptional activation by the EE. Activation curves were modeled and visualized in Matlab. A full description of the model and parameterization method is in the Supplemental Materials.
Acknowledgments
We thank Francesco Blasi, Steve Gisselbrecht, Craig Kaplan, Daniel O'Connell, and Rolf Stottmann for critical comments on the manuscript, and members of the Maas and Bulyk laboratories for helpful discussion. We are especially grateful to Neal Copeland and Francesco Blasi for gifts of Prep1 mutant mice. This work was supported by a fellowship from the Canadian Institutes of Health Research to S.R, NIH grant R01 HG003985 to M.L.B., and NIH grant R01 EY10123 to R.L.M.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1890410.
Supplemental material is available at http://www.genesdev.org.
References
- Aota S, Nakajima N, Sakamoto R, Watanabe S, Ibaraki N, Okazaki K 2003. Pax6 autoregulation mediated by direct interaction of Pax6 protein with the head surface ectoderm-specific enhancer of the mouse Pax6 gene. Dev Biol 257: 1–13 [DOI] [PubMed] [Google Scholar]
- Arnosti DN, Kulkarni MM 2005. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J Cell Biochem 94: 890–898 [DOI] [PubMed] [Google Scholar]
- Ashery-Padan R, Marquardt T, Zhou X, Gruss P 2000. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes & Dev 14: 2701–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. 2008. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133: 1266–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Phillips R 2005. Transcriptional regulation by the numbers: Models. Curr Opin Genet Dev 15: 116–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt A, Landgren H, Johansson BR, Carlsson P 2007. Foxe3 is required for morphogenesis and differentiation of the anterior segment of the eye and is sensitive to Pax6 gene dosage. Dev Biol 302: 218–229 [DOI] [PubMed] [Google Scholar]
- Davidson EH, Levine MS 2008. Properties of developmental gene regulatory networks. Proc Natl Acad Sci 105: 20063–20066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Silberman N, Kalich T, Oron-Karni V, Marquardt T, Kroeber M, Tamm ER, Ashery-Padan R 2005. Genetic dissection of Pax6 dosage requirements in the developing mouse eye. Hum Mol Genet 14: 2265–2276 [DOI] [PubMed] [Google Scholar]
- Dimanlig PV, Faber SC, Auerbach W, Makarenkova HP, Lang RA 2001. The upstream ectoderm enhancer in Pax6 has an important role in lens induction. Development 128: 4415–4424 [DOI] [PubMed] [Google Scholar]
- Donner AL, Episkopou V, Maas RL 2007. Sox2 and Pou2f1 interact to control lens and olfactory placode development. Dev Biol 303: 784–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W, Thoma G, Nüsslein-Volhard C 1989. Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity of binding sites for the bicoid morphogen. Nature 340: 363–367 [DOI] [PubMed] [Google Scholar]
- Duncan MK, Xie L, David LL, Robinson ML, Taube JR, Cui W, Reneker LW 2004. Ectopic Pax6 expression disturbs lens fiber cell differentiation. Invest Ophthalmol Vis Sci 45: 3589–3598 [DOI] [PubMed] [Google Scholar]
- Ferretti E, Schulz H, Talarico D, Blasi F, Berthelsen J 1999. The PBX-regulating protein PREP1 is present in different PBX-complexed forms in mouse. Mech Dev 83: 53–64 [DOI] [PubMed] [Google Scholar]
- Ferretti E, Villaescusa JC, Di Rosa P, Fernandez-Diaz LC, Longobardi E, Mazzieri R, Miccio A, Micali N, Selleri L, Ferrari G, et al. 2006. Hypomorphic mutation of the TALE gene Prep1 (pKnox1) causes a major reduction of Pbx and Meis proteins and a pleiotropic embryonic phenotype. Mol Cell Biol 26: 5650–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J, Mango SE 2002. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science 295: 821–825 [DOI] [PubMed] [Google Scholar]
- Gertz J, Siggia ED, Cohen BA 2009. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature 457: 215–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Siggers T, Tiana G, Caprara G, Notarbartolo S, Corona T, Pasparakis M, Milani P, Bulyk ML, Natoli G 2010. Noncooperative interactions between transcription factors and clustered DNA binding sites enable graded transcriptional responses to environmental inputs. Mol Cell 37: 418–428 [DOI] [PubMed] [Google Scholar]
- Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL 1994. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet 7: 463–471 [DOI] [PubMed] [Google Scholar]
- Gordan R, Hartemink AJ, Bulyk ML 2009. Distinguishing direct versus indirect transcription factor–DNA interactions. Genome Res 19: 2090–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, Tank DW, Wieschaus EF, Bialek W 2007. Probing the limits to positional information. Cell 130: 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley JC, Davidson DR, Hill RE 1995. The role of Pax-6 in eye and nasal development. Development 121: 1433–1442 [DOI] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BLM, Ton CCT, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V 1991. Mouse Small eye results from mutations in a paired-like homeobox-containing gene. Nature 354: 522–525 [DOI] [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, Graves BJ 2007. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes & Dev 21: 1882–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger SA, Chan ET, Berger MF, Stottmann R, Hughes TR, Bulyk ML 2010. Conservation and regulatory associations of a wide affinity range of mouse transcription factor binding sites. Genomics 95: 185–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Levine M 1993. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell 72: 741–752 [DOI] [PubMed] [Google Scholar]
- Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P 1999. Distinct cis-essential modules direct the time–space pattern of the Pax6 gene activity. Dev Biol 205: 79–97 [DOI] [PubMed] [Google Scholar]
- Kim H, O'Shea EK 2008. A quantitative model of transcription factor-activated gene expression. Nat Struct Mol Biol 15: 1192–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HD, Shay T, O'Shea EK, Regev A 2009. Transcriptional regulatory circuits: Predicting numbers from alphabets. Science 325: 429–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, Seawright A, Schedl A, Quinlan RA, Danes S, van Heyningen V 2001. Aniridia-associated translocations, DNase hypersensitivity, sequence comparison and transgenic analysis redefine the functional domain of PAX6. Hum Mol Genet 10: 2049–2059 [DOI] [PubMed] [Google Scholar]
- Lai ZC, Maxson R, Childs G 1988. Both basal and ontogenic promoter elements affect the timing and level of expression of a sea urchin H1 gene during early embryogenesis. Genes & Dev 2: 173–183 [DOI] [PubMed] [Google Scholar]
- Majka J, Speck C 2007. Analysis of protein–DNA interactions using surface plasmon resonance. Adv Biochem Eng Biotechnol 104: 13–36 [PubMed] [Google Scholar]
- Newburger D, Bulyk ML 2009. UniPROBE: An online database of protein binding microarray data on protein–DNA interactions. Nucleic Acids Res 37: D77–D82 doi: 10.1093/nar/gkn660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Conley KW, Le TT, Donner AL, Maas RL, Brown NL 2008. Notch signaling regulates growth and differentiation in the mammalian lens. Dev Biol 321: 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ 2009. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet 5: e1000511 doi: 10.1371/journal.pgen.1000511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scardigli R, Baumer N, Gruss P, Guillemot F, Le Roux I 2003. Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development 130: 3269–3281 [DOI] [PubMed] [Google Scholar]
- Schedl A, Ross A, Lee M, Engelkamp D, Rashbass P, van Heyningen V, Hastie ND 1996. Influence of PAX6 gene dosage on development: Overexpression causes severe eye abnormalities. Cell 86: 71–82 [DOI] [PubMed] [Google Scholar]
- Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U 2008. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature 451: 535–540 [DOI] [PubMed] [Google Scholar]
- Struhl G, Struhl K, Macdonald PM 1989. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell 57: 1259–1273 [DOI] [PubMed] [Google Scholar]
- van Raamsdonk CD, Tilghman SM 2000. Dosage requirement and allelic expression of PAX6 during lens placode formation. Development 127: 5439–5448 [DOI] [PubMed] [Google Scholar]
- Wang J, Ellwood K, Lehman A, Carey MF, She ZS 1999. A mathematical model for synergistic eukaryotic gene activation. J Mol Biol 286: 315–325 [DOI] [PubMed] [Google Scholar]
- Williams SC, Altmann CR, Chow RL, Hemmati-Brivanlou A, Lang RA 1998. A highly conserved lens transcriptional control element from the Pax-6 gene. Mech Dev 73: 225–229 [DOI] [PubMed] [Google Scholar]
- Yuh CH, Bolouri H, Davidson EH 1998. Genomic cis-regulatory logic: Experimental and computational analysis of a sea urchin gene. Science 279: 1896–1902 [DOI] [PubMed] [Google Scholar]
- Zhang X, Friedman A, Heaney S, Purcell P, Maas RL 2002. Meis homeoproteins directly regulate Pax6 during vertebrate lens morphogenesis. Genes & Dev 16: 2097–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Rowan S, Yue Y, Heaney S, Pan Y, Brendolan A, Selleri L, Maas RL 2006. Pax6 is regulated by Meis and Pbx homeoproteins during pancreatic development. Dev Biol 300: 748–757 [DOI] [PubMed] [Google Scholar]