Abstract
Nutrient-driven O-GlcNAcylation of key components of the transcription machinery may epigenetically modulate gene expression in metazoans. The global effects of GlcNAcylation on transcription can be addressed directly in C. elegans because knockouts of the O-GlcNAc cycling enzymes are viable and fertile. Using anti-O-GlcNAc ChIP-on-chip whole-genome tiling arrays on wild-type and mutant strains, we detected over 800 promoters where O-GlcNAc cycling occurs, including microRNA loci and multigene operons. Intriguingly, O-GlcNAc-marked promoters are biased toward genes associated with PIP3 signaling, hexosamine biosynthesis, and lipid/carbohydrate metabolism. These marked genes are linked to insulin-like signaling, metabolism, aging, stress, and pathogen-response pathways in C. elegans. Whole-genome transcriptional profiling of the O-GlcNAc cycling mutants confirmed dramatic deregulation of genes in these key pathways. As predicted, the O-GlcNAc cycling mutants show altered lifespan and UV stress susceptibility phenotypes. We propose that O-GlcNAc cycling at promoters participates in a molecular program impacting nutrient-responsive pathways in C. elegans, including stress, pathogen response, and adult lifespan. The observed impact of O-GlcNAc cycling on both signaling and transcription in C. elegans has important implications for human diseases of aging, including diabetes and neurodegeneration.
Keywords: diabetes, genomics, polycomb, epigenetics, transcription
Alterations in the hexosamine signaling pathway (HSP) have been linked to diseases of human aging, including diabetes mellitus, Alzheimer's disease, and coronary artery disease (1 –7). Genetic approaches to understanding the links between aging, human disease, and hexosamine signaling have been hampered by the fact that many of the genes important for hexosamine signaling are essential in mammals, making it difficult to analyze perturbations in the pathway in the context of the whole organism (8 –10).
The HSP leads to the dynamic and reversible Ser/Thr-O-GlcNAc modification of key nuclear and cytoplasmic proteins, and is a major cellular sensor of nutrient flux (1, 3, 5, 6). Levels of the sugar nucleotide UDP-GlcNAc are highly responsive to intracellular concentrations of glucose, amino acid, and lipids (11). In response to the levels of this nutrient-sensing substrate, O-GlcNAc transferase (OGT) posttranslationally GlcNAcylates key components of intracellular signaling pathways, mitochondrial proteins, metabolic enzymes, the proteasome, and transcription factors. O-GlcNAc modification, like other forms of posttranslational modification, serves to regulate the function of these target proteins. The O-GlcNAcase (OGA) catalyzes cycling by promoting O-GlcNAc removal (1, 3, 5, 6). In addition to this enzymatic activity, the OGA also contains a putative chromatin-interacting histone acetyltransferase-like (HAT) domain (12).
A growing body of evidence suggests that the enzymes of O-GlcNAc cycling may play a role in maintaining chromatin structure and modulating transcription (13 –16). Unlike vertebrate and Drosophila model systems (6, 8 –10, 13, 14, 17), null mutations in the enzymes of O-GlcNAc cycling are viable and fertile in Caenorhabditis elegans (18, 19), allowing a global analysis of the consequences of altered O-GlcNAc cycling on chromatin structure and gene expression in a living organism. Here, using whole-genome chromatin immunoprecipitation (ChIP)-on-chip tiling arrays, and transcriptional profiling, we have identified global changes in gene expression associated with defects in O-GlcNAc addition and removal. The findings suggest that O-GlcNAc is strongly associated with promoters of a subset of genes and microRNAs and that loss of O-GlcNAc cycling results in a dramatic deregulation of gene expression.
Results
Genome-Wide Analysis of Chromatin-Associated O-GlcNAc Proteins.
ChIP-on-chip analysis was performed to determine the location of O-GlcNAc throughout the C. elegans genome. Synchronous L1 larvae were used as a source of chromatin for these experiments. Three O-GlcNAc specific antibodies, RL2 (20), HGAC85 (21), and CTD 110.6 (22), were tested. Based on specific signal intensity in preliminary tiling experiments, RL2 was chosen for all subsequent experiments (Fig. S1A). Chromatin derived from wild-type (N2), ogt-1 mutants, and oga-1 mutants was subjected to whole-genome ChIP-on-chip analysis. The ogt-1 mutant strain provides an important negative control; it encodes a catalytically null allele, and the strain completely lacks the O-GlcNAc modification. The oga-1 mutant strain also is a catalytically null allele and exhibits elevated levels of O-GlcNAc-modified proteins. These strains have been characterized previously (18, 19). The normalized peak intensities across each of the Caenorhabditis elegans linkage groups is shown in Fig. 1A. Thresholding of wild-type and oga-1(ok1207) was performed by subtracting normalized signal intensities of ogt-1(ok430) from those of wild type and oga-1(ok1207). Peaks with intensities of 2.2 or greater were called as O-GlcNAc-specific peaks. After thresholding, wild-type and the oga-1 mutant strain retained 1166 O-GlcNAc-specific peaks. The number and distribution of O-GlcNAc-marked peaks (active regions) per chromosome is indicated in Fig. 1B. These peaks were associated with 828 genes (Dataset S1). A nearly identical set of O-GlcNAc-marked chromatin peaks was identified in both the oga-1 mutant strain and the wild-type strains (Fig. 1B). However, the marks, with few exceptions, were always higher in oga-1(ok1207) compared with wild type (Fig. 1 A and C). This is apparent when the average peak value is plotted for the 718 peaks within ±2 kb of known transcription starts; oga-1(1207) peak values are almost invariably higher than the other strains (Fig. 2A). This finding presumably reflects active cycling of O-GlcNAc in the wild-type strain, due to the action of O-GlcNAcase. In the absence of O-GlcNAcase, cycling is prevented, leading to enhanced accumulation at nearly all of the marked loci in the oga-1 mutant.
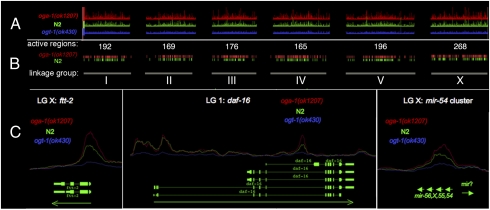
Fig. 1.
O-GlcNAc marks promoters throughout the C. elegans genome. An antibody specific for O-GlcNAc (RL2) was used for ChIP-on-chip analysis of the C. elegans genome. (A) The peak intensities across all six linkage groups: oga-1(ok1207), red; N2 (wild type), green; ogt-1(ok430), blue. Note that the O-GlcNAc-accumulating mutant oga-1(ok1207) (red) has the highest peak intensities, and the O-GlcNAc-deficient mutant ogt-1(ok430) (blue) displayed weak binding and served as a negative control. (B) A threshold was applied to peak intensities to obtain intervals for oga-1(ok1207) (red) and N2 (green). The number of peaks for each linkage group is shown. (C) Examples of the O-GlcNAc peaks at promoters are shown for two separate genes and a micro RNA. ftt-2 shows a large peak at its promoter. O-GlcNAc marks the first and third promoter of daf-16. The mir-54 cluster is also marked by O-GlcNAc. Green arrows represent direction of transcription.
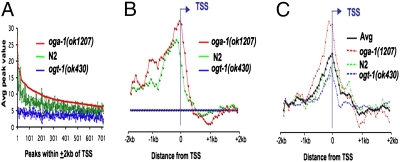
Fig. 2.
O-GlcNAc cycling occurs asymmetrically around the TSS. (A) The average chip value for all peaks within ±2 kb of TSS-associated the genes represented on the Affymetrix expression arrays is shown for each of the strains as indicated by the colored lines sorted using the peak values for oga-1(ok1207). Note that the peak values are highest in oga-1(ok1207) mutants that are unable to remove O-GlcNAc and are at background levels in ogt-1(ok430) mutants that are unable to add O-GlcNAc. (B) The position of the O-GlcNAc peaks within ±2 kb of the annotated TSS for each of the strains as indicated. Note that at this threshold, no peaks were detected in ogt-1(ok430). (C) The position of RNA Pol II peaks relative to the transcription start using antibody ab5095 for each indicated strain as described in Materials and Methods. When averaged together, the peaks for all stains exhibit similar behavior as each do individually (Avg, black line).
O-GlcNAc Marks Are Associated with Promoters.
Closer inspection of the O-GlcNAc-modified regions on chromatin showed that they were almost invariably associated with the promoters of genes. A clear example is shown with the 14-3-3 protein-encoding gene ftt-2 (Fig. 1C). Note the enhanced 5′ peak of O-GlcNAc in the oga-1(ok1207) mutant (red line) compared with wild type (green line), whereas ogt-1(ok430) (blue line) shows minimal O-GlcNAc association. These differences in the O-GlcNAc peaks are consistent with active cycling at the promoter. Many genes containing multiple promoters exhibit O-GlcNAc peaks at selective sites. For example, the daf-16 gene has three annotated promoters. Only the first and third promoters are strongly marked by O-GlcNAc; the middle promoter is not marked by the modification (Fig. 1C). In addition to protein-coding genes, O-GlcNAc marks were associated with ≈24 known microRNAs (Dataset S2), including the mir-54 cluster (Fig. 1C). Of the marked genes that are part of a multigene operon, ∼90% had O-GlcNAc marks at the promoter of the first gene. One striking example is the operon containing p38 MAP-kinase genes (pmk-1–3), where the first gene (F42G8.5) is heavily marked by O-GlcNAc (Fig. S1B).
To determine whether O-GlcNAc marks were correlated with transcriptional activity, we compared the O-GlcNAc ChIP-on-chip data set with an RNA Pol II ChIP-on-chip carried out under identical conditions. We have used two anti-RNA Pol II CTD antibodies: 5095 (Abcam) and 8WG16 (MMS-126R; Covance). The 5095 is marketed as specific for Ser-2-phosphorylation of the heptad repeat (YSPTSPS), a modification that is associated with the elongating form of the polymerase. However, 5095 was recently shown to also recognize unphosphorylated Pol II in C. elegans (23). The CTD heptad repeat-specific antibody (8WG16) is phosphorylation independent (24) and has been used previously for C. elegans ChIPs (23, 25). ChIP signals throughout the marked genes were much stronger with 5095 compared with 8WG16 (Fig. S1A). We found that O-GlcNAc marks a subset of the genes (∼40%) that were transcriptionally active as determined by 5095 Pol II ChIP. Two specific examples are shown in Fig. S1 A and B. Averaged across the genome, O-GlcNAc marks were almost exclusively associated with the 5′ end of genes, with a profile very similar, but not identical, to the distribution of RNA polymerase II (cf. Fig. 2 B and C). The asymmetric O-GlcNAc peak distribution is centered 100 bp upstream of the transcriptional start site (TSS), dropping off dramatically after crossing the TSS, whereas the RNA Pol II peak is centered near the TSS. The distribution of the O-GlcNAc peaks relative to the TSS was similar in N2 and oga-1(ok1207) thresholded samples (Fig. 2B, green and red lines), and was absent in the negative control strain ogt-1(ok430) (Fig. 2A, blue line). The distribution of elongating Pol II, relative to the TSS, was essentially identical in all of the strains (Fig. 2C), similar to previous reports (23, 25).
O-GlcNAc Marks Promoters of Genes Implicated in Growth, Aging, and the Stress Response.
To determine what kinds of proteins were encoded by genes that actively accumulate O-GlcNAc marks, we examined the O-GlcNAc promoter-marked genes bioinformatically using both traditional GO terms and the DAVID bioinformatic tools (26, 27). The O-GlcNAc marked promoters are strikingly biased toward genes involved in the biochemical pathways of PIP3 signaling (enrichment score 2.4), glyoxylate and glycerone-phosphate metabolism (enrichment score 1.2), and amine-metabolism (Table S1). Intriguingly, PIP3 signaling and glyoxylate signaling have been previously linked to OGT recruitment to target sites (28, 29), and amine-metabolism is central to the formation of UDP-GlcNAc via the hexosamine biosynthetic pathway (11). Further, DAVID functional annotation clustering analysis of the O-GlcNAc marked genes suggested that the biological pathways enriched in this data set involved reproductive behavior (enrichment score 8.5), aging (enrichment score 3.9), morphogenesis (enrichment score 3.2), neuron differentiation (enrichment score 3.0), mitotic spindle localization (enrichment score 2.7), and glycolysis (enrichment score 1.9; Table S1). Among the genes identified were the epsin family of PIP3 interacting proteins and the 14-3-3 proteins, which are known to sequester transcription factors like DAF-16/FOXO in the cytoplasm (see below). Previous work has implicated O-GlcNAc in many of these processes (1, 5).
Disruption of O-GlcNAc Cycling Leads to Changes in Transcription.
In addition to the global analysis of O-GlcNAc marks on chromatin, we examined changes in the transcriptome associated with the loss of O-GlcNAc cycling. From the same synchronous L1 larvae used for ChIP-on-chip, as well as synchronized L4 larvae, we prepared triplicate RNA samples for Affymetrix whole-genome expression arrays. The gene expression data from ogt-1 and oga-1 mutant strains were compared with wild type to determine how defects in O-GlcNAc cycling affected the transcriptome. A complete list of deregulated genes and the associated GO term analysis for both developmental stages is presented in Dataset S3.
In the oga-1(ok1207) L1 sample, 509 genes were deregulated greater than 1.5-fold relative to wild type (Fig. 3A). Of these, 291 genes (57%) were down-regulated and 218 were up-regulated. When analyzed bioinformatically using DAVID, these genes were predicted to regulate lifespan and aging, the stress response, innate immunity, and carbohydrate/lipid metabolism (Table S2). In the L4 larvae, knockout of oga-1 led to deregulation of ≈165 genes [69 down-regulated (41%); 96 up-regulated; Fig. 3B]. Though fewer genes were deregulated in the L4 stage compared with L1, bioinformatics analysis suggested that these genes were enriched for similar pathways, including aging (enrichment score 3.7), carbohydrate metabolism (enrichment score 2.7), membrane transport (enrichment score 2.5), chromatin remodeling (enrichment score 8.5), and innate immunity (Table S2). Both in L1 larvae and in L4 larvae, ≈100 representative genes altered in the oga-1(ok1207) were confirmed by a quantitative reverse transcriptase PCR (qRT-PCR; Dataset S4).
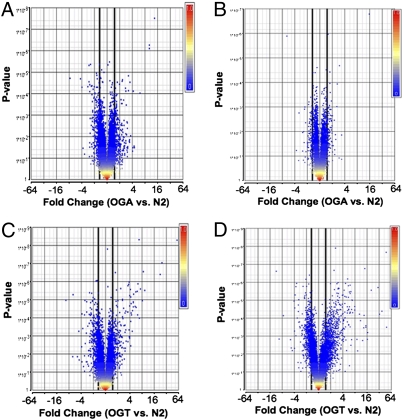
Fig. 3.
O-GlcNAc cycling mutant exhibit deregulation of gene expression. Volcano plots of gene expression, as detected by microarray analysis are shown. (A and B) oga-1(ok1207). (C and D) ogt-1(ok430). Larval stages L1 (A and C) and L4 (B and D) were used for analysis. Data are plotted as geometric fold change (x axis) vs. P value (y axis). Genes were considered significantly deregulated if the geometric fold change was greater than 1.5 (bold black line) with a P value of <0.05.
A similar transcriptional analysis was performed using the ogt-1(ok430) knockout strain. At the L1 stage, 688 genes were up- or down-regulated relative to wild type; 389 (56%) were down-regulated and 299 were up-regulated (Fig. 3C). These genes were highly enriched in pathways involved in innate immunity (C-type lectins, major facilitator superfamily, peptidoglycan catabolism), stress, and detoxification (GST; Table S3). At the L4 stage, the ogt-1(ok430) showed deregulation of 1,641 genes compared with wild type; 659 (40%) were down-regulated and 982 were up-regulated (Fig. 3D). These deregulated genes were highly enriched for loci encoding insulin and secreted peptides (enrichment score 11.7), regulators of amino acid metabolism (enrichment score 9.6), mitochondrial proteins (enrichment score 9.4), and genes involved in reproduction, stress, detoxification, and aging (Table S3). Both in L1 larvae and in L4 larvae, many of these changes were confirmed by qRT-PCR (Dataset S4). These expression data sets strongly suggest that at two developmental stages, the loss of O-GlcNAc cycling in C. elegans leads to striking deregulation of genes involved in the response to stress, adult lifespan, carbohydrate breakdown, pathogen resistance, and transcriptional regulation.
Disruption of O-GlcNAc Cycling Affects the Aging Pathway and the Stress Response.
The genomic and transcriptional analyses of the O-GlcNAc cycling mutants predicted a role for O-GlcNAc cycling in several processes, including the stress response and aging. We first examined the lifespan of the O-GlcNAc cycling mutants in both a wild-type or daf-2 mutant background (Fig. 4 A and B, respectively). DAF-2 is the C. elegans insulin-like receptor, and daf-2 mutants are long lived (30). In both the wild-type and daf-2 mutant backgrounds, ogt-1(ok430) significantly reduced the mean lifespan (by ∼20% and 40% respectively; Fig. 4 A and B). The oga-1 mutation significantly extended lifespan in the daf-2 mutant background (∼12% extension), but not in the wild-type background (Fig. 4 A and B).
Fig. 4.
O-GlcNAc cycling mutants affect aging and the stress response. The absence of O-GlcNAc modifications in ogt-1(ok430) resulted in shorter lifespan in (A) wild-type and (B) daf-2(e1370) animals. In contrast, an overabundance of O-GlcNAc modification in oga-1(ok1207) mutants had a negligible effect on wild-type lifespan (A), but extended lifespan of daf-2(e1370) animals in a daf-16-independent manner (B and C). Data are from individual representative trials; all results are presented in Dataset S5. Number of animals scored: (A) control, n = 88; ok1207, n = 83; ok430, n = 86; ok430;ok1207, n = 72; (B) control, n = 101; ok1207, n = 101; ok430, n = 95; ok430; ok1207, n = 102; (C) daf-2 daf-16 RNAi, control n = 54; ok1207, n = 59; daf-16(mu86);daf-2, control n = 88; ok1207, n = 76. Experiment in A was performed at 25 °C; similar results were obtained at 20 °C; B and C were performed at 20 °C. Animals in A and B were grown on OP50 food source; animals in C were grown on HT115 bacteria. (D) Neither mutant significantly affected daf-2 fertility at either the semipermissive (22.5 °C) or the restrictive (25 °C) temperature. Error bars represent SD for three independent experiments; P > 0.3. (E) The absence of the O-GlcNAc modification in ogt-1(ok430) decreased resistance to UV stress, whereas the abundance in oga-1(ok1207) increased stress resistance. Young adult hermaphrodites were exposed to 23 mJ of UV radiation and were followed for 3 days to determine the fraction surviving. (F) A large pool of DAF-16:GFP accumulates in the nucleus in ogt-1(ok430) L1 hermaphrodites (Left) without stress induction. In contrast, wild-type animals have very little DAF-16::GFP in the nucleus (Inset). Upon heat challenge at 37 °C for 10 min, nuclear DAF-16::GFP was detected in 100% of N2, ogt-1(ok430), and oga-1(ok1207) animals (Table S4). Quantitation of the fraction of animals with nuclear DAF-16::GFP in the absence of stress (Right).
The lifespan extension observed in a daf-2 mutant is dependent on the downstream transcription factor DAF-16/FOXO (30), so next we tested whether the lifespan extension observed with the oga-1 mutant was dependent on DAF-16. Surprisingly, we found that the lifespan extension of a daf-2;oga-1 mutant is not dependent on DAF-16. oga-1(ok1207) increased daf-2 lifespan in either a daf-16(mu86) mutant background (28% extension) or in response to daf-16 RNAi (19% extension, P < 0.0001; Fig. 4C). Thus, oga-1(1207) increased lifespan in a manner dependent upon daf-2, but independent of daf-16.
In addition to lifespan regulation and dauer formation, the DAF-2 insulin-like receptor also regulates fertility: daf-2(e1370) mutations reduce fertility in a DAF-16 and temperature-dependent manner (31) (Fig. 4D). However, although mutations in oga-1 and ogt-1 modulate dauer formation and lifespan in a daf-2(e1370) mutant, there was no statistical difference in brood size among the three strains (Fig. 4D). These findings suggest that O-GlcNAc cycling is important for control of dauer formation and lifespan regulation, but not hermaphrodite self-fertilization and reproduction.
Next, we tested whether O-GlcNAc cycling was involved in modulating the response to stress, as predicted by our tiling array and transcriptome data. We found that ogt-1(ok430) was hypersensitive to UV stress compared with wild type (Fig. 4E). In contrast, the oga-1(ok1207) strain was slightly more resistant to UV stress compared with wild type. The observed altered sensitivity to UV stress is consistent with previous reports in mammalian systems suggesting a role for O-GlcNAc in mediating the stress response (32 –34).
The FOXO homolog DAF-16 is a key mediator of the stress response and longevity in C. elegans, and is known to translocate to the nucleus in response to stress, including heat stress and starvation (35). As in the wild-type strain, a DAF-16::GFP fusion protein was normally cytoplasmic in the oga-1(ok1207) strain (Fig. 4F) and entered the nucleus upon heat stress. However, in ogt-1(ok430) animals, we found nuclear-localized DAF-16::GFP in the absence of acute heat stress (22 °C) in a significant proportion of the animals (Fig. 4F), suggesting chronic activation of DAF-16. Such chronic activation of DAF-16 does not appear to be protective, because these animals are more sensitive to acute stress (Fig. 4E). The constitutive translocation detected in ogt-1(ok430), in the absence of acute stress, is consistent with a deregulation of insulin signaling and its downstream effectors in the ogt-1 knockout.
Discussion
O-GlcNAc Cycling Occurs at Promoters of Nutrient-Responsive Genes and microRNAs.
Kelly and Hart (36) first demonstrated that glycoprotein-binding lectins are localized to Drosophila polytene chromosomes. Using whole-genome tiling array technology, we have identified over 800 genes as being linked to sites of active O-GlcNAc cycling in C. elegans. A comparison of genes marked by either O-GlcNAc or by Pol II (5095) shows a substantial overlap (∼40%). Thus O-GlcNAc marks provide a clear and convenient means of identifying promoters in C. elegans, many of which are actively transcribing, even in growth-arrested L1s.
The chromatin-associated O-GlcNAc marks share several properties with the chromatin marks of the histone variant HTZ-1, the H2A.Z-like protein in C. elegans (25). Both are located just upstream of the transcriptional start and tend to mark the first, but not subsequent, starts sites for genes within operons. Both also show a strong, but not absolute, correlation with transcriptional activity as defined by RNA Pol II ChIP (pan-Pol II CTD antibody 8WG16) (25). However, there are also differences between the HTZ-1 and O-GlcNAc data. For example, HTZ-1 is underincorporated on the X chromosome, whereas the X chromosome has the most abundant number of O-GlcNAc peaks over threshold (Fig. 1A and Dataset S1). A comparison of these data suggests that O-GlcNAc and HTZ-1 are reflective of overlapping, yet distinct, biological processes.
Like HTZ-1 chromatin marks, loci marked by O-GlcNAc are not easily correlated to direct transcriptional changes at those loci as revealed by expression array analysis. This could be due to several factors. First, the influence of O-GlcNAc on signaling cascades could lead to transcriptional changes independent of marked loci. Second, many transcription factors are modified by O-GlcNAc (e.g., daf-16), and changes in these key regulators could influence the transcription of many downstream target genes. Additionally, the O-GlcNAc-marked miRNAs highlight the possible role of posttranscriptional regulation of message stability as a confounding factor in correlating the steady-state mRNA analysis provided by expression arrays with chromatin-associated factors. The role of O-GlcNAcylated, chromatin-associated proteins and transcriptional networks will have to be much better understood before we can elucidate the molecular mechanisms linking O-GlcNAc marks to transcriptional effects on individual genes.
Despite the imperfect correlation between our O-GlcNAc ChIP data and expression array results gene by gene, a more global approach was informative. Bioinformatic examination of the O-GlcNAc marked genes revealed a significant enrichment for genes involved in hexosamine synthesis, phosphoinositol signaling, lipid and carbohydrate metabolism, and chromatin remodeling. These observed enrichments are consistent with a number of the proposed biological functions of the O-GlcNAc modification (11, 18, 19, 28, 37–40). The predictive ability of the O-GlcNAc ChIP-on-chip data set suggests that these data can be mined to identify new O-GlcNAc-responsive pathways.
O-GlcNAc Marks, Transcriptional Repression, and Polycomb.
Previous studies have hinted at a role for O-GlcNAc in transcriptional repression (29, 41, 42). Two recent papers have demonstrated that Drosophila ogt is allelic with super sex combs (sxc), a gene involved in repression of homeotic genes (13, 14). Flies deficient in catalytically active Ogt develop through all larval stages, but die as pharate adults that have a variety of homeotic transformations (13, 14, 43). Polyhomeotic (Ph), a Polycomb group (PcG) protein, was shown to be O-GlcNAc modified and was suggested to be the mediator of OGT-dependent repression. The most highly labeled 1% of the O-GlcNAc-marked genes in the fly is associated with polycomb response elements (PREs), which are correlated with homeotic repression and bound by Ph and/or the Polycomb repressive complex (PhoRC) (14). Interestingly, the most highly labeled ∼1% of the O-GlcNAc-marked genes in C. elegans also have a connection with transcriptional repression, polycomb, and homeotic gene expression (44–50).
Although Drosophila O-GlcNAc is essential for PcG repression and normal development, O-GlcNAc cycling does not appear to be essential for normal C. elegans development. However, many homeotic transformations in C. elegans manifest as cell lineage defects that are often subtle and not obvious upon casual examination (51). Further investigation will be required to determine what role O-GlcNAc cycling may play in C. elegans HOX gene expression. The robustness of the C. elegans developmental program may be due to the complex interplay between Wnt signaling and HOX gene expression (52).
O-GlcNAc Cycling Modulates the C. elegans Insulin-Like Signaling Pathway Impacting Longevity and the Stress Response.
We have provided several lines of evidence pointing to deregulation of cellular signaling and transcriptional pathways in the mutants of O-GlcNAc cycling. Phenotypically, the mutants have altered adult lifespan and sensitivity to stress. The ogt-1 null mutants also show an aberrant, constitutive nuclear accumulation of the critical transcription factor DAF-16. Among the genes most deregulated by interference with O-GlcNAc cycling are genes implicated in the stress and longevity pathways (reviewed in ref. 53). These findings are consistent with our previous work implicating O-GlcNAc cycling in the dauer diapause (18, 19). The modulation of O-GlcNAc in the insulin-signaling pathway can be thought of as a fine-tuning mechanism, not an on/off switch.
The enhancement of daf-2(e1370) longevity by oga-1(ok1207) was daf-16 independent. Combined with the ogt-1 lifespan results, these findings suggest that O-GlcNAc cycling influences lifespan via both the insulin-like signaling pathway (daf-2 dependent; daf-16 dependent) and additional signaling pathways (daf-16 independent).
Two of the three daf-16 promoters are heavily marked by O-GlcNAc in the tiling array, and the mammalian DAF-16 homolog FOXO1 protein is modified by O-GlcNAc (54). DAF-16 is known to directly or indirectly regulate hundreds of transcriptional targets (44, 55, 56), and O-GlcNAc cycling could represent a modulator that distinguishes between these, as it regulates the DAF-16-dependent lifespan extension of daf-2 mutants but not the DAF-16-dependent fertility suppression of daf-2 mutants. These findings point to considerable complexity in the interaction between O-GlcNAc cycling and the well-studied insulin-like signaling pathway in C. elegans.
DAF-16 constitutively localized to the nucleus in an ogt-1 mutant (Fig. 4F); however, given that ogt-1 mutants are sensitive to acute stress (Fig. 4E) and display longevity defects (Fig. 4 A–C), the nuclear localized DAF-16 in the ogt-1 background is further evidence of massive deregulation of converging signaling pathways that may include the insulin-signaling pathway and the p38 MAPK pathways (6, 57, 58). OGT-1 and PMK-1 physically interact in C. elegans (59), and the absence of OGT-1 could cause the deregulation of pmk-1, leading to dramatic changes in longevity and the stress response that are unrelated to daf-16-dependent transcription. Thus the nuclear localization of daf-16 could reflect a constitutive, unregulated activation of several stress-induced pathways leading to decreased lifespan and increased sensitivity to stress. In mammals, p38 MAPK physically interacts with the C terminus of OGT-1 and is known to activate O-GlcNAcylation of some substrates during starvation (60). Our data are also consistent with results from mammalian cells demonstrating that nuclear localization of the DAF-16 homolog FOXO1 is not sufficient to stimulate transcription in the presence of insulin signaling (61). Mutants in smk-1 (a presumptive phosphatase) affect stress, longevity, and innate immunity downstream of DAF-16 nuclear localization (62).
O-GlcNAc Cycling and Diseases of Aging.
C. elegans is a widely used model for aging, implicating the insulin-like signaling pathway in determination of adult lifespan (63). A growing body of evidence suggests that altered O-GlcNAc cycling may be associated with the diseases of aging, including obesity, type II diabetes mellitus, cardiovascular disease, cancer, and neurodegenerative disease (3, 6). Our data derived from a global analysis of O-GlcNAc promoters and genes deregulated by the C. elegans O-GlcNAc cycling mutants are consistent with these proposed associations. The alterations in dauer and longevity observed for the O-GlcNAc cycling mutant alleles suggest that the insulin-like signaling pathway is normally responsive to the nutrient-sensing O-GlcNAc signaling pathway. In addition, the multiple pathways leading to induction of the stress and pathogen response programs are deregulated in the O-GlcNAc cycling mutants. Thus, the genetic and molecular analysis presented here reinforces the growing body of evidence that O-GlcNAc cycling is critical for maintaining the delicate balance between the progrowth and the prosurvival functions of the insulin and MAPK pathways during senescence. Because O-GlcNAc has emerged as a potential regulator of epigenetic phenomena (13, 14), we are pursuing the hypothesis that O-GlcNAc could also function in metabolic reprogramming in the intrauterine environment.
Materials and Methods
Strains.
The following alleles were used in these studies: wild-type N2 Bristol (discovered by Sydney Brenner in 1964), ogt-1(ok430) (18), ogt-1(ok1474), ogt-1(tm1046) (National Bioresource Project of Japan), oga-1(ok1207) (19), the temperature-sensitive insulin like receptor mutant daf-2(e1370), daf-16(mu86), and TJ356 [daf-16::GFP (zIs356)]. Unless otherwise noted, all strains were obtained from the Caenorhabditis Genetics Center. For details of allele conformation and genetic crosses, see SI Materials and Methods.
Chromatin Preparation and ChIP-on-Chip.
Synchronized C. elegans L1-stage animal pellets were collected, immediately frozen in water, and shipped on dry ice for processing by Genpathway. Genomic DNA regions of interest were isolated using antibodies against the following epitopes: the unmodified and Ser-5-P-modified heptad repeat (YSPTSPS) of the C-terminal domain (CTD) of RNA PolII (8WG16 MMS-126R; Covance); the Ser-2-phosphorylated and nonmodified heptad repeat of the CTD domain (ab5095, Abcam); O-linked N-acetylglucosamine modified peptides (RL2; ab2739; Abcam); HGAC85-derived O-linked N-acetylglucosamine (21) (MA1-076; Thermo Scientific); and Ser/Thr-O-linked N-acetylglucosamine (CTD 110.6; MMS-248R; Covance). Standard chromatin immunoprecipitation methods were used (25). For more detailed methods, see SI Materials and Methods. Bioinformatic analysis (Genpathway) was used to identify genes associated with RL2 peaks above threshold and located within 2 kb of the transcriptional start site (genome version WS195). All data were submitted to the Gene Expression Omnibus (GEO) as SuperSeries GSE18132.
Gene Expression Analysis.
Embryos were obtained by hypochlorite-NaOH lysis of gravid hermaphrodite adults. The embryos were hatched in M9 medium by continuous overnight shaking at 20 °C. Growth-arrested L1 larvae were lysed using glass beads with Minibeadbeater 8 (Biospec Products) at 4 °C. L4 animals were processed as previously described (64). For further details, see SI Materials and Methods. All data were submitted to GEO as SuperSeries GSE18132.
Longevity and Fertility Assays.
Longevity assays were conducted as described (65). The fertility assays were conducted as described (31). Modifications to either assay are provided in SI Materials and Methods. RNAi feeding vector L4440 and daf-16 (RNAi) were obtained from the RNAi feeding library (66).
UV Treatment and DAF-16::GFP Translocation.
UV stress performed essentially as described (67). At least 20 animals were used for each allele tested. Any changes are detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, NIDDK, and NIA.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (SuperSeries no. GSE18132).
This article contains supporting information online at www.pnas.org/cgi/content/full/0911857107/DCSupplemental.
References
- 1.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 2.Copeland RJ, Bullen JW, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: Roles in insulin resistance and glucose toxicity. Am J Physiol Endocrinol Metab. 2008;295:E17–E28. doi: 10.1152/ajpendo.90281.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butkinaree C, Park K, Hart GW. O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta. 2010;1800:96–106. doi: 10.1016/j.bbagen.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanover JA. Glycan-dependent signaling: O-linked N-acetylglucosamine. FASEB J. 2001;15:1865–1876. doi: 10.1096/fj.01-0094rev. [DOI] [PubMed] [Google Scholar]
- 5.Love DC, Hanover JA. The hexosamine signaling pathway: Deciphering the “O-GlcNAc code”. Sci STKE. 2005;2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 6.Hanover JA, Krause MW, Love DC. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta. 2010;1800:80–95. doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshall S. Role of insulin, adipocyte hormones, and nutrient-sensing pathways in regulating fuel metabolism and energy homeostasis: A nutritional perspective of diabetes, obesity, and cancer. Sci STKE. 2006;2006:re7. doi: 10.1126/stke.3462006re7. [DOI] [PubMed] [Google Scholar]
- 8.Shafi R, et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci USA. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanover JA, et al. Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch Biochem Biophys. 2003;409:287–297. doi: 10.1016/s0003-9861(02)00578-7. [DOI] [PubMed] [Google Scholar]
- 10.O'Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol Cell Biol. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991;266:4706–4712. [PubMed] [Google Scholar]
- 12.Toleman C, Paterson AJ, Whisenhunt TR, Kudlow JE. Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J Biol Chem. 2004;279:53665–53673. doi: 10.1074/jbc.M410406200. [DOI] [PubMed] [Google Scholar]
- 13.Sinclair DA, et al. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc) Proc Natl Acad Sci USA. 2009;106:13427–13432. doi: 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gambetta MC, Oktaba K, Müller J. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- 15.Cheung WD, Sakabe K, Housley MP, Dias WB, Hart GW. O-linked beta-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J Biol Chem. 2008;283:33935–33941. doi: 10.1074/jbc.M806199200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujiki R, et al. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459:455–459. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
- 17.Webster DM, et al. O-GlcNAc modifications regulate cell survival and epiboly during zebrafish development. BMC Dev Biol. 2009;9:28. doi: 10.1186/1471-213X-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanover JA, et al. A Caenorhabditis elegans model of insulin resistance: Altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc Natl Acad Sci USA. 2005;102:11266–11271. doi: 10.1073/pnas.0408771102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forsythe ME, et al. Caenorhabditis elegans ortholog of a diabetes susceptibility locus: oga-1 (O-GlcNAcase) knockout impacts O-GlcNAc cycling, metabolism, and dauer. Proc Natl Acad Sci USA. 2006;103:11952–11957. doi: 10.1073/pnas.0601931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snow CM, Senior A, Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol. 1987;104:1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner JR, Tartakoff AM, Greenspan NS. Cytologic assessment of nuclear and cytoplasmic O-linked N-acetylglucosamine distribution by using anti-streptococcal monoclonal antibodies. Proc Natl Acad Sci USA. 1990;87:5608–5612. doi: 10.1073/pnas.87.15.5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comer FI, Vosseller K, Wells L, Accavitti MA, Hart GW. Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine. Anal Biochem. 2001;293:169–177. doi: 10.1006/abio.2001.5132. [DOI] [PubMed] [Google Scholar]
- 23.Baugh LR, Demodena J, Sternberg PW. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science. 2009;324:92–94. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- 24.Hirakata M, et al. Identification of autoantibodies to RNA polymerase II. Occurrence in systemic sclerosis and association with autoantibodies to RNA polymerases I and III. J Clin Invest. 1993;91:2665–2672. doi: 10.1172/JCI116505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whittle CM, et al. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet. 2008;4:e1000187. doi: 10.1371/journal.pgen.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dennis GJ, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 27.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Yang X, et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
- 29.Yao D, et al. High glucose increases angiopoietin-2 transcription in microvascular endothelial cells through methylglyoxal modification of mSin3A. J Biol Chem. 2007;282:31038–31045. doi: 10.1074/jbc.M704703200. [DOI] [PubMed] [Google Scholar]
- 30.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 31.Tissenbaum HA, Ruvkun G. An insulin-like signaling pathway affects both longevity and reproduction in Caenorhabditis elegans. Genetics. 1998;148:703–717. doi: 10.1093/genetics/148.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zachara NE, Hart GW. O-GlcNAc a sensor of cellular state: The role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim Biophys Acta. 2004;1673:13–28. doi: 10.1016/j.bbagen.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 33.Guinez C, et al. Hsp70-GlcNAc-binding activity is released by stress, proteasome inhibition, and protein misfolding. Biochem Biophys Res Commun. 2007;361:414–420. doi: 10.1016/j.bbrc.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 34.Chatham JC, Nöt LG, Fülöp N, Marchase RB. Hexosamine biosynthesis and protein O-glycosylation: The first line of defense against stress, ischemia, and trauma. Shock. 2008;29:431–440. doi: 10.1097/shk.0b013e3181598bad. [DOI] [PubMed] [Google Scholar]
- 35.Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- 36.Kelly WG, Hart GW. Glycosylation of chromosomal proteins: Localization of O-linked N-acetylglucosamine in Drosophila chromatin. Cell. 1989;57:243–251. doi: 10.1016/0092-8674(89)90962-8. [DOI] [PubMed] [Google Scholar]
- 37.Kornfeld R. Studies on L-glutamine D-fructose 6-phosphate amidotransferase. I. Feedback inhibition by uridine diphosphate-N-acetylglucosamine. J Biol Chem. 1967;242:3135–3141. [PubMed] [Google Scholar]
- 38.Vosseller K, Wells L, Lane MD, Hart GW. Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proc Natl Acad Sci USA. 2002;99:5313–5318. doi: 10.1073/pnas.072072399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McClain DA, et al. Altered glycan-dependent signaling induces insulin resistance and hyperleptinemia. Proc Natl Acad Sci USA. 2002;99:10695–10699. doi: 10.1073/pnas.152346899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zachara NE, Hart GW. Cell signaling, the essential role of O-GlcNAc! Biochim Biophys Acta. 2006;1761:599–617. doi: 10.1016/j.bbalip.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Whisenhunt TR, et al. Disrupting the enzyme complex regulating O-GlcNAcylation blocks signaling and development. Glycobiology. 2006;16:551–563. doi: 10.1093/glycob/cwj096. [DOI] [PubMed] [Google Scholar]
- 42.Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: Coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;110:69–80. doi: 10.1016/s0092-8674(02)00810-3. [DOI] [PubMed] [Google Scholar]
- 43.Ingham PW. A gene that regulates the bithorax complex differentially in larval and adult cells of Drosophila. Cell. 1984;37:815–823. doi: 10.1016/0092-8674(84)90416-1. [DOI] [PubMed] [Google Scholar]
- 44.Murphy CT, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 45.Yuan J, Tirabassi RS, Bush AB, Cole MD. The C. elegans MDL-1 and MXL-1 proteins can functionally substitute for vertebrate MAD and MAX. Oncogene. 1998;17:1109–1118. doi: 10.1038/sj.onc.1202036. [DOI] [PubMed] [Google Scholar]
- 46.Chamberlin HM, Thomas JH. The bromodomain protein LIN-49 and trithorax-related protein LIN-59 affect development and gene expression in Caenorhabditis elegans. Development. 2000;127:713–723. doi: 10.1242/dev.127.4.713. [DOI] [PubMed] [Google Scholar]
- 47.Cui M, Kim EB, Han M. Diverse chromatin remodeling genes antagonize the Rb-involved SynMuv pathways in C. elegans. PLoS Genet. 2006;2:e74. doi: 10.1371/journal.pgen.0020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lehner B, et al. Loss of LIN-35, the Caenorhabditis elegans ortholog of the tumor suppressor p105Rb, results in enhanced RNA interference. Genome Biol. 2006;7:R4. doi: 10.1186/gb-2006-7-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ceol CJ, Horvitz HR. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev Cell. 2004;6:563–576. doi: 10.1016/s1534-5807(04)00065-6. [DOI] [PubMed] [Google Scholar]
- 50.Zhang T, et al. RNA-binding proteins SOP-2 and SOR-1 form a novel PcG-like complex in C. elegans. Development. 2006;133:1023–1033. doi: 10.1242/dev.02275. [DOI] [PubMed] [Google Scholar]
- 51.Yang L, Sym M, Kenyon C. The roles of two C. elegans HOX co-factor orthologs in cell migration and vulva development. Development. 2005;132:1413–1428. doi: 10.1242/dev.01569. [DOI] [PubMed] [Google Scholar]
- 52.Li X, Kulkarni RP, Hill RJ, Chamberlin HM. HOM-C genes, Wnt signaling and axial patterning in the C. elegans posterior ventral epidermis. Dev Biol. 2009;332:156–165. doi: 10.1016/j.ydbio.2009.05.567. [DOI] [PubMed] [Google Scholar]
- 53.Mukhopadhyay A, Oh SW, Tissenbaum HA. Worming pathways to and from DAF-16/FOXO. Exp Gerontol. 2006;41:928–934. doi: 10.1016/j.exger.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 54.Housley MP, et al. O-GlcNAc regulates FoxO activation in response to glucose. J Biol Chem. 2008;283:16283–16292. doi: 10.1074/jbc.M802240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 56.Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- 57.Inoue H, et al. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 2005;19:2278–2283. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Troemel ER, et al. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheung WD, Hart GW. AMP-activated protein kinase and p38 MAPK activate O-GlcNAcylation of neuronal proteins during glucose deprivation. J Biol Chem. 2008;283:13009–13020. doi: 10.1074/jbc.M801222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsai WC, Bhattacharyya N, Han LY, Hanover JA, Rechler MM. Insulin inhibition of transcription stimulated by the forkhead protein Foxo1 is not solely due to nuclear exclusion. Endocrinology. 2003;144:5615–5622. doi: 10.1210/en.2003-0481. [DOI] [PubMed] [Google Scholar]
- 62.Wolff S, et al. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 2006;124:1039–1053. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- 63.Tissenbaum HA, Guarente L. Model organisms as a guide to mammalian aging. Dev Cell. 2002;2:9–19. doi: 10.1016/s1534-5807(01)00098-3. [DOI] [PubMed] [Google Scholar]
- 64.Fukushige T, Brodigan TM, Schriefer LA, Waterston RH, Krause M. Defining the transcriptional redundancy of early bodywall muscle development in C. elegans: Evidence for a unified theory of animal muscle development. Genes Dev. 2006;20:3395–3406. doi: 10.1101/gad.1481706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilson MA, et al. Blueberry polyphenols increase lifespan and thermotolerance in Caenorhabditis elegans. Aging Cell. 2006;5:59–68. doi: 10.1111/j.1474-9726.2006.00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 67.Murakami S, Johnson TE. A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics. 1996;143:1207–1218. doi: 10.1093/genetics/143.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.