Abstract
Chronic beryllium disease (CBD) is a fibrotic lung disorder caused by beryllium (Be) exposure and is characterized by granulomatous inflammation and the accumulation of Be-responsive CD4+ T cells in the lung. Genetic susceptibility to CBD has been associated with certain alleles of the MHCII molecule HLA-DP, especially HLA-DPB1*0201 and other alleles that contain a glutamic acid residue at position 69 of the β-chain (βGlu69). The HLA-DP alleles that can present Be to T cells match those implicated in the genetic susceptibility, suggesting that the HLA contribution to disease is based on the ability of those molecules to bind and present Be to T cells. The structure of HLA-DP2 and its interaction with Be are unknown. Here, we present the HLA-DP2 structure with its antigen-binding groove occupied by a self-peptide derived from the HLA-DR α-chain. The most striking feature of the structure is an unusual solvent exposed acidic pocket formed between the peptide backbone and the HLA-DP2 β-chain α-helix and containing three glutamic acids from the β-chain, including βGlu69. In the crystal packing, this pocket has been filled with the guanidinium group of an arginine from a neighboring molecule. This positively charged moiety forms an extensive H-bond/salt bridge network with the three glutamic acids, offering a plausible model for how Be-containing complexes might occupy this site. This idea is strengthened by the demonstration that mutation of any of the three glutamic acids in this pocket results in loss of the ability of DP2 to present Be to T cells.
Keywords: antigen presentation, lung disease, metal hypersensitivity
Particular alleles of the three major human MHC class II (MHCII) isotypes, HLA-DR, -DP, and -DQ, have been associated with many chronic inflammatory diseases (1). For example, the major genetic contribution to rheumatoid arthritis involves DRB1 alleles such as DRB1*0401 and *0404 (2), whereas HLA-DQ alleles, especially DQ2 and DQ8, provide the major genetic contribution to type 1 diabetes (3). In addition, certain HLA-DP alleles have been associated with pauci-articular juvenile chronic arthritis (4), Graves' disease (5), Takayasu's arteritis (6), and hard metal lung disease (7).
HLA-DP alleles have also been strongly associated with chronic beryllium disease (CBD), an inflammatory T-cell-mediated lung disease caused by hypersensitivity to beryllium (Be) (8–12). The disease is characterized by granulomatous inflammation and the influx of beryllium-responsive, Th1-polarized CD4+ T cells to the lung, resulting in the development of lung fibrosis (13). Exposure to Be in the workplace continues to be a public health concern with ≈1 million individuals exposed and potentially at risk for developing CBD (14). Of those exposed, CBD develops in up to 18% of workers, depending on the nature of the exposure (15–17) and the genetic susceptibility of the individual (8–12). Genetic susceptibility to CBD has been linked to HLA-DP alleles that contain a glutamic acid at amino acid position 69 of the β-chain (βGlu69) (8–12). When Be-reactive lung T cells are tested for an in vitro response to Be, the DP alleles implicated in disease susceptibility are the same as those shown to present Be to T cells (18, 19). In addition, soluble HLA-DP molecules expressing βGlu69, but not HLA-DP molecules with a lysine at that position, can bind beryllium in vitro with high affinity (20, 21). Thus, these findings strongly suggest that the ability to bind and present Be is the reason for the association of certain HLA-DP alleles and disease susceptibility.
The structures of a number of disease-associated DR and DQ alleles have been reported, but the DP isotype has been recalcitrant to attempts at crystallization. Therefore, the unique structural features of βGlu69-containing HLA-DP molecules that explain the disease association have remained unknown despite multiple attempts to model the protein (22–25). Because HLA-DP2 is the most prevalent of the βGlu69-containing HLA-DP alleles, we crystallized DP2 (DPA1*0103, DPB1*0201) with a bound self-peptide derived from the HLA-DR α-chain (pDRA) (26) and solved its structure to a resolution of 3.25 Å. Although the overall structure of the DP2-pDRA complex is similar to that of other MHCII/peptide complexes, the structure reveals a unique solvent-exposed acidic pocket containing three glutamic acids from the β-chain, including βGlu69, which can explain its ability to present Be to T cells. In the crystal, this pocket was filled by the guanidinium group of an arginine from a neighboring molecule, which formed an extensive H-bond/salt bridge network with the three glutamic acid residues. This network suggests a plausible model for how Be-containing complexes might occupy this site and subsequently be recognized by Be-reactive T cells.
Results
Properties of the DP2 Peptide-Binding Groove.
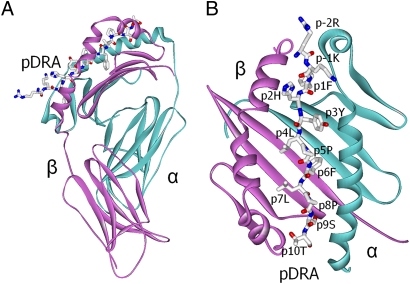
To help understand how DP2 contributes to disease susceptibility, we expressed a soluble version (27) of DP2 (Fig. S1 A and B) with a covalently linked peptide derived from the HLA-DR α-chain (pDRA). This peptide is well represented among processed self-peptides found naturally bound to DP2 (26). We purified, crystallized, and solved the structure of the DP2-pDRA complex as described in Materials and Methods. The structure was refined to a resolution of 3.25 Å. The statistics for the data collection and refined model are shown in Table 1. The overall structure of the DP2-pDRA complex is similar to that of other MHCII-peptide complexes (Fig. 1 A and B). The α1 and β1 domains of DP2 form the peptide-binding groove, and the peptide takes an extended polyproline-like helical course through the groove. Four pockets within the peptide binding groove of DP2 are positioned to accept the side chains of peptide amino acids at p1, p4, p6, and p9 (Fig. 2A).
Table 1.
Data collection and refinement statistics
| Data set 1 | Data set 2 | Merged | |
| Data collection | |||
| Space group | P3121 | P3121 | P3121 |
| Cell dimensions | |||
| a, b, c (Å) | 157.5, 157.5, 61.8 | 157.2, 157.2, 61.8 | 157.3, 157.3, 61.8 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 50–3.5 (3.69–3.5) | 50–3.25 (3.43–3.25) | 50–3.25 (3.43–3.25) |
| Rpim (%) | 13.5 (51.3) | 6.9 (33.5) | 11.2 (33.5) |
| Rmerge (%) | 18.5 (99.3) | 13.9 (98.8) | |
| I/σI | 8.3 (1.9) | 11.2 (2.3) | 12.3 (2.3) |
| Completeness (%) | 100 (100) | 87.1 (89.1) | 100 (89.1) |
| Redundancy | 7.1 (7.3) | 6.0 (6.0) | 12.3 (6.0) |
| Refinement | |||
| Resolution (Å) | 50–3.25 (3.33–3.25) | ||
| No. reflections | 12,130 (1,940) | ||
| Rwork/Rfree* | 22.6/25.1 (33.1/35.2) | ||
| No. molecules in ASU | 1 | ||
| No. atoms | |||
| Protein | 3,115 | ||
| Carbohydrate | 42 | ||
| Water | 0 | ||
| B factors | |||
| Overall | 102.8 | ||
| Peptide | 101.1 | ||
| Carbohydrates | 116.3 | ||
| Rmsd | |||
| Bond lengths (Å) | 0.007 | ||
| Bond angles (°) | 1.15 |
Rpim, redundancy-independent merging R factor; ASU, asymmetric units. Data for outer shell shown in parentheses.
*Rfree test set size 5%, 704.
Fig. 1.
Overview of the DP2-pDRA structure. (A) Ribbon representations of the full DP2 α (cyan) and β (magenta) extracellular domains and a wireframe representation of the pDRA peptide with Corey–Pauling–Koltun (CPK) coloring viewed from the β-chain side. (B) Top view as in A, but without the α2 and β2 domains. Individual amino acids in the peptide are labeled.
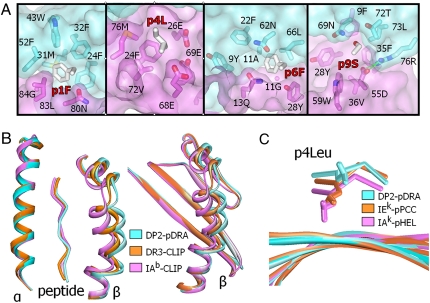
Fig. 2.
Unusual properties of the DP2 peptide-binding groove. (A) Details of the p1 (first panel), p4 (second panel), p6 (third panel), and p9 (last panel) binding pockets are shown. In each panel, the semitransparent water-accessible surfaces of the DP2 α1 (cyan) and β1 (magenta) domains are shown, as well as wireframe representations of the side chain of the pDRA amino acid at that position (CPK coloring) and the DP2 amino acids lining the pocket (CPK coloring, except for carbons: DP2 α1 carbons, dark cyan; and DP2 β1 carbons, dark magenta). (B) Three MHCII structures were overlaid on the basis of their α1 domains: DP2-pDRA (cyan), HLA-DR3 bound to the invariant chain CLIP peptide (PDB ID code 1A6A, orange) and mouse IAb bound to CLIP peptide (PDB ID code 1MUJ, magenta). (Left) MHCII α-helices as ribbons and the peptide from p1 to p9 as a CA stick. (Right) Full β1 domains as ribbons. (C) Three MHCII structure were overlaid on the basis of the four central β-strands of the floor of the peptide-binding groove: DP2-pDRA (cyan), mouse IEk bound to a peptide from pigeon cytochrome c (PCC; PDB ID code 1KTD, orange) and mouse IAk bound to a peptide from hen egg lysozyme (HEL; PDB ID code 1IAK). The figure shows the four central β-strands as ribbons, the peptide from p3 to p6 as a CA stick, and a wireframe representation of the side chain of the p4Leu in each structure.
The hydrophobic p1 pocket (Fig. 2A, panel 1) is similar to that found in a number of MHCII molecules, such as HLA-DR1 and HLA-DR4, that have a Gly at β86 (corresponding to position 84 in DPβ) (28). This deep, hydrophobic pocket can accommodate virtually all hydrophobic residues, including large aromatic amino acids such as the p1Phe of pDRA. The p4 binding pocket (Fig. 2A, panel 2) of DP2 is large, but shallow, shaped on one side by βGlu26, βGlu68, and βGlu69, and on the other side by βPhe24, βVal72, and βMet76. In agreement with DP2 peptide-binding studies (23, 26), the structure suggests that the p4 pocket could accept either positively charged amino acids, such as Arg or Lys, or hydrophobic amino acids, such as the p4Leu of pDRA. However, the side chain of this Leu does not fill the pocket.
In most MHCII molecules in mouse and human, the p6 pocket selects for small, sometimes polar or even charged, but rarely aromatic, amino acids. However, the p6 pocket of DP2 (Fig. 2A, panel 3) is large and deep and can accommodate aromatic amino acids such as the p6Phe of pDRA. It is lined with hydrophobic and polar residues, and its depth is due to the amino acids αAla11 and βGly11, whose small side chains open up the bottom of the pocket (Fig. 2A). Thus, the structure explains the preference for hydrophobic, aromatic amino acids at the p1 and p6 positions of peptides that bind to DP2 (23, 26), giving DP2 a unique peptide-binding motif. The p9 binding pocket of DP2 (Fig. 2A, panel 4) is occupied by the p9Ser of pDRA, but its size and shape predict that it would accept larger aliphatic, polar, or even charged residues. Within the p9 pocket, βAsp55 (corresponding to position 57 in most other MHCII β-chains) (28) is a strongly conserved residue among both human and mouse MHCII molecules, forming a conserved salt bridge across to αArg76 that is important in the formation of the antigen-binding cleft (29).
The most unusual feature of the DP2 peptide-binding groove is the position of the β-chain α-helix in relation to the peptide and the rest of the molecule (Fig. 2B). We compared dimensions of the DP2 binding groove with those of 28 published human and mouse MHCII structures in the Protein Data Bank (PDB) database (Table S1), measuring the distance from α62-CA to β71-CA (β69-CA of DP2) as an indication of the width of the groove. We also measured the distance from the p5-CA to β71-CA (β69-CA of DP2) as an indication of the distance between the peptide and the β-chain α-helix. The DP2 peptide-binding groove is among the widest (16.18 Å). In the other structures, the width averaged 15.01 Å and varied from 13.59 Å to 16.73 Å. Only two structures had wider grooves than DP2 (PDB ID code 1H15, HLA-DR51 and PDB 1BX2, HLA-DR2). For both DP2 and the other structures, most of the variation in width was in the gap between the peptide and the β-chain α-helix. DP2 had the largest gap of any of the structures (10.94 Å). In the other structures, this gap averaged 8.89 Å, with a range of 7.49 Å to 10.88 Å. These points are illustrated in Fig. 2B, which shows an overlay of DP2 and two other MHCII structures, human HLA-DR3 bound to the invariant chain CLIP peptide (PDB 1A6A) and mouse IAb MHCII also bound to CLIP (PDB IMUJ). These two structures were representative of structures with an average sized groove and the narrowest groove, respectively. The structures were superimposed based on the α1 domain. The left panel shows ribbon representations of the α-helices and peptide Cα backbone, illustrating the progressive increase in the width of the binding groove by movement of the β-chain α-helix away from both the α-chain and the peptide. The right panel shows ribbon representations of the β-chains and documents that the movement of the β-chain is confined to its α-helix, because the β-strands at the floor of the groove remain superimposed for all three structures. These findings suggest that the DP2 β-chain α-helix has rolled out, pulling away from the peptide and the floor of the binding groove. A recent modeling study of HLA-DR1 with and without a bound peptide has suggested that this region of the β-chain α-helix may be quite flexible (30), contributing to the variation in the width of this part of the binding groove. This may be a general feature of other MHCII molecules, such as DP2.
A second unusual feature of the DP2 peptide-binding groove is how high the peptide rides in the groove. Again, comparing DP2 with the 28 other MHCII structures (Table S1), we measured the distance from β13-CA in the floor of the binding groove to p4-CA. This distance was greatest for DP2 (8.06 Å) and varied from 5.25 Å to 7.67 Å in the other structures. This position of the peptide and the large gap between the peptide and the β-chain α-helix explains why the p4Leu of the peptide does not occupy the p4 pocket, but rather has risen to the surface of the molecule. Figure 2C illustrates this point, showing an overlay of DP2 (based on the four central β-strands of the floor of its binding groove) with two (PDB 1KTD, IEk and PDB 1IAK, IAk) of the five other MHCII-peptide structures with a p4Leu. Though the DP2 p4Leu is out of the p4 pocket and surface-exposed, this amino acid in the other structures points down into the p4 pocket.
Formation of a Unique Exposed Acidic Pocket in the DP2-pDRA Complex.
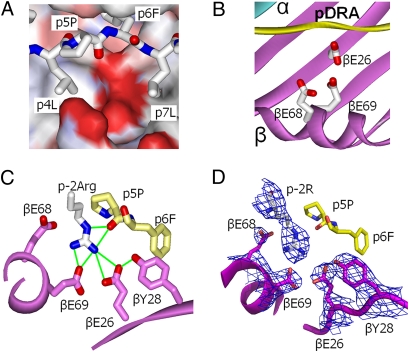
The net effect of these structural changes is to expose to the solvent an acidic pocket opposite p5Pro and flanked by p4Leu, p7Leu, and the β-chain α-helix (Fig. 3A). The acidity of this pocket is due to the presence of three DP2 β-chain amino acids: βGlu68 and βGlu69 from the β-chain α-helix and βGlu26 from the floor of the peptide-binding groove (Fig. 3B). The fact that βGlu69 is associated with genetic susceptibility to CBD and that the solved structures of other proteins associated with Be compounds generally show Be coordination by acidic amino acids (31) strongly suggest that this acidic pocket is the site of Be binding to DP2.
Fig. 3.
βGlu69 lies in a solvent-exposed acidic pocket. (A) The water-accessible surface of the DP2 molecule (without bound pDRA) is shown in the area between p5Pro and the DP2 β-chain α-helix colored by the relative charge of the surface atoms (red, negative; blue, positive) (Swiss PdbViewer). A wireframe representation of pDRA is also shown with CPK coloring. (B) Same view as A but with ribbon representations of DP2 α (cyan), DP2 β (magenta), and pDRA (yellow). Also shown are wireframe representations of the side chains of βGlu26, βGlu68, and βGlu69 with CPK coloring. (C) View of the acidic pocket looking down the peptide-binding groove from the C terminus of pDRA. Portions of the DP2 β-chain α-helix and β-sheet are shown as magenta ribbons. Wireframe representations of the side chains of βGlu26, βTyr28, βGlu68, and βGlu69 are shown with magenta carbon and red oxygen. A wireframe representation of p4 to p6 of pDRA is shown with yellow carbon, red oxygen, and blue nitrogen. Also shown is a wireframe representation (CPK coloring) of the side chain of the pDRA p-2Arg from a neighboring symmetry-related DP2-pDRA molecule. Green lines connect oxygens in the acidic pocket that potentially could form H bonds or salt bridges to each other or to the nitrogens of the arginine guanidinium group. (D) The same view as in C. In this case, the 2Fo-Fc electron density map around βGlu26, βTry28, βGlu68, and βGlu69 and p-2 Arg is shown, contoured to 1σ.
The molecular packing in the DP2-pDRA crystal offers insight into how a Be moiety could be coordinated within this acidic pocket. Within the crystal, the side chain of the N-terminal pDRA amino acid (p-2Arg; see Fig. 1B) from one DP2-pDRA molecule occupies the acidic pocket of a neighboring symmetry-related molecule (Fig. 3C), forming a daisy chain of interactions through-out the crystal (Fig. S2). The side chain carboxyl groups of βGlu68 and βGlu69 and the peptide backbone oxygen of p5Pro coordinate the guanidinium moiety of the Arg residue. The electron density within this pocket is well defined (Fig. 3D). Thus, our structure clearly establishes the presence of this acidic pocket with many potential electron donors for a Be-containing compound.
βGlu26, βGlu68, and βGlu69 Are Crucial for Be Reactive T-Cell Activation.
In addition to βGlu69, the DP2 crystal structure suggests that βGlu26 and βGlu68 may also be involved in Be coordination and presentation. These two amino acids are invariant among DP alleles. As a consequence, these residues were not identified in genetic analyses of the linkage of DP2 alleles to CBD, and their presence is not sufficient for Be presentation in the absence of βGlu69. Therefore, to determine whether they are required in addition to βGlu69, we compared the effect of their mutation on DP2 presentation of Be with Be-specific CD4+ T cells. Three mutations were tested: βGlu68 to Ala, βGlu26 to Ala, and βGlu26 to Gln. The side chain of βGlu68 is totally solvent exposed, so we assumed that the mutation to Ala would not affect DP2 assembly and expression. However, β26 is deep within the acidic pocket and only partially exposed; therefore, we mutated βGlu26 to both Ala and Gln. Because Glu and Gln are nearly isomorphous, we reasoned that this mutation was least likely to dramatically change the internal structure of the DP2 molecule, but that Gln would lack the negative charge that could be required for coordination of Be compounds. For comparison, we used wild-type DP2 and DP2 in which βGlu69 was mutated to Lys, the amino acid at β69 in DP4 and a mutation previously shown to eliminate Be presentation by DP2 (31).
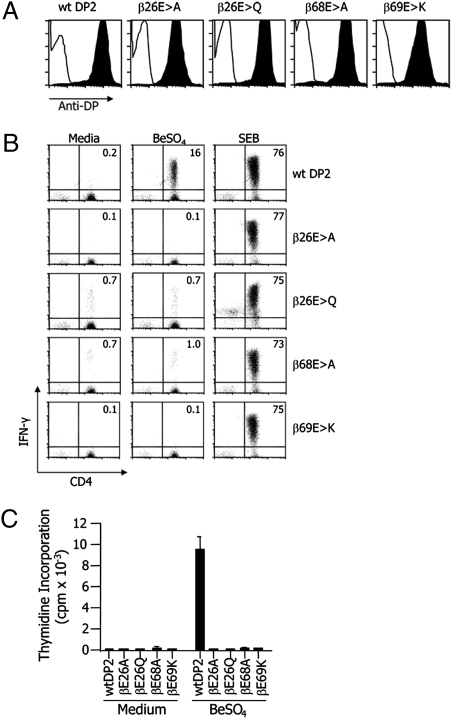
Fibroblasts transfected with each of the mutant DP2 molecules expressed DP2 equally well on the cell surface, as detected by staining with a conformation-dependent anti-DP mAb, B7.21 (Fig. 4A). When the wild-type DP2 fibroblasts were pulsed with BeSO4 and tested for stimulation of a Be-reactive T-cell line established from a DP2+ CBD patient, 16% of the T cells responded with production of IFN-γ versus only 0.2% when using unpulsed fibroblasts (Fig. 4B). By comparison, all four of the mutant DP2 molecules were unable to present Be to the T-cell line. As an additional check on the structural integrity of the various DP2 constructs, all four mutant DP2 molecules presented the MHCII-dependent superantigen, staphylococcal enterotoxin B (SEB), just as well as wild-type DP2. Similar findings were seen when Be-induced proliferation was used as a measure of T-cell response (Fig. 4C), suggesting that these three glutamic acid residues are critical for Be coordination and T-cell activation.
Fig. 4.
DP2 βGlu26, βGlu68, and βGlu69 are essential for T-cell recognition of Be. (A) Staining of DAP.3 L cells for surface expression of HLA-DP2. DAP.3 L cells transfected to express either WT DP2 or a DP2 mutant (DP2-β26E > A, DP2-β26E > Q, DP2-β68E > A, and DP2-β69E > K) were stained with either an IgG1 isotype control (open) or an anti-HLA-DP-specific mAb, B7.21 (filled). (B) Intracellular IFN-γ expression in a Be-responsive T-cell line in response to BeSO4 or SEB presentation by fibroblasts expressing either WT DP2 or a DP2 mutant is shown. The percentage of CD4+ T cells expressing intracellular IFN-γ is shown in the upper right quadrant of each density plot. (C) T-cell proliferation of the same T-cell line using BeSO4-pulsed mitomycin C-treated fibroblasts expressing either WT DP2 molecule or DP2 mutants is shown. The data are expressed as the mean cpm ± SEM.
Discussion
The reason for the genetic linkage between CBD and particular HLA-DP alleles has been a mystery for a decade (8–12). Genetic analyses that pinpointed βGlu69 as a key amino acid in this linkage have favored the hypothesis that the carboxylate of this amino acid is involved in binding the relevant Be moiety for presentation to T cells (32). This amino acid is highly polymorphic in mouse and human MHCII. In previous MHCII crystal structures, the side chains of the various amino acids that occur at this position are often partially buried under the peptide, raising the question of how Be compounds might gain access to a glutamic acid at this position.
As the first structure of an HLA-DP molecule and the first structure of an MHCII molecule with βGlu69, our structure of DP2-pDRA offers a plausible answer to this question. The widening of the peptide-binding groove between the peptide backbone and the DP2 β-chain α-helix opens a gap that exposes βGlu69 as part of an acidic pocket that could easily accommodate a compound the size of a Be-containing complex. For example, in the packing in our crystals, this pocket has been filled by the guanidinium group of an arginine from a neighboring symmetry-related molecule. The demonstration that mutation of βGlu69 or either of the other two glutamic acid residues present in this pocket, βGlu26 and βGlu68, eliminates Be presentation, provides additional support for this idea.
Genetic analyses also suggested that βAsp55 and βGlu56 might be involved in Be presentation (8). However, from the DP2 structure, it is clear that βAsp55 and βGlu56 do not lie within the usual TCR-binding interface on MHCII (Fig. S3). Furthermore, in our previous studies, mutation of βAsp55 and βGlu56 of DP2 had no effect on Be-induced proliferation and Th1-type cytokine expression from Be-responsive T cells (32). Thus, these two amino acids are unlikely to play a role in Be-induced T-cell activation.
Approximately 15% of CBD patients do not possess a βGlu69-containing HLA-DP allele, suggesting the importance of other MHCII molecules in the genetic susceptibility to Be-induced disease (9, 10). In this population of CBD patients, an increased frequency of HLA-DR13 alleles, which possess an acidic cluster composed of βAsp28, βAsp70, and βGlu71, was seen (9). To date, no structural information exists for HLA-DR13. Importantly, we have previously shown that fibroblasts expressing a mutated version of HLA-DR1302 with an arginine (R) at position 71 are unable to present Be to DR1302-restricted, Be-specific T cells (32), lending further support for the critical nature of this acidic pocket in Be coordination and subsequent T-cell activation.
In vitro, Be-specific T cells respond to a variety of added Be salts, including BeSO4 and BeCl2 (33). These salts are predicted to fully dissociate in water to yield a Be++ cation, but the extremely high charge density of this very small cation leads to rapid stable coordination by up to four electron donors, such as water, hydroxyl ion, carbonate ion, and organic acids, leading to a variety of large complexes (34, 35). The exact Be moiety required for T-cell activation is unknown. It is also possible that T cells may not directly recognize the bound Be complex, but rather recognize some common DP2 conformational change induced by any of a variety of Be compounds bound to βGlu69 in the DP2 acidic pocket.
In our structure, an arginine guanidinium group is held in the acidic pocket via interactions with βGlu26, βGlu68, βGlu69, and the peptide backbone, but not with any of the peptide side chains, suggesting that many different peptides bound to DP2 might be permissive for Be binding. However, it remains possible that the side chain of certain amino acids of the peptide, such as p4, may also participate in Be coordination (24). In addition, the position of the DP2 acidic pocket immediately adjacent to the peptide p5 amino acid makes it unlikely that a TCR could independently recognize a bound Be moiety without also having to interact with particular amino acids of the peptide. Therefore, the actual Be-responsive T-cell receptor repertoire could be very complex, perhaps with each T cell recognizing both Be and a different specifically bound peptide. This idea is consistent with our failed attempts to use DP2 with covalently attached single peptides to present Be to T cells. DP2 with the usual set of naturally bound peptides readily presents Be to a variety of Be-reactive T cells. However, DP2 expressed with a number of fixed covalent peptides, including pDRA (Fig. S4), is unable to present Be to the Be-responsive T cells that we have tested. Therefore, to understand fully Be recognition at the structural level, the relevant peptide(s) for individual T cells will likely have to be identified.
Finally, peptides naturally bound to DP2 often have an arginine or lysine at p4 (26). Depending on the conformation of these amino acids, their positively charged long side chains could reach and fill this acidic pocket (Fig. S5), similar to the arginine in our structure and potentially block Be binding. However, in the case of DP2 on the cell surface with peptides, such as pDRA, that lack an Arg or Lys at p4, one might imagine that the repulsion among three solvent-exposed acidic amino acids could destabilize the molecule unless they are neutralized by some other positively charged species, such as divalent cations (e.g., Ca2+, Mg2+, etc.). This acidic pocket might also be able to bind other metal ions, for example, that of cobalt (36), explaining the association of DP2 with hard metal lung disease (7). Taken together, we provide the first structure of an HLA-DP molecule that has been strongly linked to the development of a human disease. The unique structural features of DP2, which include the surface exposure of the key polymorphic amino acid residue (βGlu69) of that molecule, provide an explanation of the genetic linkage of DP2 to CBD.
Materials and Methods
Expression and Purification of DP2-pDRA for Crystallization.
A baculovirus construct encoding a soluble form of HLA-DP2 with a covalently attached peptide derived from the HLA-DR α-chain (pDRA) was constructed as shown in Fig. S1 using a previously described two-promoter baculovirus transfer vector (27). The construct was designed to produce an acid:base zipper stabilized version of soluble DP2 with the peptide-binding groove uniformly occupied by tethered pDRA. The construct was incorporated into Sapphire baculovirus DNA (Orbigen) by standard homologous recombination techniques to produce a stock of baculovirus encoding DP2. The DP2-pDRA protein was purified from the culture supernatant by immunoaffinity and size exclusion chromatography (see SI Materials and Methods for methodological details). The leucine zipper was partially removed by overnight treatment with papain at room temperature at a ratio of enzyme to DP2-pDRA of ∼1:50. The reaction was stopped with iodoacetamide (5 mM) for 30 min on ice. Reactants were removed by buffer exchange, and the digested DP2-pDRA was concentrated in 10 mM Hepes (pH 7), 5 mM NaN3.
Crystallization of DP2-pDRA.
DP2 was crystallized at room temperature by hanging drop vapor diffusion against 1 mL of mother liquor containing 20% PEG4000, 300 mM sodium chloride, Hepes buffer (pH 7.5), and 10% isopropanol. In all crystallization setups, 0.5 μL of protein solution (7 mg/mL) was mixed with 0.5 μL of reservoir solution. Crystals were flash cooled in liquid nitrogen after a 10- to 30-s soak in a cryoprotection solution consisting of the reservoir solution with the PEG4000 concentration increased to 27% (wt/vol).
Data Collection and Structure Determination.
Initial data were collected on a crystal that diffracted to 3.5 Å resolution at beamline 8.2.2 at the Advanced Light Source at Lawrence Berkeley National Laboratory. The data were indexed, integrated, scaled, and merged using HKL2000 (37). The structure of DP2 was determined by molecular replacement using the program CNS (38) by using HLA-DR3 (PDB 1A6A) as the search model. The model was subjected to several rounds of alternating simulated annealing/positional refinement in CNS followed by B-factor refinement in CNS. Modeling, including the pDRA and amino acids unique to DP2, was performed using the program O (39). Subsequently, data were collected at beamline 19-ID-D at the Advanced Photon Source at Argonne National Laboratory from a second crystal that diffracted to 3.25-Å resolution. The two data sets were reprocessed with Mosflm and later merged with Scala to 3.25-Å resolution. The overall completeness, I/σ, and redundancy were improved after merging. The precision-indicating merging R factor, Rpim, was used because it describes the precision of the averaged measurements, which are the quantities normally used in crystallography as observables (40, 41).
The electron density map significantly improved with the merged data, allowing the identification of 12 additional residues. Using the merged data and CNS 1.2, the model was first refined to 3.5 Å, and then simulated annealing with a starting temperature of 3,000 K was used to remove model bias and extend the resolution to 3.25 Å. Finally, a combination of restrained refinement and TLS refinement (with isotropic B-factor refining) was performed using refmac5. The final model, consisting of DP2, the DRA peptide, and three carbohydrate molecules, yielded values of Rwork and Rfree of 22.6% and 25.1%, respectively. The critical residues of the DP2 acidic pocket (Fig. 3D) and the pDRA (Fig. S6) had unequivocal electron density associated with them. Ramachandran statistics (%): most favored (86.3); additionally allowed (11.3); generously allowed (2.1); disallowed (0.3).
The structure showed relatively high overall B factors (Table 1), consistent with the rapid decrease of the diffraction intensities at higher angles. High B-factors at this resolution are not uncommon, where a Wilson plot is not able to estimate sensible B-factors. The high B-factors are probably due to the extremely high solvent content and daisy chain-like loose packing in this crystal (Fig. S2). Of more than 300 crystals analyzed, only the two used here diffracted to better than ∼5 Å, again indicating the unstable packing. However, the simulated annealing omit maps and the final 2Fo-Fc maps (Fig. S6) were of good quality for the structural determination.
Representations of the model in the figures were produced with WebLab ViewerPro 4 (Molecular Simulations, Inc.) and PyMOL (Delano Scientific). Molecular overlays were performed with Swiss PdbViewer v4 (42). This program was also used to analyze the dimensions of the peptide binding shown in Table 2.
Site-Directed Mutagenesis of the HLA-DP β-Chain and Transfection.
Mutagenesis of DPB1*0201 was performed as previously described (32). HindIII-XhoI flanked full-length DPB1*0201 cDNA was mutated at amino acid positions 26 and 68 using overlapping primers. The forward and reverse primers for generating the DP2-β26Ala and the DP2-β69Ala mutants are included in SI Materials and Methods. PCR fragments containing the mutated DP2 DNA were ligated into pcDNA3.1 (Invitrogen Life Technologies), and the sequence was confirmed by DNA sequencing.
Transfection into mouse DAP.3 L cells expressing DPA1*0103 was performed using Lipofectamine 2000 (Invitrogen Life Technologies), and selection in the presence of 0.5 mg/mL G418 was performed 48 h after the addition of DNA. After cloning via limiting dilution, HLA-DP expression was assessed using the HLA-DP-specific mAb, B7.21.
Generation of Beryllium-Specific T-Cell Lines and in Vitro T-Cell Assays.
The HLA-DP2-restricted, beryllium-responsive T-cell line was previously generated from bronchoalveolar lavage cells of a CBD patient (18). HLA-DP2-expressing fibroblasts were treated with 50 μg/mL mitomycin C for 1 h at 37 °C in a humidified 5% CO2 atmosphere followed by three washes with HBSS plus 5% FCS. The mitomycin C-treated cells were pulsed with medium, 10 ng/mL SEB (Toxin Technology), or 100 μM BeSO4 overnight at 37 °C. The cells were washed three times with HBSS and combined in a 1:1 ratio with T cells (1 × 105 cells of each) and cultured for 6 h with 10 μg/mL brefeldin A added after the first hour of stimulation (32). After stimulation, intracellular IFN-γ (BD Pharmingen) staining was performed as previously described (32). Lymphocyte proliferation assays were performed as previously described (32).
Supplementary Material
Acknowledgments
We thank Allison Lanham for technical help with intracellular cytokine analysis and Yuan Li for contributing to the development of the method of purification for the DP2-pDRA protein. We are also grateful to Stephan Ginell at the 19-ID-D beamline at the Advanced Photon Source and Corie Ralston at the 8.2.2 beamline at the Advanced Light Source for assistance in synchrotron data collection. This work was supported by US Public Health Service Grants HL62410, ES11810, AI17134, and AI18785.
Footnotes
The authors declare no conflict of interest.
Data Deposition: The coordinates of the HLA-DP2 structure have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3LQZ).
This article contains supporting information online at www.pnas.org/cgi/content/full/1001772107/DCSupplemental.
References
- 1.Jones EY, Fugger L, Strominger JL, Siebold C. MHC class II proteins and disease: A structural perspective. Nat Rev Immunol. 2006;6:271–282. doi: 10.1038/nri1805. [DOI] [PubMed] [Google Scholar]
- 2.Nepom GT. Major histocompatibility complex-directed susceptibility to rheumatoid arthritis. Adv Immunol. 1998;68:315–332. doi: 10.1016/s0065-2776(08)60563-5. [DOI] [PubMed] [Google Scholar]
- 3.Nepom GT, Erlich H. MHC class-II molecules and autoimmunity. Annu Rev Immunol. 1991;9:493–525. doi: 10.1146/annurev.iy.09.040191.002425. [DOI] [PubMed] [Google Scholar]
- 4.Begovich AB, et al. A specific HLA-DP beta allele is associated with pauciarticular juvenile rheumatoid arthritis but not adult rheumatoid arthritis. Proc Natl Acad Sci USA. 1989;86:9489–9493. doi: 10.1073/pnas.86.23.9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong RP, et al. HLA-A and DPB1 loci confer susceptibility to Graves’ disease. Hum Immunol. 1992;35:165–172. doi: 10.1016/0198-8859(92)90101-r. [DOI] [PubMed] [Google Scholar]
- 6.Dong RP, Kimura A, Numano F, Nishimura Y, Sasazuki T. HLA-linked susceptibility and resistance to Takayasu arteritis. Heart Vessels Suppl. 1992;7:73–80. doi: 10.1007/BF01744548. [DOI] [PubMed] [Google Scholar]
- 7.Potolicchio I, et al. Susceptibility to hard metal lung disease is strongly associated with the presence of glutamate 69 in HLA-DP beta chain. Eur J Immunol. 1997;27:2741–2743. doi: 10.1002/eji.1830271039. [DOI] [PubMed] [Google Scholar]
- 8.Richeldi L, Sorrentino R, Saltini C. HLA-DPB1 glutamate 69: A genetic marker of beryllium disease. Science. 1993;262:242–244. doi: 10.1126/science.8105536. [DOI] [PubMed] [Google Scholar]
- 9.Maier LA, et al. Influence of MHC class II in susceptibility to beryllium sensitization and chronic beryllium disease. J Immunol. 2003;171:6910–6918. doi: 10.4049/jimmunol.171.12.6910. [DOI] [PubMed] [Google Scholar]
- 10.Rossman MD, et al. Human leukocyte antigen Class II amino acid epitopes: Susceptibility and progression markers for beryllium hypersensitivity. Am J Respir Crit Care Med. 2002;165:788–794. doi: 10.1164/ajrccm.165.6.2104002. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, et al. Differential susceptibilities to chronic beryllium disease contributed by different Glu69 HLA-DPB1 and -DPA1 alleles. J Immunol. 1999;163:1647–1653. [PubMed] [Google Scholar]
- 12.McCanlies EC, Ensey JS, Schuler CR, Kreiss K, Weston A. The association between HLA-DPB1Glu69 and chronic beryllium disease and beryllium sensitization. Am J Ind Med. 2004;46:95–103. doi: 10.1002/ajim.20045. [DOI] [PubMed] [Google Scholar]
- 13.Fontenot AP, Canavera SJ, Gharavi L, Newman LS, Kotzin BL. Target organ localization of memory CD4(+) T cells in patients with chronic beryllium disease. J Clin Invest. 2002;110:1473–1482. doi: 10.1172/JCI15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontenot AP, Maier LA. Genetic susceptibility and immune-mediated destruction in beryllium-induced disease. Trends Immunol. 2005;26:543–549. doi: 10.1016/j.it.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Kreiss K, Mroz MM, Newman LS, Martyny J, Zhen B. Machining risk of beryllium disease and sensitization with median exposures below 2 micrograms/m3. Am J Ind Med. 1996;30:16–25. doi: 10.1002/(SICI)1097-0274(199607)30:1<16::AID-AJIM3>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 16.Kreiss K, Mroz MM, Zhen B, Martyny JW, Newman LS. Epidemiology of beryllium sensitization and disease in nuclear workers. Am Rev Respir Dis. 1993;148:985–991. doi: 10.1164/ajrccm/148.4_Pt_1.985. [DOI] [PubMed] [Google Scholar]
- 17.Kreiss K, Wasserman S, Mroz MM, Newman LS. Beryllium disease screening in the ceramics industry. Blood lymphocyte test performance and exposure-disease relations. J Occup Med. 1993;35:267–274. [PubMed] [Google Scholar]
- 18.Fontenot AP, Torres M, Marshall WH, Newman LS, Kotzin BL. Beryllium presentation to CD4+ T cells underlies disease-susceptibility HLA-DP alleles in chronic beryllium disease. Proc Natl Acad Sci USA. 2000;97:12717–12722. doi: 10.1073/pnas.220430797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombardi G, et al. HLA-DP allele-specific T cell responses to beryllium account for DP-associated susceptibility to chronic beryllium disease. J Immunol. 2001;166:3549–3555. doi: 10.4049/jimmunol.166.5.3549. [DOI] [PubMed] [Google Scholar]
- 20.Amicosante M, et al. Beryllium binding to HLA-DP molecule carrying the marker of susceptibility to berylliosis glutamate beta 69. Hum Immunol. 2001;62:686–693. doi: 10.1016/s0198-8859(01)00261-0. [DOI] [PubMed] [Google Scholar]
- 21.Fontenot AP, et al. Recombinant HLA-DP2 binds beryllium and tolerizes beryllium-specific pathogenic CD4+ T cells. J Immunol. 2006;177:3874–3883. doi: 10.4049/jimmunol.177.6.3874. [DOI] [PubMed] [Google Scholar]
- 22.Scott BL, Wang Z, Marrone BL, Sauer NN. Potential binding modes of beryllium with the class II major histocompatibility complex HLA-DP: A combined theoretical and structural database study. J Inorg Biochem. 2003;94:5–13. doi: 10.1016/s0162-0134(02)00628-1. [DOI] [PubMed] [Google Scholar]
- 23.Berretta F, et al. Detailed analysis of the effects of Glu/Lys beta69 human leukocyte antigen-DP polymorphism on peptide-binding specificity. Tissue Antigens. 2003;62:459–471. doi: 10.1046/j.1399-0039.2003.00131.x. [DOI] [PubMed] [Google Scholar]
- 24.Amicosante M, Berretta F, Dweik R, Saltini C. Role of high-affinity HLA-DP specific CLIP-derived peptides in beryllium binding to the HLA-DPGlu69 berylliosis-associated molecules and presentation to beryllium-sensitized T cells. Immunology. 2009;128(1, Suppl):e462–e470. doi: 10.1111/j.1365-2567.2008.03000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snyder JA, Weston A, Tinkle SS, Demchuk E. Electrostatic potential on human leukocyte antigen: Implications for putative mechanism of chronic beryllium disease. Environ Health Perspect. 2003;111:1827–1834. doi: 10.1289/ehp.6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Díaz G, Cañas B, Vazquez J, Nombela C, Arroyo J. Characterization of natural peptide ligands from HLA-DP2: New insights into HLA-DP peptide-binding motifs. Immunogenetics. 2005;56:754–759. doi: 10.1007/s00251-004-0735-5. [DOI] [PubMed] [Google Scholar]
- 27.Kozono H, White J, Clements J, Marrack P, Kappler J. Production of soluble MHC class II proteins with covalently bound single peptides. Nature. 1994;369:151–154. doi: 10.1038/369151a0. [DOI] [PubMed] [Google Scholar]
- 28.Stern LJ, et al. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 29.Todd JA, et al. A molecular basis for MHC class II-associated autoimmunity. Science. 1988;240:1003–1009. doi: 10.1126/science.3368786. [DOI] [PubMed] [Google Scholar]
- 30.Painter CA, Cruz A, Lopez GE, Stern LJ, Zavala-Ruiz Z. Model for the peptide-free conformation of class II MHC proteins. PLoS ONE. 2008;3:e2403. doi: 10.1371/journal.pone.0002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho H, et al. BeF(3)(-) acts as a phosphate analog in proteins phosphorylated on aspartate: Structure of a BeF(3)(-) complex with phosphoserine phosphatase. Proc Natl Acad Sci USA. 2001;98:8525–8530. doi: 10.1073/pnas.131213698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bill JR, et al. Beryllium presentation to CD4+ T cells is dependent on a single amino acid residue of the MHC class II β-chain. J Immunol. 2005;175:7029–7037. doi: 10.4049/jimmunol.175.10.7029. [DOI] [PubMed] [Google Scholar]
- 33.Rossman MD, et al. Proliferative response of bronchoalveolar lymphocytes to beryllium. A test for chronic beryllium disease. Ann Intern Med. 1988;108:687–693. doi: 10.7326/0003-4819-108-5-687. [DOI] [PubMed] [Google Scholar]
- 34.Schmidbaur H. Recent contributions to the aqueous coordination chemistry of beryllium. Coord Chem Rev. 2001;215:223–242. [Google Scholar]
- 35.Sutton M, Burastero SR. Beryllium chemical speciation in elemental human biological fluids. Chem Res Toxicol. 2003;16:1145–1154. doi: 10.1021/tx0256477. [DOI] [PubMed] [Google Scholar]
- 36.Roderick SL, Matthews BW. Structure of the cobalt-dependent methionine aminopeptidase from Escherichia coli: A new type of proteolytic enzyme. Biochemistry. 1993;32:3907–3912. doi: 10.1021/bi00066a009. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Brünger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 39.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 40.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 41.Weiss M. Global indicators of X-ray data quality. J Appl Cryst. 2001;34:130–135. [Google Scholar]
- 42.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.