Abstract
We executed a genome-wide association scan for age-related macular degeneration (AMD) in 2,157 cases and 1,150 controls. Our results validate AMD susceptibility loci near CFH (P < 10−75), ARMS2 (P < 10−59), C2/CFB (P < 10−20), C3 (P < 10−9), and CFI (P < 10−6). We compared our top findings with the Tufts/Massachusetts General Hospital genome-wide association study of advanced AMD (821 cases, 1,709 controls) and genotyped 30 promising markers in additional individuals (up to 7,749 cases and 4,625 controls). With these data, we identified a susceptibility locus near TIMP3 (overall P = 1.1 × 10−11), a metalloproteinase involved in degradation of the extracellular matrix and previously implicated in early-onset maculopathy. In addition, our data revealed strong association signals with alleles at two loci (LIPC, P = 1.3 × 10−7; CETP, P = 7.4 × 10−7) that were previously associated with high-density lipoprotein cholesterol (HDL-c) levels in blood. Consistent with the hypothesis that HDL metabolism is associated with AMD pathogenesis, we also observed association with AMD of HDL-c—associated alleles near LPL (P = 3.0 × 10−3) and ABCA1 (P = 5.6 × 10−4). Multilocus analysis including all susceptibility loci showed that 329 of 331 individuals (99%) with the highest-risk genotypes were cases, and 85% of these had advanced AMD. Our studies extend the catalog of AMD associated loci, help identify individuals at high risk of disease, and provide clues about underlying cellular pathways that should eventually lead to new therapies.
Keywords: genome-wide association study, single nucleotide polymorphism
Age-related macular degeneration (AMD) is a progressive neurodegenerative disease and a common cause of blindness in the elderly population, particularly in developed countries (1). The disease affects primarily the macular region of the retina, which is necessary for sharp central vision. An early hallmark of AMD is the appearance of drusen, which are extracellular deposits of proteins and lipids under the retinal pigment epithelium (RPE). As the disease progresses, drusen grow in size and number. In advanced stages of AMD, atrophy of the RPE (geographic atrophy) and/or development of new blood vessels (neovascularization) result in death of photoreceptors and central vision loss.
Multiple genetic linkage studies provided strong evidence of susceptibility loci, notably on chromosomes 1q31 and 10q36 (2). Disease-associated variants near CFH (1q31) and in a cluster of genes near ARMS2 (10q26) were first identified both through genome-wide association studies (GWASs) (3, 4) and fine mapping of linkage signals (5—8). Discovery of association between AMD and the CFH locus led researchers to discovery of association signals near other complement genes, including C2/CFB, C3, and CFI (9—12).
Results and Discussion
The execution of progressively larger GWAS typically results in the gradual discovery of new susceptibility loci [see the examples of Crohn’s disease (13), type 2 diabetes (14), obesity (15), and lipids (16, 17)]. To identify additional susceptibility loci and biochemical pathways contributing to AMD, we performed GWAS in a large collection of cases and controls (Table S1) using a genotyping platform that captures >90% of common variants in European ancestry samples.
We genotyped study samples, including 75 blind duplicates, together with HapMap controls at the Center for Inherited Disease Research (Johns Hopkins) with Illumina Human370 chips. After genotyping, we excluded 18 individuals with an unexpected first- or second-degree relative in the dataset and 13 individuals with evidence for a non-European ancestry component (18), resulting in a total of 2,157 unrelated cases and 1,150 unrelated controls for analyses. We excluded markers with <95% call rate, minor allele frequency <1%, or evidence for deviation from Hardy-Weinberg equilibrium at P < 10−6, resulting in a total of 324,067 autosomal SNPs for analysis. The average call rate for analyzed markers and samples was 99.94%. We identified short stretches of haplotype shared between individuals in our study and those in the HapMap CEU (19) and used these to impute missing genotypes, expanding the number of analyzed SNPs to approximately 2.5 million imputed or genotyped SNPs. Complete GWAS data and results are available from Database of Genotypes and Phenotypes accession no. phs000182.v1.p1.
An initial comparison of allele frequencies between cases and controls resulted in a genomic control parameter (20) of 1.056; adjustment for the first two principal components of ancestry (PCA) (18) reduced this to 1.007. PCA can account for subtle differences among European ancestry samples (such as North–South or East–West gradients in allele frequency; see ref. 21) and provide a useful safeguard against population stratification. All results reported here refer to this PCA-adjusted analysis.
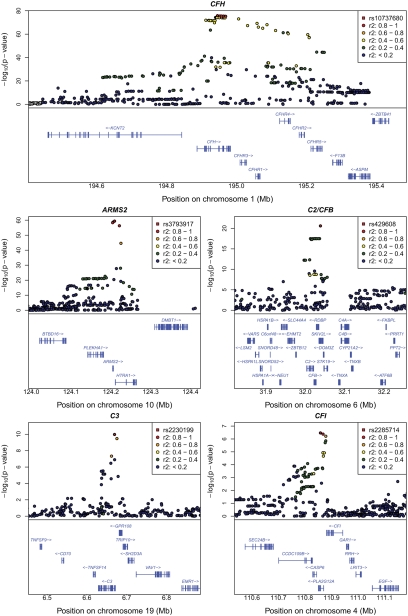
Reassuringly, we observed strong evidence of association at established susceptibility loci (Figs. 1 and 2 and Table 1); near CFH [strongest association at rs10737680, odds ratio (OR) = 3.11 (2.76, 3.51), with P < 1.6 × 10−75], near ARMS2 [at rs3793917, OR = 3.40 (2.94, 3.94), P = 4.1 × 10−60], near complement component 2 (C2) and complement factor B (CFB) [at rs429608, OR = 2.16 (1.84, 2.53), P = 2.5 × 10−21], and near complement component 3 (C3) [at rs2230199, OR = 1.74 (1.47, 2.06), P = 1.0 × 10−9]. Our study provides confirmation of a recently reported association between AMD and complement factor I (CFI) [at rs2285714, OR = 1.31 (1.18, 1.45), P = 3.4 × 10−7] (9). Conditioning on the strongest associated variant at each of these loci identified additional, strong association signals near CFH (at rs1329424, P = 6.4 × 10−16) and in the C2/CFB locus (at rs9380272, P = 2.3 × 10−8), consistent with previous reports of multiple disease-associated alleles at the two loci (8, 10, 22, 23). Where possible, we evaluated evidence for association at other previously suggested susceptibility loci using genotypes or imputed data. The results are summarized in Table S2; although none of these loci show P < 0.05 in our data, note that eight of nine signals trend in the same direction as the original report.
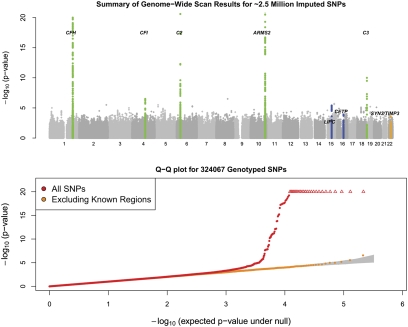
Fig. 1.
Summary of genome-wide association scan results. (Upper) Summary of the significance of the association signal at each examined SNP in the discovery samples. The five known loci are highlighted in green. The three strongest loci after follow (TIMP3, LIPC, and CETP) are highlighted in blue. (Lower) Quantile—quantile plot for test statistics. Shaded region corresponds to a 90% confidence interval for the test statistics.
Fig. 2.
Regional plots for association signals in five previously reported loci. Detailed plots of association in the discovery samples in five confirmed regions (CFH, ARMS, C2/CFB, C3, and CFI) are shown. The most significant SNP in each region is highlighted in a red square, and other SNPs are drawn as colored circles reflecting linkage disequilibrium with the top selected SNP. Exons and transcript direction for genes in each region are indicated at bottom of each panel.
Table 1.
Confirmation of previously reported association signals in the discovery samples
| SNP | Chromosome | Position (bp) | Notable nearby genes | Alleles(risk/nonrisk) | Frequency (risk allele) |
OR | P value | λsib | |
| Cases | Controls | ||||||||
| rs10737680* | 1 | 194,946,078 | CFH | A/C | 0.801 | 0.566 | 3.11 (2.76, 3.51) | 1.6 × 10−76 | 1.24 |
| rs3793917* | 10 | 124,209,265 | ARMS2/HTRA1 | G/C | 0.371 | 0.164 | 3.40 (2.94, 3.94) | 4.1 × 10−60 | 1.45 |
| rs429608 | 6 | 32,038,441 | C2/CFB | G/A | 0.920 | 0.842 | 2.16 (1.84, 2.53) | 2.5 × 10−21 | 1.05 |
| rs2230199* | 19 | 6,669,387 | C3 | C/G | 0.224 | 0.163 | 1.74 (1.47, 2.06) | 1.0 × 10−10 | 1.06 |
| rs2285714 | 4 | 110,858,259 | CFI | T/C | 0.464 | 0.395 | 1.31 (1.18, 1.45) | 3.4 × 10−7 | 1.02 |
| rs1329424* | 1 | 194,912,799 | CFH | T/G | 0.603 | 0.351 | 1.88 (1.68, 2.10) | 6.4 × 10−16 | 1.11 |
| rs9380272* | 6 | 32,013,989 | C2/CFB | A/G | 0.016 | 0.012 | 4.31 (2.76, 6.87) | 2.3 × 10−8 | 1.12 |
Association peaks at previously reported loci. For two of these loci (near CFH and C2/CFB), we found significant secondary signals after adjusting for the strongest initial signal. At C2/CFB locus, rs9380272 shows no significant association before adjusting for the primary signal because its risk allele is in linkage disequilibrium with the protective allele at rs429608. Conditioning on either of these two SNPs enhances the signal at the other SNP. The recurrence risk ratio λsib quantifies the increase in risk to siblings of affected individuals attributable to a specific allele. For example, a λsib of 1.24 implies that alleles at the first locus are responsible for 24% increase in risk to siblings of AMD patients compared with the general population.
*Imputation r2 > 0.95.
To identify previously uncharacterized AMD susceptibility loci, we conditioned on the seven strongly associated SNPs (Table 1) and repeated the genome-wide analysis. No single SNP was significant at P < 5 × 10−8 in this conditional analysis. Next, we exchanged initial results with the Tufts/Massachusetts General Hospital (MGH) GWAS for 1,358 SNPs that could be assayed directly with Affymetrix 6.0 genotyping arrays and that were significant at P < 0.001 in either study. Tufts/MGH GWAS results were adjusted for possible population stratification using genomic control (20), consistent with the analysis presented in the companion article (24). After excluding 158 samples from The Age-Related Eye Disease Study (AREDS), that were genotyped in both studies, this allowed us to examine promising SNPs in an additional 821 cases with geographic atrophy or neovascularization and 1,709 controls. Twenty-five SNPs showing consistent evidence of association in both groups of participants and five other SNPs with strong evidence for association in our data alone were genotyped in additional samples (Table S1). Summary results from follow-up experiments are presented in Table S3. Detailed results for the three most strongly associated loci (near TIMP3, CETP, and LIPC) and two other loci (LPL and ABCA1) are provided in Tables S4, S5, S6, S7, and S8. At each of these loci, Hardy-Weinberg equilibrium P values in cases, in controls, and in the combined dataset were all >0.20, suggesting no data quality problems. Furthermore, we found no evidence for heterogeneity (all Cochran's Q heterogeneity P values >0.20). Finally, when we genotyped a subset of the 1,161 samples for six of the imputed SNPs near TIMP3 (our strongest previously uncharacterized locus), we observed >99.4% concordance between imputed and genotyped alleles. Association results for this set of individuals were essentially the same whether imputed or actual genotypes were used for analysis. A comparison of results with genotyped and imputed SNPs at each locus is given in Table S9.
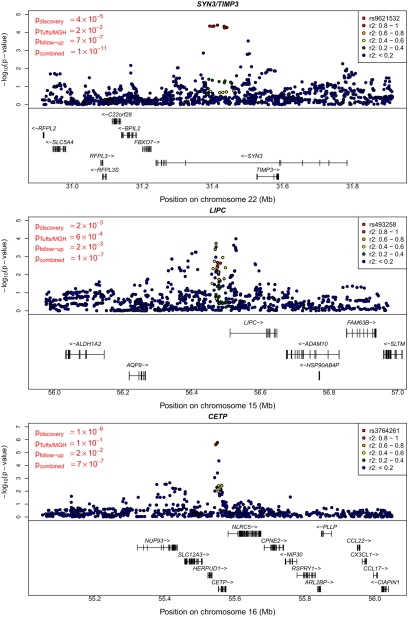
Our strongest previously uncharacterized locus maps near TIMP3 and SYN3 on chromosome 22 (Fig. 3, Top, and Table 2). There, we found that very common alleles (frequency of ≈0.94 in controls) at rs9621532 and nearby markers were associated with increased risk of AMD [OR = 1.41 (1.27, 1.57), overall P = 1.1 × 10−11, one-sided P value in newly genotyped follow-up samples Pfollow-up = 3.3 × 10−7). Consistent with the expectation that GWAS tend to estimate effect sizes (the “winner's” curse effect), we found that OR estimates in the discovery samples were larger than in the follow-up samples (25). Results at the TIMP3 locus were robust to a variety of analysis models (including different combinations of PCA, adjustment for previously known loci, and inclusion of age and sex as covariates; Table S10), are supported by nearby directly genotyped SNPs (Table S9), and remain significant when data from the companion article are excluded from analysis (overall P = 7 × 10−11 excluding all Tufts/MGH data).
Fig. 3.
Regional plot for association signals in the three previously uncharacterized loci. Detail plots for the regions surrounding the SYN3/TIMP3, LIPC, and CETP regions. Original, follow-up, and combined P values for the SNP selected for replication are indicated on the left. Discovery sample P values for the index SNP and other nearby SNPs are plotted.
Table 2.
Previously uncharacterized locus with confirmed association to AMD (P < 5×10−8)
| SNP | Chromosome | Position (bp) | Notable nearby genes | Alleles (risk/nonrisk) | Frequency (risk allele) |
OR | P value* | λsib | |
| Cases | Controls | ||||||||
| rs9621532 | 22 | 31,414,511 | SYN3/TIMP3 | A/C | |||||
| Discovery sample (2,157 cases, 1,150 controls) … | 0.964 | 0.943 | 1.81 (1.42, 2.29) | 3.9 × 10−5 | 1.011 | ||||
| Tufts/MGH sample (821 cases, 1,709 controls) … | 0.959 | 0.947 | 1.31 (0.98, 1.74) | 0.008† | 1.004 | ||||
| De novo replication sample (7,071 cases, 4,289 controls) … | 0.959 | 0.947 | 1.33 (1.17, 1.52) | 3.3 × 10−7 | 1.008 | ||||
| Combined sample (10,049 cases, 7,148 controls) … | 0.960 | 0.946 | 1.41 (1.27, 1.57) | 1.1 × 10−11 | 1.008 | ||||
| Cochran's Q heterogeneity test P = 0.245 | |||||||||
Table summarizes results for a previously uncharacterized confirmed association signal near TIMP3 (overall P < 5 × 10−8; corresponding to an adjustment for ≈1 million independent tests).
*P values for the discovery and combined samples are two sided. P values for the Tufts/MGH and de novo replication samples are one sided.
†Excluding overlapping AREDS samples in the Tufts/MGH study.
Two other loci also exhibited strong evidence for association. Near LIPC on chromosome 15, the common allele at rs493258 (frequency of ≈0.53 in controls) was associated with increased risk of AMD [OR = 1.14 (1.09, 1.20), overall P = 1.3 × 10−7, Pfollow-up = 0.0012). Near CETP on chromosome 16, the rare allele at rs3764261 (frequency ≈0.36 in controls) was associated with increased risk of AMD [OR = 1.19 (1.12, 1.27), overall P = 7.4 × 10−7, Pfollow-up = 0.009]. The signals near CETP and LIPC do not reach P < 5 × 10−8. However, note that (i) both LIPC and CETP show nominally significant association in follow-up samples alone; (ii) less than 0.3 loci per scan are expected to reach P < 3 × 10−7 by chance, suggesting that one or both of these signals are real; (iii) LIPC association with AMD reaches genome-wide significance in a companion study (24); and (iv) in a sample of Japanese individuals, top SNPs at CETP (P = 0.001), LIPC (P = 0.10), and TIMP3 (P = 0.09) trend in the right direction (Tables S4, S5, S6, S7, and S8).
Additional experiments will be required to identify the functional alleles at each locus and the genes/pathways they impact. The challenges in identifying functional alleles are illustrated by the controversy over causal alleles near ARMS2 [where the PLEKHA1, ARMS2, and HTRA1 genes have been implicated (4–6, 26)] and CFH [where noncoding variants may contribute to disease (22, 23) independently of the Y402H coding variant that was the initial focus of attention]. Despite these caveats, the previously uncharacterized loci reported here suggest biologic pathways influencing disease susceptibility and possibly new therapies.
Our top previously uncharacterized signal maps to a large intron of the synapsin III (SYN3) gene involved in neurotransmission and synapse formation (27). The SNP is located approximately 100 kb upstream of TIMP3, a metalloproteinase encoded within the same intron of SYN3. TIMP3 is involved in degradation of the extracellular matrix and mutated in Sorby's fundus dystrophy (28), an early-onset macular degenerative disease that shares clinical features with AMD but typically presents before age 40 years. Sorby's is extremely rare, presents with a highly penetrant autosomal dominant family history, and is unlikely to be misclassified as AMD. When we excluded all patients with age of onset <60 years from our sample, evidence for association at TIMP3 was essentially unchanged. Linkage of AMD to the TIMP3/SYN3 region has been reported previously (29). The effects of the common alleles reported here are too small to account for the observed linkage signal, but it is possible that missed rare high penetrance alleles could reside in the same locus and explain the linkage.
Outside of known loci and TIMP3, our two strongest signals are located near the hepatic lipase (LIPC) gene on chromosome 15q22 (initial evidence of association at rs493258 came from Tufts/MGH GWAS) and the cholesterylester transfer protein (CETP) gene on 16q21. The AMD associated alleles at these loci have been associated with high-density lipoprotein cholesterol (HDL-c) levels in blood (16, 17). This prompted us to examine whether other common HDL-c—associated polymorphisms might contribute to AMD risk. The three common alleles showing strongest association to blood HDL-c levels in an analysis of 19,840 individuals (17) also reveal evidence of association with AMD in our discovery cohort (rs173539 near CETP with P = 2.4 × 10−6; rs12678919 near LPL with P = 0.0016; rs10468017 near LIPC with P = 0.0018). Table 3 and Fig. S1 show that the same clusters of SNPs (colored) associated with HDL-c (each cluster has lead SNP with P < 5 × 10−8) are associated with macular degeneration; association signals are sharper for HDL-c given the much larger sample sizes (and greater power) of that analysis. Multiple common alleles near CETP and LIPC are associated independently with HDL-c levels (17). In our sample, we find modest association of the secondary HDL-associated alleles in each of these loci with AMD (rs289714 near CETP with P = 0.062; rs2070895 near LIPC with P = 0.051). Finally, HDL-associated alleles near ABCA1 also show evidence of association with AMD (rs1883025, P = 0.0026). The probability that 4 or more of the 14 reported HDL-associated alleles (17) would show association with AMD with P < 0.0026 is extremely low (4 × 10−8), and the probability that the top three HDL-associated alleles would reveal association with P < 0.0018 is 6 × 10−9 (the probability of P < 0.14 or better, as in the replication samples alone, is 0.003). Importantly, because we found association specifically for alleles with the largest impact on HDL levels, it seems likely that additional signals may have been missed owing to lack of power. Just as for CETP and LIPC, association signals at LPL and ABCA1 were consistent in follow-up samples and discovery samples; combining all available data we observed association with P = 3.0 × 10−3 near LPL and 5.6 × 10−4 near ABCA1 (Table 3).
Table 3.
Association of HDL-C loci with AMD
| SNP | Chromosome | Position (bp) | Notable nearby genes | Alleles (risk/nonrisk) | Frequency (risk allele) |
OR | P value* | ||
| Cases | Controls | ||||||||
| rs493258 | 15 | 56,475,172 | LIPC | C/T | |||||
| Discovery sample (2,157 cases, 1,150 controls) … | 0.564 | 0.528 | 1.21 (1.10, 1.34) | 0.002 | |||||
| Tufts/MGH sample (821 cases, 1,709 controls) … | 0.579 | 0.524 | 1.25 (1.11, 1.41) | 2.8 × 10−4† | |||||
| De novo replication sample (5,914 cases, 3,775 controls) … | 0.562 | 0.575 | 1.10 (1.03, 1.16) | 0.001 | |||||
| Combined sample (8,892 cases, 6,634 controls) … | 0.563 | 0.564 | 1.14 (1.09, 1.20) | 1.3 × 10−7 | |||||
| Cochran's Q heterogeneity test P = 0.64 | |||||||||
| rs3764261 | 16 | 55,550,825 | CETP | A/C | |||||
| Discovery sample (2,157 cases, 1,150 controls) … | 0.364 | 0.314 | 1.36 (1.26, 1.46) | 1.7 × 10−6 | |||||
| Tufts/MGH sample (821 cases, 1,709 controls) … | 0.356 | 0.329 | 1.13 (1.00, 1.28) | 0.070 | |||||
| De novo replication sample (4,945 cases, 1,960 controls) … | 0.347 | 0.317 | 1.10 (1.00, 1.22) | 0.009 | |||||
| Combined sample (7,923 cases, 4,819 controls) … | 0.354 | 0.316 | 1.19 (1.12, 1.27) | 7.4 × 10−7 | |||||
| Cochran's Q heterogeneity test P = 0.65 | |||||||||
| rs12678919 | 8 | 19,888,502 | LPL | G/A | |||||
| Discovery sample (2,157 cases, 1,150 controls) … | 0.115 | 0.096 | 1.38 (1.17, 1.63) | 0.002 | |||||
| De novo replication sample (3,333 cases, 1,288 controls) … | 0.113 | 0.108 | 1.11 (0.92, 1.35) | 0.140 | |||||
| Combined sample (5,490 cases, 2,438 controls) … | 0.114 | 0.102 | 1.26 (1.11, 1.43) | 0.003 | |||||
| Cochran's Q heterogeneity test P = 0.893 | |||||||||
| rs1883025 | 9 | 106,704,122 | ABCA1 | C/T | |||||
| Discovery sample (2,157 cases, 1,150 controls) … | 0.739 | 0.705 | 1.25 (1.12, 1.40) | 0.003 | |||||
| De novo replication sample (4,982 cases, 3,022 controls) … | 0.752 | 0.741 | 1.10 (1.00, 1.19) | 0.019 | |||||
| Combined sample (7,139 cases, 4,172 controls)… | 0.747 | 0.731 | 1.15 (1.07, 1.23) | 5.6 × 10−4 | |||||
| Cochran's Q heterogeneity test P = 0.51 | |||||||||
*P values for the discovery and combined samples are two sided. P values for the Tufts/MGH and de novo replication samples are one sided.
†Excluding overlapping AREDS samples in the Tufts/MGH study. Before excluding these samples, Tufts/MGH results differ slightly (for example, P value at rs493258 was 2.2 × 10−5).
Cholesterol and lipids accumulate underneath the RPE with age and are present in the drusen that characterize early AMD (30, 31). Genetic variants that impact cholesterol levels in the macula and RPE might impact drusen formation and thus modulate the risk of AMD. Because alleles near CETP and LPL associated with decreased HDL-c levels in blood seem to increase the risk of AMD, but alleles near LIPC and ABCA1 associated with decreased HDL-c levels in blood seem to decrease the risk of AMD, we speculate that some alleles impact cholesterol levels in blood and in the macula in opposite directions. For example, a variant that impacts cholesterol transport between tissues could facilitate transport of HDL-c from the macula to the blood (or vice versa). CETP and LPL play major roles in the synthesis and degradation of HDL-c, whereas LIPC and ABCA1 are involved in mediating the uptake of HDL-c at the cell surface. Previously, epidemiologic studies have indicated a link between cardiovascular risk factors (including HDL-c) and incidence of AMD (32, 33), but these findings have not been definitive. Our data therefore suggest an important role for HDL-c metabolism in AMD pathogenesis but also that (i) blood HDL-c levels may be a poor surrogate for the impact of HDL-c on disease risk and that (ii) further work is needed to characterize the relationship between AMD and HDL-c—associated alleles. It would be particularly interesting to examine samples with information on AMD classification and direct measurements of HDL-c levels in the retina.
To investigate whether identified risk alleles contributed preferentially to one disease subtype, we carried out a series of subgroup comparisons (Table S11). When we compared different case subgroups, we found ARMS2 risk alleles were more common in cases with neovascular disease than in cases with large drusen [OR = 1.79 (1.50, 2.13), P = 4.3 × 10−11] or with geographic atrophy [OR = 1.36 (1.13, 1.63), P = 0.0009]. In contrast, CFH risk alleles were more common in cases with geographic atrophy than in those with large drusen [OR = 1.38 (1.11, 1.73), P = 0.0012] or neovascular disease [OR = 1.32 (1.08, 1.64), P = 0.009]. Risk alleles near other complement genes seemed to be somewhat more common in cases with geographic atrophy than in those with neovascularization, whereas the reverse was true for risk alleles near TIMP3 (differences not significant). In our discovery sample, we tested for, but did not find, evidence of interactions between associated alleles at the seven loci listed in Tables 1 and 2. We also tested for, but did not find, significant interactions of risk alleles with sex and smoking.
A companion GWAS article (24) identifies variants with P < 5 × 10−8 near LIPC. Although their original GWAS did not identify TIMP3, targeted follow-up of the markers identified in our scan confirms our findings. The difference in the initial results of the two studies derives from different choices of markers to follow up after the initial GWAS: a costly experiment with maximum power would involve genotyping all discovery and follow-up samples for all markers. Practical considerations meant that each study could only examine a subset of interesting markers in available follow-up samples. Ultimately, we expect that further genotyping of follow-up samples and metaanalysis of our results with those of future GWAS will identify more disease susceptibility loci. The variants identified here have only a modest impact on the risk of AMD. However, they do point to potentially important biologic targets (such as the TIMP3 gene and HDL-c), whose effectiveness for therapeutic intervention remains to be evaluated. We note that genes like IL23 and HMGCR2 are extremely effective drug targets (for the treatment of psoriasis and for low-density lipoprotein cholesterol—lowering medications, respectively) despite the fact that naturally occurring common variants in the corresponding loci account for only small changes in the risk of psoriasis (34) and in blood lipid levels (17), respectively.
Genetic susceptibility variants may be used to predict individual risk of AMD (35). To evaluate the effectiveness of the approach, we fitted a simple logistic regression model to the data. The model included the SNPs listed in Tables 1–3 as predictors. For each SNP, a single variable encoding the number of risk alleles was modeled; no interaction terms or dominance effects were considered. In effect, the model calculates a weighted sum of risk alleles for each individual (with weights proportional to the log OR for each allele) and assigns individuals with large weighted sums the largest risk. Among the 331 individuals (10% of our sample) with the highest-risk genotypes, only 2 are controls and 329 are cases (see Fig. S2, Top, for information on other genotype risk bands). Assuming a disease prevalence of 20% at age ≈75 years, we predict that ≈80% of individuals with genotypes in the top decile of risk will develop AMD, but <5% of individuals in the bottom three deciles will develop disease (Fig. S2, Bottom). Furthermore, we find that, among cases, individuals with high-risk genotypes will present with severe disease more often (in the top risk decile for our sample, 15% of our cases have large drusen, 22% geographic atrophy, and 63% have neovascularization) than individuals with lower-risk genotypes (in the bottom risk decile, 51% of cases have large drusen, 19% geographic atrophy only, and 30% have neovascularization). A productive strategy to identify rare alleles that impact disease susceptibility might involve a detailed examination of DNA sequences in individuals with severe disease but whose common variant genotypes predict low disease risk.
Despite these encouraging contrasts between individuals with low- and high-risk genotypes, AMD susceptibility alleles must be evaluated in population-based cohorts before genotypes can become routine diagnostic tools (35). Altough trends pointing to increased frequency of severe disease in individuals with high-risk genotypes should hold, the absolute risk of developing severe disease is difficult to estimate accurately in samples collected in tertiary clinics. In the meantime, our results point to new molecular pathways and encourage new directions in the search for treatment and prevention of this common blinding disease.
Methods
All participants provided written informed consent, and the protocols were reviewed and approved by local institutional review boards.
Study Design.
Cases were classified according to AMD diagnosis in the worse eye. Controls were examined by an ophthalmologist and exhibited no signs of AMD in either eye.
Genotyping and Imputation.
Genotyping used Illumina Human370 Bead Chips and the Illumina Infinium II assay protocol. Standard quality filters were applied. See SI Methods for details. To expand the genome coverage, we performed a genome-wide imputation using haplotypes from the HapMap CEU samples as templates (release 22). Imputation was performed using MACH (www.sph.umich.edu/csg/abecasis/Mach/).
Statistical Analyses.
To investigate the association between each genotyped or imputed SNP and AMD, we first carried out a logistic regression for each SNP assuming an additive model and adjusting for the top two PCA. For samples including related individuals, the data were analyzed with the test of Thornton and McPeek (36).
Analysis for Follow-up Study.
To combine the statistics across different groups for replication, we selected an arbitrary reference allele and then calculated a z statistic summarizing the evidence for association in each study. An overall z statistic was defined as a weighted average of the individual statistics. Weights were proportional to the square root of the effective sample size for each study.
Conditional Analyses.
For conditional analyses, allele counts for the markers listed in Table 2 were used as covariates. For follow-up samples, genotypes at CFH and ARMS2 were included as covariates where available.
Risk Prediction Approach.
To evaluate the cumulative contribution of the alleles identified here to disease risk, we fitted a simple logistic regression model to the data. The effect of each genotype was modeled on a log-additive scale, with no interaction terms between genotypes. For additional details, see SI Methods.
Supplementary Material
Acknowledgments
We thank numerous clinicians, clinical staff, patients, and their families for their participation in and dedication to the project; the Kellogg Eye Center AMD group, the Complications of Age-Related Macular Degeneration Prevention Trial consortium, the Spanish AMD research group, staff at the National Center for Biotechnology Information, and Dr. Hemin Chin; and the Tufts Medical Center/Massachusetts General Hospital group for sharing genome-wide association results. This research was supported in part by the intramural program of the National Eye Institute and by National Institutes of Health Grants EY016862, EY007758, EY09859, EY012118, P30-EY014801, EY-014458, EY014467, HL084729, and HG002651, by the Foundation Fighting Blindness, the Macula Vision Research Foundation, the American Health Assistance Foundation, Research to Prevent Blindness, the Pew Charitable Trusts, the Mayo Clinic Foundation, the Casey Macular Degeneration Center Fund, the Marion W. and Edward F. Knight AMD Fund, the Harold and Pauline Price Foundation, National Genotyping Centre of Spain, and the Elmer and Sylvia Sramek Foundation. The Center for Inherited Disease Research, fully funded through a federal contract (HHSN268200782096C) from National Institutes of Health to The Johns Hopkins University, performed genotyping of the discovery cohort.
Footnotes
Conflict of interest statement: A.O.E., J.J., M.A.P.-V., Y.P.C., A.K., W.K.S., A.A., E.A.P., M.M.D., D.E.W., J.M.S., J.L.H., M.B.G., G.R.A. and A.S. are listed as inventors on patents relating to the use of genetic data in age-related macular degeneration. S.G.S. has previously received research funding from Genentech and owns equity in Pfizer.
A complete listing of The CAPT Investigative Group is available at http://amd.nei.nih.gov/gwas/CAPTAuthorList.pdf.
Data deposition: Genotype and phenotype data described in this article have been deposited in the Database of Genotypes and Phenotypes (accession no. phs000182.v1.p1).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912702107/DCSupplemental.
Contributor Information
Robert N. Johnson, THE CAPT INVESTIGATIVE GROUP.
Everett Ai, THE CAPT INVESTIGATIVE GROUP.
H. Richard McDonald, THE CAPT INVESTIGATIVE GROUP.
Margaret Stolarczuk, THE CAPT INVESTIGATIVE GROUP.
Peter Reed Pavan, THE CAPT INVESTIGATIVE GROUP.
Karina K. Billiris, THE CAPT INVESTIGATIVE GROUP.
Mohan Iyer, THE CAPT INVESTIGATIVE GROUP.
Matthew M. Menosky, THE CAPT INVESTIGATIVE GROUP.
Scott E. Pautler, THE CAPT INVESTIGATIVE GROUP.
Sharon M. Millard, THE CAPT INVESTIGATIVE GROUP.
Baker Hubbard, III, THE CAPT INVESTIGATIVE GROUP.
Thomas Aaberg, Sr., THE CAPT INVESTIGATIVE GROUP.
Lindy DuBois, THE CAPT INVESTIGATIVE GROUP.
Alice Lyon, THE CAPT INVESTIGATIVE GROUP.
Susan Anderson-Nelson, THE CAPT INVESTIGATIVE GROUP.
Lee M. Jampol, THE CAPT INVESTIGATIVE GROUP.
David V. Weinberg, THE CAPT INVESTIGATIVE GROUP.
Annie Muñana, THE CAPT INVESTIGATIVE GROUP.
Zuzanna Rozenbajgier, THE CAPT INVESTIGATIVE GROUP.
David Orth, THE CAPT INVESTIGATIVE GROUP.
Jack Cohen, THE CAPT INVESTIGATIVE GROUP.
Matthew MacCumber, THE CAPT INVESTIGATIVE GROUP.
Matthew MacCumber, THE CAPT INVESTIGATIVE GROUP.
Celeste Figliulo, THE CAPT INVESTIGATIVE GROUP.
Liz Porcz, THE CAPT INVESTIGATIVE GROUP.
James Folk, THE CAPT INVESTIGATIVE GROUP.
H. Culver Boldt, THE CAPT INVESTIGATIVE GROUP.
Stephen R. Russell, THE CAPT INVESTIGATIVE GROUP.
Rachel Ivins, THE CAPT INVESTIGATIVE GROUP.
Connie J. Hinz, THE CAPT INVESTIGATIVE GROUP.
Charles C. Barr, THE CAPT INVESTIGATIVE GROUP.
Steve Bloom, THE CAPT INVESTIGATIVE GROUP.
Ken Jaegers, THE CAPT INVESTIGATIVE GROUP.
Brian Kritchman, THE CAPT INVESTIGATIVE GROUP.
Greg Whittington, THE CAPT INVESTIGATIVE GROUP.
Jeffrey Heier, THE CAPT INVESTIGATIVE GROUP.
Albert R. Frederick, Jr., THE CAPT INVESTIGATIVE GROUP.
Michael G. Morley, THE CAPT INVESTIGATIVE GROUP.
Trexler Topping, THE CAPT INVESTIGATIVE GROUP.
Heather L. Davis, THE CAPT INVESTIGATIVE GROUP
Susan B. Bressler, THE CAPT INVESTIGATIVE GROUP.
Neil M. Bressler, THE CAPT INVESTIGATIVE GROUP.
Warren Doll, THE CAPT INVESTIGATIVE GROUP.
Michael Trese, THE CAPT INVESTIGATIVE GROUP.
Antonio Capone, THE CAPT INVESTIGATIVE GROUP.
Bruce R. Garretson, THE CAPT INVESTIGATIVE GROUP.
Tarek S. Hassan, THE CAPT INVESTIGATIVE GROUP.
Alan J. Ruby, THE CAPT INVESTIGATIVE GROUP.
Tammy Osentoski, THE CAPT INVESTIGATIVE GROUP.
Colin A. McCannel, THE CAPT INVESTIGATIVE GROUP.
Margaret J. Ruszczyk, THE CAPT INVESTIGATIVE GROUP.
Gilbert Grand, THE CAPT INVESTIGATIVE GROUP.
Kevin Blinder, THE CAPT INVESTIGATIVE GROUP.
Nancy M. Holekamp, THE CAPT INVESTIGATIVE GROUP.
Daniel P. Joseph, THE CAPT INVESTIGATIVE GROUP.
Gaurav Shah, THE CAPT INVESTIGATIVE GROUP.
Ginny S. Nobel, THE CAPT INVESTIGATIVE GROUP.
Andrew N. Antoszyk, THE CAPT INVESTIGATIVE GROUP.
David J. Browning, THE CAPT INVESTIGATIVE GROUP.
Alison H Stallings, THE CAPT INVESTIGATIVE GROUP.
Lawrence J. Singerman, THE CAPT INVESTIGATIVE GROUP.
David Miller, THE CAPT INVESTIGATIVE GROUP.
Michael Novak, THE CAPT INVESTIGATIVE GROUP.
Scott Pendergast, THE CAPT INVESTIGATIVE GROUP.
Hernando Zegarra, THE CAPT INVESTIGATIVE GROUP.
Stephanie A. Schura, THE CAPT INVESTIGATIVE GROUP.
Sheila Smith-Brewer, THE CAPT INVESTIGATIVE GROUP.
Frederick H. Davidorf, THE CAPT INVESTIGATIVE GROUP.
Robert Chambers, THE CAPT INVESTIGATIVE GROUP.
Louis Chorich, THE CAPT INVESTIGATIVE GROUP.
Jill Salerno, THE CAPT INVESTIGATIVE GROUP.
Richard F. Dreyer, THE CAPT INVESTIGATIVE GROUP.
Colin Ma, THE CAPT INVESTIGATIVE GROUP.
Marcia R. Kopfer, THE CAPT INVESTIGATIVE GROUP.
Michael L. Klein, THE CAPT INVESTIGATIVE GROUP.
David J. Wilson, THE CAPT INVESTIGATIVE GROUP.
Susan K. Nolte, THE CAPT INVESTIGATIVE GROUP
Juan E. Grunwald, THE CAPT INVESTIGATIVE GROUP.
Alexander J. Brucker, THE CAPT INVESTIGATIVE GROUP.
Josh Dunaief, THE CAPT INVESTIGATIVE GROUP.
Stuart L. Fine, THE CAPT INVESTIGATIVE GROUP.
Albert M. Maguire, THE CAPT INVESTIGATIVE GROUP.
Robert A. Stoltz, THE CAPT INVESTIGATIVE GROUP.
Monique N. McRay, THE CAPT INVESTIGATIVE GROUP
Gary Edd Fish, THE CAPT INVESTIGATIVE GROUP.
Rajiv Anand, THE CAPT INVESTIGATIVE GROUP.
Rand Spencer, THE CAPT INVESTIGATIVE GROUP.
Jean Arnwine, THE CAPT INVESTIGATIVE GROUP.
Suresh R. Chandra, THE CAPT INVESTIGATIVE GROUP.
Michael Altaweel, THE CAPT INVESTIGATIVE GROUP.
Barbara Blodi, THE CAPT INVESTIGATIVE GROUP.
Justin Gottlieb, THE CAPT INVESTIGATIVE GROUP.
Michael Ip, THE CAPT INVESTIGATIVE GROUP.
T. Michael Nork, THE CAPT INVESTIGATIVE GROUP.
Jennie Perry-Raymond, THE CAPT INVESTIGATIVE GROUP.
Stuart L. Fine, THE CAPT INVESTIGATIVE GROUP.
Maureen G. Maguire, THE CAPT INVESTIGATIVE GROUP.
Mary Brightwell-Arnold, THE CAPT INVESTIGATIVE GROUP.
Sandra Harkins, THE CAPT INVESTIGATIVE GROUP.
Ellen Peskin, THE CAPT INVESTIGATIVE GROUP.
Gui-Shuang Ying, THE CAPT INVESTIGATIVE GROUP.
Natalie Kurinij, THE CAPT INVESTIGATIVE GROUP.
References
- 1.Swaroop A, Chew EY, Rickman CB, Abecasis GR. Unravelling a late-onset multifactorial disease: From genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu Rev Genomics Hum Genet. 2009;10:19–43. doi: 10.1146/annurev.genom.9.081307.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fisher SA, et al. Meta-analysis of genome scans of age-related macular degeneration. Hum Mol Genet. 2005;14:2257–2264. doi: 10.1093/hmg/ddi230. [DOI] [PubMed] [Google Scholar]
- 3.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dewan A, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989–992. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 5.Rivera A, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14:3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 6.Jakobsdottir J, et al. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am J Hum Genet. 2005;77:389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haines JL, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 8.Edwards AO, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 9.Fagerness JA, et al. Variation near complement factor I is associated with risk of advanced AMD. Eur J Hum Genet. 2009;17:100–104. doi: 10.1038/ejhg.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gold B, et al. AMD Genetics Clinical Study Group. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maller JB, et al. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39:1200–1201. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- 12.Yates JR, et al. Genetic Factors in AMD Study Group. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 13.Barrett JC, et al. NIDDK IBD Genetics Consortium; Belgian-French IBD Consortium; Wellcome Trust Case Control Consortium. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeggini E, et al. Wellcome Trust Case Control Consortium. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willer CJ, et al. Wellcome Trust Case Control Consortium; Genetic Investigation of ANthropometric Traits Consortium. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willer CJ, et al. Genome-wide association scans identify novel loci that influence lipid lvels and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 19.Frazer KA, et al. International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 21.Tian C, et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet. 2008;4:e4. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38:1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maller J, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38:1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 24.Neale B, et al. A Genome-wide scan of advanced age-related macular degeneration suggests a novel role of lipase-C. Proc Natl Acad Sci USA. 2010;107:7395–7400. doi: 10.1073/pnas.0912019107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Göring HH, Terwilliger JD, Blangero J. Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet. 2001;69:1357–1369. doi: 10.1086/324471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanda A, et al. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci USA. 2007;104:16227–16232. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng J, et al. Regulation of neurotransmitter release by synapsin III. J Neurosci. 2002;22:4372–4380. doi: 10.1523/JNEUROSCI.22-11-04372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber BH, Vogt G, Pruett RC, Stöhr H, Felbor U. Mutations in the tissue inhibitor of metalloproteinases-3 (TIMP3) in patients with Sorsby's fundus dystrophy. Nat Genet. 1994;8:352–356. doi: 10.1038/ng1294-352. [DOI] [PubMed] [Google Scholar]
- 29.Abecasis GR, et al. Age-related macular degeneration: A high-resolution genome scan for susceptibility loci in a population enriched for late-stage disease. Am J Hum Genet. 2004;74:482–494. doi: 10.1086/382786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mullins RF, Russell SR, Anderson DH, Hageman GS. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J. 2000;14:835–846. [PubMed] [Google Scholar]
- 31.Curcio CA, et al. Esterified and unesterified cholesterol in drusen and basal deposits of eyes with age-related maculopathy. Exp Eye Res. 2005;81:731–741. doi: 10.1016/j.exer.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 32.Klein R, Klein BE, Franke T. The relationship of cardiovascular disease and its risk factors to age-related maculopathy. The Beaver Dam Eye Study. Ophthalmology. 1993;100:406–414. doi: 10.1016/s0161-6420(93)31634-9. [DOI] [PubMed] [Google Scholar]
- 33.Tomany SC, et al. Risk factors for incident age-related macular degeneration: Pooled findings from 3 continents. Ophthalmology. 2004;111:1280–1287. doi: 10.1016/j.ophtha.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 34.Nair RP, et al. Collaborative Association Study of Psoriasis. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakobsdottir J, Gorin MB, Conley YP, Ferrell RE, Weeks DE. Interpretation of genetic association studies: Markers with replicated highly significant odds ratios may be poor classifiers. PLoS Genet. 2009;5:e1000337. doi: 10.1371/journal.pgen.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thornton T, McPeek MS. Case-control association testing with related individuals: A more powerful quasi-likelihood score test. Am J Hum Genet. 2007;81:321–337. doi: 10.1086/519497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.