Abstract
Dendritic spines are actin-rich structures that accommodate the postsynaptic sites of most excitatory synapses in the brain. Although dendritic spines form and mature as synaptic connections develop, they remain plastic even in the adult brain, where they can rapidly grow, change, or collapse in response to normal physiological changes in synaptic activity that underlie learning and memory. Pathological stimuli can adversely affect dendritic spine shape and number, and this is seen in neurodegenerative disorders and some forms of mental retardation and autism as well. Many of the molecular signals that control these changes in dendritic spines act through the regulation of filamentous actin (F-actin), some through direct interaction with actin, and others via downstream effectors. For example, cortactin, cofilin, and gelsolin are actin-binding proteins that directly regulate actin dynamics in dendritic spines. Activities of these proteins are precisely regulated by intracellular signaling events that control their phosphorylation state and localization. In this review, we discuss how actin-regulating proteins maintain the balance between F-actin assembly and disassembly that is needed to stabilize mature dendritic spines, and how changes in their activities may lead to rapid remodeling of dendritic spines.
1. INTRODUCTION
Excitatory postsynaptic sites in the brain are usually formed on dendritic spines, small protrusions on the surface of dendrites that are enriched in actin [1–5]. Dendritic spines form and mature as synaptic connections develop in the brain, and their morphogenesis directly correlates with synapse formation and maturation. The most widely-held view of dendritic spine formation suggests that dendritic spines originate from dendritic filopodia-like protrusions both in vitro and in vivo [6–15]. Although mature dendritic spines can also emerge from the dendritic shaft, it is widely accepted that long, thin filopodia-like morphologies are features of immature pre-cursors to spines, while mature spines are characterized by mushroom-like or stubby shapes [16–18]. Several studies have demonstrated a correlation between dendritic spine morphology and synaptic function [19–26]. While immature thin spines with smaller heads are flexible and can rapidly enlarge or shrink in response to changes in synaptic activity [19–22], mature mushroom-shaped spines with larger heads are less likely to change, but show higher sensitivity to glutamate than immature thin spines [23–26]. These differences in synaptic strength are suggested to relate to the number of neurotransmitter receptors. Mature spines have a large spine head area that is proportional to postsynaptic density (PSD) and synapse size, as well as receptor complement (Fig. 1). Two-photon glutamate uncaging studies have shown that the mature mushroom-like spines have large PSDs with a high number of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors [23]. Indeed, numbers of AMPA receptors and N-methyl-D-aspartate (NMDA) receptors directly correlate with PSD size [27, 28]. Spine head volume and PSD size are also proportional to the number of neurotransmitter-containing vesicles at the presynaptic bouton [29] and the amount of neurotransmitter released at the synapse [30]. Mature dendritic spines also contain elements of endoplasmic reticulum, called the spine apparatus, and provide means for compartmentalization within the neuron, which results in localized control over factors such as intracellular Ca2+ concentration [31] and local protein synthesis [32].
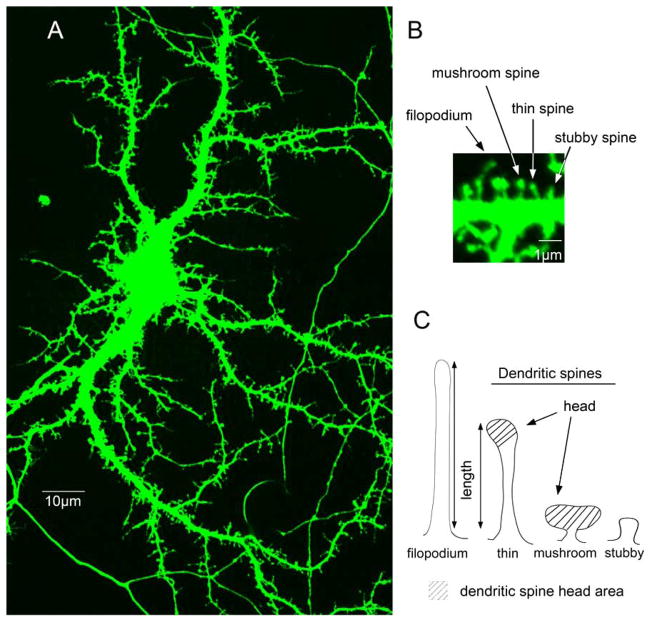
Fig. 1.
(A) A GFP-expressing hippocampal neuron at day 14 in vitro displays dendritic filopodia-like protrusions and spines with different shapes and sizes (B, C) The high magnification image of the dendrite (B) and a drawing show examples of main categories of dendritic protrusions: filopodia-like protrusions, mushroom spine, thin spine, and stubby spine. (C) Filopodia-like protrusions are precursors of dendritic spines. Mature mushroom spines display the largest heads and thin necks.
Dendritic spines remain plastic in the adult brain and can rapidly grow, change, or collapse in response to normal physiological changes in synaptic activity that underlies learning and memory. Long-term potentiation (LTP) can result in spine head enlargement [19, 20], whereas long-term depression (LTD) has been reported to induce shrinkage of dendritic spine heads and spine elimination [33]. Synaptic activity was also shown to shape dendritic spines in area CA1 of the hippocampus [8], and long-lasting synaptic enhancement leads to new spine development [34]. Although changes in synaptic activity may not always lead to structural remodeling [35], experience-dependent motility of spines and change in their morphology has been reported using various learning paradigms in developing rat barrel cortex [36], motor cortex [37], and hippocampus [38, 39]. Pathological stimuli can also lead to changes in dendritic spine shape and number in the developing and adult brain. Immature dendritic spine profiles are found in subjects with Fragile X, Down and Rett syndromes, whose brains exhibit many long, thin spines and filopodia-like protrusions [40, 41]. Moreover, dendritic spine loss is a hallmark of several neurodegenerative diseases, and may also contribute to impaired brain function in these diseases [42]. The ability of spines to rapidly turnover can lead to recovery from damage [43] and stressors such as sleep deprivation [44].
2. ACTIN DYNAMICS IN DENDRITIC SPINES
Many molecular signals that control the changes in dendritic spine morphology act through the regulation of F-actin, some through direct interaction with actin, and others via downstream effectors [5]. Actin dynamics at the leading edge in motile cells [45] and in growth cones [46] control cell locomotion and axon pathfinding, respectively. There is also a tight regulation of actin dynamics within dendritic spines and filopodia-like protrusions. While the stabilization of mature spines is required for synapse maintenance, F-actin remodeling underlies synaptic plasticity and may result in spine turnover, triggering both formation of new spines and filopodia, as well as pruning of existing spines [47, 48]. As globular actin (G-actin) monomers join together to form actin filaments (F-actin), there is a bidirectional polymerization that is biased toward what is known as the plus end or barbed end [49]. In this way, the F-actin barbed end is fast-growing, while the opposite pole of the filament, the minus end or pointed end, elongates more slowly. The constant turnover of the actin filaments in spines results from a steady-state of actin treadmilling, in which G-actin monomers are added quickly to the barbed end and are disassembled from the pointed end of F-actin filaments, while exchanging ATP for ADP [50–53]. However, this F-actin treadmilling may or may not lead to changes in dendritic spine shape and size.
3. PROTEINS REGULATING ACTIN DYNAMICS IN DENDRITIC SPINES
Actin assembly regulatory proteins are responsible for shifting the balance between spine assembly and disassembly that is required for normal synaptic function. Some regulate spine morphology through direct interaction with actin, while others control actin dynamics indirectly [54–56]. It is the interplay among many of these proteins that defines the rate of growth or disassembly of actin, and ultimately the morphology of dendritic spines and development of synapses. In this review, we discuss the role of actin-binding proteins that are found in dendritic spines and influence dendritic spine formation, maintenance or remodeling, including proteins that regulate elongation, branching, and bundling of actin filaments and those that trigger actin severing and depolymerization.
3.1. Actin Polymerization and Elongation
3.1.1. Arp2/3 Complex Promotes Nucleation and Branching of Actin Filaments
The Actin-Related Protein (Arp) 2/3 complex is comprised of seven proteins [57–59] and is responsible for nucleating branches from the sides of existing actin filaments and capping the pointed ends, thus creating additional fast-growing barbed ends for further actin polymerization and elongation (Fig. 2) [60–62]. The Arp2/3 complex contributes to the cytoskeletal rearrangements necessary for a variety of cellular functions, including cell migration and outgrowth [63]. It can be found in protruding lamellipodia of fibroblasts [64] and at the leading edge of carcinoma cells, where its branching activity is needed for lamellipodia extension [65]. In dendritic spines, Arp2/3 is sparsely distributed in the PSD, and more concentrated in a specific domain that lies midway between the plasma membrane, the spine center, the PSD, and the spine neck [66]. This specificity of Arp2/3 localization suggests a restricted spine region dedicated to actin branching. In cooperation with members of the Ena/VASP family, Arp2/3 nucleates actin polymerization and branching for pathogenic bacteria motility [67] and growth cone protrusion [68]. Other cell functions that rely on F-actin assembly, including endocytosis [69], internal receptor trafficking [70], integrin-mediated cell adhesion [71], and phagocytosis [72] are also mediated by the Arp2/3 complex.
Fig. 2.
Arp2/3 nucleates new branches from existing actin filaments, creating fast-growing barbed ends.
In maintaining the balance between assembly and disassembly of actin filaments, there are some proteins and factors that compete against the Arp2/3 complex, while others work in cooperation (Fig. 3). Tropomyosin inhibits the branching activity of Arp2/3 [73], and ATP hydrolysis also acts to antagonize its activity by de-branching older actin filaments [74]. Conversely, the Arp2/3 complex acts in cooperation with gelsolin as it polymerizes actin in the presence of gelsolin-capped filaments [75]; and cortactin activates Arp2/3 and stabilizes new branch points on actin filaments [76]. The actin-severing protein ADF/cofilin dissociates Arp2/3 from actin filaments [77], but also uses its severing activity to increase the number of preferred ends for Arp2/3 nucleation [78]. Moreover, the Arp2/3 complex acts in opposition to ADF/cofilin in lamellipodia by cross-linking actin filaments and capping pointed ends, thus stabilizing actin filaments and preventing depolymerization [79].
Fig. 3.
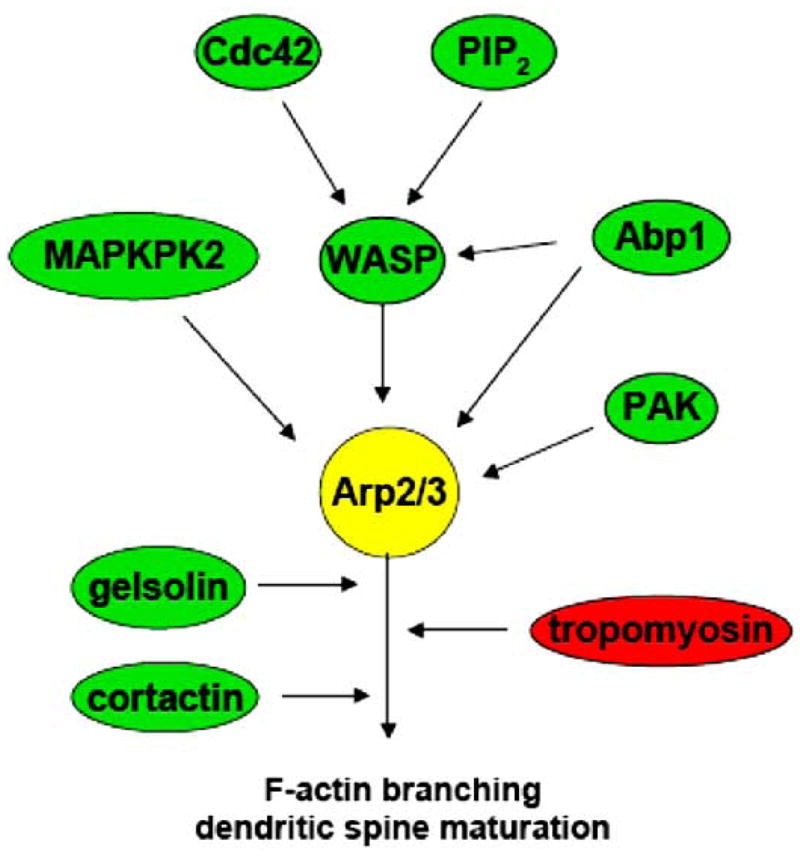
Regulatory proteins affecting the activity of Arp2/3.
Phosphorylation of the Arp2/3 complex is necessary for its nucleating activity and cellular localization [80]. Phosphorylation of Arp2/3 can be achieved by such proteins as MAPK-activated protein kinase 2 (MAPKPK2) [81] and the p21-activated kinase (PAK) [82]. Proteins of the Wiskott-Aldrich Syndrome protein (WASP) family, Neural-WASP (N-WASP), Scar, and the WASP-family verprolin-homologous protein (WAVE), are well-known to bind to and activate the Arp2/3 complex [83], each stimulating a different rate of actin nucleation [84]. Cdc42, a member of the Rho family of small GTPases, and Phosphatidylinositol 4,5 bisphosphate (PIP2) stimulate Arp2/3 nucleation activity through WASP activation [85–88]. Moreover, the overexpression of actin-binding protein Abp1, which is similar to cortactin in structure [89] and can activate N-WASP [90] or phosphorylate Arp2/3 directly, increased the length and density of mature spines and synapses, whereas RNA interference (RNAi)-mediated knock-down of Abp1 reduced the number of mushroom-shaped spines and synapses in cultured hippocampal neurons [91]. In addition, EphB receptors were found to promote dendritic spine maturation through the Cdc42-N-WASP pathway, suggesting an involvement of the Arp2/3 complex in dendritic spine development [92].
The role of N-WASP in regulating spine morphogenesis was further supported by findings that show a decrease in the number of dendritic spines in hippocampal neurons with reduced N-WASP expression [93]. The changes in spine number and morphology following N-WASP knock-down were similar to those of the RNAi-mediated decrease of the Arp3 protein [93], suggesting that activation of the Arp2/3 complex is a mechanism by which N-WASP promotes the formation of mature spines and synapses. A decrease in number of mature dendritic spines was also seen with knockdown of WAVE1 by RNAi in primary hippocampal neurons, and in mouse striatal sections of WAVE1 knock-out mice [94]. Synaptic plasticity, as well as learning and memory, were also impaired in mice with a targeted WAVE1 mutation and in WAVE1 knock-out mice [95, 96], suggesting a role of the WAVE1 pathway in regulating synaptic plasticity.
Knock-down of Arp2/3 through siRNA-targeting of the p34 subunit was recently shown to promote filopodia-like protrusions, while decreasing the number of mushroom, thin, and stubby spines and the overall protrusion density in hippocampal neurons [97]. This is similar to the effect seen when Arp2/3 localization was altered by expression of the Scar1-WA fragment, which further suggests an important role of the Arp2/3 complex in promotion of mature dendritic spine phenotype [97]. It will be interesting to determine whether the Arp2/3 complex and its regulatory proteins are also involved in the assembly of the postsynaptic scaffold and in glutamate receptor trafficking. Taken together, these results suggest that the Arp2/3 complex is tightly regulated in dendritic spines and contributes to spine morphogenesis, inducing formation of mature dendritic spines and synapses, as well as regulating synaptic plasticity.
3.1.2. Cortactin Promotes Branching and Stabilization of Actin Filaments
Another protein that can bind to and activate the Arp2/3 complex is cortactin [76, 98]. It was discovered as a cortical protein that binds to actin [99] and promotes branching and stabilization of actin filaments [100]. Cortactin is located at sites of actin assembly in lamellipodia [101], and is likely involved in endosomal movement in fibroblasts [102]. It is also implicated in controlling shape-changes of folliculostellate cells of the anterior pituitary [103], and in aberrant cell motility, such as cancer cell migration and invasion [104, 105].
In the brain, cortactin is enriched in dendritic spines, where it co-localizes with F-actin. There is a large cortactin pool in the actin core within the dendritic spine, which is implicated in regulating its shape, and a smaller pool near the PSD that may be involved in the regulation of synaptic function [106]. Cortactin has been implicated in spine morphogenesis in studies using gain- and loss-of-function approaches [107]. While overexpression of cortactin in hippocampal cultures led to spine elongation, knock-down of cortactin with small-interfering RNA (siRNA) resulted in loss of spines [107]. Cortactin interaction with the large GTPase dynamin-3 variant, Dyn3baa, also promoted filopodial extension in cultured rat hippocampal neurons [108]. The ability of cortactin to use its actin-branching and elongation activity for filopodia formation and outgrowth suggests its important role in spine morphogenesis.
In addition, cortactin regulates spine morphology by mediating the interaction between actin and microtubules. While stable microtubules that express microtubule-associated protein 2 (MAP2) are concentrated in dendritic shafts [109], dynamic microtubules can enter dendritic spines and affect actin dynamics [110]. Microtubule-associated protein EB3, which is located on the plus-end of dynamic microtubules, may act through a p140Cap-Src pathway to promote cortactin activity and mature dendritic spines. Knock-down of EB3 with shRNA induced a loss of F-actin in dendritic protrusions, as well as a reduction of mature mushroom spines and an increase in filopodia-like structures [110], which is similar to the result seen with RNAi-mediated loss of p140Cap or cortactin [107]. These effects of EB3 knock-down were reversed by overexpression of p140Cap or cortactin. In this way, dynamic microtubules may contribute to the arrangement and activity of actin-regulating proteins such as cortactin, thus shaping dendritic spines.
Cortactin influences actin organization through interactions with several proteins that promote F-actin assembly, and these interactions are negatively regulated by cortactin phosphorylation [111, 112]. For example, cortactin’s ability to activate WASP and induce actin branching through Arp2/3 is inhibited by its phosphorylation, which is mediated by Src non-receptor tyrosine kinase, and results in loss of cortactin from postsynaptic sites [113]. δ-Catenin can interact with cortactin, and this interaction was shown to promote F-actin elongation without branching, which was also inhibited by Src-mediated phosphorylation of cortactin [114]. Cortactin has the ability to link actin dynamics to the cell membrane through interaction with several PSD scaffolding proteins. In addition to Dyn3, cortactin interacts with the Shank family of proteins that are localized to the PSD of excitatory synapses [115]. Shank binds to Homer, which has been shown to anchor metabotropic glutamate receptors (mGluR) 1 and 5 [116]. It seems that cortactin’s association with Shank and Homer could act to stabilize postsynaptic clusters of glutamate receptors during synaptogenesis.
Cortactin can be regulated by synaptic activity, as a decrease in overall cortactin level was observed in rat hippocampus 4 hours following Morris water maze learning [117]. This decrease in cortactin is dependent on NMDA receptor activity, and may mediate the actin reorganization that is necessary for learning-induced spine remodeling. In contrast, rats exposed to a novel environment exhibited an increase in cortactin levels in the hippocampus 12 hours later, but this upregulation of cortactin was abolished by 48 hours of Rapid Eye Movement Sleep (REMS) deprivation [118].
Cortactin localization within neurons is regulated by NMDA receptor activity and brain-derived neurotrophic factor (BDNF). NMDA receptor activation induces cortactin redistribution from dendritic spines to the shaft [107], which is mediated by Src non-receptor tyrosine kinases in hippocampal cultures [119]. Activation of Src kinases induces cortactin phosphorylation and cortactin depletion from the postsynaptic sites. Conversely, BDNF application stimulates a mitogen-activated protein (MAP) kinase-dependent redistribution of cortactin from the dendritic shaft to the spines [119, 120]. This shift in cortactin localization may play an important role in activity-dependent spine remodeling. A reduction in the level of cortactin in spines may allow for actin remodeling triggered by NMDA receptor activation, whereas BDNF-induced accumulation of cortactin in spines may increase actin stability necessary for the maintenance of mature spines and synapses. As BDNF itself is regulated by synaptic activity [121], it seems that these converse pathways indeed work together to regulate transient changes in dendritic spine morphology in response to changes in synaptic activity. The control exerted over cortactin activity by regulating its localization within neurons seems to have a critical role in spine development and synaptogenesis, as well as synaptic plasticity.
3.1.3. α-Actinin Promotes Bundling and Extension of Actin
α-Actinin induces bundling and cross-linking of actin networks in a concentration-dependent manner [122, 123] by forming anti-parallel homodimers that exhibit an actin-binding domain at each end [124–126]. This dimer formation may also target α-actinin to the cell membrane, as phospholipids and cytoplasmic regions of transmembrane receptors have an affinity for the exposed acidic surface of α-actinin rods [123, 127, 128]. α-Actinin can form short branched actin filaments or elongate existing ones, depending on certain conditions and factors, such as the α-actinin/actin ratio, Ca2+ concentration, and binding partners [129]. α-Actinin was first recognized as a protein that enhances the contraction of actomyosin fibers [130, 131], and several muscular pathologies have been linked to α-actinin [132, 133, 134]. It is implicated in maintaining cellular shape and adhesion in epithelial cells [135] and human neutrophils [136]. α-Actinin was also found in leading lamella of smooth muscle cells (A10) [137], in growth cones of PC12 cells [138] and in spinal cord neurons [139], and is suggested to play a role in cell and growth cone motility.
The muscle form of α-actinin is Ca2+-insensitive, but the activity of the non-muscle form is controlled by Ca2+ concentration [140, 141]. At low Ca2+ levels, the actin-bundling activity of α-actinin is high, whereas an increase in Ca2+ levels inhibits the cross-linking activity by inducing a structural change in α-actinin [142]. In the brain, this has implications for the control of α-actinin by synaptic activity that regulates Ca2+ influx through NMDA receptors and voltage-gated Ca2+ channels. α-Actinin is also regulated by other actin-binding and signaling proteins. For example, tropomyosin and dre-brin competitively inhibit α-actinin binding to actin [143], whereas focal adhesion kinase (FAK) reduces α-actinin affinity for actin by phosphorylation [144]. Conversely, PIP2 binds to α-actinin and increases its gelation activity [145], and the protein actophorin promotes α-actinin bundling activity by severing and positioning actin filaments favorably for α-actinin interaction [146]. Moreover, α-actinin acts in cooperation with profilin, as it promotes the actin-polymerizing activity of the profilin-actin complex (profilactin; [147]).
The brain isoform of α-actinin [148] is enriched at the PSD of excitatory synapses and promotes the elongation of dendritic spines [149]. α-Actinin depends on F-actin for its spine localization, as actin depolymerization with latrunculin A disrupts its postsynaptic localization in cultured hippocampal neurons [150]. There is evidence that α-actinin may also be involved in the assembly of the spine apparatus via interaction with the postsynaptic protein synaptopodin [151]. Synaptopodin was shown to shift α-actinin activity from branching to elongation of existing actin filaments, and synaptopodin-deficient mice lack a spine apparatus and exhibit deficiencies in activity-dependent LTP [152].
In dendritic spines, α-actinin was shown to promote both actin filament elongation and branching depending on its interacting proteins. For example, α-actinin has been shown to interact with the Spine-Associated Rap GTPase-activating protein (SPAR), which promotes mature dendritic spines. α-Actinin2 appears to cooperate with SPAR in creating dendritic spines with large heads [153]. This study supports the role for α-actinin in promoting actin branching in the heads of mature dendritic spines. α-Actinin could also indirectly contribute to actin-branching activity not only in spines, but in dendrites and axons as well, through its interaction with the guanine nucleotide exchange factor EFA6A [154]. EFA6A was shown to activate ADP ribosylation factor 6 (ARF6), which regulates axonal and dendritic branching in cultured rat hippocampal neurons [155]. Conversely, α-actinin can promote filopodia-like extensions through its actin-elongating activity, as overexpression of α-actinin2 increased the length and number of dendritic filopodia-like protrusions in cultured hippocampal neurons [156]. The ability of α-actinin to promote dendritic filopodia-like extensions may be accomplished through its co-localization and interaction with telencephalin (TLCN), a telencephalon-specific cell adhesion molecule, which is known to promote development of filopodia-like protrusions and to slow the maturation of dendritic spines [157,158].
α-Actinin was also shown to interact with the NMDA receptor in dendritic spines, promote its anchoring to the postsynaptic membrane [159], and regulate NMDA receptor activity [160, 161]. Both Ca2+/calmodulin and Ca2+/calmodulin-dependent protein kinase II (CaMKII) compete with α-actinin for binding of the NR1 subunit of the NMDA receptor [162, 163]. While α-actinin binding to the NMDA receptor increases the receptor open probability, Ca2+/calmodulin interaction with the NMDA receptor reduces it, resulting in NMDA receptor inactivation [164–167]. The role of α-actinin in enhancing NMDA receptor activity is consistent with its ability to induce dendritic filopodia-like protrusions, as NMDA receptor activation also leads to spine remodeling. It would be interesting to see if NMDA receptor anchoring is also affected by α-actinin binding to actin filaments or whether α-actinin can regulate NMDA receptor interactions with postsynaptic scaffolding proteins, such as PSD-95 [168]. Perhaps α-actinin can contribute to NMDA receptor anchoring by stabilizing the dendritic spines through its actin-branching and bundling activity.
There is evidence that may link α-actinin to several neural pathologies. For example, α-actinin2 levels were shown to be decreased in an Alzheimer’s disease mouse model expressing mutant hAPP [169]. α-Actinin was accumulated in Hirano bodies, neuronal inclusions that increase with aging and are found in patients with Alzheimer’s disease [170]. It is possible that the cross-linking activity of α-actinin contributes to the formation of these structures, which may deplete the postsynaptic pool of α-actinin. A decrease in postsynaptic α-actinin was also seen in the Huntington’s disease mouse model R6/2 [171]. Through its actin branching and elongating activity, as well as its ability to regulate NMDA receptor activity, α-actinin is well positioned for modulating synaptic plasticity.
3.1.4. Drebrin Regulates Actin Assembly in Dendritic Spines
Drebrin is a developmentally regulated neuron-specific protein that competes with tropomyosin, fascin, α-actinin, gelsolin, cofilin, and myosin for actin-binding [172–175]. Drebrin was shown to bundle actin filaments into thick winding fibers in fibroblasts [176]. The embryonic drebrin isoform, drebrin E, exists in several cell types including neurons, while adult drebrin A is exclusively expressed in neurons [177], and was found to be accumulated in the head area of mature mushroom-shaped dendritic spines with large PSDs and a high number of NMDA receptors in adult mouse cerebral cortex [178]. Interestingly, drebrin A is specifically localized at postsynaptic sites of excitatory synapses and barely detectable in presynaptic terminals, neuronal cell bodies, or axons [174, 179]. Deletion of the actin-binding domain of drebrin A [180] or depolymerization of F-actin with latrunculin A [150] leads to drebrin A re-distribution throughout various domains of dendrites, suggesting that it is dependent on F-actin for its postsynaptic localization.
Drebrin A has been implicated in the development of dendritic spines and synapses [179, 181, 182]. In support of this, it was found in submembranous zones of dendrites prior to the assembly of PSDs, formation of dendritic spine heads, and aggregation of presynaptic vesicles [179]. Furthermore, drebrin A overexpression in immature neurons induced accumulation of F-actin and PSD-95 at postsynaptic sites [181], whereas suppression of drebrin A expression decreased spine width and the density of spines and filopodia in developing hippocampal neurons in cultures [183]. Drebrin clustering was found to precede PSD-95 accumulation at postsynaptic sites and suppression of drebrin A expression disrupted PSD-95 postsynaptic localization [184]. Moreover, drebrin can also regulate NMDA receptor localization, as membrane targeting of NMDA receptors in cultured hippocampal neurons after AP5 treatment was abolished following reduction of drebrin A expression by antisense treatment [183].
Drebrin A postsynaptic localization is regulated by synaptic activity, and drebrin was shown to change its localization upon AMPA and NMDA receptor activation [182, 185]. While NMDA receptor activation induced a loss of drebrin from dendritic spines, an increase in drebrin A-immunoreactive spines was observed following NMDA receptor blockade in vivo, which occurred in the large mushroom-shaped spines [186]. On the other hand, drebrin clustering at postsynaptic sites was induced by AMPA receptor activation during spine morphogenesis and diminished with AMPA receptor blockade [182]. Together, these studies show that drebrin A induces postsynaptic differentiation, accumulates in mature dendritic spines, and stabilizes existing actin filaments. Therefore, the re-localization of drebrin seen upon NMDA receptor activation may be necessary to allow for spine remodeling.
Besides promoting dendritic spine formation and postsynaptic differentiation, drebrin was also implicated in learning and memory and the regulation of dendritic spine plasticity [180, 183]. For example, drebrin overexpression was shown to induce dendritic spine elongation in cultured cortical neurons [180]. Drebrin overexpression may promote spine elongation by inhibiting myosin association with actin filaments, thereby reducing actin contractility [174] or by competing with actin-stabilizing proteins for actin binding. Recent study has also suggested that drebrin-induced dendritic spine remodeling may involve activation of Ras family of small GTPases [187]. The significance of drebrin in normal synapse development and synaptic plasticity is further supported by observations that decreased levels of both drebrin isoforms were found in the hippocampi of Alzheimer’s patients [188] and following cerebral ischemia [189], as well as in the frontal and temporal cortex of patients with Down syndrome [190]. In the future, it would be interesting to determine the interplay between drebrin and other actin-binding proteins in promoting postsynaptic differentiation and spine maturation, or in regulating spine remodeling and synaptic plasticity.
3.1.5. Opposing Effects of Profilin on Actin Polymerization
Profilin was first identified as an actin monomer-sequestering protein that can inhibit actin polymerization and elongation [191–193] by forming a 1:1 complex with G-actin, thus decreasing its free concentration available to form F-actin polymers [194–197]. Conversely, profilin or the profilin-actin complex (profilactin) can promote actin polymerization by binding the barbed ends of actin filaments and directly elongating it (Fig. 4) [198, 199]. A dose-dependent decrease in the amount of F-actin and lamellipodial size were seen upon microinjection of profilin into normal rat kidney (NRK) cells, whereas profilactin increased F-actin and lamellipodial ruffling [200]. Further, while profilin-bound actin monomers may be removed from the free actin pool and cannot nucleate new filaments, they can be added to F-actin barbed but not pointed ends, contributing to actin filament elongation [201]. While profilin can promote depolymerization by binding and sequestering actin monomers, it induces polymerization by lowering the critical concentration of ATP-actin and decreasing actin monomer affinity for its bound nucleotide, thereby favoring ATP-actin formation [202]. Profilin can also promote the stability of actin filaments, as F-actin concentration was shown to increase with stable expression of profilin in Chinese hamster ovary (CHO) cells [203].
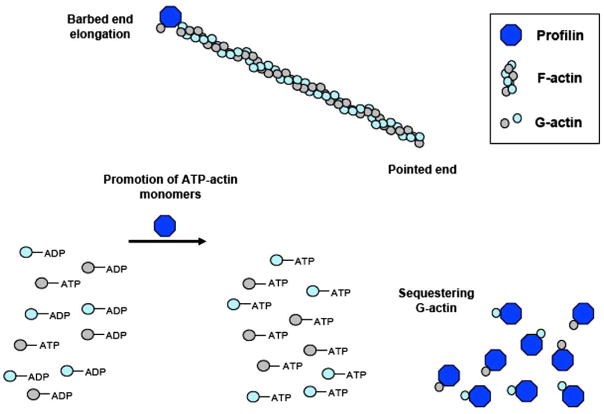
Fig. 4.
Opposing actions of profilin on actin. G-actin-sequestering promotes F-actin depolymerization, while profilin-actin complex induces polymerization by binding to F-actin barbed ends and promoting formation of ATP-actin monomers.
The ability of profilin to promote or inhibit actin polymerization under different circumstances can result in diverse cellular responses [204]. Profilin is implicated in cell motility, as it is localized with dynamic actin in fibroblast lamellipodia [205] and it was shown to regulate the movement of Listeria monocytogenes in infected host cells [206]. Profilin I mRNA was up-regulated in reactive hippocampal microglia following entorhinal deafferentation [207], which could also indicate a role for profilin in microglial migration following injury. Profilin regulates cell morphology, as it is involved in thrombin-induced platelet shape change [208] and endocytosis [209, 210]. Moreover, several abnormalities, including slow growth and loss of normal cell shape were seen with disruption or deletion of the profilin gene in yeast [211], whereas profilin gene deletion in Drosophila was lethal [212]. Profilin II was detected in neurons and its localization in dendritic spines was shown to be regulated by Ca2+ levels [213].
Profilin activity is also regulated by phosphorylation and its interaction with PIP2. Profilin interaction with PIP2 [214–216] inhibits PIP2 hydrolysis by phospholipase C (PLC) [217], and dissociates the profilin-actin complex [218], which could lead to release of free G-actin monomers and polymerization near the cell membrane. Conversely, PIP2 binding to cofilin can stimulate profilin phosphorylation by protein kinase C (PKC) [219, 220]. Profilin phosphorylation, which can also be achieved by phosphatidylinositol 3-kinase (PI3-K), increases its affinity for G-actin and poly (L-proline) [221]. While PIP2 binding can lead to increased actin polymerization, profilin phosphorylation would increase its association with G-actin and inhibit actin polymerization. Further, an increased affinity for poly (L-proline) may lead to enhanced profilin binding to membrane-associated proteins of the Mena/VASP family, resulting in F-actin elongation [213, 222]. It would be interesting to determine how profilin-PIP2 binding and PKC phosphorylation of profilin regulate F-actin polymerization in dendritic spines.
Profilin expression is developmentally regulated in rat cerebellum [223] and is detected at synaptic sites in the adult cerebellar cortex [224], suggesting that it may play a role in synaptogenesis. Indeed, profilin associates with gephyrin and Mena to form a postsynaptic cortical scaffolding complex [225]. Upon NMDA receptor activation and Ca2+ influx, profilin is recruited to and stabilizes dendritic spines in a VASP-dependent manner [213, 226, 227]. Moreover, electrical stimulation that is known to produce LTP promoted similar recruitment of profilin to dendritic spines and spine stabilization [213], suggesting that profilin may be involved in regulation of dendritic spine and synapse dynamics underlying experience-dependent plasticity.
Besides the role of profilin II in normal dendritic spine development and plasticity, several studies have suggested that profilin II may mediate cytoskeletal changes in response to pathological conditions and disease processes. For example, profilin interacts with the Huntington Disease protein Huntingtin (Htt), and could be involved in causing the pathological aggregation of Htt. [228]. Inhibition of Rho-kinase (ROCK), which forms a complex with profilin II [229], reduces Htt aggregation [230]. On the other hand, profilin depletion can lead to neuronal deficits, as some patients with Miller-Dieker syndrome have been shown to exhibit partial profilin gene deletions [231]. The Fragile X mental retardation protein in Drosophila (dFMRP) was shown to regulate the neuronal cytoskeleton through profilin, as dFMRP binds mRNA of the profilin homolog and decreases its protein expression [232]. Profilin II knock-out mice demonstrated a lack of an actin polymerizing response to depolarization, which led to increased synaptic excitability due to increased vesicle exocytosis in glutamatergic neurons [233]. These studies correlate well, as some Fragile X phenotypes could also be explained by increased synaptic excitability, perhaps from decreased profilin levels. These studies show the remarkably diverse functions of profilin in controlling actin dynamics in dendritic spines. Depending on cellular signals and interacting proteins, profilin may promote actin polymerization or depolymerization, which could lead to formation, stabilization, or remodeling of dendritic spines.
3.1.6. Spinophilin (Neurabin II) and Neurabin I Promote cross-Linking of Actin
Spinophilin derives its name from its localization in the heads of dendritic spines [234], and neurabin I is a neural tissue–specific actin–binding protein [235]. Both proteins are structurally related and display actin cross-linking and bundling activity. Spinophilin is expressed ubiquitously, but most abundantly in the brain, where it is localized in PSDs [236]. Neurabin I is accumulated in dendritic spines, as well as in growth cone lamellipodia of developing neurons.
A multitude of binding partners have been identified for spinophilin [237], including those with implications for synaptic function, such as protein phosphatase 1 (PP1). Spinophilin participates in targeting PP1 to the postsynaptic membrane (Fig. 5) [234, 238], which allows PP1 to dephosphorylate its synaptic substrates. In this way, spinophilin can indirectly modify the activities of AMPA and NMDA receptors, two known substrates of PP1. In vivo, spinophilin takes part in the anchoring of AMPA receptors to the plasma membrane and promotes its dephosphorylation through PP1 [239]. Spinophilin knock-out mice exhibited an increased density of dendritic filopodia and immature spines, abnormal regulation of AMPA receptor activity by PP1, and impaired LTD, suggesting that spinophilin may regulate excitatory synaptic transmission and spine morphogenesis through PP1-mediated regulation of AMPA receptor activity [240]. Further, phosphorylation of spinophilin by protein kinase A (PKA) [241] or CaMKII [242] reduces the affinity of spinophilin for F-actin and causes a change in its cellular localization, which may also alter its binding with PP1 due to spatial segregation. Spinophilin phosphorylation by CaMKII is Ca2+-dependent and may play a role in synaptic plasticity. In addition, a downregulation of the expression of spinophilin and GluR1 were observed during sleep following unilateral LTP induction in the prefrontal cortex of rats [243]. Taken together, these studies demonstrate that phosphorylation of spinophilin regulates its expression and specific localization within dendritic spines, where it is positioned to regulate synaptic activity and spine morphogenesis.
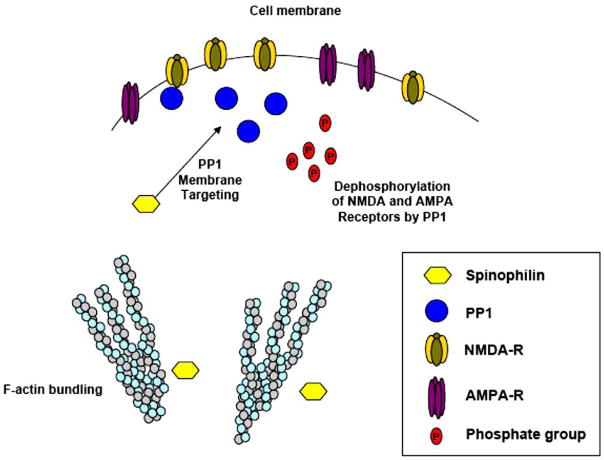
Fig. 5.
Spinophilin participates in localization of PP1 to the cell membrane, where it dephosphorylates NMDA and AMPA receptors, down-regulating their activity. The actin-bundling activity of spinophilin prevents outgrowth of filopodia-like protrusions.
Although spinophilin and neurabin I are structurally similar, are localized in dendritic spines, and bind some of the same partners, they may have variable effects on dendritic spine morphology. While the actin-bundling activity of spinophilin seems to suppress outgrowth of immature dendritic filopodia-like protrusions [240], overexpression of neurabin I in immature neuronal cultures induced these protrusions [244]. Neurite formation was also reversibly abolished by neurabin I suppression following antisense oligonucleotide treatment of rat hippocampal neurons [236]. In contrast to the effects of spinophilin depletion, knock-out of neurabin I led to impaired LTP and enhanced AMPA receptor-mediated synaptic transmission; however, LTD was not altered [245]. Neurabin I may act as a competitive inhibitor of spinophilin, resulting in opposing effects on dendritic spines and synaptic activity, as they are structurally similar and share the same binding partners.
3.2. Actin Filaments Severing and De-Polymerization
3.2.1. Cofilin Regulates Actin Filament Treadmilling
Cofilin is a member of the ADF/cofilin family of actin-depolymerizing proteins that preferentially bind to ADP-actin subunits in a minor twisted conformation of F-actin and sever filaments (Fig. 6). This increases the pool of G-actin monomers used by actin polymerizing factors and also creates free barbed ends that can nucleate filament growth [246–248]. Cofilin is expressed at high levels in the adult brain [249] and is more abundant in mammalian neurons than ADF [250]; therefore in this review, we will primarily discuss the role of cofilin in actin dynamics. Low levels of cofilin activity are detected in resting cells and contribute to F-actin depolymerization at the “pointed” ends and F-actin polymerization at the “barbed” ends, resulting in F-actin turnover at a slow rate [251]. On the other hand, enhanced cofilin activity is often detected in ruffling membranes at the leading edge of migrating cells and in neuronal growth cones, leading to fast F-actin dynamics [252–254]. The local concentration of cofilin in specific subcellular compartments determines the cofilin/actin ratio, thus influencing its effects on actin dynamics. While a low cofilin/actin ratio was shown to result in the highest severing of actin filaments, at high ratios cofilin can induce dissociation of the Arp2/3 complex and de-branching, resulting in formation of long, unbranched filaments [77, 255]. Therefore, the actions of cofilin in the cell largely depend on its localization and the level of activity [255].
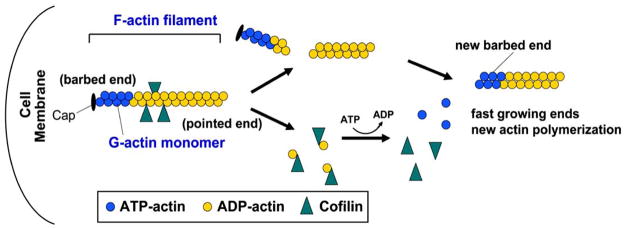
Fig. 6.
Actin-severing activity of cofilin promotes F-actin and spine remodeling.
Cofilin activity is regulated by phosphorylation. LIM kinase (LIMK) suppresses cofilin activity by phosphorylating cofilin at Serine 3, which inhibits its binding to F-actin [256, 257]. The cofilin-specific phosphatases slingshot (SSH) and chronophin (CIN) dephosphorylate and activate cofilin [258–260]. The importance of cofilin phosphorylation in synaptic structural plasticity during LTP induction and protective effects of the inactive phosphomimetic cofilinS3D mutant against Aβ-mediated spine loss have been recently reported [261, 262]. Our studies have also shown that over-expression of inactive cofilinS3D promoted stabilization of mature dendritic spines, whereas constitutively-active cofilinS3A induced remodeling and elongation of actin-rich stable dendritic spines, and extension of new filopodia-like protrusions [263]. Moreover, depletion of cofilin-1 in cultured hippocampal neurons by overexpressing cofilin-1 siRNA was shown to decrease the number of mature dendritic spines and to induce the formation of abnormal filopodia-like branches [97]. Immunoelectron microscopy studies have found that cofilin accumulates near the PSD in the “shell” area of dendritic spines, a specific region containing a dynamic F-actin pool, while avoiding the spine “core” with a stable pool of F-actin [264]. The effect of actin-remodeling activity of cofilin on dendritic spine morphology and its localization within spines suggest that cofilin may play a role in dendritic spine plasticity.
Several mechanisms that control cofilin activity in dendritic spines were recently proposed involving Rho GTPase-dependent regulation of LIMK activity (Fig. 7) [263, 265]. Members of Rho family GTPases such as RhoA, Cdc42, and Rac can promote activation of LIMK through Pak and ROCK, respectively [257, 266, 267]. Pak1 and ROCK2 were demonstrated to regulate dendritic spine properties and synaptic density through LIMK-mediated regulation of cofilin activity [268, 269]. Moreover, the Rac-Pak-LIMK pathway was suggested to contribute to SynGAP-mediated regulation of steady-state cofilin phosphorylation in dendritic spines [265]. Our studies suggest that EphB2-mediated dendritic spine stabilization relies on the ability of EphB2 receptors to activate the RhoA-ROCK-LIMK-1 pathway that works to suppress cofilin activity by phosphorylation [263]. LIMK-1 is primarily expressed in neurons and was shown to be involved in dendritic spine development. Neurons from LIMK knock out mice formed morphologically immature dendritic spines with smaller heads and postsynaptic densities compared to wild-type neurons [270], and the inhibition of LIMK translation by microRNA-134 also resulted in smaller spine heads [271]. Cofilin phosphorylation can also be regulated independently of Rho GTPases. NMDA receptor activation in cultured hippocampal neurons was shown to trigger transient dephosphorylation and activation of cofilin [265]. Ca2+-dependent changes in SSH activity were previously implicated in the regulation of cofilin activity [272]. Ca2+ influx through NMDA receptors can rapidly activate calcineurin [273], which in turn was shown to induce cofilin dephosphorylation through an upregulation of SSH activity [272]. In addition, the NMDA receptor-calcineurin pathway was shown to mediate reversible spine shrinkage in acute hippocampal slices following LTD by regulating cofilin activity [33]. While LTD induction depended on PP1, the change in spine morphology depended on cofilin, suggesting that these pathways may cooperate to regulate activity-dependent synapse pruning. In summary, the spine-remodeling activity of cofilin can be enhanced through a variety of signaling cascades that result in cofilin dephosphorylation through suppression of LIMK activity and/or SSH activation.
Fig. 7.
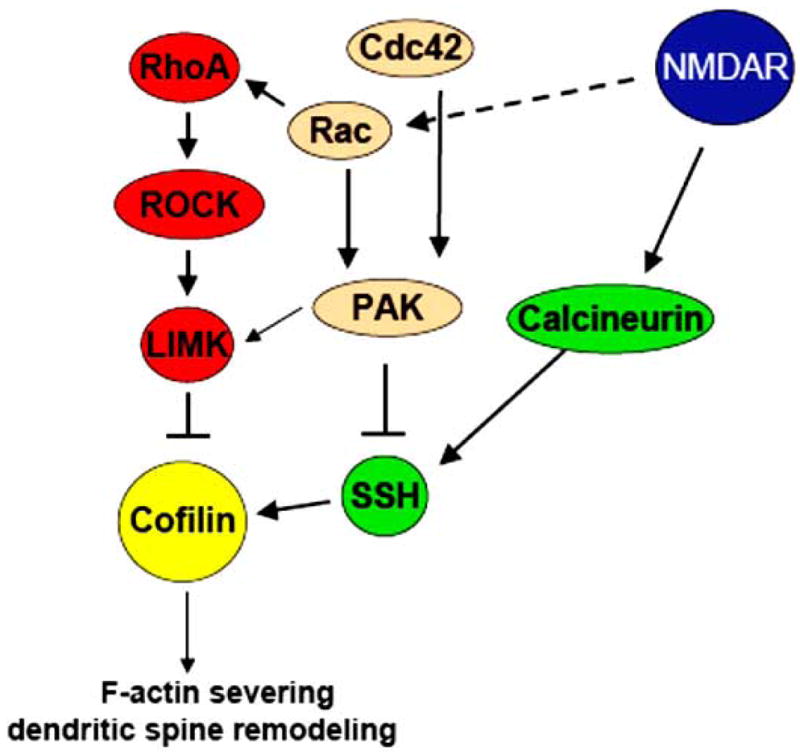
Regulatory proteins that enhance or inhibit the activity of cofilin.
Besides regulation of cofilin activity by phosphorylation, there are other mechanisms that influence the actin-severing ability of cofilin. Cofilin competes for actin binding with the actin-stabilizing protein drebrin [274]. Therefore, increased drebrin levels could lead to stabilization of actin filaments by preventing cofilin binding to actin filaments, whereas an increase in cofilin concentration could result in release of drebrin from actin filaments and enhance cofilin-mediated actin depolymerization. Cofilin-mediated actin depolymerization can also be inhibited by phosphoinositides, especially PIP2, which interacts with the actin-binding domain of cofilin and triggers its association with the plasma membrane [275, 276]. On the other hand, the EphA4 receptor was shown to regulate cofilin activity in spines through activation of phospholipase Cγ, followed by hydrolysis of PIP2 and release of active cofilin from the membrane [277]. PIP2 was also shown to inhibit cofilin’s ability to form cofilin-actin rods in myotubes [278]. In neurons, formation of cofilin-actin rods has been observed in response to neuronal stress [279, 280]. Treatment of hippocampal neurons with amyloid beta (Aβ)1-42 peptide also led to the formation of cofilin-actin rods in dendrites and axons of neurons in both dissociated hippocampal cultures [281] and organotypic hippocampal slices [280]. Similar filamentous structures were detected in brains of subjects with Alzheimer’s disease [282]. Therefore, it is possible that recruitment of a large number of cofilin molecules to actin rods would deplete the cofilin pool in dendritic spines, thus affecting dendritic spine maintenance and synaptic plasticity. Taken together, these studies indicate that cofilin is regulated by multiple mechanisms, which appear to be critical for dendritic spine maintenance and plasticity.
3.2.2. Gelsolin Severing Activity is Regulated by Ca2+
Gelsolin derives its name from the process of “gelsol” transformation of F-actin in the cortical cytoplasm of motile cells, which reversibly transitions from a highly cross-linked state (“gelation”) to a disassembled state (“solation”) in order to promote cell movement [283]. Gelsolin has a variety of functions, such as binding and severing F-actin and capping fast-growing barbed ends [284–287], as well as nucleating new filament polymerization [288]. Gelsolin binds selectively to ADP-actin [289, 290], introducing a conformational change, thus severing actin filaments [291, 292]. In combination with its capping activity, this leads to an increase in the number of pointed ends and ADP-actin dissociation. The structure of gelsolin has been delineated [293, 294], as well as its interaction with actin [295–300]. Structural provisions for gelsolin actin-severing [301–303] and polymerization activities [304] have also been well-studied in non-neuronal cells. Gelsolin is involved in the regulation of cell motility [305, 306] and phagocytosis [307], as well as inhibition of axonal transport of membranous organelles [308]. Mutated gelsolin was shown to inhibit fibroblast growth [309] and Rac-dependent motility was reduced in gelsolin-null fibroblasts [310].
Gelsolin activity can be regulated by different factors, including protein interactions and pH [311]. Tropomyosin competes with gelsolin for actin binding [312]. Moreover, phosphate analogs and activated GTP-Rac1 can reduce F-actin susceptibility to severing by promoting gelsolin dissociation from actin filaments [313, 314]. The actin severing and nucleating activities of gelsolin are inhibited by PIP2 [315, 316]. Conversely, gelsolin can promote the phosphorylation of PIP2 by activating PI3-K [317], and inhibit the hydrolysis of PIP2 through competitive substrate binding [318, 319]. This has implications for linking the cell membrane with actin dynamics through gelsolin activity. Gelsolin-actin binding is also regulated by micromolar concentrations of Ca2+ [283, 320–323]. Ca2+ was shown to promote gelsolin severing activity [320, 324], and to increase the efficiency of its capping activity [325]. By occupying the Ca2+-binding domain of gelsolin [326], Ca2+ ions induce structural changes in gelsolin [327–329].
Gelsolin is detected in the brain, spinal cord, and in cultured neurons [330], where it is implicated in plasticity. An upregulation in gelsolin mRNA [331] and gelsolin protein levels [332] was found in the rat hippocampus following entorhinal deafferentation, suggesting that gelsolin may play a role in brain remodeling following hippocampal injury. Filopodia and lamellipodia are enriched with gelsolin [333, 334], and gelsolin knock-out mice exhibit slower filopodial retraction [335], further supporting the role of gelsolin in cell-protrusive activities. Gelsolin knock-out mice exhibit increased Ca2+ influx following glutamate exposure [336] and fail to display an NMDA receptor-dependent decrease in the actin turnover rate [337]. These studies implicate gelsolin’s severing and capping activity in mediating NMDA receptor-induced spine stabilization.
Mutations in the gelsolin gene were found in subjects with familial amyloidosis [338–341]. This could partially explain the neuronal pathologies of the disease [342], including signs of de-myelination reported in these patients [343], as high levels of gelsolin are detected in myelin-forming cells and implicated in lamellipodial movement toward the axons during myelination [344–346]. Gelsolin mutations are also linked to Alzheimer’s and Parkinson’s diseases [347]. In addition, subjects with Down syndrome exhibited an increase in gelsolin levels in developing frontal cortex, and levels of gelsolin were shown to rise with aging in control subjects [348]. The ability of gelsolin to bind and sever F-actin and to cap fast-growing barbed ends, as well as to nucleate filament polymerization, supports its role in mediating actin remodeling in dendritic spines.
3.3. Myosins Influence Actin Dynamics at Synapses
Although F-actin assembly/disassembly determines actin dynamics, myosin motors posses ATPase activity and can also influence actin dynamics in dendritic spines. The myosin II, V, and VI isoforms were found in dendritic spines [349], and are suggested to regulate dendritic spine shape and synaptic plasticity, at least in part through interaction with actin and modulation of NMDA and AMPA receptor membrane insertion and function. Actin-myosin interaction is known to regulate dendritic spine shape and is inhibited by drebrin, which is enriched in mature dendritic spines [174, 178]. Although myosin IIB immunoreactivity was identified throughout neuronal cell bodies and various dendritic domains that lack drebrin immunoreactivity, a small pool of myosin IIB was also detected within the drebrin-positive fraction of F-actin in dendritic spines [350]. Drebrin overexpression, on the other hand, has been shown to elongate dendritic spines, so it may function here to inhibit actin-myosin contractility [174]. Myosin motors have been suggested to contribute to the formation of mature short mushroom-shaped and stubby spines. In support of this, myosin IIB depletion with RNAi causes a decrease in the number of short mushroom-shaped spines and an increase in long filopodia-like protrusions [351].
Myosins can also interact with NR1 and NR2 subunits of NMDA receptors through the myosin regulatory light chain [352] and control the trafficking of the GluR1 subunit of AMPA receptors from the dendritic shaft to dendritic spines in a Ca2+-dependent manner [353]. Myosin Vb associates with recycling endosomes and mobilizes them for AMPA receptor insertion and spine growth upon LTP induction, whereas depletion of myosin Vb using RNAi prevents LTP-induced AMPA receptor insertion into the postsynaptic membrane and spine growth [354]. Further, the PSD-95 interacting protein guanylate kinase domain-associated protein (GKAP) interacts with the dynein light chain (DLC) of myosin V, which may implicate myosin V in NMDA receptor regulation [355].
Myosin VI is also enriched in the brain and localized to PSDs in synapses. Spine loss was seen upon myosin VI disruption through the expression of a dominant-negative mutant or in hippocampal cultures of myosin VI deficient mice [356]. In addition, increase in AMPA receptor internalization following AMPA or insulin stimulation was not observed in myosin VI deficient neurons [356]. Myosin VI promotes the formation of mature spines and synapses, but also allows for LTP termination by receptor internalization. Although myosin is perhaps best studied for its role in generation of contractile forces in non-neuronal cells, myosin II, V, and VI also exhibit important roles in promoting mature dendritic spine morphology and regulating synaptic plasticity through NMDA and AMPA receptor trafficking at synapses.
4. INTEGRATION OF ACTIN-BINDING PROTEINS IN THE REGULATION OF DENDRITIC SPINES AND SYNAPSES
There are many actin-binding proteins within the cell that directly regulate actin polymerization, branching, cross-linking and bundling, or severing and depolymerization. By affecting actin dynamics, actin-binding proteins have the ability to form, re-arrange, stabilize, or remodel dendritic spines. These proteins also aid in clustering postsynaptic proteins and in regulating neurotransmitter receptor activities to create functional synaptic connections. Some actin-regulatory proteins like profilin have multiple functions in dendritic spines and pose opposing effects on actin, depending on several factors and conditions, such as its subcellular localization, Ca2+ levels, interactions with other proteins, and phosphorylation state. Other proteins such as spinophilin and neurabin I are similar in structure and activity, but exhibit opposing effects on dendritic spines through yet unknown mechanisms. Arp2/3 and cortactin work together, each to promote the activity of the other, contributing to actin polymerization. While α-actinin elongates dendritic spines, myosins induce the formation of short mushroom-shaped spines by enhancing actomyosin contractility. Meanwhile, ADF/cofilin and gelsolin work by severing F-actin filaments, which can promote spine elongation and remodeling by increasing actin filament turnover, or lead to spine stabilization through gelsolin capping activity.
These actin-binding proteins are regulated by signaling cascades that are initiated at the cell surface through trans-synaptic interactions, neuronglia communications, and contacts with the extracellular matrix [5]. Cell surface receptors, such as glutamate receptors, EphB receptors and ephrins, neuroligins and neurexins, integrins, cell adhesion molecules, growth factor receptors, and some proteoglycans, mediate these interactions linking extracellular events to the actin cytoskeleton in dendritic spines by initiating cytoplasmic signaling cascades. Cytoplasmic signaling proteins such as PIP2 exhibit diverse control through a variety of mechanisms, from promoting α-actinin activity through direct binding, to activating Arp2/3 through WASP, to dissociating the profilactin complex from actin. The Rho family of small GTPases, such as Rac1, Cdc42, and RhoA, are also key regulators of actin-binding proteins in dendritic spines and play an important role in dendritic spine formation, maintenance, and remodeling. In addition to the structural role of actin in dendritic spines, actin assembly was recently suggested to influence neuronal motility through a regulation of gene transcription [357]. This may have implications for a role of actin-binding proteins in regulating gene transcription and protein synthesis in spines.
Dendritic spine dynamics have been implicated in processes of learning and memory [358, 359], and abnormalities in the shape and number of spines are seen in neurodegenerative diseases, as well as some forms of mental retardation and autistic spectrum disorders [41, 42]. While the role of various actin-binding proteins in regulating spine dynamics has been studied, a challenge for future research is to understand the interplay between different actin-regulating factors under an array of physiological and pathological conditions.
Acknowledgments
The authors thank members of the laboratories of Drs. Iryna Ethell and Douglas Ethell for insightful discussions and comments. The work in the authors’ laboratory is supported by grants from the NIMH, the FRAXA Foundation, and the Department of Defense.
ABBREVIATIONS
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid
- ARF6
ADP ribosylation factor 6
- Arp2/3
Actin-Related Protein
- BDNF
brain-derived neurotrophic factor
- CaMKII
Ca2+ calmodulin-dependent protein kinase II
- CIN
chronophin
- dFMRP
Drosophila Fragile X mental retardation protein
- DLC
dynein light chain
- F-actin
filamentous actin
- FAK
focal adhesion kinase
- G-actin
globular actin
- GKAP
guanylate kinase domain-associated protein
- Htt
Huntingtin
- LIMK
LIM kinase
- LTD
long-term depression
- LTP
long-term potentiation
- MAP
mitogen-activated protein
- MAPKPK2
MAPK-activated protein kinase 2
- mGluR
metabotropic glutamate receptors
- NMDA
N-methyl-D-aspartate
- N-WASP
Neural-WASP
- PAK
p21-activated kinase
- PI3-K
phosphatidylinositol 3-kinase
- PIP2
Phosphatidylinositol 4,5 bisphosphate
- PKA
protein kinase A
- PKC
protein kinase C
- PLC
phospholipase C
- PP1
protein phosphatase 1
- PSD
postsynaptic density
- REMS
Rapid Eye Movement Sleep
- RNAi
RNA interference
- ROCK
Rho-kinase
- siRNA
small-interfering RNA
- SPAR
Spine-Associated Rap GTPase-activating protein
- SSH
slingshot
- TLCN
telenceplalin
- WASP
Wiskott-Aldrich Syndrome protein
- WAVE
WASP-family verprolin-homologous protein
Footnotes
This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
References
- 1.Rao A, Craig AM. Signaling between the actin cytoskeleton and the postsynaptic density of dendritic spines. Hippocampus. 2000;10:527–41. doi: 10.1002/1098-1063(2000)10:5<527::AID-HIPO3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 2.Sorra KE, Harris KM. Overview on the structure, composition, function, development, and plasticity of hippocampal dendritic spines. Hippocampus. 2000;10:501–11. doi: 10.1002/1098-1063(2000)10:5<501::AID-HIPO1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 3.Hering H, Sheng M. Dendritic spines: structure, dynamics and regulation. Nat Rev Neurosci. 2001;2:880–8. doi: 10.1038/35104061. [DOI] [PubMed] [Google Scholar]
- 4.Yuste R, Bonhoeffer T. Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat Rev Neurosci. 2004;5:24–34. doi: 10.1038/nrn1300. [DOI] [PubMed] [Google Scholar]
- 5.Ethell IM, Pasquale EB. Molecular mechanisms of dendritic spine development and remodeling. Prog Neurobiol. 2005;75:161–205. doi: 10.1016/j.pneurobio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Ziv NE, Smith SJ. Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron. 1996;17:91–102. doi: 10.1016/s0896-6273(00)80283-4. [DOI] [PubMed] [Google Scholar]
- 7.Dailey ME, Smith SJ. The dynamics of dendritic structure in developing hippocampal slices. J Neurosci. 1996;16:2983–94. doi: 10.1523/JNEUROSCI.16-09-02983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maletic-Savatic M, Malinow R, Svoboda K. Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science. 1999;283:1923–7. doi: 10.1126/science.283.5409.1923. [DOI] [PubMed] [Google Scholar]
- 9.Chen BE, Lendvai B, Nimchinsky EA, Burbach B, Fox K, Svoboda K. Imaging high-resolution structure of GFP-expressing neurons in neocortex in vivo. Learn Mem. 2000;7:433–41. doi: 10.1101/lm.32700. [DOI] [PubMed] [Google Scholar]
- 10.Marrs GS, Green SH, Dailey ME. Rapid formation and remodeling of postsynaptic densities in developing dendrites. Nat Neurosci. 2001;4:1006–13. doi: 10.1038/nn717. [DOI] [PubMed] [Google Scholar]
- 11.Okabe S, Miwa A, Okado H. Spine formation and correlated assembly of presynaptic and postsynaptic molecules. J Neurosci. 2001;21:6105–14. doi: 10.1523/JNEUROSCI.21-16-06105.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trachtenberg JT, Chen BE, Knott GW, et al. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–94. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- 13.Portera-Cailliau C, Pan DT, Yuste R. Activity-regulated dynamic behavior of early dendritic protrusions: evidence for different types of dendritic filopodia. J Neurosci. 2003;23:7129–42. doi: 10.1523/JNEUROSCI.23-18-07129.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziv NE, Garner CC. Cellular and molecular mechanisms of presynaptic assembly. Nat Rev Neurosci. 2004;5:385–99. doi: 10.1038/nrn1370. [DOI] [PubMed] [Google Scholar]
- 15.Knott GW, Holtmaat A, Wilbrecht L, Welker E, Svoboda K. Spine growth precedes synapse formation in the adult neocortex in vivo. Nat Neurosci. 2006;9:1117–24. doi: 10.1038/nn1747. [DOI] [PubMed] [Google Scholar]
- 16.Matus A. Growth of dendritic spines: a continuing story. Curr Opin Neurobiol. 2005;15:67–72. doi: 10.1016/j.conb.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Lippman J, Dunaevsky A. Dendritic spine morphogenesis and plasticity. J Neurobiol. 2005;64:47–57. doi: 10.1002/neu.20149. [DOI] [PubMed] [Google Scholar]
- 18.Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16:95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–6. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang C, Barco A, Zablow L, Kandel ER, Siegelbaum SA, Zakharenko SS. Transient expansion of synaptically connected dendritic spines upon induction of hippocampal long-term potentiation. Proc Natl Acad Sci USA. 2004;101:16665–70. doi: 10.1073/pnas.0407581101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nimchinsky EA, Yasuda R, Oertner TG, Svoboda K. The number of glutamate receptors opened by synaptic stimulation in single hippocampal spines. J Neurosci. 2004;24:2054–64. doi: 10.1523/JNEUROSCI.5066-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashby MC, Maier SR, Nishimune A, Henley JM. Lateral diffusion drives constitutive exchange of AMPA receptors at dendritic spines and is regulated by spine morphology. J Neurosci. 2006;26:7046–55. doi: 10.1523/JNEUROSCI.1235-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuzaki M, Ellis-Davies GC, Nemoto T, Miyashita Y, Iino M, Kasai H. Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat Neurosci. 2001;4:1086–92. doi: 10.1038/nn736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy VN, Schikorski T, Stevens CF, Zhu Y. Inactivity produces increases in neurotransmitter release and synapse size. Neuron. 2001;32:673–82. doi: 10.1016/s0896-6273(01)00500-1. [DOI] [PubMed] [Google Scholar]
- 25.Smith MA, Ellis-Davies GCR, Magee JG. Mechanism of the distance-dependent scaling of Schaffer collateral synapse in CA1 pyramidal neurons. J Physiol. 2003;548:245–58. doi: 10.1113/jphysiol.2002.036376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicholson DA, Trana R, Katz Y, Kath WL, Spruston N, Geinisman Y. Distance-dependent differences in synapse number and AMPA receptor expression in hippocampal CA1 pyramidal neurons. Neuron. 2006;50:431–42. doi: 10.1016/j.neuron.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 27.Takumi Y, Ramírez-León V, Laake P, Rinvik E, Ottersen OP. Different modes of expression of AMPA and NMDA receptors in hippocampal synapses. Nat Neurosci. 1999;2:618–24. doi: 10.1038/10172. [DOI] [PubMed] [Google Scholar]
- 28.Racca C, Stephenson FA, Streit P, Roberts JD, Somogyi P. NMDA receptor content of synapses in stratum radiatum of the hippocampal CA1 area. J Neurosci. 2000;20:2512–22. doi: 10.1523/JNEUROSCI.20-07-02512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schikorski T, Stevens CF. Quantitative ultrastructural analysis of hippocampal excitatory synapses. J Neurosci. 1997;17:5858–67. doi: 10.1523/JNEUROSCI.17-15-05858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murthy VN, Sejnowski TJ, Stevens CF. Heterogeneous release properties of visualized individual hippocampal synapses. Neuron. 1997;18:599–612. doi: 10.1016/s0896-6273(00)80301-3. [DOI] [PubMed] [Google Scholar]
- 31.Llinás R, Sugimori M, Silver RB. Microdomains of high calcium concentration in a presynaptic terminal. Science. 1992;256:677–9. doi: 10.1126/science.1350109. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka J, Horiike Y, Matsuzaki M, Miyazaki T, Ellis-Davies GC, Kasai H. Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science. 2008;319:1683–7. doi: 10.1126/science.1152864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Q, Homma KJ, Poo MM. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44:749–57. doi: 10.1016/j.neuron.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 34.Engert F, Bonhoeffer T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature. 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- 35.Sorra KE, Harris KM. Stability in synapse number and size at 2 hr after long-term potentiation in hippocampal area CA1. J Neurosci. 1998;18:658–71. doi: 10.1523/JNEUROSCI.18-02-00658.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lendvai B, Stern EA, Chen B, Svoboda K. Experience-dependent plasticity of dendritic spines in the developing rat barrel cortex in vivo. Nature. 2000;404:876–81. doi: 10.1038/35009107. [DOI] [PubMed] [Google Scholar]
- 37.Kleim JA, Barbay S, Nudo RJ. Functional reorganization of the rat motor cortex following motor skill learning. J Neurophysiol. 1998;80:3321–5. doi: 10.1152/jn.1998.80.6.3321. [DOI] [PubMed] [Google Scholar]
- 38.Leuner B, Falduto J, Shors TJ. Associative memory formation increases the observation of dendritic spines in the hippocampus. J Neurosci. 2003;23:659–65. doi: 10.1523/JNEUROSCI.23-02-00659.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geinisman Y, Disterhoft JF, Gundersen HJ, et al. Remodeling of hippocampal synapses after hippocampus-dependent associative learning. J Comp Neurol. 2000;417:49–59. [PubMed] [Google Scholar]
- 40.Rudelli RD, Brown WT, Wisniewski K, et al. Adult fragile X syndrome. Clinico-neuropathologic findings. Acta Neuropathol. 1985;67:289–95. doi: 10.1007/BF00687814. [DOI] [PubMed] [Google Scholar]
- 41.Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10:981–91. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- 42.Halpain S, Spencer K, Graber S. Dynamics and pathology of dendritic spines. Prog Brain Res. 2005;147:29–37. doi: 10.1016/S0079-6123(04)47003-4. [DOI] [PubMed] [Google Scholar]
- 43.Cotman CW, Nieto-Sampedro M. Cell biology of synaptic plasticity. Science. 1984;225:1287–94. doi: 10.1126/science.6382610. [DOI] [PubMed] [Google Scholar]
- 44.Chen JR, Wang TJ, Huang HY, et al. Fatigue reversibly reduced cortical and hippocampal dendritic spines concurrent with compromise of motor endurance and spatial memory. Neuroscience. 2009;161:1104–13. doi: 10.1016/j.neuroscience.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 45.Pollard TD, Blanchoin L, Mullins RD. Actin dynamics. J Cell Sci. 2001;114:3–4. doi: 10.1242/jcs.114.1.3. [DOI] [PubMed] [Google Scholar]
- 46.Lowery LA, Van Vactor D. The trip of the tip: understanding the growth cone machinery. Nat Rev Mol Cell Biol. 2009;10:332–43. doi: 10.1038/nrm2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smart FM, Halpain S. Regulation of dendritic spine stability. Hippocampus. 2000;10:542–54. doi: 10.1002/1098-1063(2000)10:5<542::AID-HIPO4>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 48.Kolb B, Cioe J, Comeau W. Contrasting effects of motor and visual spatial learning tasks on dendritic arborization and spine density in rats. Neurobiol Learn Mem. 2008;90:295–300. doi: 10.1016/j.nlm.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 49.Woodrum DT, Rich SA, Pollard TD. Evidence for biased bidirectional polymerization of actin filaments using heavy meromyosin prepared by an improved method. J Cell Biol. 1975;67:231–7. doi: 10.1083/jcb.67.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kirschner MW. Implications of treadmilling for the stability and polarity of actin and tubulin polymers in vivo. J Cell Biol. 1980;86:330–4. doi: 10.1083/jcb.86.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stossel TP. On the crawling of animal cells. Science. 1993;260:1086–94. doi: 10.1126/science.8493552. [DOI] [PubMed] [Google Scholar]
- 52.Bindschadler M, Osborn EA, Dewey CF, Jr, McGrath JL. A mechanistic model of the actin cycle. Biophys J. 2004;86:2720–39. doi: 10.1016/S0006-3495(04)74326-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okamoto K, Nagai T, Miyawaki A, Hayashi Y. Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat Neurosci. 2004;7:1104–12. doi: 10.1038/nn1311. [DOI] [PubMed] [Google Scholar]
- 54.Zhang W, Benson DL. Development and molecular organization of dendritic spines and their synapses. Hippocampus. 2000;10:512–26. doi: 10.1002/1098-1063(2000)10:5<512::AID-HIPO2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 55.Sekino Y, Kojima N, Shirao T. Role of actin cytoskeleton in dendritic spine morphogenesis. Neurochem Int. 2007;51:92–104. doi: 10.1016/j.neuint.2007.04.029. [DOI] [PubMed] [Google Scholar]
- 56.Sala C, Cambianica I, Rossi F. Molecular mechanisms of dendritic spine development and maintenance. Acta Neurobiol Exp (Wars) 2008;68:289–304. doi: 10.55782/ane-2008-1696. [DOI] [PubMed] [Google Scholar]
- 57.Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol. 1997;138:375–84. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gournier H, Goley ED, Niederstrasser H, Trinh T, Welch MD. Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol Cell. 2001;8:1041–52. doi: 10.1016/s1097-2765(01)00393-8. [DOI] [PubMed] [Google Scholar]
- 59.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–77. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 60.Mullins RD, Stafford WF, Pollard TD. Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. J Cell Biol. 1997;136:331–43. doi: 10.1083/jcb.136.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pantaloni D, Boujemaa R, Didry D, Gounon P, Carlier MF. The Arp2/3 complex branches filament barbed ends: functional antagonism with capping proteins. Nat Cell Biol. 2000;2:385–91. doi: 10.1038/35017011. [DOI] [PubMed] [Google Scholar]
- 62.Amann KJ, Pollard TD. The Arp2/3 complex nucleates actin filament branches from the sides of pre-existing filaments. Nat Cell Biol. 2001;3:306–10. doi: 10.1038/35060104. [DOI] [PubMed] [Google Scholar]
- 63.Schmidt KL, Marcus-Gueret N, Adeleye A, Webber J, Baillie D, Stringham EG. The cell migration molecule UNC-53/NAV2 is linked to the ARP2/3 complex by ABI-1. Development. 2009;136:563–74. doi: 10.1242/dev.016816. [DOI] [PubMed] [Google Scholar]
- 64.Machesky LM, Reeves E, Wientjes F, et al. Mammalian actin-related protein 2/3 complex localizes to regions of lamellipodial protrusion and is composed of evolutionarily conserved proteins. Biochem J. 1997;328:105–12. doi: 10.1042/bj3280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bailly M, Ichetovkin I, Grant W, et al. The F-actin side binding activity of the Arp2/3 complex is essential for actin nucleation and lamellipod extension. Curr Biol. 2001;11:620–5. doi: 10.1016/s0960-9822(01)00152-x. [DOI] [PubMed] [Google Scholar]
- 66.Rácz B, Weinberg RJ. Organization of the Arp2/3 complex in hippocampal spines. J Neurosci. 2008;28:5654–9. doi: 10.1523/JNEUROSCI.0756-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laurent V, Loisel TP, Harbeck B, et al. Role of proteins of the Ena/VASP family in actin-based motility of Listeria monocytogenes. J Cell Biol. 1999;144:1245–58. doi: 10.1083/jcb.144.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goldberg DJ, Foley MS, Tang D, Grabham PW. Recruitment of the Arp2/3 complex and mena for the stimulation of actin polymerization in growth cones by nerve growth factor. J Neurosci Res. 2000;60:458–67. doi: 10.1002/(SICI)1097-4547(20000515)60:4<458::AID-JNR4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 69.Qualmann B, Kessels MM, Kelly RB. Molecular links between endocytosis and the actin cytoskeleton. J Cell Biol. 2000;150:F111–6. doi: 10.1083/jcb.150.5.f111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lynch DK, Winata SC, Lyons RJ, et al. A Cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J Biol Chem. 2003;278:21805–13. doi: 10.1074/jbc.M211407200. [DOI] [PubMed] [Google Scholar]
- 71.DeMali KA, Barlow CA, Burridge K. Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion. J Cell Biol. 2002;159:881–91. doi: 10.1083/jcb.200206043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.May RC, Caron E, Hall A, Machesky LM. Involvement of the Arp2/3 complex in phagocytosis mediated by FcgammaR or CR3. Nat Cell Biol. 2000;2:246–8. doi: 10.1038/35008673. [DOI] [PubMed] [Google Scholar]
- 73.Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol. 2001;11:1300–4. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- 74.Le Clainche C, Pantaloni D, Carlier MF. ATP hydrolysis on actin-related protein 2/3 complex causes debranching of dendritic actin arrays. Proc Natl Acad Sci USA. 2003;100:6337–42. doi: 10.1073/pnas.1130513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ressad F, Didry D, Egile C, Pantaloni D, Carlier MF. Control of actin filament length and turnover by actin depolymerizing factor (ADF/cofilin) in the presence of capping proteins and ARP2/3 complex. J Biol Chem. 1999;274:20970–6. doi: 10.1074/jbc.274.30.20970. [DOI] [PubMed] [Google Scholar]
- 76.Weaver AM, Karginov AV, Kinley AW, et al. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr Biol. 2001;11:370–4. doi: 10.1016/s0960-9822(01)00098-7. [DOI] [PubMed] [Google Scholar]
- 77.Chan C, Beltzner CC, Pollard TD. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr Biol. 2009;19:537–45. doi: 10.1016/j.cub.2009.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ichetovkin I, Grant W, Condeelis J. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr Biol. 2002;12:79–84. doi: 10.1016/s0960-9822(01)00629-7. [DOI] [PubMed] [Google Scholar]
- 79.Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–26. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.LeClaire LL, 3rd, Baumgartner M, Iwasa JH, Mullins RD, Barber DL. Phosphorylation of the Arp2/3 complex is necessary to nucleate actin filaments. J Cell Biol. 2008;182:647–54. doi: 10.1083/jcb.200802145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Singh S, Powell DW, Rane MJ, et al. Identification of the p16-Arc subunit of the Arp 2/3 complex as a substrate of MAPK-activated protein kinase 2 by proteomic analysis. J Biol Chem. 2003;278:36410–7. doi: 10.1074/jbc.M306428200. [DOI] [PubMed] [Google Scholar]
- 82.Vadlamudi RK, Li F, Barnes CJ, Bagheri-Yarmand R, Kumar R. p41-Arc subunit of human Arp2/3 complex is a p21-activated kinase-1-interacting substrate. EMBO Rep. 2004;5:154–60. doi: 10.1038/sj.embor.7400079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 84.Zalevsky J, Lempert L, Kranitz H, Mullins RD. Different WASP family proteins stimulate different Arp2/3 complex-dependent actin-nucleating activities. Curr Biol. 2001;11:1903–13. doi: 10.1016/s0960-9822(01)00603-0. [DOI] [PubMed] [Google Scholar]
- 85.Ma L, Rohatgi R, Kirschner MW. The Arp2/3 complex mediates actin polymerization induced by the small GTP-binding protein Cdc42. Proc Natl Acad Sci USA. 1998;95:15362–7. doi: 10.1073/pnas.95.26.15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mullins RD, Pollard TD. Rho-family GTPases require the Arp2/3 complex to stimulate actin polymerization in Acanthamoeba extracts. Curr Biol. 1999;9:405–15. doi: 10.1016/s0960-9822(99)80187-0. [DOI] [PubMed] [Google Scholar]
- 87.Rohatgi R, Ma L, Miki H, et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–31. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 88.Higgs HN, Pollard TD. Activation by Cdc42 and PIP(2) of Wiskott-Aldrich syndrome protein (WASp) stimulates actin nucleation by Arp2/3 complex. J Cell Biol. 2000;150:1311–20. doi: 10.1083/jcb.150.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Olazabal IM, Machesky LM. Abp1p and cortactin, new “handholds” for actin. J Cell Biol. 2001;154:679–82. doi: 10.1083/jcb.200105061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goode BL, Rodal AA, Barnes G, Drubin DG. Activation of the Arp2/3 complex by the actin filament binding protein Abp1p. J Cell Biol. 2001;153:627–34. doi: 10.1083/jcb.153.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Haeckel A, Ahuja R, Gundelfinger ED, Qualmann B, Kessels MM. The actin-binding protein Abp1 controls dendritic spine morphology and is important for spine head and synapse formation. J Neurosci. 2008;28:10031–44. doi: 10.1523/JNEUROSCI.0336-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Irie F, Yamaguchi Y. EphB receptors regulate dendritic spine development via intersectin, Cdc42 and N-WASP. Nat Neurosci. 2002;5:1117–8. doi: 10.1038/nn964. [DOI] [PubMed] [Google Scholar]
- 93.Wegner AM, Nebhan CA, Hu L, et al. N-wasp and the arp2/3 complex are critical regulators of actin in the development of dendritic spines and synapses. J Biol Chem. 2008;283:15912–20. doi: 10.1074/jbc.M801555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim Y, Sung JY, Ceglia I, et al. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature. 2006;442:814–7. doi: 10.1038/nature04976. [DOI] [PubMed] [Google Scholar]
- 95.Soderling SH, Langeberg LK, Soderling JA, et al. Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc Natl Acad Sci USA. 2003;100:1723–8. doi: 10.1073/pnas.0438033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Soderling SH, Guire ES, Kaech S, et al. A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J Neurosci. 2007;27:355–65. doi: 10.1523/JNEUROSCI.3209-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hotulainen P, Llano O, Smirnov S, et al. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol. 2009;185:323–39. doi: 10.1083/jcb.200809046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Uruno T, Liu J, Zhang P, et al. Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat Cell Biol. 2001;3:259–266. doi: 10.1038/35060051. [DOI] [PubMed] [Google Scholar]
- 99.Wu H, Parsons JT. Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J Cell Biol. 1993;120:1417–26. doi: 10.1083/jcb.120.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ammer AG, Weed SA. Cortactin branches out: roles in regulating protrusive actin dynamics. Cell Motil Cytoskeleton. 2008;65:687–707. doi: 10.1002/cm.20296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weed SA, Karginov AV, Schafer DA, et al. Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J Cell Biol. 2000;151:29–40. doi: 10.1083/jcb.151.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kaksonen M, Peng HB, Rauvala H. Association of cortactin with dynamic actin in lamellipodia and on endosomal vesicles. J Cell Sci. 2000;113:4421–6. doi: 10.1242/jcs.113.24.4421. [DOI] [PubMed] [Google Scholar]
- 103.Zheng G, Solinet S, Pelletier RM, Vitale ML. Actin cytoskeleton remodelling in the anterior pituitary folliculostellate cell line TtT/GF: participation of the actin-binding protein cortactin. J Mol Histol. 2005;36:461–74. doi: 10.1007/s10735-006-9021-1. [DOI] [PubMed] [Google Scholar]
- 104.Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta. 2007;1773:642–52. doi: 10.1016/j.bbamcr.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Buday L, Downward J. Roles of cortactin in tumor pathogenesis. Biochim Biophys Acta. 2007;1775:263–73. doi: 10.1016/j.bbcan.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 106.Rácz B, Weinberg RJ. The subcellular organization of cortactin in hippocampus. J Neurosci. 2004;24:10310–7. doi: 10.1523/JNEUROSCI.2080-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hering H, Sheng M. Activity-dependent redistribution and essential role of cortactin in dendritic spine morphogenesis. J Neurosci. 2003;23:11759–69. doi: 10.1523/JNEUROSCI.23-37-11759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gray NW, Kruchten AE, Chen J, McNiven MA. A dynamin-3 spliced variant modulates the actin/cortactin-dependent morphogenesis of dendritic spines. J Cell Sci. 2005;118:1279–90. doi: 10.1242/jcs.01711. [DOI] [PubMed] [Google Scholar]
- 109.Matus A. Actin-based plasticity in dendritic spines. Science. 2000;290:754–8. doi: 10.1126/science.290.5492.754. [DOI] [PubMed] [Google Scholar]
- 110.Jaworski J, Kapitein LC, Gouveia SM, et al. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron. 2009;61:85–100. doi: 10.1016/j.neuron.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 111.Gunst SJ. Actions by actin: reciprocal regulation of cortactin activity by tyrosine kinases and F-actin. Biochem J. 2004;380:e7–8. doi: 10.1042/BJ20040559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lua BL, Low BC. Cortactin phosphorylation as a switch for actin cytoskeletal network and cell dynamics control. FEBS Lett. 2005;579:577–85. doi: 10.1016/j.febslet.2004.12.055. [DOI] [PubMed] [Google Scholar]
- 113.Martinez-Quiles N, Ho HY, Kirschner MW, Ramesh N, Geha RS. Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP. Mol Cell Biol. 2004;24:5269–80. doi: 10.1128/MCB.24.12.5269-5280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Martinez MC, Ochiishi T, Majewski M, Kosik KS. Dual regulation of neuronal morphogenesis by a delta-catenin-cortactin complex and Rho. J Cell Biol. 2003;162:99–111. doi: 10.1083/jcb.200211025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Naisbitt S, Kim E, Tu JC, et al. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–82. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 116.Tu JC, Xiao B, Naisbitt S, et al. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–92. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- 117.Meighan SE, Meighan PC, Choudhury P, et al. Effects of extracellular matrix-degrading proteases matrix metalloproteinases 3 and 9 on spatial learning and synaptic plasticity. J Neurochem. 2006;96:1227–41. doi: 10.1111/j.1471-4159.2005.03565.x. [DOI] [PubMed] [Google Scholar]
- 118.Davis CJ, Meighan PC, Taishi P, et al. REM sleep deprivation attenuates actin-binding protein cortactin: a link between sleep and hippocampal plasticity. Neurosci Lett. 2006;400:191–6. doi: 10.1016/j.neulet.2006.02.046. [DOI] [PubMed] [Google Scholar]
- 119.Iki J, Inoue A, Bito H, Okabe S. Bi-directional regulation of postsynaptic cortactin distribution by BDNF and NMDA receptor activity. Eur J Neurosci. 2005;22:2985–94. doi: 10.1111/j.1460-9568.2005.04510.x. [DOI] [PubMed] [Google Scholar]
- 120.Kohara K, Kitamura A, Morishima M, Tsumoto T. Activity-dependent transfer of brain-derived neurotrophic factor to postsynaptic neurons. Science. 2001;291:2419–23. doi: 10.1126/science.1057415. [DOI] [PubMed] [Google Scholar]
- 121.Zafra F, Hengerer B, Leibrock J, Thoenen H, Lindholm D. Activity dependent regulation of BDNF and NGF mRNAs in the rat hippocampus is mediated by non-NMDA glutamate receptors. EMBO J. 1990;9:3545–50. doi: 10.1002/j.1460-2075.1990.tb07564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Grazi E, Trombetta G, Guidoboni M. Binding of alpha-actinin to F-actin or to tropomyosin F-actin is a function of both alpha-actinin concentration and gel structure. J Muscle Res Cell Motil. 1991;12:579–84. doi: 10.1007/BF01738446. [DOI] [PubMed] [Google Scholar]
- 123.Sjöblom B, Salmazo A, Djinovic-Carugo K. Alpha-actinin structure and regulation. Cell Mol Life Sci. 2008;65:2688–701. doi: 10.1007/s00018-008-8080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hemmings L, Kuhlman PA, Critchley DR. Analysis of the actin-binding domain of alpha-actinin by mutagenesis and demonstration that dystrophin contains a functionally homologous domain. J Cell Biol. 1992;116:1369–80. doi: 10.1083/jcb.116.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Djinovic-Carugo K, Young P, Gautel M, Saraste M. Structure of the alpha-actinin rod: molecular basis for cross-linking of actin filaments. Cell. 1999;98:537–46. doi: 10.1016/s0092-8674(00)81981-9. [DOI] [PubMed] [Google Scholar]
- 126.Ylänne J, Scheffzek K, Young P, Saraste M. Crystal structure of the alpha-actinin rod reveals an extensive torsional twist. Structure. 2001;9:597–604. doi: 10.1016/s0969-2126(01)00619-0. [DOI] [PubMed] [Google Scholar]
- 127.Flood G, Kahana E, Gilmore AP, Rowe AJ, Gratzer WB, Critchley DR. Association of structural repeats in the alpha-actinin rod domain: alignment of inter-subunit interactions. J Mol Biol. 1995;252:227–34. doi: 10.1006/jmbi.1995.0490. [DOI] [PubMed] [Google Scholar]
- 128.Golji J, Collins R, Mofrad MR. Molecular mechanics of the alpha-actinin rod domain: bending, torsional, and extensional behavior. PLoS Comput Biol. 2009;5:e1000389. doi: 10.1371/journal.pcbi.1000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wachsstock DH, Schwartz WH, Pollard TD. Affinity of alpha-actinin for actin determines the structure and mechanical properties of actin filament gels. Biophys J. 1993;65:205–14. doi: 10.1016/S0006-3495(93)81059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ebashi S, Ebashi F. A new protein factor promoting contraction of actomyosin. Nature. 1964;203:645–6. doi: 10.1038/203645a0. [DOI] [PubMed] [Google Scholar]
- 131.Maruyama K, Kimura S. Does alpha-actinin promote contraction of actomyosin fibers? J Biochem. 1971;69:983–5. doi: 10.1093/oxfordjournals.jbchem.a129550. [DOI] [PubMed] [Google Scholar]
- 132.Minetti C, Ricci E, Bonilla E. Progressive depletion of fast alpha-actinin-positive muscle fibers in Duchenne muscular dystrophy. Neurology. 1991;41:1977–81. doi: 10.1212/wnl.41.12.1977. [DOI] [PubMed] [Google Scholar]
- 133.Domazetovska A, Ilkovski B, Cooper ST, et al. Mechanisms underlying intranuclear rod formation. Brain. 2007;130:3275–84. doi: 10.1093/brain/awm247. [DOI] [PubMed] [Google Scholar]
- 134.Roulier EM, Fyrberg C, Fyrberg E. Perturbations of Drosophila alpha-actinin cause muscle paralysis, weakness, and atrophy but do not confer obvious nonmuscle phenotypes. J Cell Biol. 1992;116:911–22. doi: 10.1083/jcb.116.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Drenckhahn D, Franz H. Identification of actin-, alpha-actinin-, and vinculin-containing plaques at the lateral membrane of epithelial cells. J Cell Biol. 1986;102:1843–52. doi: 10.1083/jcb.102.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yürüker B, Niggli V. Alpha-actinin and vinculin in human neutrophils: reorganization during adhesion and relation to the actin network. J Cell Sci. 1992;101:403–14. doi: 10.1242/jcs.101.2.403. [DOI] [PubMed] [Google Scholar]
- 137.Gotlieb AI, Heggeness MH, Ash JF, Singer SJ. Mechanochemical proteins, cell motility and cell-cell contacts: the localization of mechanochemical proteins inside cultured cells at the edge of an in vitro “wound”. J Cell Physiol. 1979;100:563–78. doi: 10.1002/jcp.1041000318. [DOI] [PubMed] [Google Scholar]
- 138.Sobue K, Kanda K. Alpha-actinins, calspectin (brain spectrin or fodrin), and actin participate in adhesion and movement of growth cones. Neuron. 1989;3:311–9. doi: 10.1016/0896-6273(89)90255-9. [DOI] [PubMed] [Google Scholar]
- 139.Jockusch H, Jockusch BM. Structural proteins in the growth cone of cultured spinal cord neurons. Exp Cell Res. 1981;131:345–52. doi: 10.1016/0014-4827(81)90237-8. [DOI] [PubMed] [Google Scholar]
- 140.Burridge K, Feramisco JR. Non-muscle alpha actinins are calcium-sensitive actin-binding proteins. Nature. 1981;294:565–7. doi: 10.1038/294565a0. [DOI] [PubMed] [Google Scholar]
- 141.Noegel A, Witke W, Schleicher M. Calcium sensitive non-muscle alpha-actinin contains EF-hand structures and highly conserved regions. FEBS Lett. 1987;221:391–6. doi: 10.1016/0014-5793(87)80962-6. [DOI] [PubMed] [Google Scholar]
- 142.Witke W, Hofmann A, Kfippel B, Sehleicher M, Noegel AA. The Ca2+-binding domains in non-muscle type α-actinin: biochemical and genetic analysis. J Cell Biol. 1993;121:599–606. doi: 10.1083/jcb.121.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zeece MG, Robson RM, Bechtel PJ. Interaction of alpha-actinin, filamin and tropomyosin with F-actin. Biochim Biophys Acta. 1979;581:365–70. doi: 10.1016/0005-2795(79)90258-7. [DOI] [PubMed] [Google Scholar]
- 144.Izaguirre G, Aguirre L, Hu YP, et al. The cytoskeletal/non-muscle isoform of alpha-actinin is phosphorylated on its actin-binding domain by the focal adhesion kinase. J Biol Chem. 2001;276:28676–85. doi: 10.1074/jbc.M101678200. [DOI] [PubMed] [Google Scholar]
- 145.Fukami K, Furuhashi K, Inagaki M, Endo T, Hatano S, Takenawa T. Requirement of phosphatidylinositol 4,5-bisphosphate for alpha-actinin function. Nature. 1992;359:150–2. doi: 10.1038/359150a0. [DOI] [PubMed] [Google Scholar]
- 146.Maciver SK, Wachsstock DH, Schwarz WH, Pollard TD. The actin filament severing protein actophorin promotes the formation of rigid bundles of actin filaments crosslinked with alpha-actinin. J Cell Biol. 1991;115:1621–8. doi: 10.1083/jcb.115.6.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Blikstad I, Eriksson S, Carlsson L. alpha-Actinin promotes polymerization of actin from profilactin. Eur J Biochem. 1980;109:317–23. doi: 10.1111/j.1432-1033.1980.tb04797.x. [DOI] [PubMed] [Google Scholar]
- 148.Schook W, Ores C, Puszkin S. Isolation and properties of brain alpha-actinin. Biochem J. 1978;175:63–72. doi: 10.1042/bj1750063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Walikonis RS, Jensen ON, Mann M, Provance DW, Jr, Mercer JA, Kennedy MB. Identification of proteins in the postsynaptic density fraction by mass spectrometry. J Neurosci. 2000;20:4069–80. doi: 10.1523/JNEUROSCI.20-11-04069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Allison DW, Chervin AS, Gelfand VI, Craig AM. Postsynaptic scaffolds of excitatory and inhibitory synapses in hippocampal neurons: maintenance of core components independent of actin filaments and microtubules. J Neurosci. 2000;20:4545–54. doi: 10.1523/JNEUROSCI.20-12-04545.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kremerskothen J, Plaas C, Kindler S, Frotscher M, Barnekow A. Synaptopodin, a molecule involved in the formation of the dendritic spine apparatus, is a dual actin/alpha-actinin binding protein. J Neurochem. 2005;92:597–606. doi: 10.1111/j.1471-4159.2004.02888.x. [DOI] [PubMed] [Google Scholar]
- 152.Asanuma K, Kim K, Oh J, et al. Synaptopodin regulates the actin-bundling activity of alpha-actinin in an isoform-specific manner. J Clin Invest. 2005;115:1188–98. doi: 10.1172/JCI23371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hoe HS, Lee JY, Pak DT. Combinatorial morphogenesis of dendritic spines and filopodia by SPAR and alpha-actinin2. Biochem Biophys Res Commun. 2009;384:55–60. doi: 10.1016/j.bbrc.2009.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sakagami H, Honma T, Sukegawa J, Owada Y, Yanagisawa T, Kondo H. Somatodendritic localization of EFA6A, a guanine nucleotide exchange factor for ADP-ribosylation factor 6, and its possible interaction with alpha-actinin in dendritic spines. Eur J Neurosci. 2007;25:618–28. doi: 10.1111/j.1460-9568.2007.05345.x. [DOI] [PubMed] [Google Scholar]
- 155.Hernández-Deviez DJ, Roth MG, Casanova JE, Wilson JM. ARNO and ARF6 regulate axonal elongation and branching through downstream activation of phosphatidylinositol 4-phosphate 5-kinase alpha. Mol Biol Cell. 2004;15:111–20. doi: 10.1091/mbc.E03-06-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nakagawa T, Engler JA, Sheng M. The dynamic turnover and functional roles of alpha-actinin in dendritic spines. Neuropharmacology. 2004;47:734–45. doi: 10.1016/j.neuropharm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 157.Nyman-Huttunen H, Tian L, Ning L, Gahmberg CG. alpha-Actinin-dependent cytoskeletal anchorage is important for ICAM-5-mediated neuritic outgrowth. J Cell Sci. 2006;119:3057–66. doi: 10.1242/jcs.03045. [DOI] [PubMed] [Google Scholar]
- 158.Furutani Y, Matsuno H, Kawasaki M, Sasaki T, Mori K, Yoshihara Y. Interaction between telencephalin and ERM family proteins me- diates dendritic filopodia formation. J Neurosci. 2007;27:8866–76. doi: 10.1523/JNEUROSCI.1047-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dunah AW, Wyszynski M, Martin DM, Sheng M, Standaert DG. alpha-actinin-2 in rat striatum: localization and interaction with NMDA glutamate receptor subunits. Brain Res Mol Brain Res. 2000;79:77–87. doi: 10.1016/s0169-328x(00)00102-9. [DOI] [PubMed] [Google Scholar]
- 160.Wyszynski M, Kharazia V, Shanghvi R, et al. Differential regional expression and ultrastructural localization of alpha-actinin-2, a putative NMDA receptor-anchoring protein, in rat brain. J Neurosci. 1998;18:1383–92. doi: 10.1523/JNEUROSCI.18-04-01383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rao A, Kim E, Sheng M, Craig AM. Heterogeneity in the molecular composition of excitatory postsynaptic sites during development of hippocampal neurons in culture. J Neurosci. 1998;18:1217–29. doi: 10.1523/JNEUROSCI.18-04-01217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wyszynski M, Lin J, Rao A, Nigh E, Beggs AH, Craig AM, Sheng M. Competitive binding of alpha-actinin and calmodulin to the NMDA receptor. Nature. 1997;385:439–42. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- 163.Leonard AS, Bayer KU, Merrill MA, et al. Regulation of calcium/calmodulin-dependent protein kinase II docking to N-methyl-D-aspartate receptors by calcium/calmodulin and alpha-actinin. J Biol Chem. 2002;277:48441–8. doi: 10.1074/jbc.M205164200. [DOI] [PubMed] [Google Scholar]
- 164.Rycroft BK, Gibb AJ. Regulation of single NMDA receptor channel activity by alpha-actinin and calmodulin in rat hippocampal granule cells. J Physiol. 2004;557:795–808. doi: 10.1113/jphysiol.2003.059212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang S, Ehlers MD, Bernhardt JP, Su CT, Huganir RL. Calmodulin mediates calcium-dependent inactivation of N-methyl-D-aspartate receptors. Neuron. 1998;21:443–53. doi: 10.1016/s0896-6273(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 166.Krupp JJ, Vissel B, Thomas CG, Heinemann SF, Westbrook GL. Interactions of calmodulin and alpha-actinin with the NR1 subunit modulate Ca2+-dependent inactivation of NMDA receptors. J Neurosci. 1999;19:1165–78. doi: 10.1523/JNEUROSCI.19-04-01165.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bouhamdan M, Yan HD, Yan XH, Bannon MJ, Andrade R. Brain-specific regulator of G-protein signaling 9-2 selectively interacts with alpha-actinin-2 to regulate calcium-dependent inactivation of NMDA receptors. J Neurosci. 2006;26:2522–30. doi: 10.1523/JNEUROSCI.4083-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Roche KW, Standley S, McCallum J, Dune Ly C, Ehlers MD, Wenthold RJ. Molecular determinants of NMDA receptor internalization. Nat Neurosci. 2001;4:794–802. doi: 10.1038/90498. [DOI] [PubMed] [Google Scholar]
- 169.Palop JJ, Chin J, Bien-Ly N, et al. Vulnerability of dentate granule cells to disruption of arc expression in human amyloid precursor protein transgenic mice. J Neurosci. 2005;25:9686–93. doi: 10.1523/JNEUROSCI.2829-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Galloway PG, Perry G, Kosik KS, Gambetti P. Hirano bodies contain tau protein. Brain Res. 1987;403:337–40. doi: 10.1016/0006-8993(87)90071-0. [DOI] [PubMed] [Google Scholar]
- 171.Luthi-Carter R, Apostol BL, Dunah AW, et al. Complex alteration of NMDA receptors in transgenic Huntington’s disease mouse brain: analysis of mRNA and protein expression, plasma membrane association, interacting proteins, and phosphorylation. Neurobiol Dis. 2003;14:624–36. doi: 10.1016/j.nbd.2003.08.024. [DOI] [PubMed] [Google Scholar]
- 172.Ishikawa R, Hayashi K, Shirao T, et al. Drebrin, a development-associated brain protein from rat embryo, causes the dissociation of tropomyosin from actin filaments. J Biol Chem. 1994;269:29928–33. [PubMed] [Google Scholar]
- 173.Sasaki Y, Hayashi K, Shirao T, Ishikawa R, Kohama K. Inhibition by drebrin of the actin-bundling activity of brain fascin, a protein localized in filopodia of growth cones. J Neurochem. 1996;66:980–8. doi: 10.1046/j.1471-4159.1996.66030980.x. [DOI] [PubMed] [Google Scholar]
- 174.Hayashi K, Ishikawa R, Ye LH, et al. Modulatory role of drebrin on the cytoskeleton within dendritic spines in the rat cerebral cortex. J Neurosci. 1996;16:7161–70. doi: 10.1523/JNEUROSCI.16-22-07161.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ishikawa R, Katoh K, Takahashi A, et al. Drebrin attenuates the interaction between actin and myosin-V. Biochem Biophys Res Commun. 2007;359:398–401. doi: 10.1016/j.bbrc.2007.05.123. [DOI] [PubMed] [Google Scholar]
- 176.Shirao T, Hayashi K, Ishikawa R, et al. Formation of thick, curving bundles of actin by drebrin A expressed in fibroblasts. Exp Cell Res. 1994;215:145–53. doi: 10.1006/excr.1994.1326. [DOI] [PubMed] [Google Scholar]
- 177.Shirao T, Obata K. Immunochemical homology of 3 developmentally regulated brain proteins and their developmental change in neuronal distribution. Brain Res. 1986;394:233–44. doi: 10.1016/0165-3806(86)90099-4. [DOI] [PubMed] [Google Scholar]
- 178.Kobayashi C, Aoki C, Kojima N, Yamazaki H, Shirao T. Drebrin a content correlates with spine head size in the adult mouse cerebral cortex. J Comp Neurol. 2007;503:618–26. doi: 10.1002/cne.21408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Aoki C, Sekino Y, Hanamura K, et al. Drebrin A is a postsynaptic protein that localizes in vivo to the submembranous surface of dendritic sites forming excitatory synapses. J Comp Neurol. 2005;483:383–402. doi: 10.1002/cne.20449. [DOI] [PubMed] [Google Scholar]
- 180.Hayashi K, Shirao T. Change in the shape of dendritic spines caused by overexpression of drebrin in cultured cortical neurons. J Neurosci. 1999;19:3918–25. doi: 10.1523/JNEUROSCI.19-10-03918.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mizui T, Takahashi H, Sekino Y, Shirao T. Overexpression of drebrin A in immature neurons induces the accumulation of F-actin and PSD-95 into dendritic filopodia, and the formation of large abnormal protrusions. Mol Cell Neurosci. 2005;30:630–8. [PubMed] [Google Scholar]
- 182.Takahashi H, Yamazaki H, Hanamura K, Sekino Y, Shirao T. Activity of the AMPA receptor regulates drebrin stabilization in dendritic spine morphogenesis. J Cell Sci. 2009;122:1211–9. doi: 10.1242/jcs.043729. [DOI] [PubMed] [Google Scholar]
- 183.Takahashi H, Mizui T, Shirao T. Down-regulation of drebrin A expression suppresses synaptic targeting of NMDA receptors in developing hippocampal neurones. J Neurochem. 2006;97:110–5. doi: 10.1111/j.1471-4159.2005.03536.x. [DOI] [PubMed] [Google Scholar]
- 184.Takahashi H, Sekino Y, Tanaka S, Mizui T, Kishi S, Shirao T. Drebrin-dependent actin clustering in dendritic filopodia governs synaptic targeting of postsynaptic density-95 and dendritic spine morphogenesis. J Neurosci. 2003;23:6586–95. doi: 10.1523/JNEUROSCI.23-16-06586.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sekino Y, Tanaka S, Hanamura K, et al. Activation of N-methyl-D-aspartate receptor induces a shift of drebrin distribution: disappearance from dendritic spines and appearance in dendritic shafts. Mol Cell Neurosci. 2006;31:493–504. doi: 10.1016/j.mcn.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 186.Fujisawa S, Shirao T, Aoki C. In vivo, competitive blockade of N-methyl-D-aspartate receptors induces rapid changes in filamentous actin and drebrin A distributions within dendritic spines of adult rat cortex. Neuroscience. 2006;140:1177–87. doi: 10.1016/j.neuroscience.2006.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Biou V, Brinkhaus H, Malenka RC, Matus A. Interactions between drebrin and Ras regulate dendritic spine plasticity. Eur J Neurosci. 2008;27:2847–59. doi: 10.1111/j.1460-9568.2008.06269.x. [DOI] [PubMed] [Google Scholar]
- 188.Harigaya Y, Shoji M, Shirao T, Hirai S. Disappearance of actin-binding protein, drebrin, from hippocampal synapses in Alzheimer’s disease. J Neurosci Res. 1996;43:87–92. doi: 10.1002/jnr.490430111. [DOI] [PubMed] [Google Scholar]
- 189.Arai H, Sato K, Uto A, Yasumoto Y. Effect of transient cerebral ischemia in mongolian gerbils on synaptic vesicle protein (SVP-38) and developmentally regulated brain protein (drebrin) Neurosci Res Commun. 1991;9:143–150. [Google Scholar]
- 190.Shim KS, Lubec G. Drebrin, a dendritic spine protein, is manifold decreased in brains of patients with Alzheimer’s disease and Down syndrome. Neurosci Lett. 2002;324:209–12. doi: 10.1016/s0304-3940(02)00210-0. [DOI] [PubMed] [Google Scholar]
- 191.Carlsson L, Nyström LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115:465–83. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- 192.Nishida E, Maekawa S, Sakai H. Characterization of the action of porcine brain profilin on actin polymerization. J Biochem. 1984;95:399–404. doi: 10.1093/oxfordjournals.jbchem.a134620. [DOI] [PubMed] [Google Scholar]
- 193.Kaiser DA, Sato M, Ebert RF, Pollard TD. Purification and characterization of two isoforms of Acanthamoeba profilin. J Cell Biol. 1986;102:221–6. doi: 10.1083/jcb.102.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tobacman LS, Korn ED. The regulation of actin polymerization and the inhibition of monomeric actin ATPase activity by Acanthamoeba profilin. J Biol Chem. 1982;257:4166–70. [PubMed] [Google Scholar]
- 195.Ozaki K, Hatano S. Mechanism of regulation of actin polymerization by Physarum profilin. J Cell Biol. 1984;98:1919–25. doi: 10.1083/jcb.98.6.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sanders MC, Wang YL. Exogenous nucleation sites fail to induce detectable polymerization of actin in living cells. J Cell Biol. 1990;110:359–65. doi: 10.1083/jcb.110.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Perelroizen I, Marchand JB, Blanchoin L, Didry D, Carlier MF. Interaction of profilin with G-actin and poly(L-proline) Biochemistry. 1994;33:8472–8. doi: 10.1021/bi00194a011. [DOI] [PubMed] [Google Scholar]
- 198.Tilney LG, Bonder EM, Coluccio LM, Mooseker MS. Actin from Thyone sperm assembles on only one end of an actin filament: a behavior regulated by profilin. J Cell Biol. 1983;97:112–24. doi: 10.1083/jcb.97.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pring M, Weber A, Bubb MR. Profilin-actin complexes directly elongate actin filaments at the barbed end. Biochemistry. 1992;31:1827–36. doi: 10.1021/bi00121a035. [DOI] [PubMed] [Google Scholar]
- 200.Cao LG, Babcock GG, Rubenstein PA, Wang YL. Effects of profilin and profilactin on actin structure and function in living cells. J Cell Biol. 1992;117:1023–9. doi: 10.1083/jcb.117.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Pollard TD, Cooper JA. Quantitative analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry. 1984;23:6631–41. doi: 10.1021/bi00321a054. [DOI] [PubMed] [Google Scholar]
- 202.Pantaloni D, Carlier MF. How profilin promotes actin filament assembly in the presence of thymosin beta 4. Cell. 1993;75:1007–14. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- 203.Finkel T, Theriot JA, Dise KR, Tomaselli GF, Goldschmidt-Clermont PJ. Dynamic actin structures stabilized by profilin. Proc Natl Acad Sci USA. 1994;91:1510–4. doi: 10.1073/pnas.91.4.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Theriot JA, Mitchison TJ. The three faces of profilin. Cell. 1993;75:835–8. doi: 10.1016/0092-8674(93)90527-w. [DOI] [PubMed] [Google Scholar]
- 205.Buss F, Temm-Grove C, Henning S, Jockusch BM. Distribution of profilin in fibroblasts correlates with the presence of highly dynamic actin filaments. Cell Motil Cytoskeleton. 1992;22:51–61. doi: 10.1002/cm.970220106. [DOI] [PubMed] [Google Scholar]
- 206.Theriot JA, Rosenblatt J, Portnoy DA, Goldschmidt-Clermont PJ, Mitchison TJ. Involvement of profilin in the actin-based motility of L. monocytogenes in cells and in cell-free extracts. Cell. 1994;76:505–17. doi: 10.1016/0092-8674(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 207.Dong JH, Ying GX, Zhou CF. Entorhinal deafferentation induces the expression of profilin mRNA in the reactive microglial cells in the hippocampus. Glia. 2004;47:102–8. doi: 10.1002/glia.10355. [DOI] [PubMed] [Google Scholar]
- 208.Hartwig JH, Chambers KA, Hopcia KL, Kwiatkowski DJ. Association of profilin with filament-free regions of human leukocyte and platelet membranes and reversible membrane binding during platelet activation. J Cell Biol. 1989;109:1571–9. doi: 10.1083/jcb.109.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Witke W, Podtelejnikov AV, Di Nardo A, et al. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 1998;17:967–76. doi: 10.1093/emboj/17.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gareus R, Di Nardo A, Rybin V, Witke W. Mouse profilin 2 regulates endocytosis and competes with SH3 ligand binding to dynamin 1. J Biol Chem. 2006;281:2803–11. doi: 10.1074/jbc.M503528200. [DOI] [PubMed] [Google Scholar]
- 211.Haarer BK, Lillie SH, Adams AE, Magdolen V, Bandlow W, Brown SS. Purification of profilin from Saccharomyces cerevisiae and analysis of profilin-deficient cells. J Cell Biol. 1990;110:105–14. doi: 10.1083/jcb.110.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Verheyen EM, Cooley L. Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development. 1994;120:717–28. doi: 10.1242/dev.120.4.717. [DOI] [PubMed] [Google Scholar]
- 213.Ackermann M, Matus A. Activity-induced targeting of profilin and stabilization of dendritic spine morphology. Nat Neurosci. 2003;6:1194–200. doi: 10.1038/nn1135. [DOI] [PubMed] [Google Scholar]
- 214.Sohn RH, Chen J, Koblan KS, Bray PF, Goldschmidt-Clermont PJ. Localization of a binding site for phosphatidylinositol 4,5-bisphosphate on human profilin. J Biol Chem. 1995;270:21114–20. doi: 10.1074/jbc.270.36.21114. [DOI] [PubMed] [Google Scholar]
- 215.Lambrechts A, Verschelde JL, Jonckheere V, Goethals M, Vandekerckhove J, Ampe C. The mammalian profilin isoforms display complementary affinities for PIP2 and proline-rich sequences. EMBO J. 1997;16:484–94. doi: 10.1093/emboj/16.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Krishnan K, Holub O, Gratton E, Clayton AHA, Cody S, Moens PDJ. Profilin interaction with phosphatidylinositol (4,5)-bisphosphate destabilizes the membrane of giant unilamellar Vesicles. Biophys J. 2009;96:5112–21. doi: 10.1016/j.bpj.2009.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Goldschmidt-Clermont PJ, Van Alstyne EL, Day JR, et al. Group-specific component (vitamin D binding protein) prevents the interaction between G-actin and profilin. Biochemistry. 1986;25:6467–72. doi: 10.1021/bi00369a019. [DOI] [PubMed] [Google Scholar]
- 218.Lassing I, Lindberg U. Specificity of the interaction between phosphatidylinositol 4,5-bisphosphate and the profilin:actin complex. J Cell Biochem. 1988;37:255–67. doi: 10.1002/jcb.240370302. [DOI] [PubMed] [Google Scholar]
- 219.Hansson A, Skoglund G, Lassing I, Lindberg U, Ingelman-Sundberg M. Protein kinase C-dependent phosphorylation of profilin is specifically stimulated by phosphatidylinositol bisphosphate (PIP2) Biochem Biophys Res Commun. 1988;150:526–31. doi: 10.1016/0006-291x(88)90425-1. [DOI] [PubMed] [Google Scholar]
- 220.Vemuri B, Singh SS. Protein kinase C isozyme-specific phosphorylation of profilin. Cell Signal. 2001;13:433–9. doi: 10.1016/s0898-6568(01)00164-4. [DOI] [PubMed] [Google Scholar]
- 221.Sathish K, Padma B, Munugalavadla V, et al. Phosphorylation of profilin regulates its interaction with actin and poly (L-proline) Cell Signal. 2004;16:589–96. doi: 10.1016/j.cellsig.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 222.Ferron F, Rebowski G, Lee SH, Dominguez R. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J. 2007;26:4597–606. doi: 10.1038/sj.emboj.7601874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Léna JY, Sri Widada J, Ferraz C, Liautard JP, Rabié A, Faivre-Sarrailh C. Profilin and profilin mRNA in the cerebellum of the developing rat. Neuroreport. 1991;2:117–20. doi: 10.1097/00001756-199103000-00001. [DOI] [PubMed] [Google Scholar]
- 224.Faivre-Sarrailh C, Lena JY, Had L, Vignes M, Lindberg U. Location of profilin at presynaptic sites in the cerebellar cortex; implication for the regulation of the actin-polymerization state during axonal elongation and synaptogenesis. J Neurocytol. 1993;22:1060–72. doi: 10.1007/BF01235749. [DOI] [PubMed] [Google Scholar]
- 225.Giesemann T, Schwarz G, Nawrotzki R, et al. Complex formation between the postsynaptic scaffolding protein gephyrin, profilin, and Mena: a possible link to the microfilament system. J Neurosci. 2003;23:8330–9. doi: 10.1523/JNEUROSCI.23-23-08330.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Neuhoff H, Sassoè-Pognetto M, Panzanelli P, Maas C, Witke W, Kneussel M. The actin-binding protein profilin I is localized at synaptic sites in an activity-regulated manner. Eur J Neurosci. 2005;21:15–25. doi: 10.1111/j.1460-9568.2004.03814.x. [DOI] [PubMed] [Google Scholar]
- 227.Birbach A, Verkuyl JM, Matus A. Reversible, activity-dependent targeting of profilin to neuronal nuclei. Exp Cell Res. 2006;312:2279–87. doi: 10.1016/j.yexcr.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 228.Shao J, Welch WJ, Diprospero NA, Diamond MI. Phosphorylation of profilin by ROCK1 regulates polyglutamine aggregation. Mol Cell Biol. 2008;28:5196–208. doi: 10.1128/MCB.00079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Da Silva JS, Medina M, Zuliani C, Di Nardo A, Witke W, Dotti CG. RhoA/ROCK regulation of neuritogenesis via profilin IIa-mediated control of actin stability. J Cell Biol. 2003;162:1267–79. doi: 10.1083/jcb.200304021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Bauer PO, Wong HK, Oyama F, et al. Inhibition of Rho kinases enhances the degradation of mutant huntingtin. J Biol Chem. 2009;284:13153–64. doi: 10.1074/jbc.M809229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kwiatkowski DJ, Aklog L, Ledbetter DH, Morton CC. Identification of the functional profilin gene, its localization to chromosome subband 17p13.3, and demonstration of its deletion in some patients with Miller-Dieker syndrome. Am J Hum Genet. 1990;46:559–67. [PMC free article] [PubMed] [Google Scholar]
- 232.Reeve SP, Bassetto L, Genova GK, et al. The Drosophila fragile X mental retardation protein controls actin dynamics by directly regulating profilin in the brain. Curr Biol. 2005;15:1156–63. doi: 10.1016/j.cub.2005.05.050. [DOI] [PubMed] [Google Scholar]
- 233.Pilo Boyl P, Di Nardo A, Mulle C, et al. Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. EMBO J. 2007;26:2991–3002. doi: 10.1038/sj.emboj.7601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Allen PB, Ouimet CC, Greengard P. Spinophilin, a novel protein phosphatase 1 binding protein localized to dendritic spines. Proc Natl Acad Sci USA. 1997;94:9956–61. doi: 10.1073/pnas.94.18.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Nakanishi H, Obaishi H, Satoh A, et al. Neurabin: a novel neural tissue-specific actin filament-binding protein involved in neurite formation. J Cell Biol. 1997;139:951–61. doi: 10.1083/jcb.139.4.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Satoh A, Nakanishi H, Obaishi H, et al. Neurabin-II/spinophilin an actin filament-binding protein with one pdz domain localized at cadherin-based cell-cell adhesion sites. J Biol Chem. 1998;273:3470–5. doi: 10.1074/jbc.273.6.3470. [DOI] [PubMed] [Google Scholar]
- 237.Sarrouilhe D, di Tommaso A, Métayé T, Ladeveze V. Spinophilin: from partners to functions. Biochimie. 2006;88:1099–113. doi: 10.1016/j.biochi.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 238.Hsieh-Wilson LC, Allen PB, Watanabe T, Nairn AC, Greengard P. Characterization of the neuronal targeting protein spinophilin and its interactions with protein phosphatase-1. Biochemistry. 1999;38:4365–73. doi: 10.1021/bi982900m. [DOI] [PubMed] [Google Scholar]
- 239.Yan Z, Hsieh-Wilson L, Feng J, et al. Protein phosphatase 1 modulation of neostriatal AMPA channels: regulation by DARPP-32 and spinophilin. Nat Neurosci. 1999;2:13–7. doi: 10.1038/4516. [DOI] [PubMed] [Google Scholar]
- 240.Feng J, Yan Z, Ferreira A, et al. Spinophilin regulates the formation and function of dendritic spines. Proc Natl Acad Sci USA. 2000;97:9287–92. doi: 10.1073/pnas.97.16.9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hsieh-Wilson LC, Benfenati F, Snyder GL, Allen PB, Nairn AC, Greengard P. Phosphorylation of spinophilin modulates its interaction with actin filaments. J Biol Chem. 2003;278:1186–94. doi: 10.1074/jbc.M205754200. [DOI] [PubMed] [Google Scholar]
- 242.Grossman SD, Futter M, Snyder GL, et al. Spinophilin is phosphorylated by Ca2+/calmodulin-dependent protein kinase II resulting in regulation of its binding to F-actin. J Neurochem. 2004;90:317–24. doi: 10.1111/j.1471-4159.2004.02491.x. [DOI] [PubMed] [Google Scholar]
- 243.Romcy-Pereira RN, Erraji-Benchekroun L, Smyrniotopoulos P, et al. Sleep-dependent gene expression in the hippocampus and prefrontal cortex following long-term potentiation. Physiol Behav. 2009;98:44–52. doi: 10.1016/j.physbeh.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Oliver CJ, Terry-Lorenzo RT, Elliott E, et al. Targeting protein phosphatase 1 (PP1) to the actin cytoskeleton: the neurabin I/PP1 complex regulates cell morphology. Mol Cell Biol. 2002;22:4690–701. doi: 10.1128/MCB.22.13.4690-4701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wu LJ, Ren M, Wang H, Kim SS, Cao X, Zhuo M. Neurabin contributes to hippocampal long-term potentiation and contextual fear memory. PLoS One. 2008;3:1407. doi: 10.1371/journal.pone.0001407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 247.Condeelis J. How is actin polymerization nucleated in vivo? Trends Cell Biol. 2001;11:288–93. doi: 10.1016/s0962-8924(01)02008-6. [DOI] [PubMed] [Google Scholar]
- 248.Sarmiere PD, Bamburg JR. Regulation of the neuronal actin cytoskeleton by ADF/cofilin. J Neurobiol. 2004;58:103–17. doi: 10.1002/neu.10267. [DOI] [PubMed] [Google Scholar]
- 249.Moriyama K, Matsumoto S, Nishida E, Sakai H, Yahara I. Nucleotide sequence of mouse cofilin cDNA. Nucleic Acids Res. 1990;18:3053. doi: 10.1093/nar/18.10.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Garvalov BK, Flynn KC, Neukirchen D, et al. Cdc42 regulates cofilin during the establishment of neuronal polarity. J Neurosci. 2007;27:13117–29. doi: 10.1523/JNEUROSCI.3322-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Bamburg JR, Wiggan OP. ADF/cofilin and actin dynamics in disease. Trends Cell Biol. 2002;12:598–605. doi: 10.1016/s0962-8924(02)02404-2. [DOI] [PubMed] [Google Scholar]
- 252.DesMarais V, Ghosh M, Eddy R, Condeelis J. Cofilin takes the lead. J Cell Sci. 2005;118:19–26. doi: 10.1242/jcs.01631. [DOI] [PubMed] [Google Scholar]
- 253.Gungabissoon RA, Bamburg JR. Regulation of growth cone actin dynamics by ADF/cofilin. J Histochem Cytochem. 2003;51:411–20. doi: 10.1177/002215540305100402. [DOI] [PubMed] [Google Scholar]
- 254.Gehler S, Shaw AE, Sarmiere PD, Bamburg JR, Letourneau PC. Brain-derived neurotrophic factor regulation of retinal growth cone filopodial dynamics is mediated through actin depolymerizing factor/cofilin. J Neurosci. 2004;24:10741–9. doi: 10.1523/JNEUROSCI.2836-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 256.Arber S, Barbayannis FA, Hanser H, et al. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–9. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- 257.Yang N, Higuchi O, Ohashi K, et al. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–12. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- 258.Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 2002;108:233–46. doi: 10.1016/s0092-8674(01)00638-9. [DOI] [PubMed] [Google Scholar]
- 259.Gohla A, Birkenfeld J, Bokoch GM. Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat Cell Biol. 2005;7:21–9. doi: 10.1038/ncb1201. [DOI] [PubMed] [Google Scholar]
- 260.Huang TY, DerMardirossian C, Bokoch GM. Cofilin phosphatases and regulation of actin dynamics. Curr Opin Cell Biol. 2006;18:26–31. doi: 10.1016/j.ceb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 261.Chen LY, Rex CS, Casale MS, Gall CM, Lynch G. Changes in synaptic morphology accompany actin signaling during LTP. J Neurosci. 2007;27:5363–72. doi: 10.1523/JNEUROSCI.0164-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Shankar GM, Bloodgood BL, Townsend M, Walsh DM, Selkoe DJ, Sabatini BL. Natural oligomers of the Alzheimer amyloid-beta protein induce reversible synapse loss by modulating an NMDA-type glutamate receptor-dependent signaling pathway. J Neurosci. 2007;27:2866–75. doi: 10.1523/JNEUROSCI.4970-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Shi Y, Pontrello CG, DeFea KA, Reichardt LF, Ethell IM. Focal adhesion kinase acts downstream of EphB receptors to maintain mature dendritic spines by regulating cofilin activity. J Neurosci. 2009;29:8129–42. doi: 10.1523/JNEUROSCI.4681-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Rácz B, Weinberg RJ. Spatial organization of cofilin in dendritic spines. Neuroscience. 2006;138:447–56. doi: 10.1016/j.neuroscience.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 265.Carlisle HJ, Manzerra P, Marcora E, Kennedy MB. SynGAP regulates steady-state and activity-dependent phosphorylation of cofilin. J Neurosci. 2008;28:13673–83. doi: 10.1523/JNEUROSCI.4695-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat Cell Biol. 1999;1:253–9. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 267.Maekawa M, Ishizaki T, Boku S, et al. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science. 1999;285:895–8. doi: 10.1126/science.285.5429.895. [DOI] [PubMed] [Google Scholar]
- 268.Asrar S, Meng Y, Zhou Z, Todorovski Z, Huang WW, Jia Z. Regulation of hippocampal long-term potentiation by p21-activated protein kinase 1 (PAK1) Neuropharmacology. 2009;56:73–80. doi: 10.1016/j.neuropharm.2008.06.055. [DOI] [PubMed] [Google Scholar]
- 269.Zhou Z, Meng Y, Asrar S, Todorovski Z, Jia Z. A critical role of Rho-kinase ROCK2 in the regulation of spine and synaptic function. Neuropharmacology. 2009;56:81–9. doi: 10.1016/j.neuropharm.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 270.Meng Y, Zhang Y, Tregoubov V, et al. Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron. 2002;35:121–33. doi: 10.1016/s0896-6273(02)00758-4. [DOI] [PubMed] [Google Scholar]
- 271.Schratt GM, Tuebing F, Nigh EA, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–9. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 272.Wang Y, Shibasaki F, Mizuno K. Calcium signal-induced cofilin dephosphorylation is mediated by Slingshot via calcineurin. J Biol Chem. 2005;280:12683–9. doi: 10.1074/jbc.M411494200. [DOI] [PubMed] [Google Scholar]
- 273.Quinlan EM, Halpain S. Postsynaptic mechanisms for bidirectional control of MAP2 phosphorylation by glutamate receptors. Neuron. 1996;16:357–68. doi: 10.1016/s0896-6273(00)80053-7. [DOI] [PubMed] [Google Scholar]
- 274.Kojima N, Shirao T. Synaptic dysfunction and disruption of postsynaptic drebrin-actin complex: a study of neurological disorders accompanied by cognitive deficits. Neurosci Res. 2007;58:1–5. doi: 10.1016/j.neures.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 275.Yonezawa N, Homma Y, Yahara I, Sakai H, Nishida E. A short sequence responsible for both phosphoinositide binding and actin binding activities of cofilin. J Biol Chem. 1991;266:17218–21. [PubMed] [Google Scholar]
- 276.Kusano K, Abe H, Obinata T. Detection of a sequence involved in actin-binding and phosphoinositide-binding in the N-terminal side of cofilin. Mol Cell Biochem. 1999;190:133–41. [PubMed] [Google Scholar]
- 277.Zhou L, Martinez SJ, Haber M, et al. EphA4 signaling regulates phospholipase Cgamma1 activation, cofilin membrane association, and dendritic spine morphology. J Neurosci. 2007;27:5127–38. doi: 10.1523/JNEUROSCI.1170-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Nagaoka R, Kusano K, Abe H, Obinata T. Effects of cofilin on actin filamentous structures in cultured muscle cells. Intracellular regulation of cofilin action. J Cell Sci. 1995;108:581–93. doi: 10.1242/jcs.108.2.581. [DOI] [PubMed] [Google Scholar]
- 279.Minamide LS, Striegl AM, Boyle JA, Meberg PJ, Bamburg JR. Neurodegenerative stimuli induce persistent ADF/cofilin-actin rods that disrupt distal neurite function. Nat Cell Biol. 2000;2:628–36. doi: 10.1038/35023579. [DOI] [PubMed] [Google Scholar]
- 280.Davis RC, Maloney MT, Minamide LS, Flynn KC, Stonebraker MA, Bamburg JR. Mapping cofilin-actin rods in stressed hippocampal slices and the role of cdc42 in amyloid-beta-induced rods. J Alzheimers Dis. 2009 doi: 10.3233/JAD-2009-1122. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Maloney MT, Minamide LS, Kinley AW, Boyle JA, Bamburg JR. Beta-secretase-cleaved amyloid precursor protein accumulates at actin inclusions induced in neurons by stress or amyloid beta: a feedforward mechanism for Alzheimer’s disease. J Neurosci. 2005;25:11313–21. doi: 10.1523/JNEUROSCI.3711-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Bamburg JR, Bloom GS. Cytoskeletal pathologies of Alzheimer disease. Cell Motil Cytoskeleton. 2009;66:635–49. doi: 10.1002/cm.20388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Yin HL, Stossel TP. Control of cytoplasmic actin gelsol transformation by gelsolin, a calcium-dependent regulatory protein. Nature. 1979;281:583–6. doi: 10.1038/281583a0. [DOI] [PubMed] [Google Scholar]
- 284.Harris HE, Weeds AG. Plasma gelsolin caps and severs actin filaments. FEBS Lett. 1984;177:184–8. doi: 10.1016/0014-5793(84)81280-6. [DOI] [PubMed] [Google Scholar]
- 285.Selve N, Wegner A. Rate constant for capping of the barbed ends of actin filaments by the gelsolin-actin complex. Eur J Biochem. 1986;155:397–401. doi: 10.1111/j.1432-1033.1986.tb09504.x. [DOI] [PubMed] [Google Scholar]
- 286.Sun HQ, Wooten DC, Janmey PA, Yin HL. The actin side-binding domain of gelsolin also caps actin filaments. Implications for actin filament severing. J Biol Chem. 1994;269:9473–9. [PubMed] [Google Scholar]
- 287.Kinosian HJ, Selden LA, Estes JE, Gershman LC. Kinetics of gelsolin interaction with phalloidin-stabilized F-actin. Rate constants for binding and severing. Biochemistry. 1996;35:16550–6. doi: 10.1021/bi961891j. [DOI] [PubMed] [Google Scholar]
- 288.Ditsch A, Wegner A. Nucleation of actin polymerization by gelsolin. Eur J Biochem. 1994;224:223–7. doi: 10.1111/j.1432-1033.1994.tb20015.x. [DOI] [PubMed] [Google Scholar]
- 289.Laham LE, Lamb JA, Allen PG, Janmey PA. Selective binding of gelsolin to actin monomers containing ADP. J Biol Chem. 1993;268:14202–7. [PubMed] [Google Scholar]
- 290.Laham LE, Way M, Yin HL, Janmey PA. Identification of two sites in gelsolin with different sensitivities to adenine nucleotides. Eur J Biochem. 1995;234:1–7. doi: 10.1111/j.1432-1033.1995.001_c.x. [DOI] [PubMed] [Google Scholar]
- 291.Hesterkamp T, Weeds AG, Mannherz HG. The actin monomers in the ternary gelsolin: 2 actin complex are in an antiparallel orientation. Eur J Biochem. 1993;218:507–13. doi: 10.1111/j.1432-1033.1993.tb18403.x. [DOI] [PubMed] [Google Scholar]
- 292.Khaitlina S, Hinssen H. Conformational changes in actin induced by its interaction with gelsolin. Biophys J. 1997;73:929–37. doi: 10.1016/S0006-3495(97)78125-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.McLaughlin PJ, Gooch J. Crystallization of human gelsolin. FEBS Lett. 1992;302:253–5. doi: 10.1016/0014-5793(92)80453-n. [DOI] [PubMed] [Google Scholar]
- 294.Burtnick LD, Koepf EK, Grimes J, et al. The crystal structure of plasma gelsolin: implications for actin severing, capping, and nucleation. Cell. 1997;90:661–70. doi: 10.1016/s0092-8674(00)80527-9. [DOI] [PubMed] [Google Scholar]
- 295.Bryan J, Kurth MC. Actin-gelsolin interactions. Evidence for two actin-binding sites. J Biol Chem. 1984;259:7480–7. [PubMed] [Google Scholar]
- 296.Porte F, Harricane MC. Interactions of plasma gelsolin with actin. Isolation and characterization of binary and ternary plasma-gelsolin-actin complexes. Eur J Biochem. 1986;154:87–93. doi: 10.1111/j.1432-1033.1986.tb09362.x. [DOI] [PubMed] [Google Scholar]
- 297.Pope B, Way M, Weeds AG. Two of the three actin-binding domains of gelsolin bind to the same subdomain of actin. Implications of capping and severing mechanisms. FEBS Lett. 1991;280:70–4. doi: 10.1016/0014-5793(91)80206-i. [DOI] [PubMed] [Google Scholar]
- 298.Schoepper B, Wegner A. Rate constants and equilibrium constants for binding of actin to the 1:1 gelsolin-actin complex. Eur J Biochem. 1991;202:1127–31. doi: 10.1111/j.1432-1033.1991.tb16480.x. [DOI] [PubMed] [Google Scholar]
- 299.Mannherz HG, Gooch J, Way M, Weeds AG, McLaughlin PJ. Crystallization of the complex of actin with gelsolin segment 1. J Mol Biol. 1992;226:899–901. doi: 10.1016/0022-2836(92)90641-v. [DOI] [PubMed] [Google Scholar]
- 300.McGough A, Chiu W, Way M. Determination of the gelsolin binding site on F-actin: implications for severing and capping. Biophys J. 1998;74:764–72. doi: 10.1016/S0006-3495(98)74001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Chaponnier C, Janmey PA, Yin HL. The actin filament-severing domain of plasma gelsolin. J Cell Biol. 1986;103:1473–81. doi: 10.1083/jcb.103.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.McLaughlin PJ, Gooch JT, Mannherz HG, Weeds AG. Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature. 1993;364:685–92. doi: 10.1038/364685a0. [DOI] [PubMed] [Google Scholar]
- 303.Fujita H, Allen PG, Janmey PA, et al. Characterization of gelsolin truncates that inhibit actin depolymerization by severing activity of gelsolin and cofilin. Eur J Biochem. 1997;248:834–9. doi: 10.1111/j.1432-1033.1997.00834.x. [DOI] [PubMed] [Google Scholar]
- 304.Feinberg J, Mery J, Heitz F, Benyamin Y, Roustan C. Conformational and functional studies of three gelsolin subdomain-1 synthetic peptides and their implication in actin polymerization. Biopolymers. 1997;41:647–55. doi: 10.1002/(SICI)1097-0282(199705)41:6<647::AID-BIP5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 305.Wang E, Yin HL, Krueger JG, Caliguiri LA, Tamm I. Unphosphorylated gelsolin is localized in regions of cell-substratum contact or attachment in Rous sarcoma virus-transformed rat cells. J Cell Biol. 1984;98:761–71. doi: 10.1083/jcb.98.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Chen P, Murphy-Ullrich JE, Wells A. A role for gelsolin in actuating epidermal growth factor receptor-mediated cell motility. J Cell Biol. 1996;134:689–98. doi: 10.1083/jcb.134.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Yin HL, Albrecht JH, Fattoum A. Identification of gelsolin, a Ca2+-dependent regulatory protein of actin gelsol transformation, and its intracellular distribution in a variety of cells and tissues. J Cell Biol. 1981;91:901–6. doi: 10.1083/jcb.91.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Brady ST, Lasek RJ, Allen RD, Yin HL, Stossel TP. Gelsolin inhibition of fast axonal transport indicates a requirement for actin microfilaments. Nature. 1984;310:56–8. doi: 10.1038/310056a0. [DOI] [PubMed] [Google Scholar]
- 309.Ishizaki A, Fujita H, Kuzumaki N. Growth-inhibitory functions of a mutated gelsolin (His321) in NIH/3T3 mouse fibroblasts. Exp Cell Res. 1995;217:448–52. doi: 10.1006/excr.1995.1108. [DOI] [PubMed] [Google Scholar]
- 310.Azuma T, Witke W, Stossel TP, Hartwig JH, Kwiatkowski DJ. Gelsolin is a downstream effector of rac for fibroblast motility. EMBO J. 1998;17:1362–70. doi: 10.1093/emboj/17.5.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Selve N, Wegner A. pH-dependent rate of formation of the gelsolin-actin complex from gelsolin and monomeric actin. Eur J Biochem. 1987;168:111–5. doi: 10.1111/j.1432-1033.1987.tb13394.x. [DOI] [PubMed] [Google Scholar]
- 312.Ishikawa R, Yamashiro S, Matsumura F. Differential modulation of actin-severing activity of gelsolin by multiple isoforms of cultured rat cell tropomyosin: potentiation of protective ability of tropomyosins by 83-kDa nonmuscle caldesmon. J Biol Chem. 1989;264:7490–7. [PubMed] [Google Scholar]
- 313.Allen PG, Laham LE, Way M, Janmey PA. Binding of phosphate, aluminum fluoride, or beryllium fluoride to F-actin inhibits severing by gelsolin. J Biol Chem. 1996;271:4665–70. doi: 10.1074/jbc.271.9.4665. [DOI] [PubMed] [Google Scholar]
- 314.Arcaro A. The small GTP-binding protein Rac promotes the dissociation of gelsolin from actin filaments in neutrophils. J Biol Chem. 1998;273:805–13. doi: 10.1074/jbc.273.2.805. [DOI] [PubMed] [Google Scholar]
- 315.Janmey PA, Stossel TP. Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature. 1987;325:362–4. doi: 10.1038/325362a0. [DOI] [PubMed] [Google Scholar]
- 316.Yu FX, Sun HQ, Janmey PA, Yin HL. Identification of a poly-phosphoinositide-binding sequence in an actin monomer-binding domain of gelsolin. J Biol Chem. 1992;267:14616–21. [PubMed] [Google Scholar]
- 317.Singh SS, Chauhan A, Murakami N, Chauhan VP. Profilin and gelsolin stimulate phosphatidylinositol 3-kinase activity. Biochemistry. 1996;35:16544–9. doi: 10.1021/bi9609634. [DOI] [PubMed] [Google Scholar]
- 318.Banno Y, Nakashima T, Kumada T, Ebisawa K, Nonomura Y, Nozawa Y. Effects of gelsolin on human platelet cytosolic phosphoinositide-phospholipase C isozymes. J Biol Chem. 1992;267:6488–94. [PubMed] [Google Scholar]
- 319.Sun H, Lin K, Yin HL. Gelsolin modulates phospholipase C activity in vivo through phospholipid binding. J Cell Biol. 1997;138:811–20. doi: 10.1083/jcb.138.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Yin HL, Stossel TP. Purification and structural properties of gelsolin, a Ca2+-activated regulatory protein of macrophages. J Biol Chem. 1980;255:9490–3. [PubMed] [Google Scholar]
- 321.Janmey PA, Chaponnier C, Lind SE, Zaner KS, Stossel TP, Yin HL. Interactions of gelsolin and gelsolin-actin complexes with actin. Effects of calcium on actin nucleation, filament severing, and end blocking. Biochemistry. 1985;24:3714–23. doi: 10.1021/bi00335a046. [DOI] [PubMed] [Google Scholar]
- 322.Coué M, Korn ED. Interaction of plasma gelsolin with G-actin and F-actin in the presence and absence of calcium ions. J Biol Chem. 1985;260:15033–41. [PubMed] [Google Scholar]
- 323.Pope B, Maciver S, Weeds A. Localization of the calcium-sensitive actin monomer binding site in gelsolin to segment 4 and identification of calcium binding sites. Biochemistry. 1995;34:1583–8. doi: 10.1021/bi00005a014. [DOI] [PubMed] [Google Scholar]
- 324.Allen PG, Janmey PA. Gelsolin displaces phalloidin from actin filaments: a new fluorescence method shows that both Ca2+ and Mg2+ affect the rate at which gelsolin severs F-actin. J Biol Chem. 1994;269:32916–23. [PubMed] [Google Scholar]
- 325.Kilhoffer MC, Mely Y, Gérard D. The effect of plasma gelsolin on actin filaments. Ca2+-dependency of the capping and severing activities. Biochem Biophys Res Commun. 1985;131:1132–8. doi: 10.1016/0006-291x(85)90208-6. [DOI] [PubMed] [Google Scholar]
- 326.Ditsch A, Wegner A. Two low-affinity Ca(2+)-binding sites of gelsolin that regulate association with actin. Eur J Biochem. 1995;229:512–6. doi: 10.1111/j.1432-1033.1995.0512k.x. [DOI] [PubMed] [Google Scholar]
- 327.Hwo S, Bryan J. Immuno-identification of Ca2+-induced conformational changes in human gelsolin and brevin. J Cell Biol. 1986;102:227–36. doi: 10.1083/jcb.102.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Rouayrenc JF, Fattoum A, Méjean C, Kassab R. Characterization of the Ca2+-induced conformational changes in gelsolin and identification of interaction regions between actin and gelsolin. Biochemistry. 1986;25:3859–67. doi: 10.1021/bi00361a018. [DOI] [PubMed] [Google Scholar]
- 329.Hellweg T, Hinssen H, Eimer W. The Ca(2+)-induced conformational change of gelsolin is located in the carboxyl-terminal half of the molecule. Biophys J. 1993;65:799–805. doi: 10.1016/S0006-3495(93)81121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Petrucci TC, Thomas C, Bray D. J Isolation of a Ca2+-dependent actin-fragmenting protein from brain, spinal cord, and cultured neurones. J Neurochem. 1983;40:1507–16. doi: 10.1111/j.1471-4159.1983.tb08119.x. [DOI] [PubMed] [Google Scholar]
- 331.Ying GX, Liu X, Wang WY, et al. Regulated transcripts in the hippocampus following transections of the entorhinal afferents. Biochem Biophys Res Commun. 2004;322:210–6. doi: 10.1016/j.bbrc.2004.07.099. [DOI] [PubMed] [Google Scholar]
- 332.Dong JH, Ying GX, Liu X, et al. Lesion-induced gelsolin upregulation in the hippocampus following entorhinal deafferentation. Hippocampus. 2006;16:91–100. doi: 10.1002/hipo.20134. [DOI] [PubMed] [Google Scholar]
- 333.Tanaka J, Kira M, Sobue K. Gelsolin is localized in neuronal growth cones. Brain Res Dev Brain Res. 1993;76:268–71. doi: 10.1016/0165-3806(93)90217-x. [DOI] [PubMed] [Google Scholar]
- 334.Neely MD, Macaluso E. Motile areas of leech neurites are rich in microfilaments and two actin-binding proteins: gelsolin and profilin. Proc Biol Sci. 1997;264:1701–6. doi: 10.1098/rspb.1997.0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Lu M, Witke W, Kwiatkowski DJ, Kosik KS. Delayed retraction of filopodia in gelsolin null mice. J Cell Biol. 1997;138:1279–87. doi: 10.1083/jcb.138.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Furukawa K, Fu W, Li Y, Witke W, Kwiatkowski DJ, Mattson MP. The actin-severing protein gelsolin modulates calcium channel and NMDA receptor activities and vulnerability to excitotoxicity in hippocampal neurons. J Neurosci. 1997;17:8178–86. doi: 10.1523/JNEUROSCI.17-21-08178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Star EN, Kwiatkowski DJ, Murthy VN. Rapid turnover of actin in dendritic spines and its regulation by activity. Nat Neurosci. 2002;5:239–46. doi: 10.1038/nn811. [DOI] [PubMed] [Google Scholar]
- 338.Haltia M, Levy E, Meretoja J, Fernandez-Madrid I, Koivunen O, Frangione B. Gelsolin gene mutation--at codon 187--in familial amyloidosis, finnish: DNA-diagnostic assay. Am J Med Genet. 1992;42:357–9. doi: 10.1002/ajmg.1320420321. [DOI] [PubMed] [Google Scholar]
- 339.de la Chapelle A, Kere J, Sack GH, Jr, Tolvanen R, Maury CP. Familial amyloidosis, finnish type: G654- a mutation of the gelsolin gene in finnish families and an unrelated American family. Genomics. 1992;13:898–901. doi: 10.1016/0888-7543(92)90182-r. [DOI] [PubMed] [Google Scholar]
- 340.Maury CP, Nurmiaho-Lassila EL, Rossi H. Amyloid fibril formation in gelsolin-derived amyloidosis: definition of the amyloidogenic region and evidence of accelerated amyloid formation of mutant Asn-187 and Tyr-187 gelsolin peptides. Lab Invest. 1994;70:558–64. [PubMed] [Google Scholar]
- 341.Paunio T, Kangas H, Kiuru S, Palo J, Peltonen L, Syvänen AC. Tissue distribution and levels of gelsolin mRNA in normal individuals and patients with gelsolin-related amyloidosis. FEBS Lett. 1997;406:49–55. doi: 10.1016/s0014-5793(97)00237-8. [DOI] [PubMed] [Google Scholar]
- 342.Westberg JA, Zhang KZ, Andersson LC. Regulation of neural differentiation by normal and mutant (G654A, amyloidogenic) gelsolin. FASEB J. 1999;13:1621–6. doi: 10.1096/fasebj.13.12.1621. [DOI] [PubMed] [Google Scholar]
- 343.Kiuru S, Seppäläinen AM. Neuropathy in familial amyloidosis, Finnish type (FAF): electrophysiological studies. Muscle Nerve. 1994;17:299–304. doi: 10.1002/mus.880170307. [DOI] [PubMed] [Google Scholar]
- 344.Tanaka J, Sobue K. Localization and characterization of gelsolin in nervous tissues: gelsolin is specifically enriched in myelin-forming cells. J Neurosci. 1994;14:1038–52. doi: 10.1523/JNEUROSCI.14-03-01038.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Léna JY, Legrand C, Faivre-Sarrailh C, Sarliève LL, Ferraz C, Rabié A. High gelsolin content of developing oligodendrocytes. Int J Dev Neurosci. 1994;12:375–86. doi: 10.1016/0736-5748(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 346.Vouyiouklis DA, Brophy PJ. A novel gelsolin isoform expressed by oligodendrocytes in the central nervous system. J Neurochem. 1997;69:995–1005. doi: 10.1046/j.1471-4159.1997.69030995.x. [DOI] [PubMed] [Google Scholar]
- 347.Haltia M, Ghiso J, Wisniewski T, Kiuru S, Miller D, Frangione B. Gelsolin variant and beta-amyloid co-occur in a case of Alzheimer’s with Lewy bodies. Neurobiol Aging. 1991;12:313–6. doi: 10.1016/0197-4580(91)90007-7. [DOI] [PubMed] [Google Scholar]
- 348.Ji L, Chauhan A, Muthaiyah B, Wegiel J, Chauhan V. Gelsolin levels are increased in the brain as a function of age during normal development in children that are further increased in down syndrome. Alzheimer Dis Assoc Disord. 2009 doi: 10.1097/WAD.0b013e31819d494e. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Morales M, Fifková E. In situ localization of myosin and actin in dendritic spines with the immunogold technique. J Comp Neurol. 1989;279:666–74. doi: 10.1002/cne.902790412. [DOI] [PubMed] [Google Scholar]
- 350.Cheng XT, Hayashi K, Shirao T. Non-muscle myosin IIB-like immunoreactivity is present at the drebrin-binding cytoskeleton in neurons. Neurosci Res. 2000;36:167–73. doi: 10.1016/s0168-0102(99)00123-6. [DOI] [PubMed] [Google Scholar]
- 351.Ryu J, Liu L, Wong TP, et al. A critical role for myosin IIb in dendritic spine morphology and synaptic function. Neuron. 2006;49:175–82. doi: 10.1016/j.neuron.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 352.Amparan D, Avram D, Thomas CG, et al. Direct interaction of myosin regulatory light chain with the NMDA receptor. J Neurochem. 2005;92:349–61. doi: 10.1111/j.1471-4159.2004.02869.x. [DOI] [PubMed] [Google Scholar]
- 353.Correia SS, Bassani S, Brown TC, et al. Motor protein-dependent transport of AMPA receptors into spines during long-term potentiation. Nat Neurosci. 2008;11:457–66. doi: 10.1038/nn2063. [DOI] [PubMed] [Google Scholar]
- 354.Wang Z, Edwards JG, Riley N, et al. Myosin Vb mobilizes recycling endosomes and AMPA receptors for postsynaptic plasticity. Cell. 2008;135:535–48. doi: 10.1016/j.cell.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Naisbitt S, Valtschanoff J, Allison DW, et al. Interaction of the postsynaptic density-95/guanylate kinase domain-associated protein complex with a light chain of myosin-V and dynein. J Neurosci. 2000;20:4524–34. doi: 10.1523/JNEUROSCI.20-12-04524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Osterweil E, Wells DG, Mooseker MS. A role for myosin VI in postsynaptic structure and glutamate receptor endocytosis. J Cell Biol. 2005;168:329–38. doi: 10.1083/jcb.200410091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Stern S, Debre E, Stritt C, Berger J, Posern G, Knöll B. A nuclear actin function regulates neuronal motility by serum response factor-dependent gene transcription. J Neurosci. 2009;29:4512–8. doi: 10.1523/JNEUROSCI.0333-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Segal M. Dendritic spines and long-term plasticity. Nat Rev Neurosci. 2005;6:277–84. doi: 10.1038/nrn1649. [DOI] [PubMed] [Google Scholar]
- 359.Newpher TM, Ehlers MD. Spine microdomains for postsynaptic signaling and plasticity. Trends Cell Biol. 2009;19:218–27. doi: 10.1016/j.tcb.2009.02.004. [DOI] [PubMed] [Google Scholar]