Abstract
BAG3, a member of the BAG family of heat shock protein (HSP) 70 cochaperones, is expressed in response to stressful stimuli in a number of normal cell types and constitutively in a variety of tumors, including pancreas carcinomas, lymphocytic and myeloblastic leukemias, and thyroid carcinomas. Down-regulation of BAG3 results in cell death, but the underlying molecular mechanisms are still elusive. Here, we investigated the molecular mechanism of BAG3-dependent survival in human osteosarcoma (SAOS-2) and melanoma (M14) cells. We show that bag3 overexpression in tumors promotes survival through the NF-κB pathway. Indeed, we demonstrate that BAG3 alters the interaction between HSP70 and IKKγ, increasing availability of IKKγ and protecting it from proteasome-dependent degradation; this, in turn, results in increased NF-κB activity and survival. These results identify bag3 as a potential target for anticancer therapies in those tumors in which this gene is constitutively expressed. As a proof of principle, we show that treatment of a mouse xenograft tumor model with bag3siRNA-adenovirus that down-regulates bag3 results in reduced tumor growth and increased animal survival.
Keywords: BAG3, IKK-gamma, apoptosis
The bag3 gene belongs to a highly conserved gene family identified in yeast (Saccharomyces cerevisiae and Schizosaccharomyces pombe), invertebrates (Caenorhabditis elegans, Ciona intestinalis, and Drosophila), amphibians (Xenopus laevis), mammals (Homo sapiens and Mus musculus), and plants (Oryza sativa and Arabidopsis thaliana) (1–9). All BAG proteins are characterized by a conserved domain of 110–124 amino acids (BAG domain) (3, 10) through which they bind to the HSC70/HSP70 ATPase domain regulating the heat shock protein (HSP) polypeptide folding activity (2, 11). In addition, BAG3 contains a WW domain and a proline-rich repeat (PXXP) through which it interacts with other partners, such as phospholipase C (PLC)-γ (12) and Akt kinase (13).
BAG3 is a 74-kDa cytoplasmic protein mainly localized in the rough endoplasmic reticulum; a slightly different molecular weight or a doublet form can be observed in some cell types and/or following cell exposition to stressors (11, 14, 15). In humans, bag3 is constitutively expressed in myocytes and a few other normal cell types and in several tumors (leukemia and lymphoma, myeloma, and pancreas and thyroid carcinomas) (11, 16–20). In addition, its expression is induced in different normal cell types (leukocytes and epithelial and glial cells) in response to cell stressors, such as oxidants, high temperature, heavy metals, and HIV-1 infection (15, 21–23). Interestingly, induction of bag3 in response to stress is under the control of HSF1 (24), a member of the heat shock factor family of transcription factors (25, 26) involved in tumor initiation and maintenance (27, 28). Actually, several lines of evidence suggest that overexpression of BAG3 in tumors plays a role in survival of these cells. Indeed down-modulation of BAG3 in primary samples of B-cell chronic lymphocytic leukemia and acute lymphoblastic leukemia results in a dramatic increase of basal as well as drug-induced apoptosis (17, 18). Furthermore, BAG3 is overexpressed in thyroid carcinomas, where higher levels of expression are reached in anaplastic tumors compared with well-differentiated forms. In these tumors, down-modulation of BAG3 results in sensitization to TNF-related apoptosis-inducing ligand-dependent apoptosis (20).
The influence of BAG3 on cell survival is potentially mediated, at least in part, by regulation of HSC70/HSP70 function (11). Some of the apoptosis-regulating proteins that are known to interact with HSP70 are therefore potential targets of BAG3 regulatory activity. Among these regulating proteins, the IKKγ subunit of the IKK complex is of particular interest. Indeed, HSP70 has been shown to interact with IKKγ (29, 30) and to suppress NF-κB activity. Interference with the HSP70-IKKγ interaction through a dominant negative mutant restores NF-κB activity (30). In this paper, we show that BAG3 affects NF-κB activity by interfering with the binding between HSP70 and IKKγ, thus favoring IKK complex formation and preventing the proteosomal degradation of IKKγ. Moreover, we show that down-regulation of bag3 in vivo can be a potential anticancer therapeutical strategy.
Results
Apoptosis Modulation by BAG3 Is Dependent on NF-κB Activity.
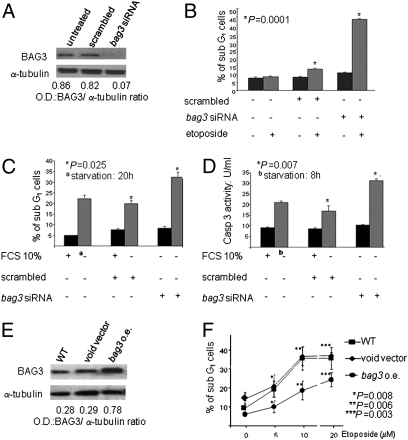
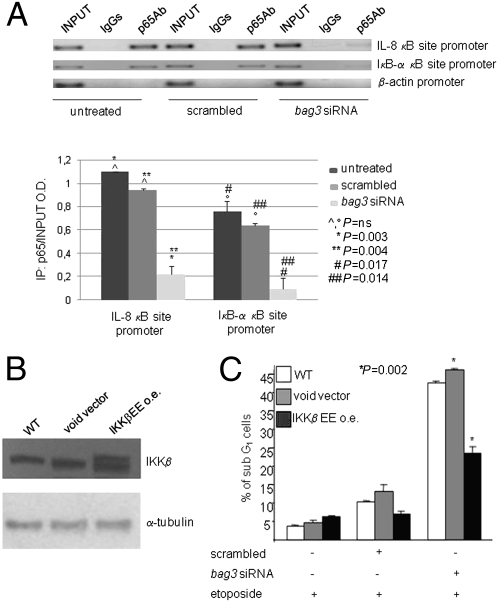
We investigated the effects of bag3 silencing in SAOS-2 human osteosarcoma cells, which express high constitutive levels of BAG3 (Fig. 1A). Our results show that down-regulation of BAG3 by a specific siRNA in these cells results in increased death (detected as sub-G1 peak of propidium iodide-stained cells) in response to etoposide (Fig. 1B) or serum deprivation (Fig. 1 C and D). Consistent overexpression of bag3 in the same cell line protects cells from etoposide-induced death (Fig. 1 E and F), thus also confirming the antiapoptotic property of BAG3 in this tumor. Because apoptosis is also known to be regulated by NF-κB factors in human osteosarcoma (31, 32) and, as mentioned before, NF-κB is a potential target of BAG3 action, we investigated if BAG3 modulation affected the NF-κB pathway. By ChIP experiments (Fig. 2A), we were able to show that down-regulation of bag3 by siRNA results in dramatically reduced binding of NF-κB to two well-known responsive elements on the IL-8 and IκB-α promoters. These results suggested that bag3 siRNA was sensitizing SAOS-2 cells to apoptosis by altering NF-κB activity; we therefore tested if sustaining NF-κB activity by expressing a constitutively active form of IKKβ (IKKβ EE) (33) protected cells from the effect of bag3 siRNA. Indeed, as shown in Fig. 2 B and C, overexpression of IKKβ EE results in reduced etoposide-dependent death in cells in which BAG3 was down-regulated. Altogether, the evidence indicates an involvement of the NF-κB pathway in BAG3-mediated modulation of apoptosis.
Fig. 1.
BAG3 protein levels influence starvation or etoposide-induced apoptosis in SAOS-2 cells. (A) SAOS-2 cells (30% confluency) were transfected with bag3siRNA or control scrambled RNA. After 72 h, whole-cell extracts were analyzed by immunoblotting with anti-BAG3 TOS-2 polyclonal antibody or control anti-α-tubulin antibody. (B) Cells were transfected as in A. After 72 h, cells were washed and incubated with medium alone or in the presence of etoposide (20 μM). Twenty-four hours later, apoptosis was evaluated as the percentage of sub-G1 cells by cell permeabilization and propidium iodide staining in flow cytometry. (C) Cells were transfected as in A. After 72 h, cells were washed and incubated with complete medium or in starvation medium (absence of FCS). Twenty hours later, the sub-G1 cell percentage was analyzed. (D) Transfected cells, treated as described in C, were assessed for caspase-3 activity after 8 h of treatment with starvation medium. (E) SAOS-2 cells, WT or transfected (12 weeks with stable transfectants), with a bag3 cDNA construct in either the pcDNA3.1 vector [bag3 overexpressing (o.e.)] or the void vector were analyzed for their content of BAG3 protein by immunoblotting with TOS-2 polyclonal antibody. (F) Cells were transfected as described in E and cultured for 48 h with the indicated concentrations (μM) of etoposide; cell apoptosis was then analyzed by cell permeabilization and PI staining in flow cytometry.
Fig. 2.
BAG3 protein levels influence NF-κB activity. (A) SAOS-2 cells at 30% confluency were transfected with bag3 or control scrambled siRNA. Chromatin was immunoprecipitated after 72 h with p65 antibody or control IgGs, and purified DNA was subjected to PCR assay to amplify a segment spanning the κB-responsive elements on the IL-8 and IκB-α promoters. β-actin promoter primers were used as a negative control. The graph depicts band densitometry values from three separate experiments. ns, not significant. (B) Lysates from SAOS-2 cells, WT or transfected (12 weeks of stable transfectants), with either a construct expressing a constitutively active form of IKKβ [IKKβ EE overexpressing (o.e.)] or the void vector were analyzed in immunoblotting with anti-α-tubulin antibody. (C) WT, void vector-transfected, and IKKβ EE-transfected cells were plated at 30% confluency and transfected with scrambled or bag3 siRNA. After 72 h, cells were washed and incubated either with medium alone or in presence of etoposide (20 μM). Twenty-four hours later, apoptosis was analyzed by flow cytometry.
BAG3 Regulates HSP70 Binding to IKKγ.
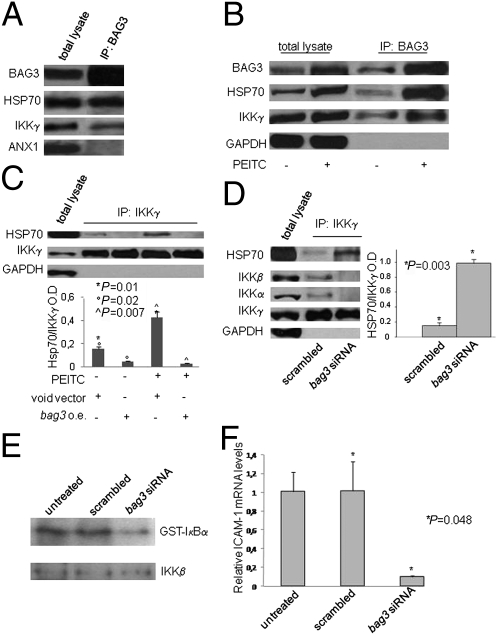
Because it has been reported that the BAG3 partner molecule HSP70 interacts with IKKγ, the regulatory subunit (also called NF-κB-essential modulator) of the IKK complex (29, 30), we investigated the possibility that BAG3 was also involved in this interaction. Fig. 3A shows that, indeed, both endogenous HSP70 and endogenous IKKγ [but not an unrelated protein (annexin1) used as a control] coimmunoprecipitated with endogenous BAG3.
Fig. 3.
BAG3 protein levels influence HSP70 association with IKKγ and IKK activity. (A) SAOS-2 cell lysate was immunoprecipitated with an anti-BAG3 polyclonal antibody and analyzed by immunoblotting with anti-HSP70 and anti-IKKγ antibodies. An antibody recognizing annexin I was used as a negative control. (B) M14 whole lysates from untreated or PEITC-treated (5 μM for 30 min) cells were immunoprecipitated with the anti-BAG3 monoclonal antibody AC-2 and analyzed by immunoblotting with anti-BAG3 TOS-2 polyclonal, anti-HSP70, or anti-GAPDH antibody. (C) M14 cells were transfected either with a bag3 overexpressing (o.e.) vector or the void vector. After 48 h, cells were treated as described in B. Immunoprecipitation was performed using an anti-IKKγ polyclonal antibody and analyzed by immunoblotting with anti-HSP70, anti-IKKγ, or anti-GAPDH antibody. The graph depicts densitometry values of bands obtained from two separate experiments. (D) M14 cells were plated at 30% confluency and transfected with scrambled or bag3 siRNA. After 48 h, cell lysates were immunoprecipitated with anti-IKKγ polyclonal antibody and analyzed by immunoblotting with anti-HSP70, anti-IKKγ, anti-IKKα, or anti-IKKβ antibody. The amount of coimmunoprecipitated HSP70 was quantified by densitometry from three separate experiments and normalized to the amount of IKKγ (OD: HSP70/IKKγ). An antibody recognizing GAPDH was used as a negative control. (E) Whole-cell lysates were prepared, and IKK kinase activity was measured after immunoprecipitation with anti-IKKα antibody using GST-IκBα (1–54) as a substrate (34). IKK recovery was determined by immunoblotting with anti-IKKβ antibody. (F) Total RNA was extracted, and ICAM-1 mRNA levels were analyzed by quantitative RT-PCR using β-actin mRNA levels for normalization.
To demonstrate that this interaction is not restricted to a particular cell line, we performed an analogous experiment in melanoma, a tumor whose growth and progression are known to be modulated by NF-κB (34–36). As shown in Fig. 3B, coimmunoprecipitation experiments performed in M14 cells confirm the interaction between BAG3, HSP70, and IKKγ. Induction of oxidative stress by treatment with phenethyl isothiocyanate (PEITC) (Fig. S1A) results in the expected increase of BAG3 and HSP70, and thus in increased amounts of immunoprecipitated proteins. These results suggest that BAG3 can modulate the HSP70-IKKγ interaction. We therefore investigated if BAG3 overexpression affected this interaction by performing immunoprecipitation experiments in the presence of overexpressed BAG3. As shown in Fig. 3C, overexpression of bag3 results in a reduction of the amount of HSP70 that coimmunoprecipitates with IKKγ. Interestingly, oxidative stress (by PEITC treatment) increases this interaction, which can still be almost completely suppressed by BAG3 overexpression. As expected, down-regulation of bag3 results in increased binding of HSP70 to IKKγ (Fig. 3D).
According to the model proposed by Ran et al. (30), increased binding of HSP70 to IKKγ hampers the formation of active IKK complex. In keeping with the model, the amounts of IKKα and IKKβ coimmunoprecipitation with IKKγ were reduced in bag3 siRNA-treated cells (Fig. 3D). To assess whether IKK activation was affected by BAG3, we measured the kinase activity after immunoprecipitation with anti-IKKα-antibody, using GST-IκBα (1–54) as a substrate (37). As shown in Fig. 3E, bag3 silencing results in reduced amounts of phosphorylated GST-IκBα. Consistently, the mRNA levels of ICAM-1, a gene highly regulated by NF-κB (38), are reduced in cells transfected with bag3 siRNA (Fig. 3F), confirming the effect of BAG3 on NF-κB activity. The above reported findings demonstrated that BAG3 protein regulates the extent of HSP70 association with IKKγ, thus relieving HSP70 inhibition of IKK (29, 30).
BAG3 Regulates IKKγ Protein Levels.
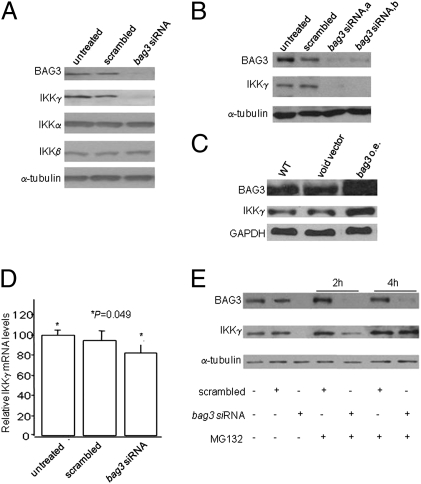
It is well known that HSP70 can control proteasome-dependent degradation of a number of proteins (39) and that this activity can, in turn, be controlled by BAG3 (13). As an example, Doong et al. (13) showed that BAG3 expression inhibited the HSP70-mediated proteasomal degradation of Akt, sustaining the intracellular levels of the kinase. It is thus possible that BAG3 is controlling IKKγ in a similar fashion. We therefore prolonged (to 96 h) the time of SAOS-2 or M14 treatment with scrambled or bag3siRNA and analyzed the intracellular levels of IKKγ protein. These were highly reduced in bag3siRNA-treated compared with control cells (Fig. 4 A and B). The bag3siRNA effect was specific for IKKγ, because the levels of the other IKK subunits were not modulated (Fig. 4A). Consistently, bag3 overexpression produced increased intracellular levels of IKKγ (Fig. 4C).
Fig. 4.
BAG3 protein influences the intracellular levels of IKKγ protein. (A) SAOS-2 cells (30% confluency) were transfected with scrambled or bag3 siRNA. After 96 h, cell lysates were obtained and analyzed by immunoblotting with anti-BAG3 or anti-α-tubulin antibody. (B) M14 cells were plated at 30% confluency and transfected with two different bag3-specific siRNAs (indicated as bag3 siRNA a and b) or with a control scrambled siRNA. After 96 h, lysates were analyzed by immunoblotting with anti-BAG3 or anti-α-tubulin antibody. (C) M14 cells, WT or transfected, with either a bag3 cDNA construct in pcDNA3.1 vector [bag3 overexpressing (o.e.)] or the void vector were analyzed for their content of BAG3 protein by immunoblotting with TOS-2 polyclonal antibody; furthermore, IKKγ levels were checked by immunoblotting and GAPDH was used to monitor equal loading conditions. (D) Total RNA was extracted, and IKKγ mRNA levels were analyzed by quantitative RT-PCR using 18s rRNA levels for normalization. (E) M14 cells were plated at 30% confluency and transfected with bag3 or control scrambled siRNA. After 96 h, cells were treated with MG132 (10 μM) for 2 and 4 h. Cell lysates were analyzed by immunoblotting with anti-BAG3, anti-IKKγ, or anti-α-tubulin antibody.
To verify whether the effect of BAG3 down-regulation on IKKγ levels was exerted at a posttranscriptional level, we measured IKKγ mRNA levels by real-time PCR and showed that they were not affected by bag3 silencing (Fig. 4D). Furthermore, IKKγ protein levels were restored in cultures in which a proteasome inhibitor was added to bag3 siRNA-treated cells (Fig. 4E and Fig. S1B). These findings indicated that BAG3 influenced the extent of IKKγ degradation. To demonstrate further that the effect of BAG3 on IKKγ protein levels is dependent on HSP70- mediated delivery of client proteins to proteasome (11, 13), we down-regulated HSP70 by siRNA in the same cells in which we had down-regulated BAG3. As shown in the Western blot in Fig. S1C, down-modulation of BAG3 results in the expected reduction of IKKγ protein levels; however, contemporaneous reduction of HSP70 rescues this phenotype. These findings indicated that BAG3 influenced the extent of IKKγ degradation, in agreement with the known interference of the cochaperone in HSP70-mediated delivery of client proteins to proteasome (11, 13).
Therefore, BAG3 protein was apparently able to modulate both the inhibition and the proteasomal delivery of IKKγ mediated by HSP70. The effects on IKK activation were detectable already a short time after bag3 down-modulation, whereas the effects on intracellular IKKγ levels required a longer time.
BAG3 Down-Modulation Inhibits Tumor Growth in Vivo.
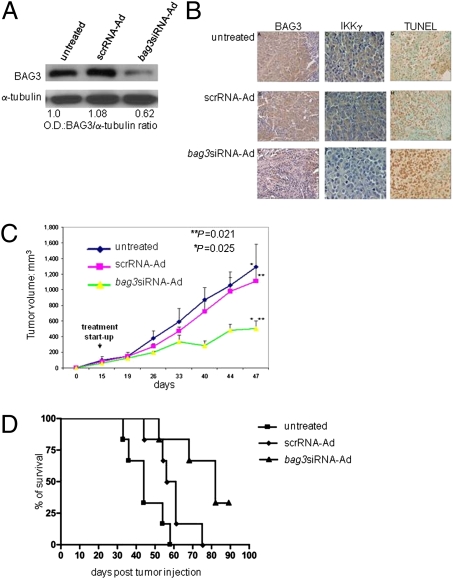
Because our original hypothesis was that BAG3 overexpression in tumors is responsible for their reduced death, we investigated the effects of BAG3 down-modulation on in vivo tumor growth. To this end, we implanted M14 cells in nude mice and treated the xenografts with a bag3siRNA-carrying adenovirus (bag3siRNA-Ad) or a control Ad that carried a scrambled sequence (scrRNA-Ad). Analyses of tumors treated for 2 weeks with bag3siRNA-Ad showed decreased BAG3 and IKKγ levels paralleled by increased cell death detected by TUNEL staining (Fig. 5B). Consistently, treatment with bag3siRNA-Ad resulted in reduced tumor growth, up to 75% after 47 days of treatment (Fig. 5C). More importantly, although treatment with scrRNA-Ad had a slight, if any, effect on animal survival, >70% of bag3siRNA-Ad-treated animals survived after the death of both control animals and mice inoculated with scrRNA-Ad (day 75) (P < 0.0016) (Fig. 5D).
Fig. 5.
Knockdown of BAG3 protein results in diminishing tumor IKKγ levels, increasing tumor cell apoptosis, inhibiting tumor growth, and enhancing animal survival in M14-xenografted mice. (A) M14 cells (5 × 106) were injected s.c. onto the back of 6-week-old female BALB/c nu/nu mice. Two weeks later, animals were randomized into three groups (10 animals per group) and control PBS (100 μL), bag3siRNA-Ad, or scrRNA-Ad (108 pfu per 100 μL) was injected into the tumors twice a week. After 2 weeks of treatment, four tumors per animal group were excised and BAG3 protein content was analyzed by Western blot. (B) BAG3 and IKKγ protein were detected by immunohistochemistry, whereas apoptosis was analyzed by TUNEL assay, in tumor sections. Representative results are shown. (C) Tumor sizes in bag3siRNA-Ad, scrRNA-Ad, treated, or untreated remaining mice (6 per group) were measured every week using calipers. (D) Evaluation of animal survival was carried out according to Kaplan–Meier analysis. Differences among the treatment groups were analyzed by ANOVA.
Discussion
The observation that bag3 is constitutively expressed in a number of different tumors (11, 16–18, 20, 21, 40–42) prompted us to investigate the potential role played by this protein in cancer. Our results show that overexpression of bag3 promotes survival, whereas its down-regulation sensitizes cells to apoptosis, both in vitro and in vivo. Indeed, treatment of mice with bag3siRNA-Ad that down-regulates bag3 results in reduced growth of xenografted tumors and increased animal survival. We show that BAG3 prosurvival activity is a result of the activation of the NF-κB pathway. In particular, we show that BAG3 interferes with HSP70 binding to IKKγ, thus releasing it and allowing it to form the IKK complex required for NF-κB activation. These results are in accordance with previous findings showing that HSP70 regulates NF-κB activity by binding to IKKγ (30). Here, we present additional complexity to the system by demonstrating that another player, BAG3, is capable of interfering with HSP70, counteracting its action and favoring survival. Moreover, our data seem to suggest a double effect of BAG3 on IKKγ: Although BAG3 initially releases it from a complex with HSP70, increasing its availability, it also results in increased IKKγ steady-state levels at longer time points. This second effect is attributable to the inhibition of HSP70-dependent delivery to the proteasome (Fig. S1D).
Altogether, these results ascribe a functional role to the finding of elevated BAG3 levels in tumors and indicate a unique potential target for anticancer treatment in this protein, or at least for sensitization of cancer cells to more conventional drugs. To this end, our in vivo data constitute a proof of principle that interfering with BAG3 can result in reduced tumor size.
In this study, the biological effect of BAG3 on tumor growth and survival has been linked to direct interaction and modulation of a specific protein: IKKγ. Indeed, it has previously been shown that BAG3 can physically associate with a number of proteins potentially involved in tumorigenesis (e.g., PLC-γ, Akt, Bcl-XL, HspB8) (12, 13, 43, 44); however, no correlation between these interactions and the prosurvival effect of BAG3 has been provided. Similarly, although it has been clearly demonstrated that BAG3 can affect the levels and/or activity of a number of other proteins potentially involved in tumor transformation (e.g., p27, Rac1, the focal adhesion proteins FAK and paxillin, the matricellular signal regulator CCN1) (45–48), neither a clear mechanism through which BAG3 regulates them nor a clear causative role of BAG3 antiapoptotic activity has been demonstrated. Clearly, we believe that some of these proteins may play a causative role in the mechanism of action of BAG3, together with the pathway elucidated in this paper, because it is likely that BAG3 interacts with and regulates a number of different proteins through its multiple protein-binding domains. This, however, might be the subject of future studies.
The importance of BAG3 in regulating NF-κB signaling is very likely not restricted to tumor cells but potentially plays an important role in other contexts. BAG3 is usually expressed in normal cells in response to stress, where it probably represents a homeostatic response that sustains NF-κB activation and cell survival after perturbation of cell equilibrium. Because hsp70 expression is also stimulated in response to stress (25), final NF-κB activity is likely the result of fine-tuning of inputs that regulate the levels, kinetics of expression, and molecular forms of the chaperone and its cochaperone, and, possibly, of other interacting partners (25). The association of HSP70 with IKKγ (30) and its displacement by BAG3 are therefore events whose onset and modulations are probably dependent on pro- and antiapoptotic stimuli under stressing events. In a few normal cell types, BAG3 is expressed in a constitutive manner and likely plays a fundamental role in the survival of these cells. As an example, in cardiomyocytes, where cell stress is physiologically connected with the contractile function and cardiac hemodynamic load, BAG3 protein is essential for cell survival; indeed, mice with homozygous disruption of the bag3 gene develop fulminant myopathy with apoptotic features postnatally (19). Similarly, mice with a cardiomyocyte-specific deletion of IKKγ develop adult-onset cardiomyopathy accompanied by apoptosis (49), suggesting that alteration of any component of this pathway that results in reduced NF-κB activity results in reduced cell survival. Moreover, BAG3 is known to play a role in other cellular processes, such as autophagy (50, 51) and cell motility (46–48), that also involve the NF-κB pathway (52–55). We therefore believe that our results very likely have additional implications in these other contexts that require further attention.
In conclusion, our findings disclose a previously unknown mechanism that can sustain NF-κB activity and cell survival both in physiological and pathological situations.
Materials and Methods
Cells and Reagents.
SAOS-2 (American Type Culture Collection) and M14 (kindly provided by G. Zupi and C. Leonetti, Regina Elena Cancer Institute, Rome, Italy) cells were grown in DMEM with 10% (vol/vol) FBS (Invitrogen). Etoposide and PEITC were purchased from Sigma.
Plasmids, siRNAs, and Adenoviral Constructs.
The bag3 siRNA a (5′-AAGGUUCAGACCAUCUUGGAA-3′) and scrambled RNA (5′-CAGUCGCGUUUGCGACUGG-3′) were synthesized by Dharmacon. The bag3 siRNA b (target sequence: 5′-ATCGAAGAGTATTTGACCAAA-3′) was purchased from Quiagen. TRANSIT-TKO (Mirus) was used for cell transfection with siRNA (200 nM). The bag3 overexpressing plasmid (bag3 o.e.) was obtained by cloning bag3 full-length cDNA from human brain in the expression vector pcDNA 3.1. Plasmid expressing constitutively active IKKβ (IKKβ EE) was produced in our laboratory. Lipofectamine (Invitrogen) was used for cell transfection with plasmidic constructs. The bag3siRNA-Ad and scrambled siRNA-Ad were made using the BD Adeno-X Expression Systems 2 PT3674-1 (Pr36024) and BD knockout RNAi Systems PT3739 (PR42756) (BD Biosciences–Clontech).
Analysis of Hypodiploid (Apoptotic) Nuclei and Caspase-3 Activity.
Apoptosis was analyzed by propidium iodide incorporation in permeabilized cells and flow cytometry as described (17). Caspase-3 activity was determined using the Caspase-3 Fluorometric Detection kit (Assay Designs).
ChIP Assay.
The ChIP assay was performed as described (56) according to the manufacturer's instructions (Upstate Cell Signaling Solutions). Briefly, cells were collected at the indicated time and nuclear extracts were obtained using the NE-PER Kit (Pierce). Chromatin from SAOS-2 cells was formaldehyde cross-linked, shared by sonication (four times, 15 s each, one-third power), and immunoprecipitated using anti-p65 antibody (anti-p65 polyclonal antibody C-20; Santa Cruz Biotechnology) and protein A-Sepharose (Sigma) saturated with salmon sperm DNA (Applied Biosystems). IgGs from rabbit serum (DAKO A/S) were used as negative controls. After reversal of the cross-linking, the purified DNA fragments were subjected to PCR to amplify a segment spanning the κB-responsive elements on IL-8 promoter and IκBα promoter. Input DNA from fragmented chromatin before immunoprecipitation was used to monitor equal starting conditions. Primers (obtained from Primm) used in the PCR assay were IκB-α promoter (FWD 5′-GACGACCCCAATTCAAATCG-3′ and REV 5′-TCAGGCTCGGGGAATTTCC-3′), IL-8 promoter (FWD 5′-GGGCCATCAGTTGCAAATC-3′ and REV 5′-TTCCTTCCGGTGGTTTCTTC-3′), and β-actin promoter (FWD 5′-TGCACTGTGCGGCGAAGC-3′ and REV 5′-TCGAGCCATAAAAGGCAA-3′). β-actin promoter primers were used as a negative control. PCR products were analyzed on 1.5% (wt/vol) agarose gels, stained with ethidium bromide, and quantified by densitometry.
Coimmunoprecipitation and Western Blotting.
Three hundred micrograms of proteins was used for immunoprecipitation assays using 3 μg of anti-IKKγ polyclonal antibody purchased from Santa Cruz Biotechnology or anti-BAG3 monoclonal antibody AC-2 (developed in our laboratory; US Patent 7,537,760) and incubated at 4 °C overnight on a tube rotator. Twenty-five microliters of protein A-Sepharose was then added, and the immunocomplexes were precipitated and washed five times with radioimmunoprecipitation assay buffer. For Western blot analyses, proteins obtained from immunoprecipitations or 30 μg of total protein were run on 10% (wt/vol) SDS/PAGE gels and transferred to nitrocellulose. Nitrocellulose blots were blocked with 5% nonfat dried milk in Tris buffer saline Tween-20 buffer (TBST) [20 mM Tris-HCl (pH 7.4), 500 mM NaCl, and 0.01% Tween 20] and incubated with primary antibodies with 5% (weight/vol) milk in TBST buffer overnight at 4 °C. Immunoreactivity was detected by sequential incubation with horseradish peroxidase-conjugated secondary antibody (Amersham Biosciences) and enhanced chemiluminescence reagents (SuperSignal West Dura Extended Duration Substrate; Pierce) following standard protocols. Anti-BAG3 polyclonal and monoclonal (AC-1) antibodies were purchased from Enzo Biochem. The anti-BAG3 monoclonal antibody AC-2 was produced and characterized in our laboratory. Anti-IKKγ, anti-IKKβ, and anti-IKKα antibodies were purchased from BD Biosciences. Anti-HSP70 was purchased from Stressgen. Anti-GAPDH antibody, anti-annexin I, and anti-α-tubulin were purchased from Santa Cruz Biotechnology. Scanning densitometry of the bands was performed with an Image Scan (SnapScan 1212; Agfa–Gevaert N.V.). The area under the curve related to each band was determined using Gimp2 software. Background was subtracted from the calculated values. Significance between the two groups was calculated by the Student's t test.
IKK Activity Assay.
Whole-cell lysates were prepared, and IKK kinase activity was measured after immunoprecipitation with anti-IKKα antibody (BD Biosciences), as described (46), using GST-IκBα (1–54) as a substrate. IKK recovery was determined by immunoblotting with anti-IKKβ antibody (Santa Cruz Biotechnology).
Immunohistochemistry.
Tumors excised after 2-week treatments were sectioned at a thickness of 6 μm. Immunohistochemistry was performed as described (20). Labeling of apoptotic cells was performed using the TUNEL ApopTag Peroxidase In Situ Apoptosis Detection kit (Intergen) following the manufacturer's instructions. Negative controls for the TUNEL reaction consisted of sections incubated with reaction buffer but without TdT enzyme.
Quantitative RT-PCR.
Total RNA was extracted using TRIzol Reagent (Invitrogen) and digested with DNase (Invitrogen); 1 μg of RNA was then retrotranscribed using random examers and treated with RNase A (Invitrogen). A quantitative RT-PCR assay was performed using the LightCycler 480 SYBR green I Master (Roche Diagnostics, GmbH) with a Roche 480 LightCycler. IKKγ mRNA levels were expressed as a ratio to 18s rRNA levels; primers used were IKKγ FWD (5′-CTTTTGGGGTAGATGCG-3′), IKKγ REV (5′-GGTTAAATACACATCGGTCTG-3′), 18s FWD (5′-ACAGGTCTGTGATGCC-3′), and 18s REV (5′-ATCGGTAGTAGCGACG-3′). ICAM-1 mRNA levels were expressed as a ratio to β-actin mRNA levels; primers used were ICAM-1 FWD (5′-CTGTCAAACGGGAGATGAATGGT-3′), ICAM-1 REV (5′-TCTGGCGGTAATAGGTGTAAATGG-3′), β-actin FWD (5′-ACCACCACAGCTGAGAGGGAAATCG-3′), and β-actin REV (5′-CTGACCGTCAGGCAGCTCATAGCTC-3′). Each sample was run in triplicate. Analysis of relative gene expression data was calculated using the 2−ΔΔCt method (57).
Xenografts.
M14 xenografts were produced on the back of 6-week-old female BALB/c nu/nu mice (Charles River Laboratories) by s.c. injection of 5 × 106 M14 cells in 500 μL of Hanks’ balanced salt solution. Two weeks after tumor cell injection, mice with similar tumor sizes were randomized into three groups (10 mice per group) and treated with control PBS (100 μL), bag3siRNA-Ad, or scrRNA-Ad (108 pfu/100 μL) via intratumoral injection twice a week. Tumor size was measured once every week by calipers, and tumor volume was calculated as length × width2 × 0.52. Mice with tumors >1.5 cm in diameter were euthanized. Differences among the treatment groups were analyzed by ANOVA using statistical software (Statistica; StatSoft). Survival was analyzed by the Kaplan–Meier method, and survival curves were compared by use of the log-rank test (58).
Supplementary Material
Acknowledgments
This work was supported, in part, by funds from the Italian Association for Cancer Research and Ministry of University.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907696107/DCSupplemental.
References
- 1.Thress K, Henzel W, Shillinglaw W, Kornbluth S. Scythe: A novel reaper-binding apoptotic regulator. EMBO J. 1998;17:6135–6143. doi: 10.1093/emboj/17.21.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274:781–786. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- 3.Briknarová K, et al. Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein. Nat Struct Biol. 2001;8:349–352. doi: 10.1038/86236. [DOI] [PubMed] [Google Scholar]
- 4.Moribe Y, Niimi T, Yamashita O, Yaginuma T, Samui A. Samui, a novel cold-inducible gene, encoding a protein with a BAG domain similar to silencer of death domains (SODD/BAG-4), isolated from Bombyx diapause eggs. Eur J Biochem. 2001;268:3432–3442. doi: 10.1046/j.1432-1327.2001.02244.x. [DOI] [PubMed] [Google Scholar]
- 5.Takayama S, Reed JC. Molecular chaperone targeting and regulation by BAG family proteins. Nat Cell Biol. 2001;3:237–241. doi: 10.1038/ncb1001-e237. [DOI] [PubMed] [Google Scholar]
- 6.Reed JC, et al. RIKEN GER Group; GSL Members. Comparative analysis of apoptosis and inflammation genes of mice and humans. Genome Res. 2003;13(6B):1376–1388. doi: 10.1101/gr.1053803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coulson M, Robert S, Saint R. Drosophila starvin encodes a tissue-specific BAG-domain protein required for larval food uptake. Genetics. 2005;171:1799–1812. doi: 10.1534/genetics.105.043265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wada S, Hamada M, Satoh N. A genomewide analysis of genes for the heat shock protein 70 chaperone system in the ascidian Ciona intestinalis. Cell Stress Chaperones. 2006;11:23–33. doi: 10.1379/CSC-137R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doukhanina EV, et al. Identification and functional characterization of the BAG protein family in Arabidopsis thaliana. J Biol Chem. 2006;281:18793–18801. doi: 10.1074/jbc.M511794200. [DOI] [PubMed] [Google Scholar]
- 10.Sondermann H, et al. Structure of a Bag/Hsc70 complex: Convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291:1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- 11.Rosati A, et al. Apoptosis inhibition in cancer cells: A novel molecular pathway that involves BAG3 protein. Int J Biochem Cell Biol. 2007;39:1337–1342. doi: 10.1016/j.biocel.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Doong H, et al. CAIR-1/BAG-3 forms an EGF-regulated ternary complex with phospholipase C-gamma and Hsp70/Hsc70. Oncogene. 2000;19:4385–4395. doi: 10.1038/sj.onc.1203797. [DOI] [PubMed] [Google Scholar]
- 13.Doong H, et al. CAIR-1/BAG-3 abrogates heat shock protein-70 chaperone complex-mediated protein degradation: Accumulation of poly-ubiquitinated Hsp90 client proteins. J Biol Chem. 2003;278:28490–28500. doi: 10.1074/jbc.M209682200. [DOI] [PubMed] [Google Scholar]
- 14.Doong H, Vrailas A, Kohn EC. What's in the ‘BAG’? A functional domain analysis of the BAG-family proteins. Cancer Lett. 2002;188:25–32. doi: 10.1016/s0304-3835(02)00456-1. [DOI] [PubMed] [Google Scholar]
- 15.Pagliuca MG, Lerose R, Cigliano S, Leone A. Regulation by heavy metals and temperature of the human BAG-3 gene, a modulator of Hsp70 activity. FEBS Lett. 2003;541:11–15. doi: 10.1016/s0014-5793(03)00274-6. [DOI] [PubMed] [Google Scholar]
- 16.Liao Q, et al. The anti-apoptotic protein BAG-3 is overexpressed in pancreatic cancer and induced by heat stress in pancreatic cancer cell lines. FEBS Lett. 2001;503:151–157. doi: 10.1016/s0014-5793(01)02728-4. [DOI] [PubMed] [Google Scholar]
- 17.Romano MF, et al. BAG3 protein controls B-chronic lymphocytic leukaemia cell apoptosis. Cell Death Differ. 2003;10:383–385. doi: 10.1038/sj.cdd.4401167. [DOI] [PubMed] [Google Scholar]
- 18.Romano MF, et al. BAG3 protein regulates cell survival in childhood acute lymphoblastic leukemia cells. Cancer Biol Ther. 2003;2:508–510. doi: 10.4161/cbt.2.5.524. [DOI] [PubMed] [Google Scholar]
- 19.Homma S, et al. BAG3 deficiency results in fulminant myopathy and early lethality. Am J Pathol. 2006;169:761–773. doi: 10.2353/ajpath.2006.060250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiappetta G, et al. The antiapoptotic protein BAG3 is expressed in thyroid carcinomas and modulates apoptosis mediated by tumor necrosis factor-related apoptosis-inducing ligand. J Clin Endocrinol Metab. 2007;92:1159–1163. doi: 10.1210/jc.2006-1712. [DOI] [PubMed] [Google Scholar]
- 21.Bonelli P, et al. BAG3 protein regulates stress-induced apoptosis in normal and neoplastic leukocytes. Leukemia. 2004;18:358–360. doi: 10.1038/sj.leu.2403219. [DOI] [PubMed] [Google Scholar]
- 22.Chen L, et al. Light damage induced changes in mouse retinal gene expression. Exp Eye Res. 2004;79:239–247. doi: 10.1016/j.exer.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Rosati A, et al. Evidence for BAG3 modulation of HIV-1 gene transcription. J Cell Physiol. 2007;210:676–683. doi: 10.1002/jcp.20865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franceschelli S, et al. Bag3 gene expression is regulated by heat shock factor 1. J Cell Physiol. 2008;215:575–577. doi: 10.1002/jcp.21397. [DOI] [PubMed] [Google Scholar]
- 25.Morimoto RI. Regulation of the heat shock transcriptional response: Cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 26.Voellmy R, Boellmann F. Chaperone regulation of the heat shock protein response. Adv Exp Med Biol. 2007;594:89–99. doi: 10.1007/978-0-387-39975-1_9. [DOI] [PubMed] [Google Scholar]
- 27.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;21:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solimini NL, Luo J, Elledge SJ. Non-oncogene addiction and the stress phenotype of cancer cells. Cell. 2007;21:986–988. doi: 10.1016/j.cell.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Agou F, et al. NEMO trimerizes through its coiled-coil C-terminal domain. J Biol Chem. 2002;277:17464–17475. doi: 10.1074/jbc.M201964200. [DOI] [PubMed] [Google Scholar]
- 30.Ran R, et al. Hsp70 promotes TNF-mediated apoptosis by binding IKK gamma and impairing NF-kappa B survival signaling. Genes Dev. 2004;18:1466–1481. doi: 10.1101/gad.1188204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Javelaud D, Wietzerbin J, Delattre O, Besançon F. Induction of p21Waf1/Cip1 by TNFalpha requires NF-kappaB activity and antagonizes apoptosis in Ewing tumor cells. Oncogene. 2000;19:61–68. doi: 10.1038/sj.onc.1203246. [DOI] [PubMed] [Google Scholar]
- 32.Campbell KJ, Witty JM, Rocha S, Perkins ND. Cisplatin mimics ARF tumor suppressor regulation of RelA (p65) nuclear factor-kappaB transactivation. Cancer Res. 2006;66:929–935. doi: 10.1158/0008-5472.CAN-05-2234. [DOI] [PubMed] [Google Scholar]
- 33.Maeda S, et al. IKKbeta is required for prevention of apoptosis mediated by cell-bound but not by circulating TNFalpha. Immunity. 2003;19:725–737. doi: 10.1016/s1074-7613(03)00301-7. [DOI] [PubMed] [Google Scholar]
- 34.Huang S, DeGuzman A, Bucana CD, Fidler IJ. Nuclear factor-kappaB activity correlates with growth, angiogenesis, and metastasis of human melanoma cells in nude mice. Clin Cancer Res. 2000;6:2573–2581. [PubMed] [Google Scholar]
- 35.McNulty SE, del Rosario R, Cen D, Meyskens FL, Jr, Yang S. Comparative expression of NFkappaB proteins in melanocytes of normal skin vs. benign intradermal naevus and human metastatic melanoma biopsies. Pigm Cell Res. 2004;17:173–180. doi: 10.1111/j.1600-0749.2004.00128.x. [DOI] [PubMed] [Google Scholar]
- 36.Amiri KI, Richmond A. Role of nuclear factor-kappa B in melanoma. Cancer Metastasis Rev. 2005;24:301–313. doi: 10.1007/s10555-005-1579-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li N, Karin M. Ionizing radiation and short wavelength UV activate NF-kappaB through two distinct mechanisms. Proc Natl Acad Sci USA. 1998;95:13012–13017. doi: 10.1073/pnas.95.22.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou J, Baichwal V, Cao Z. Regulatory elements and transcription factors controlling basal and cytokine-induced expression of the gene encoding intercellular adhesion molecule 1. Proc Natl Acad Sci USA. 1994;91:11641–11645. doi: 10.1073/pnas.91.24.11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Höhfeld J, Cyr DM, Patterson C. From the cradle to the grave: Molecular chaperones that may choose between folding and degradation. EMBO Rep. 2001;2:885–890. doi: 10.1093/embo-reports/kve206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valdez BC, et al. Altered gene expression in busulfan-resistant human myeloid leukemia. Leuk Res. 2008;32:1684–1697. doi: 10.1016/j.leukres.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu P, Xu B, Li J, Lu H. BAG3 gene silencing sensitizes leukemic cells to Bortezomib-induced apoptosis. FEBS Lett. 2009;583:401–406. doi: 10.1016/j.febslet.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 42.Wang HQ, et al. Inhibition of the JNK signalling pathway enhances proteasome inhibitor-induced apoptosis of kidney cancer cells by suppression of BAG3 expression. Br J Pharmacol. 2009;158:1405–1412. doi: 10.1111/j.1476-5381.2009.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacobs AT, Marnett LJ. HSF1-mediated BAG3 expression attenuates apoptosis in 4-hydroxynonenal-treated colon cancer cells via stabilization of anti-apoptotic Bcl-2 proteins. J Biol Chem. 2009;284:9176–9183. doi: 10.1074/jbc.M808656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carra S, Brunsting JF, Lambert H, Landry J, Kampinga HH. HspB8 participates in protein quality control by a non-chaperone-like mechanism that requires eIF2alpha phosphorylation. J Biol Chem. 2009;284:5523–5532. doi: 10.1074/jbc.M807440200. [DOI] [PubMed] [Google Scholar]
- 45.Seo YJ, et al. Bis induces growth inhibition and differentiation of HL-60 cells via up-regulation of p27. Exp Mol Med. 2005;37:624–630. doi: 10.1038/emm.2005.76. [DOI] [PubMed] [Google Scholar]
- 46.Iwasaki M, et al. BAG3 regulates motility and adhesion of epithelial cancer cells. Cancer Res. 2007;67:10252–10259. doi: 10.1158/0008-5472.CAN-07-0618. [DOI] [PubMed] [Google Scholar]
- 47.Kassis JN, Guancial EA, Doong H, Virador V, Kohn EC. CAIR-1/BAG-3 modulates cell adhesion and migration by downregulating activity of focal adhesion proteins. Exp Cell Res. 2006;312:2962–2971. doi: 10.1016/j.yexcr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 48.Kassis JN, et al. Genomic and phenotypic analysis reveals a key role for CCN1 (CYR61) in BAG3-modulated adhesion and invasion. J Pathol. 2009;218:495–504. doi: 10.1002/path.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kratsios P, et al. Antioxidant amelioration of dilated cardiomyopathy caused by conditional deletion of NEMO/IKKgamma in cardiomyocytes. Circ Res. 2010;106:133–144. doi: 10.1161/CIRCRESAHA.109.202200. [DOI] [PubMed] [Google Scholar]
- 50.Carra S, Seguin SJ, Lambert H, Landry J. HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J Biol Chem. 2008;283:1437–1444. doi: 10.1074/jbc.M706304200. [DOI] [PubMed] [Google Scholar]
- 51.Gamerdinger M, et al. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Djavaheri-Mergny M, Codogno P. Autophagy joins the game to regulate NF-kappaB signaling pathways. Cell Res. 2007;17:576–577. doi: 10.1038/cr.2007.58. [DOI] [PubMed] [Google Scholar]
- 53.Ye RD. Regulation of nuclear factor kappaB activation by G-protein-coupled receptors. J Leukoc Biol. 2001;70:839–848. [PubMed] [Google Scholar]
- 54.Palumbo R, et al. Cells migrating to sites of tissue damage in response to the danger signal HMGB1 require NF-kappaB activation. J Cell Biol. 2007;179:33–40. doi: 10.1083/jcb.200704015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niu J, et al. Keratinocyte growth factor/fibroblast growth factor-7-regulated cell migration and invasion through activation of NF-kappaB transcription factors. J Biol Chem. 2007;282:6001–6011. doi: 10.1074/jbc.M606878200. [DOI] [PubMed] [Google Scholar]
- 56.Collas P. The state-of-the-art of chromatin immunoprecipitation. Methods Mol Biol. 2009;567:1–25. doi: 10.1007/978-1-60327-414-2_1. [DOI] [PubMed] [Google Scholar]
- 57.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;4:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 58.Hosmer DW, Lemeshow S, May S. Applied Survival Analysis: Regression Modeling of Time-to-Event Data. , 2nd Ed. Hoboken, NJ: John Wiley & Sons; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.