Abstract
Kinetic control of macromolecular interactions plays key roles in biological regulation. An example of such control occurs in cotranslational protein targeting by the signal recognition particle (SRP), during which the SRP RNA and the cargo both accelerate complex assembly between the SRP and SRP receptor FtsY 102-fold. The molecular mechanism underlying these rate accelerations was unclear. Here we show that a highly conserved basic residue, Lys399, on the lateral surface of FtsY provides a novel RNA tetraloop receptor to mediate the SRP RNA- and cargo-induced acceleration of SRP–FtsY complex assembly. We propose that the SRP RNA, by using its tetraloop to interact with FtsY–Lys399, provides a transient tether to stabilize the early stage and transition state of complex formation; this accelerates the assembly of a stable SRP–FtsY complex and allows the loading of cargo to be efficiently coupled to its membrane delivery. The use of a transient tether to increase the lifetime of collisional intermediates and reduce the dimension of diffusional search represents a novel and effective mechanism to accelerate macromolecular interactions.
Keywords: catalytic RNA, macromolecular interaction, signal recognition particle, GTPases, fluroescence
Kinetic control of macromolecular interactions plays key roles in biological regulation. An example of such kinetic control is found during the cotranslational targeting of proteins to cellular membranes (1), during which the signal recognition particle (SRP) recognizes ribosome-nascent chain complexes (RNCs) containing signal sequences (1–3), and delivers the RNC to the target membrane via interactions with the SRP receptor (4, 5). The functional core of SRP is comprised of an SRP54 protein and an SRP RNA. SRP54 (Ffh in bacteria) contains a methionine-rich M domain that recognizes signal sequences and binds the SRP RNA (6–8). In addition, both Ffh and the SRP receptor (FtsY in bacteria) contain GTPase, NG-domains that directly interact with each other to mediate SRP–FtsY complex assembly (9, 10). As cotranslational protein targeting must be completed before the nascent polypeptide exceeds approximately 140 amino acids in length (11, 12), this imposes a 3–5 s time window for the targeting reaction and mandates that the SRP–FtsY interaction, which is responsible for delivering the cargo to the target membrane, must occur rapidly upon loading of cargo on the SRP.
Formation of a stable SRP–FtsY complex is a dynamic process involving at least two distinct steps (13): (i) the initial rapid association between Ffh and FtsY to form a transient, GTP-independent early intermediate (13), and (ii) the GTP-dependent rearrangement of the early intermediate to a stable complex (13). This rearrangement requires the removal of steric blocks imposed by the N-terminal helices of both Ffh and FtsY (14–16). In addition, the N domains of both proteins move closer to one another and form stabilizing interactions at the heterodimer interface (9, 10). Finally, the two bound GTP molecules need to be correctly aligned, forming a cyclic pair of hydrogen bonds with each other across the dimer interface (9, 10).
Due to the extensive rearrangements required to form a stable SRP–FtsY complex, this process is extremely slow (17, 18). This barrier is overcome by the SRP RNA, which accelerates complex formation 200-fold (17, 18). Intriguingly, the effect of the SRP RNA is purely catalytic, as it also accelerates complex disassembly without changing the equilibrium stability of the SRP–FtsY complex (17). This is the first example of an RNA molecule catalyzing a protein–protein interaction, and different models have been suggested to explain its mechanism of action. One class of models postulates that the SRP RNA preorganizes the Ffh NG domain into a conformation more conducive to stable interaction with FtsY. A second class of models suggests that the SRP RNA provides a transient tether, either directly or indirectly, that holds the two GTPases together during the transition state of their complex assembly (17). Nevertheless, no direct evidence is available to support either the “preorganization” or “transient tether” model.
To distinguish between these models and understand the precise mechanism underlying the rate acceleration provided by the SRP RNA, we need to identify the structural motifs and the molecular interactions essential for mediating the catalytic effect of SRP RNA. Thus far, the conserved GGAA tetraloop of the SRP RNA provides the strongest candidate. This tetraloop is conserved throughout bacterial, archaeal, and eukaryotic SRPs, and its mutations abolish the ability of the RNA to accelerate SRP–FtsY complex formation in vitro (13, 19) and block protein targeting in vitro and in vivo (13, 20). Hydroxyl radical probing experiments further suggested that the tetraloop is located near the Ffh–FtsY heterodimer interface (21), suggesting that it could interact with the Ffh or FtsY GTPase.
In addition to the SRP RNA, the cargo for the SRP, RNCs with strong signal sequences, further accelerate the SRP–FtsY interaction 100- to 400-fold (22). Together, the SRP RNA and the cargo raise the SRP–FtsY assembly rate constant to over 4 × 106 M-1 s-1 (22), a range appropriate to support cotranslational protein targeting. The molecular mechanism by which the cargo stimulates SRP–FtsY complex assembly is not understood. Nevertheless, the SRP RNA, through its close proximity to the signal sequence binding site (6) and the ribosome (23–26) and its ability to communicate with the GTPases, provides a likely candidate to mediate the cargo-induced stimulation. This is supported by the recent observation that a signal peptide stimulates SRP–FtsY complex formation only in the presence of the SRP RNA (27).
Here we show that a highly conserved basic residue, Lys399 on FtsY, interacts with the SRP RNA to stabilize the early intermediate and the transition state of SRP–FtsY complex assembly. These data provide direct evidence for a transient tether mechanism to explain the catalytic effect of the SRP RNA on this protein–protein interaction. Further, mutation of FtsY–Lys399 largely abolishes the ability of cargo to stimulate the SRP–FtsY interaction, indicating that interaction of the RNA tetraloop with FtsY–Lys399 provides a key contact that mediates the cargo-induced stimulation of SRP–FtsY complex assembly.
Results
Basic Residues Away from the Ffh–FtsY Dimer Interface Enable Efficient SRP-Receptor Complex Assembly.
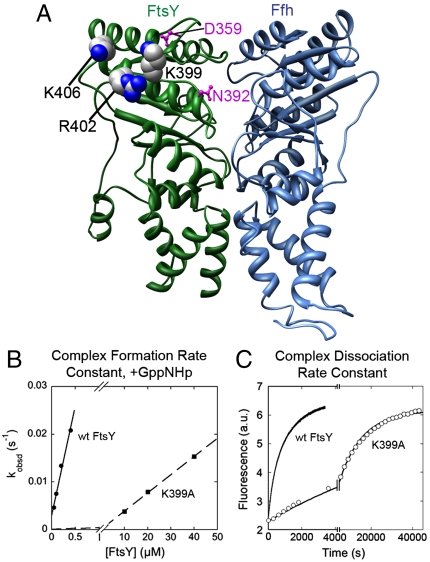
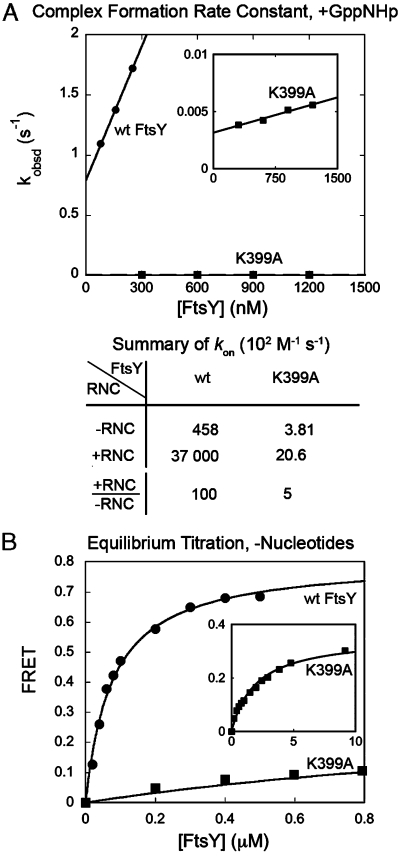
In a previous structure-function analysis, the results of biochemical analyses agreed well with the crystal structure of the Ffh–FtsY complex: Mutations of conserved residues at or near the dimer interface severely disrupted complex formation, whereas mutation of residues away from the interface had no significant effects (9, 28). A notable exception to this were the mutants K399A (9, 28), R402A, and K406A. These residues are on the lateral surface of FtsY and together form a continuous patch of basic surface on the Gα2-helix (Fig. 1A, in spacefill). Although these residues are ≥15 Å away from the heterodimer interface, their mutations severely disrupted the kinetics of SRP–FtsY complex formation. In a well-established GTPase assay, the rate constant of the reaction: SRP + FtsY → products (kcat/Km) was reduced 82-fold for mutant FtsY–K399A (Fig. S1, open vs. closed circles). FtsY–R402A and K406A also caused significant, albeit modest reductions in kcat/Km (6- and 5-fold, respectively; Fig. S1). As the kcat/Km values in this assay equal the association rate constants to form a stable SRP–FtsY complex (18), these results suggest that these basic residues, especially FtsY–Lys399, play a crucial role in SRP–FtsY complex formation.
Fig. 1.
FtsY–Lys399 plays a crucial role in SRP–FtsY complex assembly. (A) The basic residues on the FtsY Gα2-helix are highlighted in spacefill in the crystal structure of the Thermus aquaticus Ffh–FtsY NG-domain complex (PDB: 1RJ9). FtsY residues previously identified to be near the RNA tetraloop (21) are highlighted in magenta. (B) Rate constants of SRP–FtsY complex assembly, measured using FRET as described in Materials and Methods. Linear fits of data gave complex formation rate constants (kon) of 4.58 × 104 M-1 s-1 for wild-type FtsY (•) and 3.81 × 102 M-1 s-1 for mutant FtsY–K399A (▪). (C) Rate constants of SRP–FtsY complex disassembly (koff), determined by pulse-chase experiments as described (13, 22). Nonlinear fits of the time courses for loss of FRET from the complex (or gain in donor fluorescence) gave koff values of 1.46 × 10-3 s-1 for wild-type FtsY (•) and 4.05 × 10-5 s-1 for mutant FtsY–K399A (∘).
To independently test this conclusion, we directly measured SRP–FtsY complex formation using fluorescence resonance energy transfer (FRET) between donor and acceptor labeled Ffh and FtsY (13). To uncouple complex formation from GTP hydrolysis, the nonhydrolyzable GTP analog 5′-guanylylimido-diphosphate (GppNHp) was used to assemble a stable SRP–FtsY complex. The complex assembly rate constant was 120-fold slower with mutant FtsY–K399A than with wild-type FtsY (Fig. 1B), providing direct evidence that FtsY–Lys399 plays an essential role in accelerating complex formation. Mutant FtsY–K399A also had a deleterious effect on complex disassembly, reducing the dissociation rate constant 36-fold (Fig. 1C). In contrast, the equilibrium stability of the SRP–FtsY complex, either calculated from the dissociation and association rate constants or directly measured by equilibrium titration (Fig. S2), was reduced only approximately 3-fold by the FtsY–K399A mutation [Fig. S2 and (17)].
FtsY–Lys399 Functionally Interacts with the 4.5S RNA Tetraloop.
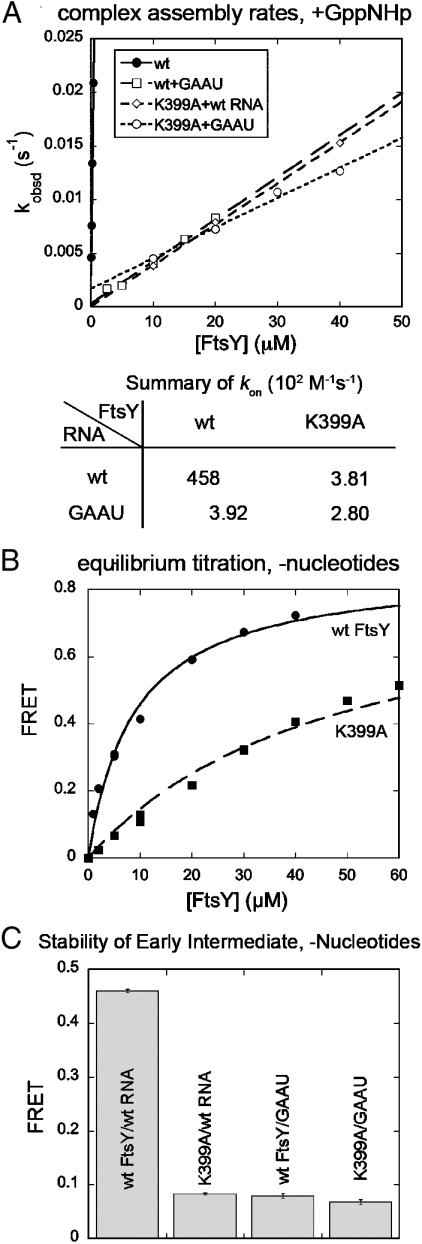
The roles of FtsY–Lys399 are reminiscent of those of the SRP RNA; i.e., accelerating both complex formation and dissociation without significantly changing the equilibrium stability of the complex. This suggests that Lys399 interacts, directly or indirectly, with the SRP RNA tetraloop to enable acceleration of complex assembly. If this were true, then the effects of FtsY–Lys399 and the RNA tetraloop should be cooperative; i.e., they contribute to complex assembly only when both motifs are present. To test this hypothesis, we compared the effect of the FtsY–K399A mutation with wild-type SRP and mutant SRP(GAAU), in which the GGAA tetraloop was replaced by GAAU and the SRP–FtsY complex formation rate constant was reduced 120-fold (Fig. 2A, squares). Indeed, FtsY–Lys399 no longer contributes to complex formation in the presence of mutant SRP(GAAU) (Fig. 2A). As a control, mutation of other residues that disrupt the SRP–FtsY interaction, such as FtsY–E475K or FtsY–T307W (9, 28), still slowed down complex assembly with SRP(GAAU) (Fig. S3). These results strongly support a functional interaction between FtsY–Lys399 and the RNA tetraloop in the transition state of complex assembly.
Fig. 2.
FtsY–Lys399 interacts with the SRP RNA tetraloop. (A) Effects of the FtsY–K399A and RNA(GAAU) mutations on the rate of stable complex formation, determined as described in Materials and Methods. (B) The FtsY–K399A mutation destabilizes the GTP-independent early intermediate. Nonlinear fits of the equilibrium titrations gave Kd values of 8.85 μM for wild-type FtsY (•), and ≥48.4 μM for mutant FtsY–K399A (▪). (C) Effects of the FtsY–K399A and RNA(GAAU) mutations on the stability of the early complex. FRET values were measured with 10 μM FtsY.
We recently found that the SRP RNA stabilizes the early intermediate that precedes the stable SRP–FtsY complex, and this stabilization directly correlates with the ability of the RNA to accelerate complex assembly (13). If FtsY–Lys399 mediates the RNA-induced acceleration of complex formation, then FtsY–Lys399 should also play an important role in stabilizing the early intermediate. To test this possibility, we isolated the early intermediate by assembling the complex in the absence of nucleotides (13), and monitored its formation using FRET. The early complex formed by FtsY–K399A had an estimated Kd value of ≥48 μM, over 6-fold weaker than that formed by wild-type FtsY (Fig. 2B). Further, whereas either the FtsY–K399A or the SRP(GAAU) mutation alone significantly destabilized the early complex, these mutations did not cause additional defects when the other mutation was already present (Fig. 2C). Thus, FtsY–Lys399 also functionally interacts with the RNA tetraloop to stabilize the early intermediate.
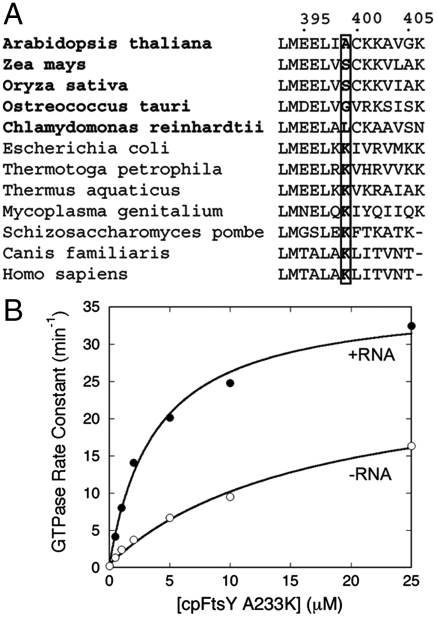
An A → K Reversal Mutant of Chloroplast FtsY Allows its Interaction with Ffh to be Stimulated by the SRP RNA.
To provide independent evidence that FtsY–Lys399 interacts with the SRP RNA, we explored an RNAless SRP pathway in chloroplast (cpSRP). This pathway uses close homologues of the SRP and FtsY GTPases (cpSRP54 and cpFtsY, respectively), but cpSRP54 does not bind the SRP RNA (29). We recently showed that the NG domains of the SRP and FtsY GTPases can interact with their heterologous binding partners across species (30). However, the interaction of Escherichia coli Ffh with cpFtsY cannot be stimulated by the SRP RNA (30), suggesting that cpFtsY has lost the structural element that responds to the SRP RNA. Sequence analysis showed that Lys399 is highly conserved among prokaryotic and eukaryotic SRP receptors, but is replaced by uncharged amino acids in cpFtsYs (Fig. 3A). We therefore reasoned that, if FtsY–Lys399 interacts with the SRP RNA, then mutation of the corresponding Ala233 to lysine in Arabidopsis thaliana cpFtsY should allow its interaction with Ffh to be stimulated by the SRP RNA.
Fig. 3.
The cpFtsY–A233K reversal mutation allows complex formation with cpFtsY to be stimulated by the SRP RNA. (A) Sequence alignment of FtsY homologues. The residue numbering is for E. coli FtsY. Bold highlights the cpFtsYs. (B) GTPase assay to measure the interaction between cpFtsY-A233K and E. coli Ffh in the absence (∘) and presence (•) of SRP RNA. Nonlinear fits of data gave kcat/Km values of 9.85 × 106 and 1.26 × 106 M-1 min-1 with and without the SRP RNA, respectively. For comparison, the kcat/Km value of the reaction of wild-type cpFtsY with Ffh is 1.77 × 106 M-1 min-1 (30).
We therefore tested the ability of the reversal mutant cpFtsY–A233K to interact with E. coli Ffh with and without the SRP RNA (Fig. 3B). Values of kcat/Km in the GTPase assay were used as indices for the rate of stable complex formation. Mutant cpFtsY–A233K interacted with and stimulated the GTPase activity of Ffh, with rate constants comparable to that of wild-type cpFtsY (Fig. 3B, open circles) (30). Interestingly, whereas the reaction of wild-type cpFtsY with Ffh is RNA-independent (30), the SRP RNA stimulated the reaction of mutant cpFtsY–A233K with Ffh 8-fold (Fig. 3B, closed circles). Although we did not restore the full extent of stimulation by the SRP RNA, this was not unexpected given the heterologous nature of the Ffh–cpFtsY interaction and the evolutionary divergence in the precise location or orientation of cpFtsY residue 233 that could have occurred in the RNAless cpSRP pathway. The ability of the SRP RNA to stimulate the interaction of Ffh with the cpFtsY–A233K reversal mutant provided independent evidence that the SRP RNA interacts with FtsY–Lys399 to stimulate complex formation.
Electrostatic Interactions Drive RNA-Stimulated Complex Assembly.
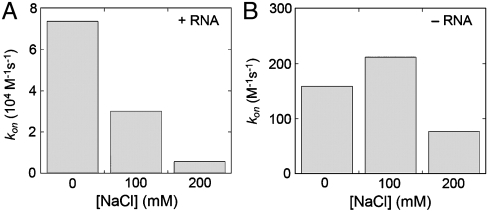
The high density of positive charge on and surrounding FtsY–Lys399 (Fig. 1A) raises the possibility that electrostatic interactions of this basic site with the backbone phosphates of the SRP RNA play a major role in accelerating SRP–FtsY complex assembly. A hallmark of macromolecular interactions driven by electrostatic attractions is that their rate constants are highly sensitive to ionic strength (31). We therefore tested the effect of ionic strength on the SRP–FtsY complex assembly kinetics using the FRET assay (Fig. 4 and Fig. S4). Indeed, the complex assembly rate constants decreased 15-fold when the ionic strength was increased from 50 to 250 mM (Fig. 4A and Fig. S4A). Although we could not vary the ionic strength above 250 mM without disrupting protein structure, the ionic strength dependence of complex assembly kinetics that we observed within this range was comparable to those found in systems where electrostatics plays a major role in macromolecular association (31). In contrast, complex assembly in the absence of the SRP RNA was affected less than 2-fold (Fig. 4B and Fig. S4B). These results strongly suggest that the rate-limiting step for complex assembly is strongly dictated by electrostatic interactions with the SRP RNA.
Fig. 4.
Effect of ionic strength on the Ffh–FtsY complex assembly kinetics in the presence (A) or absence (B) of the SRP RNA, determined using the FRET assay. All the reactions also contain 50 mM K+ and 2 mM Mg2+, therefore, the ionic strength was increased from 50 to 250 mM. The complex assembly rate constants were obtained from the data in Fig. S4.
FtsY–Lys399 Mediates the Cargo-Induced Stimulation of SRP–FtsY Complex Assembly.
A model cargo for SRP, RNC exposing the FtsQ signal sequence (RNCFtsQ), accelerates SRP–FtsY complex formation another 100- to 400-fold (22). Importantly, in the presence of RNCFtsQ mutant FtsY–K399A had an even greater deleterious effect, reducing the complex assembly rate constant 1800-fold (Fig. 5A). The stimulatory effect from the cargo was reduced to only approximately 5-fold in the presence of mutant FtsY–K399A, in contrast to the 100-fold stimulatory effect of RNCFtsQ in the presence of wild-type FtsY (Fig. 5A, Lower) (22). Thus FtsY–Lys399 plays a crucial role in mediating the cargo-induced stimulation of SRP–FtsY complex assembly.
Fig. 5.
Mutation of FtsY–Lys399 diminishes the stimulatory effect of RNC on SRP–FtsY complex assembly. (A) Effect of FtsY–K399A on the rate constants of complex formation with cargo-loaded SRP. The inset shows the data with FtsY–K399A on an expanded scale. (B) Effect of FtsY–K399A on the equilibrium stability of the RNC–SRP–FtsY early targeting complex. The inset shows the data with FtsY–K399A on an expanded scale. Nonlinear fits of data gave Kd values and FRET end points of 76.5 nM and 0.72 for wild-type FtsY (•), and approximately 2 μM and 0.35 for mutant FtsY–K399A (▪).
We showed that RNCFtsQ substantially stabilizes the early intermediate (22), and this stabilization is a key to the cargo-induced stimulation of SRP–FtsY complex assembly. We therefore tested whether FtsY–Lys399 is required to stabilize the RNC–SRP–FtsY early targeting intermediate. With wild-type FtsY, cargo-loaded SRP formed a stabilized early complex with a Kd value of 76 nM (Fig. 5B, circles) (22). In contrast, the early targeting intermediate formed by FtsY–K399A was 26-fold less stable than that by wild-type FtsY (Fig. 5B, squares). Moreover, the FRET value of the RNC–SRP–FtsY early intermediate was approximately 0.3 with mutant FtsY–K399A (Fig. 5B, Inset), much lower than that with wild-type FtsY (approximately 0.7; Fig. 5B, circles) (22). This observation, together with the slower complex assembly kinetics with FtsY–K399A, suggests that the early targeting intermediate formed by FtsY–K399A is in a different conformation and likely mispositioned. Thus FtsY–Lys399 is crucial for stabilizing and properly orienting the GTPases in the early targeting intermediate. Together, these results strongly suggest that FtsY–Lys399 is essential for mediating many of the cargo-induced allosteric regulations on the SRP–FtsY GTPase complex. Consistent with its crucial roles, mutant FtsY–K399A severely inhibited the targeting and translocation of SRP-dependent protein substrates across the microsomal membrane (Fig. S5) (32).
Discussion
The SRP RNA is universally conserved and essential for cotranslational protein targeting. An important role of this RNA is to accelerate the interaction between the SRP and SRP receptor GTPases, allowing them to form a stable complex at rates suitable for cotranslational protein targeting. This is the first example of an RNA accelerating a protein–protein interaction, and the precise mechanism underlying this unprecedented catalytic effect was not completely understood. Here we showed that FtsY–Lys399 on the lateral surface of the FtsY G-domain provides a key site that mediates the SRP RNA-induced stimulation of complex assembly. Further, this site also provides a key link that couples the binding of cargo to efficient SRP–receptor interactions.
How does FtsY–Lys399 mediate the stimulatory effect of the SRP RNA? Although several mechanisms are possible, a direct interaction between the RNA tetraloop and FtsY–Lys399 provides the simplest and most likely model. This is supported by multiple evidences. (i) Our results here established a functional link between the SRP RNA tetraloop and FtsY–Lys399, as their effects were only observed when both sites are present and functional. Further, both sites affect the same stages of the SRP–FtsY interaction, stabilizing the early intermediate and the transition state for complex assembly, without affecting other stages. (ii) Structural probing experiments showed that the RNA tetraloop can gain close proximity to the FtsY G-domain in the SRP–FtsY complex (21). Residue 359, adjacent to the ϵ-amino group of Lys399 (Fig. 1A), cleaved the RNA tetraloop when tethered with Fe-EDTA. Residue 392 (Fig. 1A) cleaved nucleotides immediately preceding the tetraloop. (iii) Comparisons with the RNAless cpSRP system further support a direct Lys399–RNA interaction. In cpSRP, the M-domain of cpSRP54 replaces the SRP RNA to accelerate its complex assembly with cpFtsY (30). Intriguingly, the SRP RNA and the cpSRP54 M-domain only stimulate complex assembly with their homologous SRP receptors (30). This specificity strongly suggests that specific sites have coevolved in FtsY (or cpFtsY) that allow each receptor to interact with the SRP RNA or the M-domain in their respective pathway. The findings here that Lys399 is conserved among cytosolic SRP receptors but diverged in the cpFtsYs, and that an A → K reversal mutation of the corresponding residue in cpFtsY restores the RNA-stimulation of complex assembly strongly supports FtsY–Lys399 as the RNA-interaction site. Finally, the high density of positive charges on and surrounding Lys399 provides an attractive site for electrostatic interactions with the RNA backbone, and the rate constants of SRP–FtsY complex assembly exhibits a strong ionic strength dependence consistent with a major role of electrostatic interactions in complex assembly. Alternative models to explain the effects of FtsY–Lys399 are possible, but such models would invoke a role of FtsY–Lys399 in mediating rearrangements that, in turn, allows the SRP RNA to interact with another site.
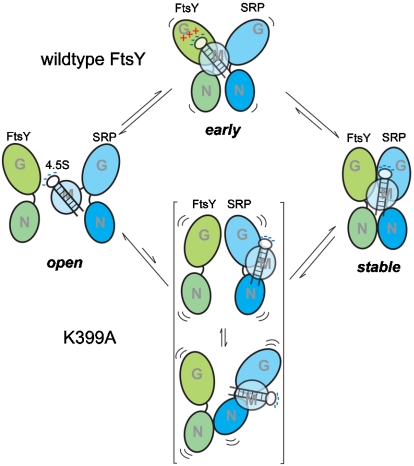
A direct interaction between the SRP RNA and FtsY–Lys399 provides strong support for the transient tether model, and suggests a simple and elegant mechanism to account for the catalytic effect of the SRP RNA on SRP–FtsY complex formation (Fig. 6). Because free Ffh and FtsY by themselves exist in conformations suboptimal for stable complex assembly (28, 33), their initial association to form the early intermediate, though rapid (13), is not sufficient to give a stable complex. The conformational changes required to form the stable complex occur on a much slower time scale (0.5–1 s) than the lifetime of the early intermediate (< 16 ms) (13). The SRP RNA, by interacting with FtsY–Lys399 in the early intermediate, can temporarily hold both proteins together and prevent their premature dissociation (Fig. 6, upper vs. lower pathways). This prolongs the lifetime of the intermediate, thus increasing the probability that a successful rearrangement takes place before the intermediate disassembles. This tethering interaction might also restrict the translational and rotational degrees of freedom with which the two GTPases explore different conformations (Fig. 6, upper vs. lower pathways) and thus facilitates their subsequent rearrangement. Once the stable complex is formed, the interaction between the RNA tetraloop and Lys399 likely dissolves (Fig. 6), as the RNA tetraloop does not significantly affect the stability of the final stable complex.
Fig. 6.
Model for the role of RNA tetraloop and FtsY–Lys399 on SRP–FtsY complex assembly, as described in the text. The upper panel depicts the complex assembly reaction with the assistance from the transient interaction between the RNA tetraloop and FtsY–Lys399, and the lower panel depicts the process in the absence of such a tethering interaction.
Macromolecular assemblies often begin with transient intermediates formed by inelastic collisions in which both binding partners engage in relative rotatory diffusions to bring the correct interacting surfaces into appropriate opposition. The principle that formation of such intermediates could reduce the dimension of diffusional search and thus provide significant rate enhancements was supported by theoretical and experimental work (34–37). Our finding here, that a transient tether can be used to increase the lifetime of transient intermediates and thereby accelerate protein–protein interactions, represents a natural extension of this principle and a simple and effective mechanism to enhance the kinetics of macromolecular recognition. This mechanism bears some resemblance to facilitated target site binding by the lac repressor and other transcriptional factors (38), in that in both cases the initial interactions are low affinity but serves to effectively reduce the dimension of “search” to achieve the final, correct interaction. In principle, such a transient tether can be provided by either a nucleic acid or protein molecule (see below). Nevertheless, the polyanionic nature of nucleic acids could allow them to engage in relatively long-range electrostatic interactions that do not have highly stringent stereochemical requirements, and hence might make them particularly suitable for providing transient tethering interactions that need to be broken at later stages.
The SRP and SRP receptor belong to a unique family of GTPases activated by nucleotide-dependent dimerization (GADs) (39). Intriguingly, although direct interaction between the GTPase sites occur or have been proposed in almost all members of this family, dimerization of these GTPases are often mediated at least in part by motifs away from the GTPase module. For example, dimerization of MnmE is driven primarily by interactions between its N-terminal domains whereas the GTPase sites transiently contact one another during the transition state of GTP hydrolysis (40). In a bacterial dynamin-like protein, motifs away from the GTPase domain—the paddle and tip domains—engage in more extensive intermolecular interactions than the GTPase modules at the dimer interface (41). We speculate that the use of tethering interactions from sites away from the GTPase active site could be an important mechanism that facilitates “kissing” between G domains in the GAD family of proteins.
The SRP–FtsY interaction is further accelerated 100- to 400-fold by the cargo (22). Together, the combined effect of the cargo and the SRP RNA brings the SRP–FtsY interaction kinetics to a range (> 106 M-1 s-1) appropriate for cotranslational protein targeting in the cell. Here we found that disruption of the RNA tetraloop–Lys399 interaction largely abolishes the cargo-induced stimulation of the GTPase interactions, demonstrating that this interaction provides a key contact that mediates the cargo-induced acceleration of SRP–FtsY complex assembly. Further, with cargo-loaded SRP, FtsY–Lys399 has an even greater effect in stabilizing a productive early intermediate and in accelerating stable SRP–FtsY complex assembly, suggesting that the intrinsic energetic contribution of the RNA tetraloop–Lys399 interaction is significantly larger than that observed with free SRP. It is possible that the cargo could preorganize the SRP such that the RNA tetraloop is prepositioned to interact with FtsY–Lys399, thus activating the SRP for complex formation. In this way, the RNA tetraloop–Lys399 interaction provides a key link that transmits the information about cargo-binding in the SRP M-domain to the SRP and FtsY GTPases, thus turning on their GTPase cycles and driving the rapid delivery of cargo to the target membrane.
Materials and Methods
Material.
E. coli Ffh, FtsY, and SRP RNA were expressed and purified using established procedures (18). Mutant proteins and SRP RNA were constructed using the QuickChange mutagenesis protocol (Stratagene). Fluorescent dyes were from Invitrogen. RNCFtsQ was prepared and purified as described (42).
GTPase Assay.
The assay to measure the stimulated GTP hydrolysis reaction between SRP and FtsY have been described (18). In general, reactions contained 100 nM Ffh, 200 nM 4.5S RNA (where applicable), 100–200 μM GTP (doped with γ-32P-GTP), and varying concentrations of wild-type or mutant FtsY.
Fluorescence Experiments.
Single cysteine mutants of Ffh and FtsY were labeled with N-(7-dimethylamino-4-methylcoumarin-3-yl)maleimide (DACM) and BODIPY-FL N-(2-aminoethyl)maleimide, respectively, and purified as described (13). Fluorescence measurements were carried out on a FluoroLog-3-22 spectrofluorometer (Jobin-Yvon) in SRP buffer [50 mM KHEPES (pH 7.5), 150 mM KOAc, 2 mM Mg(OAc)2, 2 mM DTT, and 0.01% Nikkol]. The rate and equilibrium constants for SRP–FtsY complex formation and dissociation were measured as described (13, 22).
Translocation Assay.
The protein targeting efficiency of wild-type FtsY and mutant FtsY-K399A were determined by a cotranslational translocation assay using preprolactin (pPL) as the model substrate, as described previously (32, 43). Reactions were carried out using 1 μM FtsY and 2 eq of trypsin-digested, salt-washed ER microsomal membrane.
Supplementary Material
Acknowledgments.
We thank William M. Clemons, Nenad Ban, Christiane Schaffitzel, and members of the Shan group for helpful comments on the manuscript. This work was supported by National Institutes of Health Grant GM078024 (S.S.). S.S. was supported by a career award from the Burroughs Welcome Foundation, the Beckman Young Investigator award, the Packard and Lucile Award in Science and Engineering, and the Henry Dreyfus Teacher-Scholar Award.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002968107/-/DCSupplemental.
References
- 1.Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic-reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 2.Pool MR, et al. Distinct modes of signal recognition particle interaction with the ribosome. Science. 2002;297:1345–8. doi: 10.1126/science.1072366. [DOI] [PubMed] [Google Scholar]
- 3.Walter P, Ibrahimi I, Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981;91:545–50. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilmore R, Blobel G, Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol. 1982;95:463–9. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilmore R, Walter P, Blobel G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J Cell Biol. 1982;95:470–7. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batey RT, et al. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–1239. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- 7.Keenan RJ, et al. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–91. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- 8.Zopf D, et al. The methionine-rich domain of the 54 kd protein subunit of the signal recognition particle contains an RNA binding site and can be crosslinked to a signal sequence. EMBO J. 1990;9:4511–7. doi: 10.1002/j.1460-2075.1990.tb07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egea PF, et al. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- 10.Focia PJ, et al. Heterodimeric GTPase core of the SRP targeting complex. Science. 2004;303:373–7. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flanagan JJ, et al. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J Biol Chem. 2003;278:18628–37. doi: 10.1074/jbc.M300173200. [DOI] [PubMed] [Google Scholar]
- 12.Siegel V, Walter P. The affinity of signal recognition particle for presecretory proteins is dependent on nascent chain length. EMBO J. 1988;7:1769–75. doi: 10.1002/j.1460-2075.1988.tb03007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Kung S, Shan SO. Demonstration of a multistep mechanism for assembly of the SRP. SRP receptor complex: Implications for the catalytic role of SRP RNA. J Mol Biol. 2008;381:581–593. doi: 10.1016/j.jmb.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Focia PJ, et al. Structure of a GDP:AlF4 complex of the SRP GTPases Ffh and FtsY, and identification of a peripheral nucleotide interaction site. J Mol Biol. 2006;360:631–43. doi: 10.1016/j.jmb.2006.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gawronski-Salerno J, Freymann DM. Structure of the GMPPNP-stabilized NG domain complex of the SRP GTPases Ffh and FtsY. J Struct Biol. 2007;158:122–8. doi: 10.1016/j.jsb.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neher SB, et al. SRP RNA controls a conformational switch regulating the SRP–SRP receptor interaction. Nat Struct Mol Biol. 2008;15:916–923. doi: 10.1038/nsmb.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peluso P, et al. Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science. 2000;288:1640–1643. doi: 10.1126/science.288.5471.1640. [DOI] [PubMed] [Google Scholar]
- 18.Peluso P, et al. Role of SRP RNA in the GTPase cycles of ffh and FtsY. Biochemistry. 2001;40:15224–15233. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- 19.Jagath JR, et al. Important role of the tetraloop region of 4.5S RNA in SRP binding to its receptor FtsY. RNA. 2001;7:293–301. doi: 10.1017/s1355838201002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siu FY, Spanggord RJ, Doudna JA. SRP RNA provides the physiologically essential GTPase activation function in cotranslational protein targeting. RNA. 2007;13:240–250. doi: 10.1261/rna.135407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spanggord RJ, et al. RNA-mediated interaction between the peptide-binding and GTPase domains of the signal recognition particle. Nat Struct Mol Biol. 2005;12:1116–1122. doi: 10.1038/nsmb1025. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, et al. Multiple conformational switches in a GTPase complex control co-translational protein targeting. Proc Natl Acad Sci USA. 2009;106:1754–1759. doi: 10.1073/pnas.0808573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halic M, et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–14. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- 24.Halic M, et al. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006;444:507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- 25.Rinke-Appel J, et al. Crosslinking of 4.5S RNA to the Escherichia coli ribosome in the presence or absence of the protein Ffh. RNA. 2002;8:612–625. doi: 10.1017/s1355838202020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaffitzel C, et al. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature. 2006;444:503–506. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- 27.Bradshaw N, et al. Signal Sequences Activate the Catalytic Switch of SRP RNA. Science. 2009;323:127–130. doi: 10.1126/science.1165971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan SO, Stroud RM, Walter P. Mechanism of association and reciprocal activation of two GTPases. PLoS Biol. 2004;2:1572–1581. doi: 10.1371/journal.pbio.0020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richter CV, Trager C, Schunemann D. Evolutionary substitution of two amino acids in chloroplast SRP54 of higher plants cause its inability to bind SRP RNA. FEBS Lett. 2008;582:3223–9. doi: 10.1016/j.febslet.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 30.Jaru-Ampornpan P, Nguyen TX, Shan SO. A distinct mechanism to achieve efficient signal recognition particle (SRP)–SRP receptor interaction by the chloroplast srp pathway. Mol Biol Cell. 2009;20:3965–73. doi: 10.1091/mbc.E08-10-0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schreiber G, Fersht AR. Rapid, electrostatically assisted association of proteins. Nat Struct Biol. 1996;3:427–31. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- 32.Shan SO, Chandrasekar S, Walter P. Conformational changes in the GTPase modules of the signal reception particle and its initiation of protein translocation. J Cell Biol. 2007;178:611–620. doi: 10.1083/jcb.200702018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shan SO, Walter P. Induced nucleotide specificity in a GTPase. Proc Natl Acad Sci USA. 2003;100:4480–4485. doi: 10.1073/pnas.0737693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–30. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 35.Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein–protein association. Nature. 2006;444:383–6. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 36.Volkov AN, et al. Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR. Proc Natl Acad Sci USA. 2006;103:18945–50. doi: 10.1073/pnas.0603551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou HX. Brownian dynamics study of the influences of electrostatic interaction and diffusion on protein–protein association kinetics. Biophys J. 1993;64:1711–26. doi: 10.1016/S0006-3495(93)81543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Hippel PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–8. [PubMed] [Google Scholar]
- 39.Gasper R, et al. It takes two to tango: Regulation of G proteins by dimerization. Nat Rev Mol Cell Biol. 2009;10:423–9. doi: 10.1038/nrm2689. [DOI] [PubMed] [Google Scholar]
- 40.Meyer S, et al. Kissing G domains of MnmE monitored by x-ray crystallography and pulse electron paramagnetic resonance spectroscopy. PLoS Biol. 2009;7:e1000212. doi: 10.1371/journal.pbio.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Low HH, Lowe J. A bacterial dynamin-like protein. Nature. 2006;444:766–9. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- 42.Schaffitzel C, Ban N. Generation of ribosome nascent chain complexes for structural and functional studies. J Struct Biol. 2007;158:463–471. doi: 10.1016/j.jsb.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 43.Powers T, Walter P. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 1997;16:4880–6. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.