Abstract
The primary sequence of proteins usually dictates a single tertiary and quaternary structure. However, certain proteins undergo reversible backbone rearrangements. Such metamorphic proteins provide a means of facilitating the evolution of new folds and architectures. However, because natural folds emerged at the early stages of evolution, the potential role of metamorphic intermediates in mediating evolutionary transitions of structure remains largely unexplored. We evolved a set of new proteins based on ∼100 amino acid fragments derived from tachylectin-2—a monomeric, 236 amino acids, five-bladed β-propeller. Their structures reveal a unique pentameric assembly and novel β-propeller structures. Although identical in sequence, the oligomeric subunits adopt two, or even three, different structures that together enable the pentameric assembly of two propellers connected via a small linker. Most of the subunits adopt a wild-type-like structure within individual five-bladed propellers. However, the bridging subunits exhibit domain swaps and asymmetric strand exchanges that allow them to complete the two propellers and connect them. Thus, the modular and metamorphic nature of these subunits enabled dramatic changes in tertiary and quaternary structure, while maintaining the lectin function. These oligomers therefore comprise putative intermediates via which β-propellers can evolve from smaller elements. Our data also suggest that the ability of one sequence to equilibrate between different structures can be evolutionary optimized, thus facilitating the emergence of new structures.
Keywords: beta-propellers, conformational diversity, lectins, oligomerization, protein evolution
Maynard-Smith’s conjecture of protein evolution dictates that transitions of function and structure occur gradually (by single mutational steps), and smoothly, namely via intermediates that are all functional (1). This restriction imposes a major challenge, because mutational steps may create large structural perturbations that need to be tolerated while retaining function (2). In single domain proteins, significant “cut and paste” structural changes perturb a well-packed hydrophobic core (3, 4). Evolutionary transitions from one folded and functional domain structure into a new one should therefore (or must, by default, if one follows Maynard-Smith’s conjecture) involve intermediates able to adopt more than one structure (5, 6). It has been also speculated that evolutionary intermediates of many existing folds, and folds that show internal symmetry in particular, were oligomeric assemblies of smaller subunits. Interestingly, most of the reported “metamorphic” proteins—proteins adopting more than one fold (7), are oliogomers in at least one of the two folds they adopt (8–10). Oligomerization may therefore serve as a means of augmenting structural metamorphism and of facilitating the emergence of new folds (11). However, metamorphic proteins are very rarely identified, and their evolutionary relevance is yet to be established. The involvement of structural metamorphism in fold transitions has been reported for a small cysteine-rich domain (12). Different folds have been observed for natural proteins that are close in sequence (40% identity) (9) or artificially designed proteins that are even closer (88% identity, up to a single amino acid difference) (13, 14). Here we show that fragments of ∼100-amino acid length that were truncated from a naturally occurring monomeric protein of 236 amino acids assemble into functional oligomers via the metamorphism of their monomeric subunits.
The evolutionary transitions described here regard β-propellers. These are highly symmetrical, single domain folds in which repeated, four-stranded β-sheets are arranged in a circular, slightly tilted fashion, like the blades of a propeller (15). β-propellers exhibit high similarity in structure and sometimes in sequence, not only between different propellers but also between blades of the same propeller. This implies evolutionary origins involving amplification, fusion, and recombination of small structural elements such as one or two blades (16). In a previous study, we have isolated a set of fragments truncated from a five-bladed β-propeller lectin (sugar-binding protein) dubbed tachylectin-2. These fragments oligomerized spontaneously to form functional homopentameric lectins (17).
How did the transition from a single domain, monomeric, 236-amino acid protein into a homopentamer of 5 × 100 amino acids occur with such ease, and while retaining lectin function? To explore the molecular details of this transition, we applied directed evolution to improve the expression, folding, and assembly of the newly emerged pentameric lectins, thus generating highly expressed, soluble, and functional lectins. Consequently, the crystal structures of two of the evolved pentameric lectins could also be determined.
Results
The homopentameric lectins were isolated from a gene library generated by random digestion of the 5′ and 3′ ends of the tachylectin-2 gene. The library was screened for binding of the glycoprotein mucin. The smallest active fragments from this library were found to be approximately 100 amino acids and to oligomerize into homopentamers as indicated by chemical crosslinking (17). The active variants largely belonged to two different topologies represented by variants Lib2-D2 and Lib1-B7. None of the selected variants corresponded to half a propeller or any other simple architectural subdivision that could yield a homooligomer (e.g., a dimer) with the size of wild-type tachylectin-2. The wild-type derived fragments expressed at very low soluble levels, were highly prone to aggregation, and could not be concentrated above 0.5 mg/ml. We therefore performed directed evolution of the two representative variants toward higher levels of soluble expression.
Directed Evolution Toward Higher Foldability.
Linkers of 3-4-amino acids were added in the library-making procedure at the N and C termini (17). These were removed, and the resulting “net” Lib1-B7 and Lib2-D2 fragments were subjected to rounds of mutagenesis and screening. The randomly mutated fragments were recloned and expressed in Escherichia coli, and the resulting bacterial lysates were screened for mucin binding. The enzyme linked lectin assay (ELLA) screen involved mucin-coated microtiter plates and detection of lectin binding with rabbit polyclonal antibodies generated against wild-type tachylectin-2. As the fragments were evolving for higher folding propensities, decreasing the volume of the bacterial lysate applied to the mucin-coated plates increased the screen’s stringency. After three or four rounds of mutation and selection, several mutations were present in nearly all variants that exhibited enhanced expression and solubility (Table 1 and Table S1). The directed evolution process affected almost exclusively the expression and solubility levels. The affinity for mucin as determined by the ELLA titers remained largely unchanged and similar to that of wild-type techylectin-2. The affinities of individual binding sites for GlcNAc were measured by isothermal calorimetry (ITC) and were also found to be similar to wild-type (Fig. S1). The best evolved variants reached expression levels of up to 100 mg per liter culture (comparing to 100-fold improvement in expression relative to ∼2 fold improvement in sugar binding as determined by ELLA of crude lysates). They could also be readily concentrated to 30 mg/ml. Amongst these, variants Lib1-B7-18 and Lib2-D2-15 produced diffracting crystals.
Table 1.
Sequence changes in variants derived from Lib1-B7 upon directed evolution for higher foldability
| Lib1-B7(net) | |||||||||||||
| Position | 2 | 11 | 20 | 21 | 27 | 31 | 41 | 44 | 45 | 55 | 80 | 95 | |
| Residue | Glu | Asn | Lys | Ile | Asn | Phe | Tyr | Ser | Lys | Gln | Phe | Phe | |
| Round 1 variants * | 1 (SE1E7) | Asp | |||||||||||
| 2 (SE1E8) | Leu | ||||||||||||
| 3 (SE1D9) | Leu | ||||||||||||
| 4 (SE1F2) | Ser | ||||||||||||
| 5 (SE1A3) | Arg | ||||||||||||
| 6 (SE1C11) | Asp | ||||||||||||
| Round 2 variants | 7 (SE2E10) | Ser | Leu | ||||||||||
| 8 (SE2C4) | Ser | Asp | |||||||||||
| 9 (SE2A11) | Ser | Leu | |||||||||||
| 10 (SE2D9) | Asp | Tyr | Asn | ||||||||||
| 11 (SE2A8) | Ser | Leu | |||||||||||
| 12 (SE2F2) | Ser | Leu | |||||||||||
| Round 3 variants | 13 (SE3C11) | Ser | Leu | ||||||||||
| 14 (SE3G8) | Asp | Tyr | Asn | Leu | |||||||||
| 15 (SE3G11) | Ser | Glu | Asp | Asn | |||||||||
| Round 4 variants | 16 (SE4B2) | Gly | Ser | Glu | Asp | Asn | Arg | Leu | |||||
| 17 (SE4H3) | Ser | Glu | Asp | His | Arg | Asn | Leu | ||||||
| 18 (SE4E11) † | Ser | Glu | Asp | Asn | |||||||||
| 19 (SE4H12) | Ser | Glu | Val | Asp | Asn | Leu | |||||||
*For simplicity, the evolved variants derived from the wild-type fragment Lib1-B7 were sequentially numbered as Lib1-B7-1 to Lib1-B7-19. The original annotations of these variants are given in parenthesis.
†Variant Lib1-B7-18, the structure of which is available and its complete amino acid sequence is: MEKGTPPTHDSDNWMGRAKEIGNGGWDQFQFLFFDPNGYLYAVSNDKLYKASPPQSDTDNWIARATEIGSGGWSGFKFLFFHPNGYLYAVRGQRFYKALPPVSNQD
3D Structures.
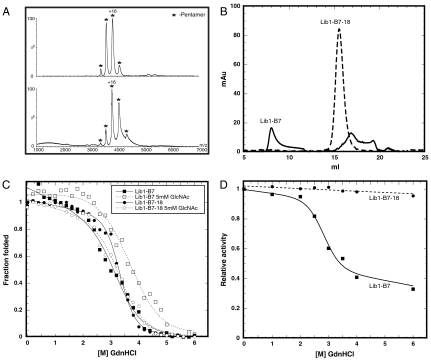
The crystal structures of these oligomeric lectins in complex with the ligand N-acetyl-D-glucosamine (GlcNAc) were determined to 2.5 Å resolution by molecular replacement using the coordinates of wild-type tachylectin-2 (18) (PDB code 1tl2; Table S2). Lib2-D2-15 crystallized with five monomers per asymmetric unit (PDB code 3KIH), whereas the Lib1-B7-18 crystals contained 10 monomers per asymmetric unit (PDB code 3KIF) comprising two pentamers. Both crystal structures were therefore consistent with the pentameric arrangement indicated by crosslinking, mass spectrometry, and gel filtration experiments (Fig. 1 A and B and Figs. S2 and S3).
Fig. 1.
Biophysical features of the pentameric lectin Lib1-B7. Similar patterns observed with Lib2-D2 are described in Fig. S1. (A) Mass spectrometry of a variant from second round of evolution (Lib1-B7-10, Upper Spectrum) and variant Lib1-B7-18 (Lower Spectrum) indicate that pentamers comprised the main oligomeric state throughout the evolution of Lib1-B7 (complementary data are provided as Fig. S2). (B) Elution patterns Lib1-B7 and its evolved variant from a Superdex 200/30 gel filtration column. Lib1-B7 was eluted in several peaks presumably corresponding to high molecular weight aggregates (8.05 ml), pentamers (15.35 ml), and smaller assemblies (≥16.9 ml). In contrast, Lib1-B7-18 was eluted as one peak at 15.5 ml corresponding to pentamers. (C) Guanidinium hydrochloride (GdnHCl) denaturation curves in the absence or presence of the sugar ligand (GlcNAc, 5 mM). Curves systematically deviated from the two-state model, and only apparent D50 values could be derived: 3.26 and 3.71 M for Lib1-B7 without and with ligand, respectively, and 3.32 and 3.34 M for Lib1-B7-18, respectively. (D) Refolding to the native state. The starting point (Lib1-B7) and its evolved variant (Lib1-B7-18) were unfolded in various concentrations of GdnHCl and refolding was induced by dilution into buffer. The fraction of refolded protein was determined by measuring the residual mucin-binding activity.
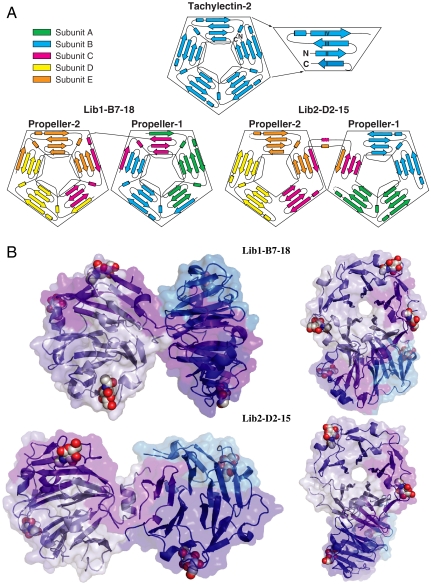
Unique structural rearrangements gave rise to the newly evolved lectins. The pentameric lectins are made from two interconnected, nearly identical five-bladed β-propellers that align almost perfectly with tachylectin-2 (Fig. 2). In Lib1-B7-18, each of the two propellers is composed of two complete subunits (intrapropeller subunits). The third subunit (subunit C) is shared between the two propellers (interpropeller subunit) so that each propeller is comprised of 2.5 monomers that together comprise five blades. As illustrated in Fig. 2A, the topology of the intrapropeller subunits (A, B, D, and E) is 3-4-1. Namely, three strands anneal with another strand from the adjacent subunit to give a complete blade, followed by one complete blade (four strands), and terminated by one strand that anneals with three strands of the next blade. In contrast, the interpropeller subunit (subunit C) exhibits a 1-3-1-3 topology (Fig. 2A). In Lib2-D2-15, the first propeller is composed of two intact subunits, each corresponding to two intact blades (A and B). The fifth blade is contributed by two bridging, interpropeller subunits (subunit C and E). The second propeller is composed of one intact subunit (D, two blades) and the two bridging subunits that contribute the remaining three blades. Thus, the subunits within Lib2-D2-15 adopt three different structures and topologies (a 4-4 topology for the intrapropeller subunits and the asymmetric 3-1-4 and 4-3-1 topologies for the interpropeller subunits C and E, respectively).
Fig. 2.
Structures of the evolved pentameric lectins. (A) Topology diagram depicting the strand rearrangements in wild-type tachylectin-2 versus the oligomeric lectins. Magnification of the first blade with N and C termini of wild-type tachylectin-2 is provided to indicate the Velcro closure that completes the first blade. The subunits of the oligomeric lectins are colored differentially for clarity and are named A to E. (B) The evolved pentamers (shown in two 180° rotated views) comprise two five-bladed propellers connected via a short linker. The individual propellers are essentially identical to each other and to wild-type tachylectin-2 (Cα RMSD values of 0.35 Å between the individual propellers of Lib1-B7-18, and 0.87 and 0.43 Å between these two propellers and wild-type tachilectin-2; 0.44 Å, 0.52, and 0.49 Å, are the respective values for Lib2-D2-15). The five sequence-identical subunits composing the pentamers are colored as follows: intrapropeller subunits in shades of blue, and bridging subunits in purple and light blue. The bound GlcNAc ligands (five in Lib1-B7-18, and four in Lib2-D2-15) are shown in spheres; details of the binding sites are provided as Fig. S5.
The Metamorphic Nature of the Oligomeric Subunits.
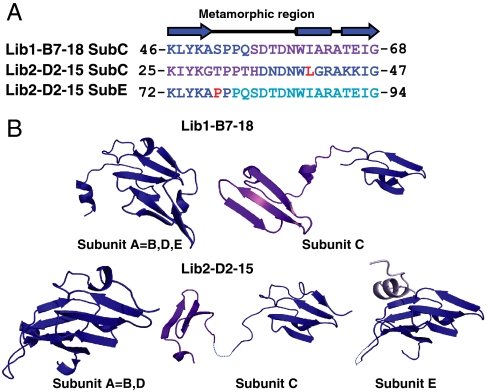
In comparison to the intrapropeller subunits, the interpropeller, bridging subunits (subunit C in Lib1-B7-18 and subunits C and E in Lib2-D2-15) adopt distinctly different topologies and structures that are necessary to complete the two five-bladed propellers and connect them. The metamorphic nature of the subunits is manifested in a structural rearrangement within a long stretch that includes the third and the fourth β-strands of the wild-type blades and a helix-turn motif that separates these strands (Fig. 3A). This resulted in “open” structures that enable the subunit to fulfill its role in bridging and completing the two five-bladed propellers (Fig. 3B). These rearrangements brake polar and hydrophobic interactions involved in the formation of the intact propellers (in both wild-type tachilectin-2, and in the intrapropeller subunits of the evolved lectins) and generate new sets of interactions with the neighboring propeller (Fig. S4). Stabilization of truncated proteins through domain swapping and oligomerization has been observed (8, 19, 20). In the case of tachylectin-2, however, the open bridging subunits (Fig. 3B) exchange strands asymmetrically (Fig. 2A) in a manner that deviates from the regular mode of domain swapping (21). The ability of the same subunit to adopt different structures and interaction modes is a prerequisite for the oligomeric assembly and formation of a functional protein. The mode of interactions between strands also differs from the regular, symmetric mode of strand exchange. In the evolved variants, Velcro closures modes of 3 + 1 are observed, as opposed to the 1 + 3 closures in wild-type tachylectin-2 (one strand from one end of the polypeptide chain assembles with three strands from another end to give a complete blade; Fig. 2A, Top). Further, in both pentameric variants, strands coming from the same blade in tachylectin-2 were asymmetrically split between two different blades (Fig. 2A, Bottom). The differences in the packing interactions of the secondary structure elements, and in the conformations of the loops connecting them, therefore represent cases of structural metamorphism that is required for correct assembly of the pentamers. As expected (22), oligomerization via Velcro closures between all blades endowed much higher thermodynamic stability (Lib1-B7-18; D50 for guanidinium hydrochloride unfolding = 3.3 M; Fig. 1C) than the mode with the Velcro closure only in the bridging subunits (Lib2-D2-15; D50 = 1.1 M; Fig. S2C).
Fig. 3.
Structural metamorphism in the evolved oligomeric lectins. (A) Sequence alignment of the metamorphic region in subunit C of Lib1-B7-18 with the two metamorphic regions observed in subunits C and E of Lib2-D2-15. This region is a stretch of 23 amino acids that in wild-type tachylectin-2 comprises the third and the fourth strands of a blade and provides part of the sugar-binding site. Regions with wild-type-like structures are colored in blue and the alternative structure is colored according to Fig. 2B. Residues mutated during rounds of evolution are marked in red. (B) Structures of the intrapropeller subunits represented by subunit A in each variant and the interpropeller bridging subunits (subunits C and E in Lib2-D2-15, and subunit C in Lib1-B7-18). A short segment within the bridging subunit C in Lib2-D2-15 (within the linker connecting the two propellers), and a short loop in subunit E, appear to be disordered and are presented as dashed lines.
Evolution of Higher Foldability.
The pentameric arrangement already appeared in the initially isolated fragments derived directly from wild-type tachilectin-2 and was maintained throughout the process of optimization for higher expression and solubility (Fig. 1 and Figs. S2 and S3). Hence, the metamorphic potential of these subunits existed in the wild-type sequence and was unraveled upon truncation into smaller fragments. However, truncation of the wild-type protein that evolved to be monomeric and monomorphic (one sequence—one structure) resulted in impaired folding and stability. The initially isolated pentamers were expressed primarily as insoluble inclusion bodies. Size exclusion chromatography and mass spectrometry indicated heterogeneity that included assemblies larger than pentamers as well as smaller forms (Fig. 1A and B). However, the mutations that followed the evolution for higher expression, solubility, and structural homogeneity do not seem to have improved the thermodynamic stability of the pentamers (i.e., the stability of the folded, oligomeric state) (Fig. 1C). Rather, they seem to have primarily optimized the kinetic stability of the evolved variants (8, 23) and thereby their ability to refold (Fig. 1D). Specifically, the relative abundance of the different conformers, and/or the energetic barrier that separates them, is likely to have been optimized.
The above hypothesis is supported by analysis of the mutations that accumulated in the evolved variants Lib1-B7-18 and Lib2-D2-15. One factor to consider is the linkage between the protein’s isoelectric point (pI) and soluble expression in E. coli (24). The evolved variant Lib1-B7-18 has a pI of 6.1, versus 8.9 for its starting point Lib1-B7. Similarly, the pI of Lib2-D2-15 dropped from to 8.3 to 7.0 in the evolved variant. However, although better expression in E. coli also correlates with increased thermodynamic stability (25, 26), higher soluble expression of the pentameric variants was not accompanied by increased stability. Concomitantly, most of the mutations do not seem to affect the packing, and thereby stability, of the folded oligomeric forms. In the evolved variant Lib1-B7-18, only mutation Lys20Glu seems to contribute to the formation of a new salt bridge with Arg94 of the adjacent subunit and may therefore improve the Velcro closure between subunits (Fig. S5A). In Lib2-D2-15, the mutation Asn48Asp could lead to a new salt bridge with the Lys25, and/or Lys45 and may thus stabilize blades within the same subunit (Fig. S5B). The other mutations observed in the evolved variants do not seem to create new contacts that can stabilize the folded, oligomeric state. Indeed, in Lib2-D2-15, the remaining two mutations are in the metamorphic region of the subunit (Fig. 3A). Moreover, equilibrium unfolding induced by guanidinium hydrochloride indicated that despite ≥100-fold improvements in the levels of expression of soluble and functional lectins, the evolved variants showed similar, or even slightly lower D50 values than those of the starting points (the fragments of wild-type tachylectin-2). Both the starting points and the evolved variants show similar increases in stability upon addition of the ligand GlcNAc (as does wild-type tachtlectin-2), further suggesting no significant changes in the folded, oligomeric state. Further, size exclusion chromatography indicated that unlike the wild-type derived fragments, the evolved variants elute as one species (Fig. 1B and Fig. S2B). The increased kinetic stability is also manifested in the fact that the evolved lectins refold fully to the native functional state after denaturation with guanidinium chloride, while the starting points do not (Fig. 1D and Fig. S2D). It is therefore likely that the major contribution of the mutations relate to the folding trajectory and the ability to achieve the right stoichiometry between the various conformers needed to assemble the oligomeric lectin. In other words, it appears that the laboratory evolution process optimized the metamorphic nature of these lectin subunits.
Discussion
Our results highlight the remarkably modular and plastic nature of the β-propeller fold, and possibly other folds exhibiting internal symmetry. This structural malleability could be one of the reasons β-propellers are so abundant and structurally variable—structures of propellers with four to 10 blades are known. These structures also support the notion that β-propellers evolved via the multiplication of smaller gene segments (16) and could assemble with different numbers of such elements. In contrast, (β/α)8-barrels are also thought to have evolved through a process of duplication and fusion of smaller units (4, 27, 28) but are known to exist with fourfold symmetry only (8 β/α modules).
Two new β-propeller arrangements (and possibly many more) that are functional could be easily derived from the same starting protein. Interestingly, one of the new structures is reminiscent of a natural oligomeric β-propeller lectin that comprises a trimer of two-blade subunits (29). The resulting six-bladed β-propeller is formed via blade-blade interaction similar to those seen in variant Lib2-D2-15. Thus, our in vitro experiment reproduced at least one type of fold exchange that occurred in nature, between oligomers comprised of two-blade subunits and monomeric multiblade propellers. The second variant (Lib1-B7) explores a different mode of oligomerization by which strand exchanges between subunits contribute to the formation of individual blades and to the completion of the overall structure, similarly to the commonly observed Velcro clasps of β-propellers (Fig. 2A). Indeed, the interpropeller subunit of Lib1-B7-18 exhibits a 1-3-1-3 topology (subunit C, Fig. 2A), which is reminiscent of the noncanonical arrangement observed in the β-propeller domain of DNA gyrase A (30).
In both variants, the metamorphic nature of the subunits underlies the emergence of the novel oligomeric structures. The five subunits of the oligomeric lectins, although identical in sequence, adopt two, or even three, distinct structures and can thus exchange strands with up to four subunits in an asymmetric manner. Thus, more than one structure can be derived from the same sequence as previously noted (9, 10, 14, 25, 31, 32). In the pentameric lectins, however, the different structures must coincide within the same oligomer to enable its assembly. The asymmetric, irregular nature of the observed exchanges of strands and blades, and subtle differences in the interactions between strands and in the conformations of their connecting loops, differ from the commonly observed mode of domain swapping or strand exchanges (20) and may thus comprise a case of backbone metamorphism (7). Asymmetric assemblies of natural oligomers are rare, but not unprecedented, as indicated by IscA (33). The newly evolved pentameric lectins may therefore represent evolutionary intermediates in which structural metamorphism facilitates changes in quaternary structure and/or functional changes and thereby provide a vivid example for the role structural plasticity plays in protein evolution. The evolution of β-propellers is likely to have proceeded through stepwise additions of smaller elements (16). Our results indicate that such elements could be functional via oligomeric assembly and that backbone plasticity can mediate the assembly of such small elements to give functional oligomers. Future work may reveal whether truncation followed by metamorphic mediated oligomerization comprises a generic mechanism by which new folds and functions may evolve.
The oligomeric subunits described here were derived directly from an existing structure, and thus their metamorphic potential was not selected for (or was possibly selected against, because it may have collided with correct folding of the monomeric, monomorphic tachylectin-2) (34). Thus, metamorphic traits can lie dormant in monomorphic or heteromorphic structures (35) and be unraveled by mutations that introduce new stop or start codons. Notably, in this case and others (7), oligomerization seems to mediate metamorphism, because the alternative structures appear to fold and be stabilized within an oligomeric context (9, 10). While truncation and oligomerization resulted in aggregation, it had little effect on the binding affinity that remained comparable to wild-type tachylectin-2. Indeed, the structures and ITC measurements indicate similar binding properties for the individual binding sites of tachylectin-2 and of the newly evolved pentamers (Figs. S1 and S6).
Refinement of the new oligomeric structures involved mutations that seem to be optimizing the transition and/or the stoichiometry between different structural conformers of the same sequence. Mutations leading to structural metamorphism have been previously reported (12, 31, 32, 36). Here, these mutations were not designed, but were selected from random repertoires by virtue of their ability to refine of the functional assembly of the new oligomeric forms. Thus, scaffold metamorphism need not be an accidental side effect but can, when necessary, become an evolvable trait. As demonstrated here, structural polymorphism facilitated the evolutionary transitions that led to the new structures, including changes in secondary, tertiary, and quaternary structure. Such structural transitions might be related to changes in binding specificity, especially in lectins, where changes in quaternary structure provide new multivalent binding modes (37). Indeed, while the original binding specificity toward GlcNAc has been maintained, the new oligomeric forms show subtle changes in specificity, in particular with respect to complex sugars containing GlcNAc (Fig. S7). Thus, as is the case with local structural plasticity within protein active sites (38), scaffold plasticity could be one of the keys to rapid evolution of new protein functions, folds, and architectures.
Materials and Methods
A detailed description of materials and methods is available in SI Text.
The “net” variants: Four amino acids (RVTK) added during the incremental truncation procedure were removed from the C terminus of Lib1-B7 and Lib2-D2 and replaced by a stop codon followed by a NotI restriction site. In Lib2-D2, the additional N terminus amino acids (VE) were similarly removed. To remove the added amino acids (VD) at the N terminus of Lib1-B7 while maintaining the NcoI site, a small combinatorial library was prepared. Screening for mucin binding indicated that the Glu right after the first Met yielded the highest activity. The resulting net variants, that were identical in sequence to wild-type tachylectin-2 with no additional aminoacids, were subjected to random mutagenesis.
Library-Making.
Mutagenesis was performed by error-prone PCR, and the conditions were adjusted to give an average of one nonsynonymous mutation per gene. The open reading frames of the net variants Lib1-B7 and Lib2-D2 were cloned into pETtr using the NcoI/NotI sites, yielding pETtrB7 or pETtrD2, respectively, and were mutated by amplifying 20–200 ng/ml plasmid templates at three different nucleotide bias ratios: 1∶5, 1∶10, 1∶20 dG/TTP to dA/CTP. Alternatively, error-prone PCR was performed by a single reaction on 20–200 ng plasmid template using GenemorphII© (Stratagene). The mutated PCR fragments were recloned back into pETtr, transformed to E. coli DH5-α cells, and the library plasmid DNAs were extracted.
Enzyme Linked Lectin Assay (ELLA) Screens.
Library plasmids were transformed into E. coli BL21 (DE3) cells for expression. The transformed cells were plated on agar plates supplemented with 100 μg/ml ampicillin and 1% glucose. Individual colonies were picked and grown in 96-well plates containing 500 μl LB and 100 μg/ml ampicillin. The starters were then transferred into new 96 plates containing LB (500 μl) and ampicillin (100 μg/ml) and grown to an OD600 nm of 0.6. IPTG was added to a final concentration of 0.4 mM and the culture shaken for 4.5 h at 37 °C. Cells were harvested, and the bacterial pellet was lysed and supplemented with protease inhibitor. The crude lysates were diluted and incubated on mucin-coated (5 μg/ml mucin in PBS) 96-well plates, in buffer containing 0.2% BSA for 1 h, and washed three times with PBS containing 0.5% tween-20. Binding was detected using polyclonal rabbit anti-tachylectin-2 antibodies, followed by goat-anti-rabbit HRP antibodies, and the TMB substrate (Dako). To increase the stringency of the screen between rounds of evolution, the amount of bacterial lysate applied to the plates was reduced to the minimal volume that enabled detection for the best clones from the previous round (5 μl at the first round, down to 0.005 μl at the fourth round).
Protein Expression and Purification.
Lectin variants were expressed in BL-21(DE-3) from pET32b (Novagen) vector from which the Trx fusion protein and peptide tags were removed (pET32-tr). The expressed proteins were purified on GlcNAc agarose as described (17). Selenomethionine Lib1-B7-18 was produced using the Van Duyne et al. protocol (39).
Crystallization, Data Collection and Refinement.
Crystals of Lib1-B7-18 were obtained by the hanging drop method. Selenomethionine Lib1-B7-18 crystals were grown from a solution of 100 mM Bis-Tris pH = 5.5, 10 mM GlcNAc, 0.2 M lithium chloride, and 19% PEG 3350. Crystals formed in a space group P32 with cell constants a = b = 80.56 Å, c = 170.68 Å, and contain 10 monomers in the asymmetric unit cell with Vm of 2.66 Å3/Dalton. The data to 2.5 Å resolution from a single crystal were collected at the European Synchrotron Radiation Facility (ESRF) beam line ID14-4. The crystals of Lib2-D2-15 obtained under oil by the microbatch method using the Oryx6 robot (Douglas Instruments Ltd., East Garston, Hungerford, Berkshire, UK), diffracted to 2.5 Å resolution. Lib2-D2-15 crystals were grown from a solution of 100 mM HEPES pH = 7.0, 10 mM GlcNAc, 0.2 M magnesium chloride, and 20% PEG 6000. Crystals formed in a space group C2 with cell constants a = 125.56 Å, b = 56.42 Å, c = 86.31 Å, and β = 122.90° and contain five monomers in the asymmetric unit cell with Vm of 2.41 Å3/Dalton. A complete dataset up to 2.5 Å was collected. The structures of Lib2-D2-15 and Lib1-B7-18 were determined by molecular replacement using the known structure of wild-type tachylectin-2 (PDB code 1TL2) (18) as a starting model without the need for a SeMet MAD experiment. Details regarding the processing of the diffraction data and refinement of the model are available in SI Text. The refinement statistics are presented in Table S2. The coordinates and structure factors have been deposited in the RCSB PDB under accession No. 3KIH and 3KIF respectively. All figures depicting structures were prepared using PyMOL (DeLano Scientific LLC).
Mass Spectrometry.
Structural mass spectrometry analysis was carried out on a nanoflow ESI QSTAR Elite (Sciex) mass spectrometer modified for transmission and isolation of high mass ions (40). Prior to MS analysis, 25 μl of up to 100 µM sample was buffer-exchanged into 0.5 M ammonium acetate pH 7.5 using microspin columns, and 1.5 μl aliquots were introduced via nanoflow capillaries prepared in-house. Throughout the analysis, conditions within the mass spectrometer were adjusted to preserve noncovalent interactions. The following experimental parameters were used: capillary voltage up to 1.2 kV, declustering potential up to 150 V, focusing potential 250 V, second declustering potential 55 V, and focusing rod offset ranging from 20 to 100 V, MCP 2350 V. All spectra were calibrated externally by use of a solution of cesium iodide. Spectra are shown here with minimal smoothing and without background subtraction.
Supplementary Material
Acknowledgments.
We thank Korina Goldin for technical assistance and Sebastian Meier, Birte Höcker, Colin Jackson, Alexey G. Murzin, and Leo James for their insightful comments. Financial support by the Israel Science Foundation and the Sasson and Marjorie Peress Philanthropic Fund is gratefully acknowledged. We thank to the Consortium of Functional Glycomics for performing the sugar-binding arrays.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912616107/DCSupplemental.
References
- 1.Maynard-Smith J. Natural selection and the concept of a protein space. Nature. 1970;225:563–564. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- 2.Andreeva A, Murzin AG. Evolution of protein fold in the presence of functional constraints. Curr Opin Struct Biol. 2006;16:399–408. doi: 10.1016/j.sbi.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Peisajovich SG, Rockah L, Tawfik DS. Evolution of new protein topologies through multistep gene rearrangements. Nat Genet. 2006;38:168–174. doi: 10.1038/ng1717. [DOI] [PubMed] [Google Scholar]
- 4.Claren J, Malisi C, Hocker B, Sterner R. Establishing wild-type levels of catalytic activity on natural and artificial (beta alpha) 8-barrel protein scaffolds. Proc Natl Acad Sci USA. 2009;106:3704–3709. doi: 10.1073/pnas.0810342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meier S, Ozbek S. A biological cosmos of parallel universes: does protein structural plasticity facilitate evolution? Bioessays. 2007;29:1095–1104. doi: 10.1002/bies.20661. [DOI] [PubMed] [Google Scholar]
- 6.Meyerguz L, Kleinberg J, Elber R. The network of sequence flow between protein structures. Proc Natl Acad Sci USA. 2007;104:11627–11632. doi: 10.1073/pnas.0701393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murzin AG. Biochemistry. Metamorphic proteins. Science. 2008;320:1725–1726. doi: 10.1126/science.1158868. [DOI] [PubMed] [Google Scholar]
- 8.Murray AJ, Head JG, Barker JJ, Brady RL. Engineering an intertwined form of CD2 for stability and assembly. Nat Struct Biol. 1998;5:778–782. doi: 10.1038/1816. [DOI] [PubMed] [Google Scholar]
- 9.Roessler CG, et al. Transitive homology-guided structural studies lead to discovery of Cro proteins with 40% sequence identity but different folds. Proc Natl Acad Sci USA. 2008;105:2343–2348. doi: 10.1073/pnas.0711589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuinstra RL, et al. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc Natl Acad Sci USA. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alva V, Koretke KK, Coles M, Lupas AN. Cradle-loop barrels and the concept of metafolds in protein classification by natural descent. Curr Opin Struct Biol. 2008;18:358–365. doi: 10.1016/j.sbi.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Meier S, et al. Continuous molecular evolution of protein-domain structures by single amino acid changes. Curr Biol. 2007;17:173–178. doi: 10.1016/j.cub.2006.10.063. [DOI] [PubMed] [Google Scholar]
- 13.Alexander PA, He Y, Chen Y, Orban J, Bryan PN. The design and characterization of two proteins with 88% sequence identity but different structure and function. Proc Natl Acad Sci USA. 2007;104:11963–11968. doi: 10.1073/pnas.0700922104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexander PA, He Y, Chen Y, Orban J, Bryan PN. A minimal sequence code for switching protein structure and function. Proc Natl Acad Sci USA. 2009;106:21149–21154. doi: 10.1073/pnas.0906408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fulop V, Jones DT. Beta propellers: Structural rigidity and functional diversity. Curr Opin Struct Biol. 1999;9:715–721. doi: 10.1016/s0959-440x(99)00035-4. [DOI] [PubMed] [Google Scholar]
- 16.Chaudhuri I, Soding J, Lupas AN. Evolution of the beta-propeller fold. Proteins. 2008;71:795–803. doi: 10.1002/prot.21764. [DOI] [PubMed] [Google Scholar]
- 17.Yadid I, Tawfik DS. Reconstruction of functional beta-propeller lectins via homo-oligomeric assembly of shorter fragments. J Mol Biol. 2007;365:10–17. doi: 10.1016/j.jmb.2006.09.055. [DOI] [PubMed] [Google Scholar]
- 18.Beisel HG, Kawabata S, Iwanaga S, Huber R, Bode W. Tachylectin-2: Crystal structure of a specific GlcNAc/GalNAc-binding lectin involved in the innate immunity host defense of the Japanese horseshoe crab Tachypleus tridentatus. EMBO J. 1999;18:2313–2322. doi: 10.1093/emboj/18.9.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Bono S, Riechmann L, Girard E, Williams RL, Winter G. A segment of cold shock protein directs the folding of a combinatorial protein. Proc Natl Acad Sci USA. 2005;102:1396–1401. doi: 10.1073/pnas.0407298102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gronenborn AM. Protein acrobatics in pairs—Dimerization via domain swapping. Curr Opin Struct Biol. 2009;19:39–49. doi: 10.1016/j.sbi.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: A mechanism for oligomer assembly. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neer EJ, Smith TF. G protein heterodimers: New structures propel new questions. Cell. 1996;84:175–178. doi: 10.1016/s0092-8674(00)80969-1. [DOI] [PubMed] [Google Scholar]
- 23.Baker D, Agard DA. Kinetics versus thermodynamics in protein folding. Biochemistry. 1994;33:7505–7509. doi: 10.1021/bi00190a002. [DOI] [PubMed] [Google Scholar]
- 24.Mehlin C, et al. Heterologous expression of proteins from Plasmodium falciparum: Results from 1000 genes. Mol Biochem Parasitol. 2006;148:144–160. doi: 10.1016/j.molbiopara.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Mayer S, Rudiger S, Ang HC, Joerger AC, Fersht AR. Correlation of levels of folded recombinant p53 in Escherichia coli with thermodynamic stability in vitro. J Mol Biol. 2007;372:268–276. doi: 10.1016/j.jmb.2007.06.044. [DOI] [PubMed] [Google Scholar]
- 26.Roodveldt C, Aharoni A, Tawfik DS. Directed evolution of proteins for heterologous expression and stability. Curr Opin Struct Biol. 2005;15:50–56. doi: 10.1016/j.sbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Bharat TA, Eisenbeis S, Zeth K, Hocker B. A beta alpha-barrel built by the combination of fragments from different folds. Proc Natl Acad Sci USA. 2008;105:9942–9947. doi: 10.1073/pnas.0802202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lang D, Thoma R, Henn-Sax M, Sterner R, Wilmanns M. Structural evidence for evolution of the beta/alpha barrel scaffold by gene duplication and fusion. Science. 2000;289:1546–1550. doi: 10.1126/science.289.5484.1546. [DOI] [PubMed] [Google Scholar]
- 29.Kostlanova N, et al. The fucose-binding lectin from Ralstonia solanacearum. A new type of beta-propeller architecture formed by oligomerization and interacting with fucoside, fucosyllactose, and plant xyloglucan. J Biol Chem. 2005;280:27839–27849. doi: 10.1074/jbc.M505184200. [DOI] [PubMed] [Google Scholar]
- 30.Corbett KD, Shultzaberger RK, Berger JM. The C-terminal domain of DNA gyrase A adopts a DNA-bending beta-pinwheel fold. Proc Natl Acad Sci USA. 2004;101:7293–7298. doi: 10.1073/pnas.0401595101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cordes MH, Burton RE, Walsh NP, McKnight CJ, Sauer RT. An evolutionary bridge to a new protein fold. Nat Struct Biol. 2000;7:1129–1132. doi: 10.1038/81985. [DOI] [PubMed] [Google Scholar]
- 32.He Y, Chen Y, Alexander P, Bryan PN, Orban J. NMR structures of two designed proteins with high sequence identity but different fold and function. Proc Natl Acad Sci USA. 2008;105:14412–14417. doi: 10.1073/pnas.0805857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morimoto K, et al. The asymmetric IscA homodimer with an exposed [2Fe-2S] cluster suggests the structural basis of the Fe-S cluster biosynthetic scaffold. J Mol Biol. 2006;360:117–132. doi: 10.1016/j.jmb.2006.04.067. [DOI] [PubMed] [Google Scholar]
- 34.Stott KM, Yusof AM, Perham RN, Jones DD. A surface loop directs conformational switching of a lipoyl domain between a folded and a novel misfolded structure. Structure. 2009;17:1117–1127. doi: 10.1016/j.str.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Alexander PA, Rozak DA, Orban J, Bryan PN. Directed evolution of highly homologous proteins with different folds by phage display: Implications for the protein folding code. Biochemistry. 2005;44:14045–14054. doi: 10.1021/bi051231r. [DOI] [PubMed] [Google Scholar]
- 36.Cordes MH, Walsh NP, McKnight CJ, Sauer RT. Evolution of a protein fold in vitro. Science. 1999;284:325–328. doi: 10.1126/science.284.5412.325. [DOI] [PubMed] [Google Scholar]
- 37.Srinivas VR, et al. Legume lectin family, the ‘natural mutants of the quaternary state’ provide insights into the relationship between protein stability and oligomerization. BBA-Gen Subjects. 2001;1527:102–111. doi: 10.1016/s0304-4165(01)00153-2. [DOI] [PubMed] [Google Scholar]
- 38.Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 39.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 40.Chernushevich IV, Thomson BA. Collisional cooling of large ions in electrospray mass spectrometry. Anal Chem. 2004;76:1754–1760. doi: 10.1021/ac035406j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.