Abstract
A fundamental challenge in gene therapy is to develop approaches for delivering nucleic acid-based gene interfering agents, such as small interfering RNAs and ribozymes, to the appropriate cells in a way that is tissue/cell specific, efficient, and safe. Using human cytomegalovirus (HCMV) infection of differentiated macrophages as the model, we showed that Salmonella can efficiently deliver RNase P-based ribozyme sequence in specific human cells, leading to substantial ribozyme expression and effective inhibition of viral infection. We constructed a functional RNase P ribozyme (M1GS RNA) that targets the overlapping mRNA region of two HCMV capsid proteins, the capsid scaffolding protein (CSP) and assemblin, which are essential for viral capsid formation. Substantial expression of ribozymes was observed in human differentiated macrophages that were treated with attenuated Salmonella strains carrying the ribozyme sequence constructs. A reduction of 87–90% in viral CSP expression and a reduction of about 5,000-fold in viral growth were observed in cells that were treated with Salmonella carrying the sequence of the functional ribozyme but not with those carrying the sequence of a control ribozyme that contained mutations abolishing the catalytic activity. To our knowledge, this study showed for the first time that ribozymes expressed following targeted gene transfer with Salmonella-based vectors are highly active and specific in blocking viral infection. Moreover, these results demonstrate the feasibility to develop Salmonella-mediated gene transfer of RNase P ribozymes as an effective approach for gene-targeting applications.
Keywords: antisense, gene targeting, antiviral, gene delivery, RNA cleavage
Human cytomegalovirus (HCMV), a common opportunistic pathogen, causes significant morbidity and mortality in immunocompromised or immunologically immature individuals, including neonates, AIDS patients, and transplant recipients (1). Macrophages and their progenitor cells, such as monocytic cells, represent the major reservoirs for HCMV. HCMV can establish latent infection in undifferentiated monocytic cells but engage in productive and lytic replication in terminally differentiated macrophages, leading to viral pathogenesis. Thus, blocking HCMV infection and replication in macrophages is central in treating and preventing HCMV-associated diseases.
Nucleic acid-based gene interference technologies, including ribozymes and small interfering RNAs, represent promising gene-targeting strategies for specific inhibition of mRNA sequences of choice (2). Altman and colleagues have previously shown that RNase P of Escherichia coli contains a catalytic RNA subunit (M1 RNA) that can be engineered to cleave tRNA substrates and other target RNAs, including specific mRNAs (3, 4). A sequence-specific ribozyme, M1GS, constructed by attaching to M1 RNA a guide sequence (GS) complementary to a target mRNA (Fig. 1 A and B), is effective in blocking mRNA expression in cultured cells (4, 5). M1GS-based strategy represents a unique nucleic acid-based interference approach because of the use of M1 RNA, one of the most efficient catalytic RNAs found in nature (3). Previous studies have shown that M1GS RNAs and RNase P are effective in cleaving both viral and cellular mRNAs and blocking their expression in cultured cells, including inhibition of gene expression of human influenza and herpes viruses (5–7).
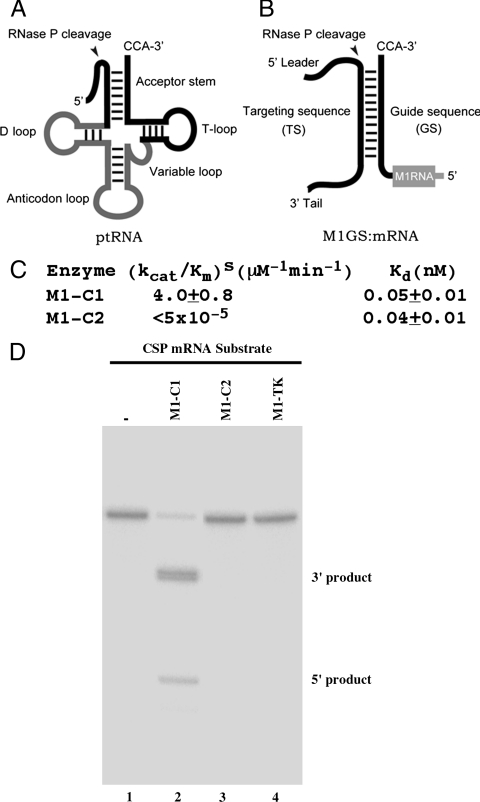
Fig. 1.
(A and B) Schematic representation of a natural substrate (ptRNA) (A) and a complex formed between a M1GS RNA and its mRNA substrate (B). (C) Overall cleavage rate [(kcat/Km)s] and binding affinity (Kd) in reactions with ribozymes (average values of triplicate experiments). (D) Cleavage of the CSP mRNA substrate by M1GS RNA. The substrate (20 nM) was incubated alone (lane 1), with 5 nM of M1-C1 (lane 2), M1-C2 (lane 3), or M1-TK (lane 4). Reactions were carried out for 40 min in buffer A (50 mM Tris.HCl, pH 7.5, 100 mM NH4Cl, and 100 mM MgCl2) at 37 °C.
A fundamental challenge in gene-targeting therapy is to develop approaches for delivering genetic material to the appropriate cells of a patient in a way that is tissue/cell specific, efficient, and safe (8, 9). Many of the vectors currently in use are based on attenuated or modified viruses, or synthetic vectors in which complexes of DNA, proteins, and/or lipids are formed in particles. Like all other nucleic acid-based interference approaches, delivery and expression of M1GS sequences in specific cell types and tissues is central in developing this technology for gene-targeting applications.
Invasive bacteria, such as Salmonella, which possess the ability to enter human cells, are capable of transferring genetic material to host cells, leading to efficient expression of the transferred genes (9). Attenuated Salmonella strains have recently been shown to function as a carrier system for delivery of nucleic acid-based vaccines and antitumor small hairpin RNAs (shRNAs) for cancer therapy (10–13). In these studies, plasmid constructs, which contained the transgenes under the control of an eukaryotic expression promoter, were introduced to the attenuated bacterial strains. These attenuated strains can target specific cells, such as dendritic cells, macrophages, and epithelial cells, and survive within the target cells to a limited extent. How the plasmid DNA from a bacterial vector is transferred to the host is not completely understood. It is generally believed that once bacteria undergo lysis intracellularlly, the plasmid content is released, leading to expression of the encoded transgenes in the construct by cellular machinery (11, 14). Macrophages represent the major in vivo reservoir for Salmonella following their systemic dissemination (15) and, therefore, are considered an optimal target for a Salmonella-based gene therapy.
Using HCMV infection of differentiated macrophages as the model, we in this study provide direct evidence that Salmonella can efficiently deliver the M1GS sequence into human differentiated macrophages, leading to substantial expression of the ribozymes. M1GS ribozymes were constructed to target the region of the mRNA encoding the HCMV capsid scaffolding protein (CSP). CSP completely overlaps with and is within the 3′ coding sequence of another viral capsid protein, assemblin (1). Both CSP and assemblin are essential for HCMV capsid formation and replication (1). Our results showed that targeted gene delivery of RNase P ribozyme by Salmonella to HCMV-infected cells resulted in effective inhibition of viral gene expression and replication. Furthermore, these results demonstrate the feasibility of developing Salmonella-based vectors for delivering RNase P ribozymes for treatment of viral diseases.
Results
In Vitro Cleavage of HCMV mRNA Sequence by M1GS Ribozyme.
Using DMS, we employed an in vivo mapping approach (5) to determine the accessibility of the region of the CSP mRNA in HCMV-infected cells, and we chose a highly accessible region as the cleavage site for M1GS RNA. A functional ribozyme, M1-C1, was constructed by covalently linking the 3′ terminus of an engineered M1GS ribozyme, V57, with a guide sequence of 18 nucleotides that is complementary to the targeted mRNA sequence. V57 is a M1 RNA variant (G190 → U190 and A258 → C258) generated from an in vitro selection procedure, and ribozymes derived from this variant are among the most active M1GS RNAs in cleaving the CSP mRNA and the thymidine kinase (TK) mRNA of herpes simplex virus 1 (HSV-1) (Fig. 1) (16). A control ribozyme, M1-C2, was derived from M1-C1 by introducing several point mutations (A347C348 → C347U348 and C353C354C355G356 → G353G354A355U356) at the catalytic P4 domain. These mutations reduced the activity of M1 RNA in cleaving a pre-tRNA by at least 10,000-fold (7). M1-C2 is therefore expected to be catalytically inactive.
Incubation of a substrate containing the CSP mRNA sequence with functional ribozyme M1-C1 yielded efficient cleavage (Fig. 1, lane 2). In contrast, cleavage by M1-C2 was barely detected (Fig. 1, lane 3). Gel-shift assays indicate that the binding affinity of M1-C2 to the CSP mRNA substrate, measured as the dissociation constant (Kd), is similar to that of M1-C1 (Fig. 1C). Since M1-C2 contains the same antisense guide sequence and exhibits similar affinity to the CSP mRNA substrate as M1-C1 but is catalytically inactive, this ribozyme was used as a control for the antisense effect in our experiments (see below).
Efficient Gene Delivery for the Expression of Ribozymes in Macrophages by Salmonella.
The DNA sequence coding for M1-C1 and M1-C2 were cloned into vector pU6, which contains a GFP expression cassette, and placed under the control of the small nuclear U6 RNA promoter. This promoter, which is transcribed by RNA polymerase III, has previously been shown to express M1GS RNA and other RNAs steadily (5, 17). To determine whether Salmonella-mediated delivery for expression of a M1GS RNA with an incorrect guide sequence could affect the CSP mRNA, the DNA sequence for ribozyme M1-TK, which was derived from V57 and targeted the HSV-1 TK mRNA (16), was also cloned into vector pU6. No cleavage of the CSP mRNA substrate by M1-TK was observed in vitro (Fig. 1, lane 4). The DNAs of constructs containing the M1GS sequence were transformed into auxotrophic Salmonella strain SL7207, which is attenuated in virulence and pathogenesis in vivo and has been shown to function efficiently as a gene delivery carrier for the expression of several transgenes in mammalian cells (12, 13).
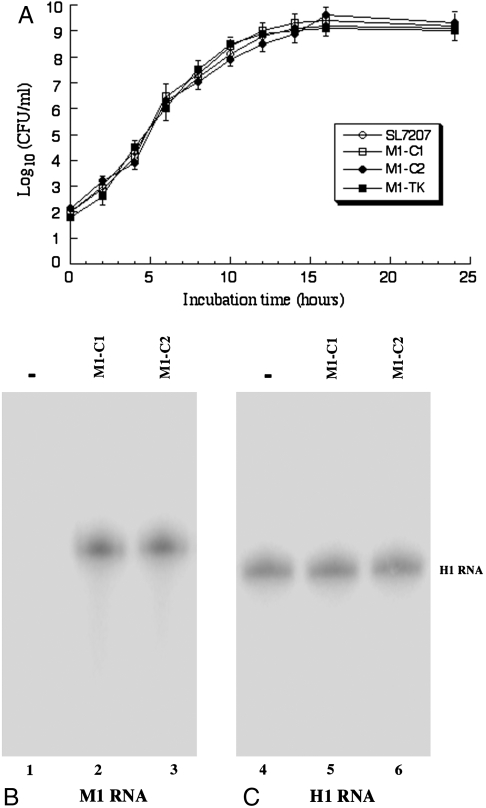
Four sets of experiments were carried out to characterize Salmonella carrying the plasmids with the ribozyme sequences. First, growth analyses of Salmonella were performed. There was no significant difference in the growth kinetics of Salmonella carrying no constructs or constructs pU6-M1-C1, pU6-M1-C2, pU6-M1-TK, and the pU6 empty vector in LB broth (Fig. 2A), indicating that the presence of the ribozyme sequence does not result in an impaired viability of the bacterial carrier. Second, Northern analysis indicated that neither the GFP nor the M1GS RNA transcript was detected in Salmonella carrying different ribozyme constructs. Furthermore, no GFP signal was observed when Salmonella was examined under a fluorescence microscope. These results suggested that M1GS RNA, which was under the control of the U6 expression cassette, was not expressed in the Salmonella vector. Third, to determine whether Salmonella can efficiently deliver the M1GS sequences into human cells, differentiated macrophage THP-1 cells were infected with Salmonella SL7207 carrying pU6-M1-C1, pU6-M1-C2, pU6-M1-TK, and the pU6 empty vector. At 24 h postinfection, more than 80% of cells were GFP positive, indicating efficient gene transfer mediated by Salmonella. Fourth, to examine the ribozyme expression after Salmonella-mediated gene transfer, total RNAs were isolated from Salmonella-infected cells at 24 h postinfection. The levels of M1GS RNAs (Fig. 2B) were assayed with Northern analyses, using H1 RNA [the RNA subunit of human RNase P (3)] as the internal control (Fig. 2C). Similar levels of ribozymes were found when cells were infected at the same multiplicity of infection (MOI) with Salmonella carrying different ribozyme sequences. The M1GS RNAs appeared to be exclusively expressed in the nuclei as they were detected only in the nuclear but not the cytoplasmic RNA fractions. This is consistent with previous observations that the transcripts expressed by the U6 promoter are primarily localized in the nuclei (7, 17).
Fig. 2.
(A) Analysis of growth in LB broth of Salmonella strain SL7207 and its derivatives that carried constructs of the sequence of M1-C1, M1-C2, and M1-TK. (B and C) Northern analyses of the expression of M1GS ribozymes in differentiated THP-1 macrophages that were treated with Salmonella carrying the empty vector (-, lane 1, 4) or with Salmonella carrying constructs that contained the sequence of M1-C1 (lanes 2 and 5) and M1-C2 (lanes 3 and 6). RNA samples (25 μg) were separated on 2% agarose gels that contained formaldehyde, transferred to nitrocellulose membranes, and hybridized to a [32P]-radiolabeled probe that contained the DNA sequence coding for M1 RNA (lanes 1–3) (B) or H1 RNA (lanes 4–6) (C), the RNA subunit of human RNase P (3).
Inhibition of Viral Gene Expression by Ribozymes Delivered via Salmonella-Based Vector.
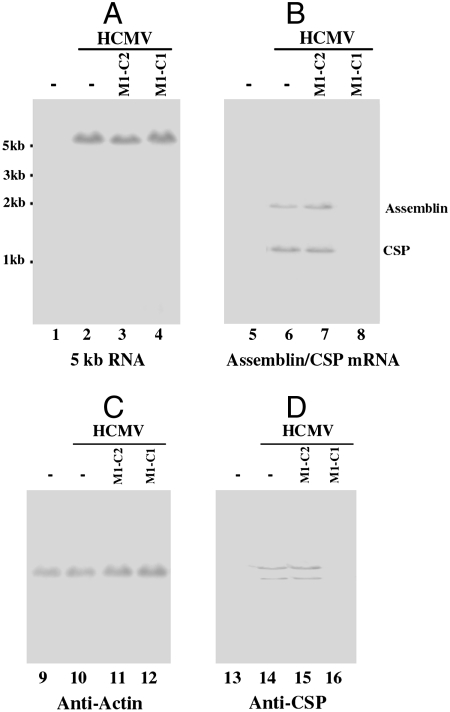
To determine the effect of the M1GS sequence delivered by Salmonella on HCMV gene expression, differentiated THP-1 cells were first treated with Salmonella carrying the U6-M1GS plasmids. The Salmonella-containing cells were then isolated by FACS analysis based on GFP expression, and infected with HCMV at a multiplicity of infection (MOI) of 0.05–1. Total RNAs were isolated from the infected cells at 8–72 h postinfection. The expression levels of CSP and assemblin mRNAs were determined by Northern analyses. The level of the 5-kb long viral immediate-early transcript (5-kb RNA), whose expression is not regulated by assemblin and CSP under the assay conditions (1), was used as an internal control for the quantitation of expression of the target mRNAs (Fig. 3A). A reduction of about 90 ± 8% and 90 ± 9% (average of three experiments) in the expression levels of CSP and assemblin mRNA was observed in cells that expressed M1-C1, respectively (Fig. 3B and Table 1). In contrast, a reduction of less than 10% in the expression levels of these two mRNAs was observed in cells that were treated with Salmonella carrying the M1-C2 or M1-TK sequences. The CSP protein levels in cells that were treated with Salmonella carrying the M1-C1 sequence-containing plasmid were also reduced. Proteins were isolated from cells at 24–72 h postinfection, separated in SDS-polyacrylamide gels, and transferred to identical membranes. One of the membranes was stained with an anti-CSP antibody, and another membrane was stained with a monoclonal antibody against human actin (Fig. 3 C and D). The latter serves as an internal control for the quantitation of CSP protein expression. A reduction of about 87% in the protein level of CSP (from three independent experiments) was observed in cells treated with Salmonella carrying the M1-C1 sequence, while a reduction of less than 10% was found in cells that were treated with Salmonella carrying the plasmids expressing the M1-C2 or M1-TK RNAs. The low level of inhibition found in cells treated with Salmonella carrying the M1-C2 sequence was presumably due to an antisense effect because M1-C2 exhibited similar binding affinity to the target sequence as M1-C1 but was catalytically inactive. These results suggest that the significant reduction of CSP expression in cells treated with the M1-C1-containing Salmonella is due to Salmonella-mediated gene delivery of the ribozyme.
Fig. 3.
(A–D) Expression levels of HCMV mRNAs (A and B) and proteins (C and D). Differentiated THP-1 cells were first treated with Salmonella carrying the empty vector (-, lanes 1–2, 5–6, 9–10, and 13–14) or with Salmonella carrying constructs that contained the sequence of M1-C2 (lanes 3, 7, 11, and 15) and M1-C1 (lanes 4, 8, 12, and 16). The cells were then either mock-infected (lanes 1, 5, 9, and 13) or infected with HCMV (lanes 2–4, 6–8, 10–12, and 14–16) and were harvested at 12–72 h postinfection. In Northern analysis (A and B), RNA samples (25 μg) were separated on agarose gels, transferred to nitrocellulose membranes, and hybridized to [32P]-radiolabeled probes that contained the sequence of the HCMV 5-kb transcript (lanes 1–4) and CSP mRNA (lanes 5–8). For Western analyses (C and D), protein samples (35 μg) were separated in SDS-polyacrylamide gels. The membranes were stained with the antibodies against human actin (C) and HCMV CSP (D).
Table 1.
Levels of inhibition of HCMV gene expression in differentiated THP-1 cells that were treated with Salmonella carrying constructs that contained the sequence of M1-C1 (M1-C1), M1-C2 (M1-C2), and M1-TK (M1-TK), as compared to that in cells treated with Salmonella carrying an empty pU6 vector construct with no ribozyme sequence (THP-1); the values shown are the means of triplicate experiments and the values of standard deviation that were less than 5% are not shown
| Viral gene class |
Ribozymes |
||||
| THP-1 |
M1-TK |
M1-C1 |
M1-C2 |
||
| IE2 mRNA | α | 0% | 0% | 0% | 1% |
| US2 mRNA | γ | 0% | 0% | 0% | 1% |
| CSP mRNA | γ | 0% | 2% | 90 ± 8% | 9% |
| Assemblin mRNA | γ | 0% | 0% | 90 ± 9% | 8% |
| UL44 protein | β,γ | 0% | 0% | 2% | 1% |
| CSP protein | γ | 0% | 0% | 87 ± 9% | 2% |
| gB protein | γ | 0% | 1% | 4% | 3% |
Inhibition of HCMV Growth by Salmonella-Mediated Gene Delivery of Ribozyme.
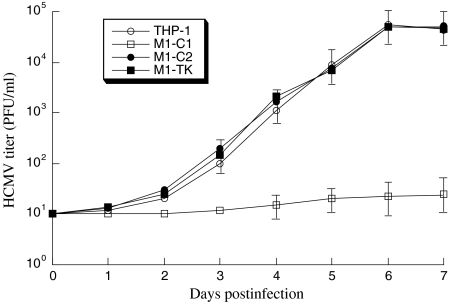
To determine whether Salmonella-mediated gene delivery of ribozymes inhibits the growth of HCMV, differentiated THP-1 cells were first treated with Salmonella carrying the ribozyme sequences. The Salmonella-containing cells were then isolated by FACS analysis based on GFP expression and infected by HCMV at an MOI of 1–5. We harvested the infected cultures (cells and culture medium together) at one-day intervals through seven days postinfection and determined the viral titers of these samples. At six days postinfection, a reduction of at least 5,000-fold in viral yield was observed in cells that were treated with Salmonella carrying the vector containing the M1-C1 sequence (Fig. 4). In contrast, no significant reduction was found in cells that were treated with Salmonella that carried the empty vector plasmid, or plasmids containing the M1-C2 and M1-TK sequence (Fig. 4).
Fig. 4.
Growth of HCMV in differentiated THP-1 cells that were treated with Salmonella carrying the empty vector construct or constructs containing the sequence of M1-C1, M1-C2, and M1-TK (see Supporting Information).
Blocking Viral Capsid Maturation by Salmonella-Mediated Gene Delivery of M1GS Ribozymes.
HCMV CSP is essential for viral capsid maturation and assembly (1). Meanwhile, it is possible that the observed reduction of viral growth in cells that were treated with M1-C1 sequence-containing Salmonella is not necessarily due to specific M1GS RNA-mediated cleavage of CSP mRNA but is due to other effects of the ribozyme or Salmonella on viral lytic replication that are unrelated to the consequence of the ribozyme cleavage or the inhibition of viral CSP expression. To further determine the antiviral mechanism of the Salmonella-mediated gene delivery of M1GS against the CSP mRNA, two sets of experiments were performed to investigate which step of the viral lytic cycle was blocked in cells treated with M1-C1 sequence-containing Salmonella. First, we examined the expression of other viral genes. Inhibition of CSP/assemblin expression is not expected to affect the expression of other viral genes, including immediate-early (α), early (β), and late (γ) genes (1). The levels of the IE2 (an α-transcript) and US2 mRNA (a β-transcript) were examined using Northern analyses, whereas the levels of viral protein UL44, a viral early late (βγ) protein and gB, a viral late (γ) protein were assayed with Western analyses. We observed no significant difference in the levels of these genes among Salmonella-treated cells (Table 1), suggesting that the Salmonella-mediated delivery of M1-C1 specifically inhibits the expression of its target and does not affect overall viral gene expression.
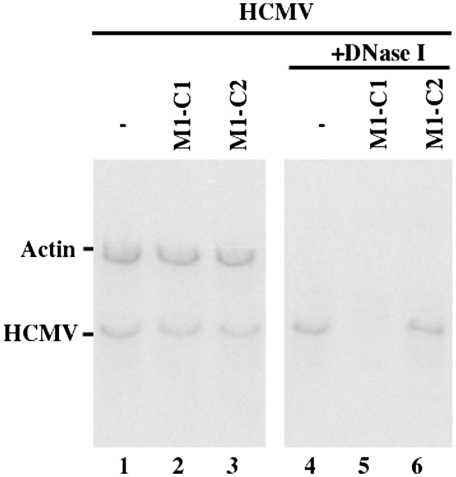
The second set of experiments was performed to determine whether viral genomic replication as well as capsid maturation is affected in cells treated with Salmonella that carried the ribozyme constructs. Cells were first treated with Salmonella, and the Salmonella-containing cells were then isolated by FACS analysis based on GFP expression and infected with HCMV. Total DNA was isolated from HCMV-infected cell lysates, and the level of intracellular viral DNA was determined by PCR detection of the HCMV IE1 sequence, using levels of β-actin DNA as the internal control (Fig. 5). The amount of intracellular viral DNA detected by the PCR assay should represent the replication level of the viral genome because HCMV replicates only in an episomal form and does not integrate its DNA into the host genome (1). To determine the level of mature capsids assembled in infected cells and examine viral capsid formation, we assayed the level of encapsidated viral DNA. DNA samples were isolated from HCMV-infected cell lysates that were treated with DNase I. The encapsidated viral DNAs are resistant to DNase I digestion, whereas those that are not packaged in the capsid will be susceptible to degradation. When the DNA samples from cell lysates that were not treated with DNase I were assayed, no significant difference in the level of total intracellular (both encapsidated and uncapsidated) viral DNA were observed (Fig. 5, lanes 1–3). However, when the samples were first treated with DNase I and then assayed, the “encapsidated” DNA was hardly detected in cells that were treated with Salmonella carrying the M1-C1 expression plasmid (Fig. 5, lane 5). These observations suggest that Salmonella-mediated gene delivery of ribozymes against the CSP mRNA does not affect the replication of viral DNA but blocks DNA encapsidation and capsid formation.
Fig. 5.
Level of total intracellular and encapsidated viral DNA as determined by semiquantative PCR. Total DNA (lanes 1–3) or DNase I-treated DNA samples (lanes 4–6) were isolated from differentiated THP-1 cells that were treated with Salmonella carrying the empty vector construct (-, lanes 1 and 4) or carrying constructs that contained the sequence of M1-C1 (lanes 2 and 5) or M1-C2 (lanes 3 and 6) and were infected with HCMV at MOI of 1. We determined the levels of viral IE1 sequence by PCR using human β-actin DNA as the internal controls. The radiolabeled PCR products were separated in 4% nondenaturing polyacrylamide gels.
Activation of Toll-Like Receptor 9 in Salmonella-Infected Cultured Cells.
It is known that bacterial DNAs, which contain unmethylated CpG motifs, can potently activate the Toll-like receptor 9 (TLR9) pathway (18, 19). To assess whether gene transfer by Salmonella vectors activates TLR9 expression, the levels of TLR9 mRNA in cells that were infected with SL7207 carrying different ribozyme constructs after 4-h infection were determined using quantitative RT-PCR, with the level of human actin mRNA as the internal control. The levels of TLR9 mRNA in cells infected with Salmonella carrying pU6-M1-C1, pU6-M1-C2, and the pU6 empty vector were similar and were at least 3-fold higher than those in uninfected cells. These results suggest that infection of the attenuated Salmonella vectors led to modest activation of TLR9 pathway in cells and that activation of TLR9 did not account for the observed antiviral effects associated with Salmonella containing M1-C1 because similar levels of TLR9 mRNA were found in cells infected with SL7207 carrying different ribozyme constructs.
Discussion
Nucleic acid-based gene interference strategies, such as antisense oligonucleotides, ribozymes or DNAzymes, and RNA interference, represent powerful research tools and promising therapeutic agents against human diseases (2). The RNase P ribozyme-based technology represents an attractive approach for gene inactivation by using M1 RNA, a highly active RNA enzyme found in nature (3). The properties and activities of RNase P ribozyme, as well as the simple design of the guide sequence, make M1GS an attractive and unique gene-targeting agent that can be generally used for antiviral as well as other in vivo applications. For nucleic acid-based gene interfering approaches including M1GS ribozyme to be successful as a therapeutic tool for practical applications, one of the most important issues is targeted gene delivery of these agents to specific types of cells and tissues. This study demonstrates targeted gene delivery of RNase P ribozymes into specific human cells by Salmonella.
In the current study, we constructed a M1GS RNA targeting the overlapping region of HCMV CSP and assemblin mRNAs. The ribozyme cleaved the target mRNAs efficiently in vitro and, furthermore, reduced their expression levels by 87–90% and inhibited viral growth by about 5,000-fold in human macrophages that were treated with Salmonella carrying the plasmid of the M1-C1 sequence. In contrast, a reduction of less than 10% in the levels of CSP expression and viral growth was observed in cells that were treated with Salmonella carrying the plasmids of the M1-C2 and M1-TK sequences. M1-TK targets an unrelated mRNA, and M1-C2 is catalytically inactive and contains an identical guide sequence to M1-C1. Thus, the observed reduction in viral gene expression and inhibition of viral growth in cells treated with the M1-C1-containing Salmonella is primarily attributed to Salmonella-mediated gene delivery of the functional ribozyme sequence for targeted cleavage of HCMV CSP mRNA.
Several lines of evidence presented in our study indicate that the Salmonella-mediated gene transfer is efficient, and ribozymes expressed following the Salmonella-mediated gene transfer are active and specific. First, targeted gene transfer of the ribozyme constructs by Salmonella yields substantial expression of the ribozymes (Fig. 2). Furthermore, more than 80% of the cells were GFP positive, suggesting efficient transfer of the ribozyme construct, which also contained the GFP expression cassette. Second, the presence of ribozyme sequences in Salmonella did not significantly affect the viability and gene transfer ability of the bacteria (Fig. 2). Third, the ribozymes expressed following transfer effectively and specifically inhibited the expression of CSP/assemblin. No reduction in the expression levels of other viral genes examined (e.g., IE2, US2, UL44, and gB) was observed in cells treated with M1GS-containing Salmonella (Table 1). Furthermore, the ribozyme expression does not affect the replication of viral genomic DNA (Fig. 5). Fourth, the inhibition of viral growth and capsid maturation appears to result from the reduction of CSP and assemblin. We observed that the level of capsid formation (DNA encapsidation) and the expression of CSP greatly decreased in cells treated with Salmonella carrying the sequence of M1-C1 but not M1-C2 or M1-TK (Figs. 3 and 5). Together, these results suggest that Salmonella can function as a gene transfer vector for efficient delivery of RNase P ribozyme in human macrophages and that the ribozymes expressed following gene transfer are active and specific in inhibiting only the expression of their target mRNA and do not affect the expression of other viral genes and genome replication.
The fundamental challenge in gene therapy is to develop approaches for delivering genetic material to the appropriate cells of the patient in a way that is tissue/cell specific, efficient, and safe (8, 9). Tissue-specific vectors have been only partially obtained by using carriers that, in nature, infect certain cell types, such as herpes simplex virus does for cells of the nervous system (8, 9). Salmonella-based vectors exhibit several unique and attractive features as a gene delivery tool. First, the Salmonella-based vector is low cost and easy to prepare, store, and transport. Second, one of the most interesting aspects associated with the use of attenuated Salmonella as a vector is the possibility of administering these bacteria via the oral route, a strategy that has proved to be successful in terms of efficacy and acceptability (20, 21). In fact, the antityphoid fever vaccine based on the attenuated strain Ty21a is one of the few live vaccines licensed for human use and has been extensively used to immunize both adults and children (21, 22). Third, it is conceivable to generate attenuated mutants with more than one independent deletion leading to an attenuated phenotype (23). This will eliminate the risk of reversion following horizontal gene transfer. Furthermore, different mutants have been identified, which are not harmful even for immunocompromised hosts (24). Fourth, while some safety considerations for the use of Gram-negative bacteria refer to the toxic effect of LPS, this concern has mostly been ruled out by oral delivery and by the fact that Salmonella strains have been widely used as vaccines both in human and veterinary medicine (9, 20). Finally, integration of bacteria-delivered DNA in host cell genome is not common (25), and oncogenesis promotion of the bacterial infection has not been known. Thus, the low-cost, safe, noninvasive administration and lifelong suitability of the therapy, combined with easy preparation of the bacterial carrier, support the notion that Salmonella represents an attractive and promising gene delivery tool for gene therapy for human diseases, including viral infections.
It is well established that bacterial infection elicits various innate immune responses, including activation of TLR4 and TLR9, which can be induced by bacterial LPS and the unmethylated CpG motifs of bacterial DNA, respectively (18, 19). While some of these responses are beneficial to the host (e.g., inhibiting bacterial infection and replication), some other responses may have unwanted side effects (e.g., induction of apoptosis), leading to potential cytotoxicity. To avoid these responses and reduce potential cytotoxicity, mutations have been introduced to bacterial vectors in order to inactivate the expression of specific bacterial components (e.g., LPS) (20). Alternatively, bacteria carrying transgenes that modulate these responses (e.g., inhibition of TLR9 expression) can be used (19). Further experiments to generate mutant Salmonella strains and examine the innate immune responses and cytotoxicity associated with these mutants should facilitate the development of better Salmonella-based vectors for gene therapy applications.
HCMV is a member of the herpesvirus family, which includes different viruses such as herpes simplex virus and Epstein-Barr virus (1). Macrophages represent the major reservoirs for HCMV as this virus can establish both primary and latent infections in these cells, leading to pathogenesis such as life-threatening complications in immunocompromised individuals. Eliminating infection in macrophages is central to the treatment and prevention of HCMV-associated diseases. Our study provides direct evidence that Salmonella-mediated gene transfer of a RNase P ribozyme can specifically block HCMV infection and replication in human differentiated macrophages. Future studies, including the construction of Salmonella strains through mutagenesis strategies, should lead to the generation of Salmonella vectors with better gene transfer efficiency and less toxicity. Moreover, investigation of the expression and activity of the ribozymes following gene transfer should provide insight into the mechanism of Salmonella-mediated gene transfer of RNase P ribozymes. These studies will facilitate the development of RNase P ribozymes as a promising gene-targeting agent for in vivo applications.
Materials and Methods
Growth of Viruses and Cells.
Human fibroblasts and THP-1 cells (American Type Culture Collection) were maintained in DMEM and RPMI medium 1640 supplemented with 10% (vol/vol) fetal bovine serum, respectively. The propagation of HCMV (AD169) in cells was carried out as described previously (7).
Ribozyme and Substrate Constructs.
The DNA sequence that encodes the CSP mRNA substrate was constructed by PCR using pGEM3zf(+) as a template and oligonucleotides AF25 (5′-GGAATTCTAATACGACTCACTATAG-3′) and sSCP3 (5′-CGGGATCCGTAACGCTCCCATCCGGACGGTGGTTCATCCTATAGTGAGTCGTATTA-3′) as 5′ and 3′ primers, respectively. Plasmid pV57 contained the DNA sequence coding for V57 RNA (16). The DNA sequence that encodes ribozyme M1-C1 was constructed by PCR using pV57 as the template and oligonucleotides AF25 as the 5′ primer and M1CSP3 (5′-CCCGCTCGAGAAAAAATGGTGTCCGGATGGGAGCGTTATGTGGAATTGTG -3′) as the 3′ primer. M1GS RNAs and the CSP mRNA substrate were synthesized in vitro by T7 RNA polymerase and cleavage and binding assays were carried out (see SI Text) (7). M1-TK was generated from pV57 as described previously (16).
Expression of Ribozymes by Salmonella-Mediated Delivery.
The auxotrophic Salmonella typhimurium aroA strain SL7207 was kindly provided by Bruce A. D. Stocker (Stanford University, Stanford, CA). Salmonella carrying different constructs were obtained by transforming SL7207 with plasmids pU6, pU6-M1-C1, pU6-M1-C2, and pU6-M1-TK. Construct pU6 contained the GFP expression cassette and, in addition, the small U6 RNA promoter, which was used for the expression of ribozymes in human cells.
To study gene transfer of ribozyme by Salmonella vectors, THP-1 cells were allowed to differentiate into adherent macrophage-like cells in the presence of 100 ng/mL tetradecanoyl phorbol acetate (Sigma) for 24 h (26) and then seeded into cultured wells at a concentration of 1 × 106 cells/mL. The differentiated cells were then infected with Salmonella at a MOI of 10–20 bacteria/cell. Cultures were centrifuged at 200 × g for 5 min and incubated at 37 °C for 30 min to allow phagocytosis to occur. Under these conditions, essentially most of cells were infected with bacteria. The medium was then replaced with fresh medium containing gentamicin (20 μg/mL) and incubated for the indicated times. Cells were harvested and the expression of ribozymes was assayed using Northern analyses (see SI Text) (7).
Studies of Viral Gene Expression, Growth, and Genome Replication.
Differentiated THP-1 cells (approximately 1–5 × 106 cells) were first incubated with Salmonella carrying different constructs at a MOI of 10–20 bacteria/cell. The medium was then replaced with fresh medium containing gentamicin (20 μg/mL) and incubated for 16 h to allow the expression of the ribozymes. The Salmonella-containing cells were then subjected to FACS using a FACSVantage SE sorter (Becton Dickinson), and a population of GFP-positive cells (usually 1–5 × 105 cells with a positive fluorescence of > 99%) was isolated. The isolated cells were incubated for 8 h and then either mock-infected or infected with HCMV as described previously (7). The multiplicity of HCMV infection (MOI) is specified in Results. The infected cells were incubated for another 8–72 h. The RNA and protein samples were isolated from infected cells, and the expression of specific mRNAs and proteins was assayed by Northern and Western analyses, respectively (see SI Text) (7). Total and encapsidated (DNase I-treated) DNAs were isolated and used as the PCR templates for a semiquantitative PCR, and inhibition of HCMV growth by ribozymes was studied as described previously (7) (see SI Text).
Supplementary Material
Acknowledgments.
We are indebted to Jiaming Zhu and Xiaohong Jiang for technical assistance and John Wu and Annette Meyer for anti-HCMV antibodies. S.U. is a recipient of a predoctoral fellowship from State of California AIDS Research Program. G.V. and Y.B. were partially supported by a predoctoral Block grant from University of California at Berkeley. This research has been supported by grants from National Institutes of Health (AI041927 and DE014842).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912813107/DCSupplemental.
References
- 1.Mocarski ES, Shenk T, Pass RF. In: Fields Virology. Knipe DM, et al., editors. Philadelphia: Lippincott-William & Wilkins; 2007. pp. 2701–2772. [Google Scholar]
- 2.Scherer LJ, Rossi JJ. Approaches for the sequence-specific knockdown of mRNA. Nat Biotechnol. 2003;21:1457–1465. doi: 10.1038/nbt915. [DOI] [PubMed] [Google Scholar]
- 3.Altman S, Kirsebom LA. In: The RNA World. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1999. pp. 351–380. [Google Scholar]
- 4.Guerrier-Takada C, Li Y, Altman S. Artificial regulation of gene expression in Escherichia coli by RNase P. Proc Natl Acad Sci USA. 1995;92:11115–11119. doi: 10.1073/pnas.92.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu F, Altman S. Inhibition of viral gene expression by the catalytic RNA subunit of RNase P from Escherichia coli. Genes Dev. 1995;9:471–480. doi: 10.1101/gad.9.4.471. [DOI] [PubMed] [Google Scholar]
- 6.Plehn-Dujowich D, Altman S. Effective inhibition of influenza virus production in cultured cells by external guide sequences and ribonuclease P. Proc Natl Acad Sci USA. 1998;95:7327–7332. doi: 10.1073/pnas.95.13.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trang P, et al. Effective inhibition of human cytomegalovirus gene expression and replication by a ribozyme derived from the catalytic RNA subunit of RNase P from Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5812–5817. doi: 10.1073/pnas.100101797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robbins PD, Ghivizzani SC. Viral vectors for gene therapy. Pharmacol Ther. 1998;80:35–47. [PubMed] [Google Scholar]
- 9.Vassaux G, Nitcheu J, Jezzard S, Lemoine NR. Bacterial gene therapy strategies. J Pathol. 2006;208:290–298. doi: 10.1002/path.1865. [DOI] [PubMed] [Google Scholar]
- 10.Darji A, et al. Oral somatic transgene vaccination using attenuated S. typhimurium. Cell. 1997;91:765–775. doi: 10.1016/s0092-8674(00)80465-1. [DOI] [PubMed] [Google Scholar]
- 11.Grillot-Courvalin C, Goussard S, Courvalin P. Bacteria as gene delivery vectors for mammalian cells. Curr Opin Biotechnol. 1999;10:477–481. doi: 10.1016/s0958-1669(99)00013-0. [DOI] [PubMed] [Google Scholar]
- 12.Paglia P, Terrazzini N, Schulze K, Guzman CA, Colombo MP. In vivo correction of genetic defects of monocyte/macrophages using attenuated Salmonella as oral vectors for targeted gene delivery. Gene Ther. 2000;7:1725–1730. doi: 10.1038/sj.gt.3301290. [DOI] [PubMed] [Google Scholar]
- 13.Yang N, Zhu X, Chen L, Li S, Ren D. Oral administration of attenuated S. typhimurium carrying shRNA-expressing vectors as a cancer therapeutic. Cancer Biol Ther. 2008;7:145–151. doi: 10.4161/cbt.7.1.5195. [DOI] [PubMed] [Google Scholar]
- 14.Loessner H, Endmann A, Rohde M, Curtiss R, 3rd, Weiss S. Differential effect of auxotrophies on the release of macromolecules by Salmonella enterica vaccine strains. FEMS Microbiol Lett. 2006;265:81–8. doi: 10.1111/j.1574-6968.2006.00470.x. [DOI] [PubMed] [Google Scholar]
- 15.Galan JE. Salmonella interactions with host cells: Type III secretion at work. Annu Rev Cell Dev Biol. 2001;17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- 16.Kilani AF, et al. RNase P ribozymes selected in vitro to cleave a viral mRNA effectively inhibit its expression in cell culture. J Biol Chem. 2000;275:10611–10622. doi: 10.1074/jbc.275.14.10611. [DOI] [PubMed] [Google Scholar]
- 17.Bertrand E, et al. The expression cassette determines the functional activity of ribozymes in mammalian cells by controlling their intracellular localization. RNA. 1997;3:75–88. [PMC free article] [PubMed] [Google Scholar]
- 18.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 19.Krieg AM. Therapeutic potential of Toll-like receptor 9 activation. Nat Rev Drug Discov. 2006;5:471–484. doi: 10.1038/nrd2059. [DOI] [PubMed] [Google Scholar]
- 20.Clairmont C, et al. Biodistribution and genetic stability of the novel antitumor agent VNP20009, a genetically modified strain of Salmonella typhimurium. J Infect Dis. 2000;181:1996–2002. doi: 10.1086/315497. [DOI] [PubMed] [Google Scholar]
- 21.Levine MM, et al. Safety, infectivity, immunogenicity, and in vivo stability of two attenuated auxotrophic mutant strains of Salmonella typhi, 541Ty and 543Ty, as live oral vaccines in humans. J Clin Invest. 1987;79:888–902. doi: 10.1172/JCI112899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hone DM, et al. A galE via (Vi antigen-negative) mutant of Salmonella typhi Ty2 retains virulence in humans. Infect Immun. 1988;56:1326–1333. doi: 10.1128/iai.56.5.1326-1333.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medina E, et al. Pathogenicity island 2 mutants of Salmonella typhimurium are efficient carriers for heterologous antigens and enable modulation of immune responses. Infect Immun. 1999;67:1093–1099. doi: 10.1128/iai.67.3.1093-1099.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dietrich G, et al. Delivery of antigen-encoding plasmid DNA into the cytosol of macrophages by attenuated suicide Listeria monocytogenes. Nat Biotechnol. 1998;16:181–185. doi: 10.1038/nbt0298-181. [DOI] [PubMed] [Google Scholar]
- 26.Yee LF, Lin PL, Stinski MF. Ectopic expression of HCMV IE72 and IE86 proteins is sufficient to induce early gene expression but not production of infectious virus in undifferentiated promonocytic THP-1 cells. Virology. 2007;363:174–188. doi: 10.1016/j.virol.2007.01.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.