Abstract
Intestinal epithelium is a rapidly self-renewing tissue in the body, and its homeostasis is tightly regulated by numerous factors including polyamines. Decreased levels of cellular polyamines increase activating transcription factor (ATF)-2, but the exact role and mechanism of induced ATF-2 in the regulation of intestinal epithelial cell (IEC) growth remain elusive. Cyclin-dependent kinase (CDK) 4 is necessary for the G1-to-S phase transition during the cell cycle, and its expression is predominantly controlled at the transcription level. Here, we reported that induced ATF-2 following polyamine depletion repressed CDK4 gene transcription in IECs by increasing formation of the ATF-2/JunD heterodimers. ATF-2 formed complexes with JunD as measured by immunoprecipitation using the ATF-2 and JunD antibodies and by glutathione S-transferase (GST) pull-down assays using GST-ATF-2 fusion proteins. Studies using various mutants of GST-ATF-2 revealed that formation of the ATF-2/JunD dimers depended on the COOH-terminal basic region-leucine zipper domain of ATF-2. Polyamine depletion increased ATF-2/JunD complex and inhibited CDK4 transcription as indicated by a decrease in the levels of CDK4-promoter activity and its mRNA. ATF-2 silencing not only prevented inhibition of CDK4 transcription in polyamine-deficient cells but also abolished repression of CDK4 expression induced by ectopic JunD overexpression. ATF-2 silencing also promoted IEC growth in polyamine-depleted cells. These results indicate that induced ATF-2/JunD association following polyamine depletion represses CDK4 transcription, thus contributing to the inhibition of IEC growth.
Keywords: activating protein-1, gene expression, cell proliferation, growth arrest, ornithine decarboxylase, α-difluoromethylornithine
activating transcription factor (ATF)-2 is a sequence-specific DNA-binding protein and belongs to the ATF/cAMP-response element (CRE)-binding protein (CREB) family of transcription factors that represent a large group of basic region-leucine zipper (b-ZIP) proteins (22, 42). These proteins can form homodimers or heterodimers via their leucine zipper motifs and bind to the consensus ATF/CRE site (TGACGTCA) within promoters of their target genes via the basic regions (20). Based on the similarity of their amino acid sequences, ATF/CREB proteins are grouped into two major subgroups: CREB consisting of CREB and CRE modulator (22) and ATF that includes ATF-2, ATF-3, ATF-4, ATF-6, ATF-7, and CRE-BPa (20, 22). ATF-2 bears a b-ZIP domain in its COOH-terminal region and also contains a trans-activation domain in the NH2-terminal region (3, 36). An increasing body of evidence indicates that ATF-2 is implicated in many aspects of cellular functions and plays diverse role in the mammalian cells (3, 4, 36, 42). For example, induced ATF-2 activates expression of various growth-promoting genes such as c-jun, cyclin A, and cyclin D in response to stress stimuli (1, 32, 36), and its overexpression also is correlated with maintenance of a cancer cell phenotype. On the other hand, several studies demonstrate an antiproliferative or apoptotic role of ATF-2 (17, 36). In an in vivo study, null ATF-2 mutant mice display symptoms of severe respiratory distress and die shortly after birth (19). Another ATF-2 mutant mice overexpressing only a fragment of ATF-2 exhibit lowered postnatal viability and growth, a defect in endochondrial ossification, and reduced numbers of cerebellar Purkinje cells (27). However, little is known about the biological role of ATF-2 in the regulation of normal intestinal mucosal growth.
The epithelium of the intestinal mucosa is a rapidly self-renewing tissue in the body, and maintenance of its integrity depends on a complex interplay among cell proliferation, growth arrest, and apoptosis (9, 24, 25). Undifferentiated epithelial cells continuously replicate in the proliferative zone within crypts and differentiate as they migrate up the luminal surface of the colon and the villous tips in the small intestine. Apoptosis occurs in both the crypt area, where this process maintains the balance in cell number between newly divided and surviving cells, and at the luminal surface of the intestine, where differentiated cells are lost (2, 5, 12, 44). This rapid dynamic turnover rate of intestinal epithelial cells (IECs) is tightly regulated and critically controlled by numerous factors including cellular polyamines (9, 18, 38, 43). The natural polyamines spermidine and spermine and their precursor putrescine are organic cations found in all eukaryotic cells (31, 37), and the regulation of cellular polyamines has been recognized for many years as a central convergence point for the multiple signaling pathways driving IEC functions. Normal IEC proliferation in the intestinal mucosa is dependent on the supply of polyamines to the dividing cells in the crypts, whereas decreasing cellular polyamines inhibits cell renewal in vivo as well as in vitro (2, 11, 15, 43, 45), although the exact mechanism underlying polyamines in this process at the molecular level remains to be fully understood.
We (42) have recently reported that depletion of cellular polyamines by inhibiting ornithine decarboxylase (ODC, the first rate-limiting enzyme in polyamine biosynthesis) with α-difluoromethylornithine (DFMO) increases the nuclear abundance of ATF-2 by stabilizing its mRNA, which is associated with a decrease in the levels of cyclin-dependent kinase 4 (CDK4) and cell proliferation. We (41) have also found that polyamine depletion increases levels of AP-1 (activating factor-1) transcriptional factor JunD and that induced JunD represses CDK4 gene transcription by interacting with the proximal region of CDK4-promoter. However, the exact relationship between JunD and ATF-2, particularly their roles in the regulation of CDK4 expression and IEC growth after polyamine depletion, remains unknown. This study was to investigate whether ATF-2 directly interacts with JunD in IECs and whether induced ATF-2 dimerization with JunD is required for repression of CDK4 transcription following polyamine depletion. The data presented herein clearly show that ATF-2 formed heterodimers with JunD via its b-ZIP domain and that induced ATF-2/JunD complex following polyamine depletion inhibited CDK4 gene transcription through its proximal promoter region. Furthermore, increased ATF-2 in polyamine-deficient cells also plays an important role in the inhibition of IEC growth.
MATERIALS AND METHODS
Chemicals and cell culture.
Tissue culture medium and dialyzed fetal bovine serum were from Invitrogen (Carlsbad, CA), and biochemicals were from Sigma (St. Louis, MO). The antibody recognizing ATF-2, JunD, and CDK4 were from Santa Cruz Biotechnology (Santa Cruz, CA). α-Difluoromethylornithine (DFMO) was from Genzyme (Cambridge, MA).
The IEC-6 cell line was purchased from the American Type Culture Collection (ATCC) at passage 13. The cell line was derived from normal rat intestine and was developed and characterized by Quaroni et al. (26). IEC-6 cells originated from intestinal crypt cells as judged by morphological and immunologic criteria. They are nontumorigenic and retain undifferentiated characteristics of intestinal crypt cells. Stock cells were maintained in T-150 flasks in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% heat-inactivated fetal bovine serum (FBS), 10 μg of insulin/ml, and 50 μg/ml gentamicin. Flasks were incubated at 37°C in a humidified atmosphere of 90% air-10% CO2 and passages 15–20 were used in experiments. IEC-6 cells at passage 15–20 exhibit a stable phenotype (14, 16). The Caco-2 cell line (a human colon carcinoma cell line) was also obtained from ATCC at passage 16. It was maintained similarly to the IEC-6 cell line except that it was maintained in an atmosphere of 95% air and 5% CO2. The medium used was Eagle's minimum essential medium with 10% heat-inactivated FBS, and passages 18–23 were used for the experiments.
Luciferase plasmid construction and transfection.
The plasmid clone (pRSV-hjD) containing the human junD gene was obtained from ATCC. Two PCR primers (sense: TACCGCTAG-CGGAGGATGGAAACACCCTTC; antisense: GTCAGGTACCCTCAGTAC-GCCGGGAC-CTG) were used to amplify the complete open reading frame of junD from pRSV-hjD. The resulting PCR product was sequenced to confirm that there were no mutations introduced by PCR and then cloned into an expression vector pcDNA3.1(+) (Invitrogen) with the cytomegalovirus (CMV) promoter as described previously (11, 41). The construct of the CDK4-promoter luciferase (Luc) reporter gene was kindly provided by Dr. Burakoff (Harvard Institutes of Medicine, Boston, MA). Transient transfection was performed with Lipofectamine Reagent from Invitrogen (Carlsbad, CA). The promoter constructs were transfected into cells along with phRL-null, a Renilla luciferase control reporter vector from Promega (Madison, WI), to monitor transfection efficiencies. The transfected cells were lysed for assays of promoter activity using the Dual Luciferase Reporter Assay System (Promega). The luciferase activity from individual constructs was normalized by Renilla-driven luciferase activity in every experiment.
RNA interference.
The small interfering RNA (siRNA) specifically targeting the coding region of ATF-2 mRNA (siATF-2) was purchased from Santa Cruz Biotechnology. Scrambled control siRNA (C-siRNA), which had no sequence homology to any known genes, was used as the control. The siATF-2 and C-siRNA were transfected into cells as described previously (42). Briefly, for each 60-mm cell culture dish, 15 μl of the 20 μM stock duplex siATF-2 or C-siRNA were mixed with 300 μl Opti-MEM medium (Invitrogen). This mixture was gently added to a solution containing 15 μl LipofectAMINE-2000 in 300 μl Opti-MEM medium. The solution was incubated for 20 min at room temperature and gently overlaid onto the monolayer of cells in 3 ml of medium, and cells were harvested for various assays after 48-h incubation.
Immunoprecipitation and immunoblotting analysis.
Cell samples, dissolved in ice-cold RIPA-buffer (50 mM Tris·HCl, pH 7.4, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, 1.0% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 2 mM phenylmethyl-sulfonyl fluoride, 20 μg/ml aprotinin, 2 μg/ml leupeptin, and 2 mM sodium orthovanadate) were sonicated and centrifuged at 4°C, and the supernatants were collected for immunoprecipitation. Equal amounts of proteins (500 μg) for each sample were incubated with the specific antibody against ATF-2 or JunD (4 μg) at 4°C for 3 h, and protein A/G-PLUS-Agarose was added and incubated overnight at 4°C. The precipitates were washed five times with ice-cold D-PBS, and the beads were resuspended in SDS sample buffer for subsequent Western blot analysis. For immunoblotting, samples were subjected to electrophoresis on PAGE gels described previously (40, 44). Briefly, after the transfer of protein onto nitrocellulose membranes, the membranes were incubated for 1 h in 5% nonfat dry milk in 1× TBS-T buffer (Tris-buffered saline, pH 7.4, with 0.1% Tween 20). Immunological evaluation was then performed overnight at 4°C in 5% nonfat dry milk/TBS-T buffer containing a specific antibody against ATF-2 or JunD protein. The membranes were subsequently washed with 1× TBS-T and incubated with the secondary antibodies conjugated with horseradish peroxidase for 1 h at room temperature. The immunocomplexes on the membranes were reacted for 1 min with Chemiluminiscence Reagent (NEL-100 DuPont NEN).
Real-time quantitative PCR analysis.
Total RNA was isolated by using the RNeasy Mini kit (Qiagen, Valencia, CA). Equal amounts of total RNA (2μg) were transcribed to synthesize single-stranded cDNA with an RT-PCR kit (Invitrogen). Real-time quantitative PCR (Q-PCR) was performed by using Applied Biosystems Instrument (Foster City, CA) using specific primers, probes, and software (Applied Biosystems) as described in our previous publications (15, 44). The levels of CDK4 mRNA were quantified by Q-PCR analysis and normalized with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels.
GST-fusion protein expression and GST pull-down assays.
The cDNA insert of the full-length ATF-2 (1–505), COOH-terminal ATF-2 (203–505), and deleted COOH-terminal ATF-2 (330–505), (376–505), and (420–505) were amplified by PCR with sense primers GGATCCATGAAATTCAAGTTACATGTG, GATCCCAGTCAGAGGAATCTCGAC, GGATCCCACACAACTCCACAGACC, GGATCCACCGCCATGCAGAAGA, GGATCCTT-AGAGAAGAAAGCTGAAGAC, respectively, and antisense primers CTCGAGTCAACTTCCTGAGGGCTGTG. All these inserts have BamHI and XhoI ends that are compatible with the linearized pGEX-4T-2 vector ends. After the inserts were cloned into pGEX-4T-2 vector, the proper orientation and sequences were screened. The established constructs were then used to express glutathione S-transferase (GST) fusion proteins in Eschericia coli BL21 induced by isopropyl-β-d-thiogalactopyranoside (IPTG) as described (33). The fusion protein expression was monitored by SDS-PAGE analysis.
GST pull-down assays with various mutants of GST-ATF-2 and in vivo translated JunD were performed by using MagneGST pull-down system (Promega) according to the manufacturer's instruction. Briefly, E. coli BL21 cultures expressing GST control or GST fusion ATF-2 proteins were harvested and incubated with equilibrated MagneGST particles for 30 min at room temperature on a rotating platform. The magnetic particles that were bound with GST or GST-fusion ATF-2 proteins were then gently washed five times. After the final washing was completed, the immobilized GST or GST-fusion ATF-2 proteins were incubated with cell lysates for another 30 min. The magnetic particles that were bound with protein complexes were washed five times and then dissolved in 1× SDS loading buffer and boiled for 5 min. SDS-PAGE was performed to analyze the pull-down results.
Chromatin immunoprecipitation.
Cells were fixed with 1% formaldehyde to cross-link chromatin, and chromatin immunoprecipitation (ChIP) assays were carried out by using the Upstate Biotechnology ChIP Assay kit (Charlottesville, VA) according to the provided protocol with minor modification. Cells were washed with ice-cold PBS, suspended in SDS lysis buffer, and then sonicated. After centrifugation, supernatant was transferred and diluted in 10-fold ChIP dilution buffer and then precleared with Salmon Sperm DNA/Protein-A Agarose-50% Slurry with agitation. Supernatant was incubated with the anti-JunD antibody or control IgG overnight with constant rotation. The immunocomplexes were captured and eluted with fresh elution buffer. The DNA-protein cross-links were reversed and deproteinized, and DNA was recovered and amplified by PCR. A pair of primers to amplify the proximal region of CDK4-promoter containing an AP-1 binding site were 5′-ATGCAGACAGGCTGAAAGAC-3′ and 5′-GATGGCAGCCA-CGTGATCTG-3′. Primers to amplify the proximal region of the GAPDH promoter (a negative control) were 5′-TACTAGCGGTTTTACGGGCG-3′ and 5′-TCGAACAGGAG-GAGCAGAGAGCGA-3′. The DNA isolated through IgG ChIP was used as a negative control. The input DNA, obtained from chromatin that was processed (cross-linked and reversed) similarly to the samples, served as a positive control for PCR effectiveness.
Statistics.
Values are means ± SE from three to six samples. Autoradiographic and immunoblotting results were repeated three times. The significance of the difference between means was determined by analysis of variance. The level of significance was determined using Duncan's multiple range test (7).
RESULTS
Induced ATF-2 physically interacts with JunD via its COOH-terminal b-ZIP domain after polyamine depletion.
ATF-2 consists of ∼505 amino acids and has a b-ZIP domain in its COOH-terminus (4, 29). To examine the physical interactions of ATF-2 with JunD in the presence or absence of cellular polyamines, the following two sets of experiments were performed. First, we determined whether induced levels of endogenous ATF-2 by polyamine depletion formed a specific complex with JunD as measured by immunoprecipitation assays using specific antibodies. Exposure of IEC-6 cells to 5 mM DFMO for 6 days completely inhibited ornithine decarboxylase (ODC) activity and almost totally depleted cellular polyamines. The levels of putrescine and spermidine were undetectable, and spermine was decreased by ∼60% (data not shown). Similar results have been reported in our previous publications (11, 12). Consistent with our previous observations (11, 42), depletion of cellular polyamines by DFMO increased the levels of both ATF-2 and JunD proteins, which was completely prevented by exogenous putrescine given together with DFMO (Fig. 1A). To examine whether induced ATF-2 interacts with JunD, whole cell lysates obtained from controls and polyamine-deficient cells were immunoprecipitated with the antibody that recognizes either JunD or ATF-2, and then these precipitates were examined by using Western blot analysis with a specific antibody against ATF-2 or JunD. As shown in Fig. 1B, ATF-2 was able to form a complex with JunD in IEC-6 cells, and this ATF-2/JunD association was increased after polyamine depletion. On the other hand, induction in the ATF-2/JunD complex in DFMO-treated cells was totally abolished by exogenous putrescine (10 μM). Spermidine at the concentration of 5 μM had an equal effect to that of putrescine on levels of ATF-2/JunD complex when it was added to cultures that contained DFMO (data not shown). Our results also showed that immunoprecipitation of cell lysates with nonspecific antibody IgG failed to precipitate JunD or ATF-2 as measured by Western blotting using anti-ATF-2 or anti-JunD antibody (data not shown). These results strongly suggest that induced levels of endogenous ATF-2 following polyamine depletion physically associate with JunD and form ATF-2/JunD heterodimers in normal IECs.
Fig. 1.
Changes in the levels of total activating transcription factor-2 (ATF-2) and JunD proteins and their interactions after polyamine depletion in intestinal epithelial cell (IEC)-6 cells. Cells were grown in control cultures, cultures in which ornithin decarboxylase (ODC) activity inhibited by 5 mM α-difluoromethylornithine (DFMO), and cultures inhibited with DFMO and supplemented with 10 μM putrescine (Put) for 6 days. A: representative immunoblots of ATF-2 and JunD proteins. Whole cell lysates were harvested, applied to each lane (20 μg) equally, and subjected to electrophoresis on a 10% acrylamide gel. Levels of ATF-2 (∼70 kDa) and JunD (∼40 kDa) proteins were identified by a specific antibody (Ab) that recognizes ATF-2 or JunD. After the blots were stripped, actin (∼42 kDa) immunoblotting was performed as an internal control for equal loading. B: representative immunoblots of ATF-2 and JunD proteins in complexes immunoprecipitated (IP) by either anti-JunD (a) or anti-ATF-2 (b) antibody (Ab). After whole cell lysates (300 μg) were incubated with a specific Ab against JunD or ATF-2, precipitates were separated by performing electrophoresis on 10% acrylamide gels. Levels of ATF-2 and JunD were examined by Western immunoblotting with a specific anti-ATF-2 or anti-JunD Ab. Three experiments were performed that showed similar results.
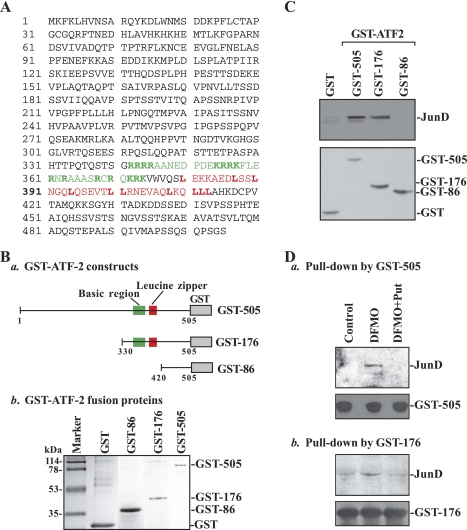
Second, we detected an in vitro association of ATF-2 with JunD by using GST pull-down assays using GST-ATF-2 fusion proteins. Three constructs expressing GST-full-length ATF-2 fusion protein (GST-505), GST-mutated ATF-2 containing a b-ZIP domain (GST-176), and GST-mutated ATF-2 lacking a b-ZIP domain (GST-86) were generated as shown in Fig. 2Ba, and their products of GST-ATF-2 proteins (Fig. 2Bb) were coupled to glutathione-Sepharose beads. Cell lysates obtained from JunD-overexpressing cells were incubated with each of these fusion proteins for 30 min, and then the levels of JunD in the materials pull-down by GST-ATF-2 fusion proteins were identified by Western blot analysis using the anti-JunD antibody. As shown in Fig. 2C, top, ATF-2 formed a heterodimer with JunD when its b-ZIP domain was present (GST-505 and GST-176, as shown in two middle lanes), but this association was disappeared when the b-ZIP domain was deleted (GST-86, as shown in right lane). Changes in the levels of detectable JunD in GST-pull down materials were not due to loading variations or integrity of fusion proteins, because the quantity and quality of all fusion proteins utilized were similar as judged by Western blot analysis using the anti-GST antibody (Fig. 2C, bottom). Results presented in Fig. 2D further show that JunD levels in the materials pull-down by GST-505 or GST-176 fusion protein increased significantly in polyamine-deficient cells, but they were restored to normal levels by exogenous putrescine. In contrast, JunD levels in the material pull-down by GST-86 fusion protein were undetectable regardless of the presence or absence of cellular polyamines (data not shown). These results indicate that the COOH-terminal b-ZIP domain of ATF-2 is necessary for its dimerization with JunD and also mediates an induction in ATF-2/JunD complex following polyamine depletion.
Fig. 2.
Interactions of ATF-2 with JunD in vitro as measured by glutathionine S-transferase (GST)-ATF-2 pull-down assays. A: human ATF-2 amino acid sequence. The basic region and leucine-zipper area were indicated by green and red colors, respectively. B: GST-ATF-2 fusion proteins: schematic diagram depicting various GST-ATF-2 constructs(a); and GST-ATF-2 fusion proteins as measured by Coomassie blue staining assays(b). Constructs were transformed into Eschericia coli BL21, and their expression was induced by treatment with isopropyl-b-d-thiogalactopyranoside (IPTG) at the concentration of 0.5 mM. Expressed GST (without ATF-2) or GST-ATF-2 fusion proteins were harvested and purified by equilibrated MagneGST particles. These fusion proteins were monitored by SDS-PAGE analysis and shown by Coomassie blue staining. C: ATF-2 association with JunD in cells overexpressing JunD. Cells were transfected by using the expression vector containing human junD cDNA by LipofectAMINE technique; whole cell lysates were harvested 48 h after the transfection. The magnetic particles bound to GST or GST-ATF-2 fusion proteins were incubated with cell lysate for 30 min, dissolved in 1× SDS loading buffer, and then subjected to SDS-PAGE. Levels of JunD in the complexes pull-down by using GST or GST-ATF-2 fusion proteins were measured by Western blot analysis with the antibody against JunD (top), whereas input GST or GST-ATF-2 fusion proteins were examined by using anti-GST antibody (bottom). Three experiments were performed that showed similar results. D: levels of JunD protein in the complexes pull-down by GST-ATF-2 fusion proteins GST-505 (a) and GST-176 (b) from control cells and cells treated with DFMO alone or DFMO plus Put for 6 days.
Induced ATF-2/JunD heterodimers repress CDK4 gene transcription by binding to the proximal region of the CDK4-promoter.
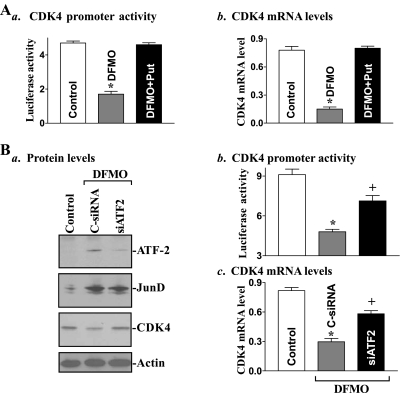
Increased levels of the ATF-2/JunD complex following polyamine depletion were associated with a repression of CDK4 transcription as indicated by a decrease in the levels of CDK4-promoter activity and its mRNA (Fig. 3A). To determine the putative role of induced ATF-2/JunD association in the inhibition of CDK4 transcription in polyamine-deficient cells, the specific siRNA targeting the siATF-2 was used to reduce ATF-2 levels. These specific siATF-2 nucleotides were designed to cleave rat ATF-2 mRNA by activating endogenous RNase H and to have a unique combination of specificity, efficiency, and reduced toxicity (42). Initially, we examined the transfection efficiency of the siRNA nucleotides in IEC-6 cells and demonstrated that >95% of cells were positive when they were transfected with a fluorescent FITC-conjugated siRNA for 24 h (data not shown). As shown in Fig. 3Ba, transfection with the siATF-2 specifically decreased ATF-2 protein but had no effect on JunD protein in polyamine-deficient cells. Induced ATF-2 protein following polyamine depletion was totally prevented when DFMO-treated cells were exposed to siATF-2 for 48 h. Interestingly, ATF-2 silencing also partially but significantly restored CDK4 transcription in the presence of DFMO (Fig. 3, Bb and Bc). The levels of CDK4-promoter activity and CDK4 mRNA in DFMO-treated cells transfected with siATF-2 were increased by ∼50% compared with those observed in DFMO-treated cells transfected with C-siRNA. Restoration of CDK4 transcription by ATF-2 silencing was also accompanied by an increase in the level of CDK4 protein in polyamine-deficient cells (Fig. 3Ba, third bottom panel). We also examined the effect of JunD inhibition on CDK4 expression and demonstrated that JunD silencing by transfection with the specific siRNA targeting JunD mRNA significantly prevented the repression of CDK4 transcription in polyamine-deficient cells (data not shown). Similar results have been published previously (41). These findings indicate that polyamine depletion represses CDK4 transcription by inducing ATF-2/JunD association.
Fig. 3.
Changes in cyclin-dependent kinase-4 (CDK4) gene expression in polyamine-deficient cells after ATF-2 silencing. A: effect of polyamine depletion on CDK4 transcription. a, levels of CDK4-promoter activity. Cells were grown in DMEM containing difluoromethylornithine (DFMO) alone or DFMO plus Put for 4 days and then transfected with the CDK4-promoter luciferase reporter construct. Luciferase activity was measured 48 h after the transfection in the presence or absence of DFMO or DFMO plus Put; data were normalized by Renilla-driven luciferase activity and expressed as means ± SE of data from 3 separate experiments. *P < 0.05 compared with control cells and cells exposed to DFMO plus Put. b, changes in mRNA levels of CDK4 in cells that were processed as described in Aa. Total cellular RNA was isolated, and levels of CDK4 mRNA were measured by using real-time quantitative PCR analysis. Values were means ± SE of data from 3 separate experiments. *P < 0.05 compared with control cells and DFMO plus Put cells. B: changes of CDK4 gene expression after ATF-2 silence in polyamine-deficient cells. a, representative immunoblots of ATF-2, JunD, and CDK4 proteins. After DFMO treatment for 4 days, cells were transfected with specific small interfering RNA (siRNA) targeting the coding region of ATF-2 mRNA (siATF-2) or control siRNA (C-siRNA), and whole cell lysates were harvested 48 h thereafter. The levels of ATF-2, JunD, and CDK4 were measured by Western blot analysis, and equal loading was monitored by actin immunoblotting. b, CDK4-promoter activity in cells described in Ba. Values are means ± SE of data from 3 separate experiments. *P < 0.05 compared with control cells; +P < 0.05 compared with DFMO-treated cells transfected with C-siRNA. c, changes in levels of CDK4 mRNA in cells described in Ba. *P < 0.05 compared with controls, +P < 0.05 compared with DFMO-treated cells transfected with C-siRNA.
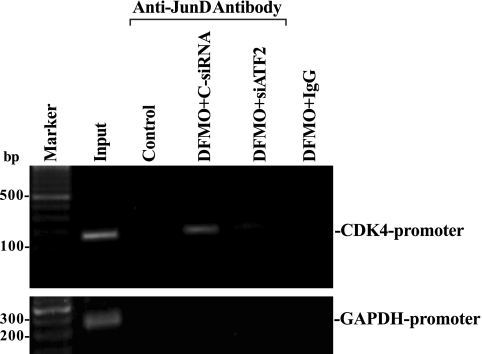
ChIP analysis also was used to examine the association of ATF-2/JunD complex with the CDK4-promoter region after ATF-2 silencing in polyamine-deficient cells. Nuclear fractions isolated from control cells and DFMO-treated cells transfected with siATF-2 or C-siRNA were immunoprecipitated using a specific anti-JunD antibody, and the associated DNA was purified. With the use of specific PCR primers, a 189-bp PCR product was obtained, which matched the sequence of the proximal region of the CDK4-promoter from -412 to 43 (containing the AP-1 binding site) relative to the transcriptional start site. ATF-2/JunD complex was bound to the CDK4-promoter in vivo, as shown using an anti-JunD antibody in polyamine-deficient cells (Fig. 4, top). The dimerization of ATF-2 with JunD is necessary for this binding activity, because ATF-2 silencing abolished this interaction without effect on JunD level (Fig. 3Ba). In addition, this association was specific for ATF-2/JunD dimers, since no PCR product was detectable when using a nonspecific antibody (IgG) or when using primers to an unrelated promoter such as the GAPDH promoter (Fig. 4, bottom). These results indicate that induced levels of ATF-2/JunD complex following polyamine depletion repress CDK4 gene transcription by interacting with the AP-1 binding site within the proximal region of the CDK4-promoter.
Fig. 4.
Activity of ATF-2/JunD complex binding to CDK4 promoter as measured by chromatin immunoprecipitation (ChIP) analysis in cells described in Fig. 3. The association of ATF-2/JunD complex with the proximal region of CDK4 promoter (between −412 and 43) was determined using anti-JunD antibody. Cross-linked chromatin isolated from control cells and DFMO-treated cells transfected with ATF-2 siRNA or C-siRNA was immunoprecipitated by anti-JunD antibody, and the associated chromosomal DNA fragments were amplified by PCR using CDK4-promoter-specific primers or GAPDH promoter-specific primers as described in the materials and methods. The expected size of the PCR product was ∼189 bp. Chromosomal DNA input was subject to the same procedure and served as a positive control. Three experiments were performed that showed similar results.
To further define the role of induced ATF-2/JunD dimers in the regulation of CDK4 transcription, the effect of ATF-2 silencing on CDK4 expression after ectopic JunD overexpression was also examined in Caco-2 cells. This line of cells was used in this study based on the following reasons: 1) polyamine depletion also increased levels of both ATF-2 and JunD that were associated with an inhibition of CDK4 expression in Caco-2 cells (41); and 2) this cell line is an excellent model for transient transfection. A JunD expression vector containing the corresponding human junD cDNA under the control of pCMV was constructed as described previously (11). Transient transfection with the JunD vector increased JunD expression (Fig. 5A, top), and the level of JunD protein was increased by approximately sixfold at 48 h after the transfection. The vector that lacked exogenous junD cDNA (Null) was used as a negative control and did not induce JunD protein levels. Ectopic JunD overexpression inhibited CDK4-promoter activity (Fig. 5Ba) and decreased levels of CDK4 mRNA (Fig. 5Bb) and protein (Fig. 5A, bottom). Importantly, ATF-2 silencing by transfection with siATF-2 prevented the repression of CDK4 transcription induced by JunD overexpression. The levels of CDK4-promoter activity and its mRNA in cells cotransfected with the JunD expression vector and siATF-2 were increased by ∼52% when they were compared with those observed in cells cotransfected with the JunD expression vector and C-siRNA. These results indicate that JunD dimerization with ATF-2 is necessary for JunD-induced repression of CDK4 transcription.
Fig. 5.
Effect of ATF-2 silencing on CDK4 gene expression in cells overexpressing JunD. A: representative immunoblots of ATF-2, JunD, and CDK4 proteins. Cells were cotransfected with the JunD expressing vector and either siATF-2 or C-siRNA, and whole cell lysates were harvested 48 h thereafter. Vector containing no junD cDNA (Null) served as a negative control. Levels of JunD, ATF-2, and CDK4 were measured by Western blot analysis, and equal loading was monitored by actin immunoblotting. B: CDK4 gene transcription in cells described in A. CDK4-promoter activity as measured by luciferase reporter gene activity (a) and changes in levels of CDK4 mRNA as measured by real-time quantitative PCR analysis (b) are shown. Values are means ± SE of data from 3 separate experiments. *P < 0.05 compared with control cells and cells transfected with Null; +P < 0.05 compared with cells cotransfected with the JunD and C-siRNA.
ATF-2 silencing promotes IEC cell growth in polyamine-deficient cells.
To elucidate whether increased accumulation of the ATF-2/JunD complex plays a role in the process of growth inhibition after polyamine depletion, we examined the effect of ATF-2 silencing on IEC-6 cell growth in polyamine-deficient cells. Cells were initially treated with 5 mM DFMO for 3 days and then transfected with either siATF-2 or C-siRNA. The changes in cell growth were measured at various times after the transfection in the presence of DFMO. As shown in Fig. 6, polyamine depletion by administration of DFMO inhibited IEC-6 cell growth, but exposure of these polyamine-deficient cells to siATF-2 significantly promoted cell growth. Cell numbers in DFMO-treated cells transfected with siATF-2 were increased by ∼60% at 48 h and ∼73% at 72 h, respectively, when compared with those observed in cells treated with DFMO alone or DFMO-treated cells transfected with C-siRNA. Transfection of DFMO-treated cells with C-siRNA at the same concentration, however, showed no significant effects on cell growth. In addition, there was no apparent loss of cell viability in cells treated with DFMO alone or DFMO-treated cells transfected with siATF-2 or C-siRNA. These results indicate that increased ATF-2/JunD association contributes to the inhibition of IEC growth following polyamine depletion.
Fig. 6.
Effect of ATF-2 silencing on cell growth in polyamine-deficient cells. A: images of intestinal epithelial cell (IEC)-6 cells after various treatments: controls (a); cells treated with DFMO alone (b); DFMO-treated cells transfected with C-siRNA (c); and DFMO-treated cells transfected with ATF-2 siRNA (d). After cells were grown in the presence of DFMO for 3 days, they were transfected with siATF-2 or C-siRNA and maintained in the same culture conditions for additional 72 h. Original magnification, ×100. B: cell numbers at different times in cells described in A. Values are means ± SE from 6 dishes. *P < 0.05 compared with cells treated with DFMO alone and DFMO-treated cells transfected with C-siRNA.
DISCUSSION
Polyamines have emerged as critical factors that are essential for the maintenance of intestinal epithelial integrity, but the exact mechanisms underlying polyamines at cellular and molecular levels remain unclear. Our previous studies have shown that polyamines regulate the stability of ATF-2 mRNA through the RNA-binding protein HuR and that depletion of cellular polyamines increases nuclear ATF-2 abundance in IECs (42). However, little is known about the role and mechanism of induced ATF-2 in the regulation of IEC proliferation. In the present study, we identified a novel function of ATF-2 in the modulation of CDK4 expression and show that induced ATF-2 represses CDK4 gene transcription, thus contributing to the inhibition of IEC growth following polyamine depletion. Studies aimed at characterizing the molecular aspects of this processes indicated that induced ATF-2 in polyamine-deficient cells physically interacts with JunD and formed ATF-2/JunD heterodimers that directly bind to the CDK4-promoter. This study also provides new evidence showing that formation of ATF-2/JunD complex is necessary for JunD-induced CDK4 repression, since ATF-2 silencing prevented inhibition of CDK4 transcription without effect on JunD levels in cells overexpressing JunD.
The results reported in the present study clearly show that the physical interaction of ATF-2 with JunD is mediated through the b-ZIP domain located in COOH-terminal region of ATF-2. ATF-2 formed heterodimers with JunD when its b-ZIP domain was present, but this association was disappeared completely when the b-ZIP domain was deleted (Fig. 2). JunD is a member of the Fos/Jun protein family and acts as an activating protein-1 (AP-1) transcription factor that activates or represses a diverse collection of target genes (8, 28). Initially, the Fos/Jun and ATF/CREB protein families were regarded as distinct sets of transcription factors that share the b-ZIP motif but have different DNA binding specificities (6, 22). ATF/CREB proteins seem to bind to the ATF/CRE site with a higher affinity than to the AP-1 site, whereas Fos/Jun proteins appear to bind to the AP-1 site with a greater affinity than to the ATF/CRE site. However, an increasing body of evidence, including the present study, indicates that members of these two families form selective cross-family heterodimers that display distinguishable DNA binding specificities from each other and from their parental homodimers, and they are now defined as a super family of b-ZIP transcription factors (20, 35). The current finding showing ATF-2 dimerization with JunD is particularly interesting and important because ATF-2 has been proposed for many years to act as a key modifier of the dynamic balance between c-Jun and the other members of the Jun family, JunB and JunD (3, 4, 36). Their interactions affect cell cycle progression positively or negatively, since c-Jun enhances G1-to-S phase transition by activating cyclin A and cyclin D after mitogenic stimulation (10, 30), whereas JunB and JunD have opposite effects (23).
Our present results also indicate that interaction of ATF-2 with JunD is regulated by cellular polyamines and that depleted polyamines by inhibiting ODC with DFMO increased formation of the ATF-2/JunD complex. Because increased levels of ATF-2 protein and ATF-2/JunD dimers in DFMO-treated cells were completely prevented by the addition of exogenous putrescine, these observed changes in levels of ATF-2/JunD complex are more likely related to polyamine depletion rather than to the nonspecific effect of DFMO. Most significantly, however, are the observations that induced ATF-2 plays a critical role in the repression of CDK4 transcription following polyamine depletion and that formation of ATF-2/JunD heterodimers is necessary for this process. Our previous studies (41, 42) demonstrate that the CDK4 is a target of JunD and that polyamine depletion increases both JunD and ATF-2 levels. Increased levels of JunD, either endogenously by polyamine depletion or exogenously by ectopic JunD overexpression, are shown to repress CDK4 transcription by directly interacting with the CDK4-promoter (41). Results presented in Fig. 3 further show that formation of ATF-2/JunD dimers is required for this repression, because ATF-2 silencing in polyamine-deficient cells restored CDK4 transcription and its protein expression without effect on JunD levels. In JunD overexpressing cells, the repression of CDK4 transcription was also prevented by ATF-2 silencing (Fig. 5), although there were no changes in levels of induced JunD.
The results reported here also suggest that ATF-2/JunD heterodimer interacts with the CDK4-promoter through an AP-1 binding site. The CDK4-promoter contains a functional AP-1 binding site but has no any typical ATF/CRE site (22, 41). When compared with the canonical AP-1 motif (TGAGTCA), this AP-1 sequence (TGAGACA) within the proximal region of the CDK4-promoter has only one nucleotide difference as indicated by underline. Our previous studies have demonstrated that JunD binds to this AP-1 site of the CDK4-promoter in vitro as well as in vivo, and this interaction mediates JunD-induced repression of CDK4 transcription in IECs (41). Deletion or point-mutation of this AP-1 sequence from the CDK4-promoter prevents CDK4 repression induced by either ectopic expression of junD gene or increased endogenous JunD following polyamine depletion. Our current observations provide additional new evidence showing that dimerization of JunD with ATF-2 is crucial for their interaction with the CDK4-promoter through this functional AP-1 site, because disruption of ATF-2/JunD complex by ATF-2 silencing blocked the binding of ATF-2/JunD to the proximal region of the CDK4-promoter containing the AP-1 site in polyamine-deficient cells (Fig. 4).
Induced ATF-2 plays a critical role in the inhibition of IEC proliferation following polyamine depletion. Increased ATF-2 levels and resultant reduction in CDK4 expression were associated with a significant increase in G1 phase growth arrest in polyamine-deficient cells (13, 37, 41), whereas decreased levels of ATF-2 by its gene silencing significantly promoted cell growth in the absence of cellular polyamines (Fig. 6). These results are consistent with others who have demonstrated an antiproliferative role of ATF-2 in several types of cells (41), although there are also reports showing a proliferative role of ATF-2 (34). These controversial effects of ATF-2 on cell proliferation are not surprising, as ATF-2 function is affected by selective dimerization with other b-ZIP proteins and since different b-ZIP content in different tissues or cells could result in either proliferative or antiproliferative ATF-2 effect (34). Because expression of other Fos/Jun members such as c-Fos, c-Jun, and JunB decreased dramatically following polyamine depletion in IECs (1, 21, 39), it is likely that the inhibition of IEC growth through repression of CDK4 transcription in DFMO-treated cells is primarily due to the ATF-2/JunD heterodimers. In support of this notion, JunD silencing promoted CDK4 expression and enhanced cell proliferation in polyamine-deficient cells (41). Given the fact that polyamines are required for maintaining intestinal epithelial integrity and their cellular levels are tightly regulated, these findings suggest that repression of CDK4 transcription by induced ATF-2/JunD heterodimers contributes to the inhibition of IEC growth following polyamine depletion and plays an important role in intestinal epithelial homeostasis.
GRANTS
This work was supported by Merit Review Grant from the Department of Veterans Affairs and by National Institutes of Health Grants DK-57819, DK-61972, DK-68491. J.-Y. Wang is a Research Career Scientist, Medical Research Service, Department of Veterans Affairs.
DISCLOSURES
No conflicts of interest are declared by the author(s).
REFERENCES
- 1.Beier F, Taylor AC, LuValle P. Activating transcription factor 2 is necessary for maximal activity and serum induction of the cyclin A promoter in chondrocytes. J Biol Chem 275: 12948–12953, 2000 [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharya S, Ray RM, Johnson LR. Prevention of TNF-α-induced apoptosis in polyamine-depleted IEC-6 cells is mediated through the activation of ERK1/2. Am J Physiol Gastrointest Liver Physiol 286: G479–G490, 2004 [DOI] [PubMed] [Google Scholar]
- 3.Bhoumik A, Jones N, Ronai Z. Transcriptional switch by activating transcription factor 2-derived peptide sensitizes melanoma cells to apoptosis and inhibits their tumorigenicity. Proc Natl Acad Sci USA 101: 4222–4227, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhoumik A, Ronai Z. ATF2: a transcription factor that elicits oncogenic or tumor suppressor activities. Cell Cycle 7: 2341–2345, 2008 [DOI] [PubMed] [Google Scholar]
- 5.Dufour G, Demers MJ, Gagne D, Dydensborg AB, Teller IC, Bouchard V, Degongre I, Beaulieu JF, Cheng JQ, Fujita N, Tsuruo T, Vallee K, Vachon PH. Human intestinal epithelial cell survival and anoikis: differentiation state-distinct regulation and roles of protein kinase B/Akt isoforms. J Biol Chem 279: 44113–44122, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA 88: 3720–3724, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harter JL. Critical values for Duncan new multiple range test. Biometrics 16: 671–685, 1960 [Google Scholar]
- 8.Hirai SI, Ryseck RP, Mechta F, Bravo R, Yaniv M. Characterization of junD: a new member of the jun proto-oncogene family. EMBO J 8: 1433–1439, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson LR. Regulation of gastrointestinal mucosal growth. Physiol Rev 68: 456–502, 1988 [DOI] [PubMed] [Google Scholar]
- 10.Leventaki V, Drakos E, Medeiros LJ, Lim MS, Elenitoba-Johnson KS, Claret FX, Rassidakis GZ. NPM-ALK oncogenic kinase promotes cell-cycle progression through activation of JNK/c-Jun signaling in anaplastic large-cell lymphoma. Blood 110: 1621–1630, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Li L, Liu L, Rao JN, Esmaili A, Strauch ED, Bass BL, Wang JY. JunD stabilization results in inhibition of normal intestinal epithelial cell growth through p21 after polyamine depletion. Gastroenterology 123: 764–779, 2002 [DOI] [PubMed] [Google Scholar]
- 12.Li L, Rao JN, Bass BL, Wang JY. NF-κB activation and susceptibility to apoptosis after polyamine depletion in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 280: G992–G1004, 2001 [DOI] [PubMed] [Google Scholar]
- 13.Liu L, Guo X, Rao JN, Zou T, Marasa BS, Chen J, Greenspon J, Casero RA, Jr, Wang JY. Polyamine-modulated c-Myc expression in normal intestinal epithelial cells regulates p21Cip1 transcription through a proximal promoter region. Biochem J 398: 257–267, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L, Li L, Rao JN, Zou T, Zhang HM, Boneva D, Bernard MS, Wang JY. Polyamine-modulated expression of c-myc plays a critical role in stimulation of normal intestinal epithelial cell proliferation. Am J Physiol Cell Physiol 288: C89–C99, 2005 [DOI] [PubMed] [Google Scholar]
- 15.Liu L, Rao JN, Zou T, Xiao L, Wang PY, Turner DJ, Gorospe M, Wang JY. Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol Biol Cell 20: 4885–4898, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Santora R, Rao JN, Guo X, Zou T, Zhang HM, Turner DJ, Wang JY. Activation of TGF-β-Smad signaling pathway following polyamine depletion in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 285: G1056–G1067, 2003 [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Wang Y, Li W, Zheng P. Activating transcription factor 2 and c-Jun-mediated induction of FoxP3 for experimental therapy of mammary tumor in the mouse. Cancer Res 69: 5954–5960, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lux GD, Marton LJ, Baylin SB. Ornithine decarboxylase is important in intestinal mucosal maturation and recovery from injury in rats. Science 210: 195–198, 1980 [DOI] [PubMed] [Google Scholar]
- 19.Maekawa T, Bernier F, Sato M, Nomura S, Singh M, Inoue Y, Tokunaga T, Imai H, Yokoyama M, Reimold A, Glimcher LH, Ishii S. Mouse ATF-2 null mutants display features of a severe type of meconium aspiration syndrome. J Biol Chem 274: 17813–17819, 1999 [DOI] [PubMed] [Google Scholar]
- 20.Manna PR, Dyson MT, Stocco DM. Role of basic leucine zipper proteins in transcriptional regulation of the steroidogenic acute regulatory protein gene. Mol Cell Endocrinol 302: 1–11, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel AR, Wang JY. Polyamine depletion is associated with an increase in JunD/AP-1 activity in small intestinal crypt cells. Am J Physiol Gastrointest Liver Physiol 276: G441–G450, 1999 [DOI] [PubMed] [Google Scholar]
- 22.Persengiev SP, Green MR. The role of ATF/CREB family members in cell growth, survival and apoptosis. Apoptosis 8: 225–228, 2003 [DOI] [PubMed] [Google Scholar]
- 23.Piechaczyk M, Farras R. Regulation and function of JunB in cell proliferation. Biochem Soc Trans 36: 864–867, 2008 [DOI] [PubMed] [Google Scholar]
- 24.Potten CS. Epithelial cell growth and differentiation II: Intestinal apoptosis. Am J Physiol Gastrointest Liver Physiol 273: G253–G257, 1997 [DOI] [PubMed] [Google Scholar]
- 25.Potten CS, Loeffler M. Stem cells: attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Development 110: 1001–1020, 1990 [DOI] [PubMed] [Google Scholar]
- 26.Quaroni A, Wands J, Trelstad RL, Isselbacher KJ. Epithelioid cell cultures from rat small intestine: characterization by morphologic and immunologic criteria. J Cell Biol 80: 248–265, 1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reimold AM, Grusby MJ, Kosaras B, Fries JW, Mori R, Maniwa S, Clauss IM, Collins T, Sidman RL, Glimcher MJ, Glimcher LH. Chondrodysplasia and neurological abnormalities in ATF-2-deficient mice. Nature 379: 262–265, 1996 [DOI] [PubMed] [Google Scholar]
- 28.Ryder K, Lau LF, Nathans D. A gene activated by growth factors is related to the oncogene v-jun. Proc Natl Acad Sci USA 85: 1487–1491, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sano Y, Akimaru H, Okamura T, Nagao T, Okada M, Ishii S. Drosophila activating transcription factor-2 is involved in stress response via activation by p38, but not c-Jun NH(2)-terminal kinase. Mol Biol Cell 16: 2934–2946, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimizu Y, Kinoshita I, Kikuchi J, Yamazaki K, Nishimura M, Birrer MJ, Dosaka-Akita H. Growth inhibition of non-small cell lung cancer cells by AP-1 blockade using a cJun dominant-negative mutant. Br J Cancer 98: 915–922, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabor CW, Tabor H. Polyamines. Annu Rev Biochem 53: 749–790, 1984 [DOI] [PubMed] [Google Scholar]
- 32.Takasawa S, Ikeda T, Akiyama T, Nata K, Nakagawa K, Shervani NJ, Noguchi N, Murakami-Kawaguchi S, Yamauchi A, Takahashi I, Tomioka-Kumagai T, Okamoto H. Cyclin D1 activation through ATF-2 in Reg-induced pancreatic beta-cell regeneration. FEBS Lett 580: 585–591, 2006 [DOI] [PubMed] [Google Scholar]
- 33.Toualbi K, Guller MC, Mauriz JL, Labalette C, Buendia MA, Mauviel A, Bernuau D. Physical and functional cooperation between AP-1 and β-catenin for the regulation of TCF-dependent genes. Oncogene 26: 3492–3502, 2007 [DOI] [PubMed] [Google Scholar]
- 34.Vale-Cruz DS, Ma Q, Syme J, LuValle PA. Activating transcription factor-2 affects skeletal growth by modulating pRb gene expression. Mech Dev 125: 843–856, 2008 [DOI] [PubMed] [Google Scholar]
- 35.Verde P, Casalino L, Talotta F, Yaniv M, Weitzman JB. Deciphering AP-1 function in tumorigenesis: fra-ternizing on target promoters. Cell Cycle 6: 2633–2639, 2007 [DOI] [PubMed] [Google Scholar]
- 36.Vlahopoulos SA, Logotheti S, Mikas D, Giarika A, Gorgoulis V, Zoumpourlis V. The role of ATF-2 in oncogenesis. Bioessays 30: 314–327, 2008 [DOI] [PubMed] [Google Scholar]
- 37.Wang JY. Polyamines and mRNA stability in regulation of intestinal mucosal growth. Amino Acids 33: 241–252, 2007 [DOI] [PubMed] [Google Scholar]
- 38.Wang JY, McCormack SA, Viar MJ, Johnson LR. Stimulation of proximal small intestinal mucosal growth by luminal polyamines. Am J Physiol Gastrointest Liver Physiol 261: G504–G511, 1991 [DOI] [PubMed] [Google Scholar]
- 39.Wang JY, McCormack SA, Viar MJ, Wang H, Tzen CY, Scott RE, Johnson LR. Decreased expression of protooncogenes c-fos, c-myc, and c-jun following polyamine depletion in IEC-6 cells. Am J Physiol Gastrointest Liver Physiol 265: G331–G338, 1993 [DOI] [PubMed] [Google Scholar]
- 40.Wang PY, Rao JN, Zou T, Liu L, Xiao L, Yu TX, Turner DJ, Gorospe M, Wang JY. Posttranscriptional regulation of MEK-1 by polyamines through the RNA-binding protein HuR modulating intestinal epithelial apoptosis. Biochem J 426: 293–306, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao L, Rao JN, Zou T, Liu L, Marasa BS, Chen J, Turner DJ, Passaniti A, Wang JY. Induced JunD in intestinal epithelial cells represses CDK4 transcription through its proximal promoter region following polyamine depletion. Biochem J 403: 573–581, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao L, Rao JN, Zou T, Liu L, Marasa BS, Chen J, Turner DJ, Zhou H, Gorospe M, Wang JY. Polyamines regulate the stability of activating transcription factor-2 mRNA through RNA-binding protein HuR in intestinal epithelial cells. Mol Biol Cell 18: 4579–4590, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang HM, Rao JN, Guo X, Liu L, Zou T, Turner DJ, Wang JY. Akt kinase activation blocks apoptosis in intestinal epithelial cells by inhibiting caspase-3 after polyamine depletion. J Biol Chem 279: 22539–22547, 2004 [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, Zou T, Rao JN, Liu L, Xiao L, Wang PY, Cui YH, Gorospe M, Wang JY. Stabilization of XIAP mRNA through the RNA binding protein HuR regulated by cellular polyamines. Nucleic Acids Res 37: 7623–7637, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou T, Rao JN, Liu L, Marasa BS, Keledjian KM, Zhang AH, Xiao L, Bass BL, Wang JY. Polyamine depletion induces nucleophosmin modulating stability and transcriptional activity of p53 in intestinal epithelial cells. Am J Physiol Cell Physiol 289: C686–C696, 2005 [DOI] [PubMed] [Google Scholar]