Abstract
One or more of the signaling lymphocytic activation molecule (SLAM) family (SLAMF) of cell surface receptors, which consists of nine transmembrane proteins, i.e., SLAMF1-9, are expressed on most hematopoietic cells. While most SLAMF receptors serve as self-ligands, SLAMF2 and SLAMF4 use each other as counter structures. Six of the receptors carry one or more copies of a unique intracellular tyrosine-based switch motif, which has high affinity for the single SH2-domain signaling molecules SLAM-associated protein and EAT-2. Whereas SLAMF receptors are costimulatory molecules on the surface of CD4+, CD8+, and natural killer (NK) T cells, they also involved in early phases of lineage commitment during hematopoiesis. SLAMF receptors regulate T lymphocyte development and function and modulate lytic activity, cytokine production, and major histocompatibility complex-independent cell inhibition of NK cells. Furthermore, they modulate B cell activation and memory generation, neutrophil, dendritic cell, macrophage and eosinophil function, and platelet aggregation. In this review, we will discuss the role of SLAM receptors and their adapters in Tcell function, and we will examine the role of these receptors and their adapters in X-linked lymphoproliferative disease and their contribution to disease susceptibility in systemic lupus erythematosus.
Keywords: XLP, SAP, SH2D1A, SLAM family, Systemic lupus erythematosus
The nine SLAM family receptors
Signaling lymphocytic activation molecule (SLAM) family (SLAMF) receptors are type I transmembrane glycoproteins, except SLAMF2, which has a glycosylphosphatidylinositol membrane anchor [1–5]. While the ectodomains of eight SLAMF receptors are comprised of an N-terminal V-Ig domain and a C-terminal C2-Ig domain, SLAMF3 consists of two V-Ig/C2-Ig sets.
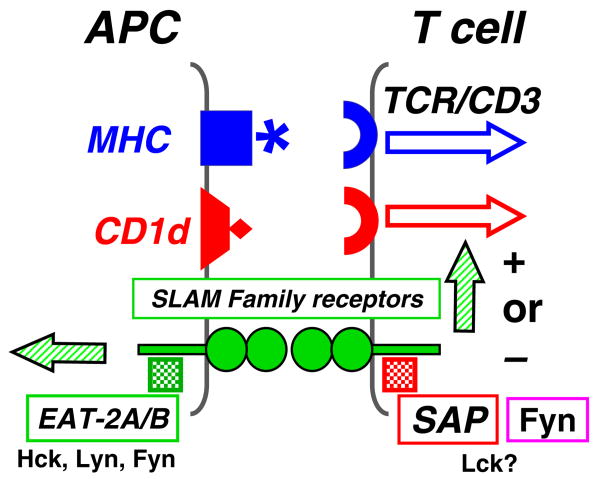
SLAMF2 is a receptor for SLAMF4, but the remaining SLAMF (1, 3, 5, 6, 7, 8, and 9) represent self-ligands and their ectodomains interact in a homophilic fashion ([6, 7] and unpublished). Interestingly, SLAMF1 also serves as one of the two known receptors for measles virus, and SLAMF2 interacts with CD2 and FimH (Table 1). The specificity of the self-ligand interactions of most of the SLAMF receptors is explained by the known structures of their extracellular domains [8, 9]. For example, at the interface of the two V-Ig in the SLAMF6 homodimer, the β-sheets contain multiple residues, which contribute to the stability of the interaction. Similarly, the structure of SLAMF5 explains the orthogonal association of two SLAMF5 molecules but precludes an interaction with SLAMF6. The orthogonal association of two SLAMF5 or of two SLAMF6 molecules enables these complexes to localize to the immunological synapse [8, 9]. Specific SLAMF members are thought to function as costimulatory molecules in the conventional T cell, natural killer (NK) T (NKT) cell, innate CD8+ T cell, or NK cell synapse through their homophilic or heterophilic interactions [10–12] (Fig. 1).
Table 1.
SLAM family receptors, their ligands, and binding partners
| SLAM family | Synonym | Ligands | Binds SAP EAT-2 | Effectors |
|---|---|---|---|---|
| SLAMF1 | CD150, SLAM | SLAMF1, measles virus | + | Fyn, Lck, SHIP1, Dok1, PKCθ, Akt |
| SLAMF2 | CD48 | SLAMF4, CD2, Enterobac. FimH1 | − | Fyn, Lck |
| SLAMF3 | CD229 | SLAMF3 | + | Fyn, Lck, ERK, AP2, Grb2 |
| SLAMF4 | CD244 | SLAMF2 | + | Fyn, Lck, LAT, PI3K, Vav1, SHIP1, cCbl, ERK, p38, SHP1, SHP2 |
| SLAMF5 | CD84 | SLAMF5 | + | Fyn, Lck |
| SLAMF6 | NTB-A (h), Ly108 (m) | SLAMF6 | + | Fyn, Lck, PLCγ, PI3K, SHP1, cCbl, Vav1 |
| SLAMF7 | CD319 | SLAMF7 | + | Fyn, Lck, PLCγ, Vav1, PI3K |
| SLAMF8 | BLAME | SLAMF8 | − | ? |
| SLAMF9 | SF2001 | SLAMF9? | − | ? |
Fig. 1.
SLAM family receptors in the immune synapse. The mostly homophilic interactions between SLAMF receptors subserve their role as costimulatory molecules. The binding of the SLAMF receptors to their ligands induces tyrosine phosphorylation of ITSM motifs in their cytoplasmic tail to which the adapters SAP or EAT-2 bind subsequently. SAP is mostly expressed by T cells, whereas EAT-2 is primarily expressed in APC
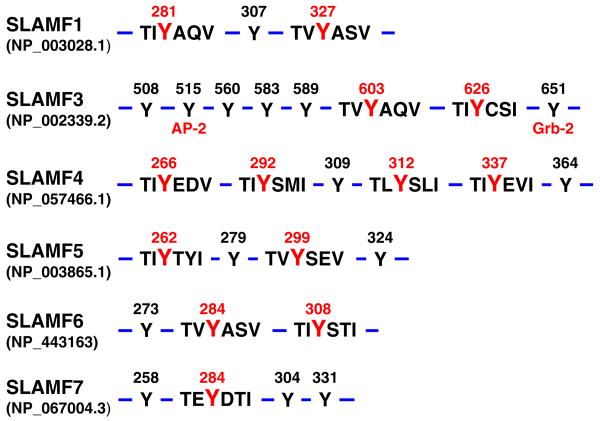
The cytoplasmic tails of SLAMF1 and SLAMF3 through SLAMF7 contain several intracellular tyrosine-based switch motifs (ITSM) as well as a number of other phosphotyrosine (pY) residues. Thus, SLAM-associated protein (SAP) or EAT-2-dependent and EAT-2-independent recruitment of intracellular molecules leads to the inhibitory and stimulatory signals, which ensue upon engagement of these receptors. Integration of signaling networks determines the outcome of several effector functions depending on the cell type and state of activation. But, SAP/EAT-2-independent signal transduction events have also been observed, e.g., Grb-2 binds to the most C-terminal pY of SLAMF3 and plays a role in endocytosis of this receptor in conjunction with TCR/CD3 [13, 14]. In addition, some receptors may play a dominant role in a given cell, e.g., SLAMF6 appears to be a major signaling entity in activation-induced cell death (AICD) and/or restimulation-induced cell death (RICD) of CD8+ T cells, while other SLAMF receptors are expressed in the same cell. The notion that both SLAMF receptors and their protein isoforms are often co-expressed in the same cell and exert partially overlapping functions remains one of the challenges of this field.
The SLAM-associated protein and the X-linked lymphoproliferative syndrome
X-linked lymphoproliferative syndrome (XLP) is an extremely rare primary immunodeficiency, which affects only males and is at first recognized as a defective immune response to Epstein–Barr virus (EBV) [15]. The majority (60%) of XLP patients infected with EBV develop fatal or fulminant infectious mononucleosis, which is caused by a dysregulated immune response, resulting in a large polyclonal proliferation of T cells and B cells. This leads to lymphoid infiltration of several organs such as the liver, kidney, thymus, and bone marrow causing organ failure. The extensive destruction of liver and bone marrow often results in fulminant hepatitis and virus-associated hemophagocytic syndrome [6]. Patients often exhibit low serum levels of IgG and increased levels of IgA and IgM. Almost all lymphomas found in XLP patients are of B cell origin and are located along the gastrointestinal tract. Of them, 53% are Burkitt's, 18% immunoblastic, 12% large noncleaved, 12% small cleaved or mixed cell, and 5% unclassifiable lymphomas [16–18]. XLP patients are usually asymptomatic prior to virus exposure, but after recurrent viral infections, they develop dysgammaglobulinemia [18–20] (Table 2).
Table 2.
Disease manifestations of XLP patients and related phenotypes in SAP−/− mice
| Disease manifestations in XLP patients | Observations in SAP−/− mice |
|---|---|
| Fatal infectious mononucleosis | |
| Increased numbers of EBV specific CD8+ T cells | Increased numbers of virus specific CTLs and IFN-γ-producing CD8+ cells |
| Reduced AICD of CD8+ T cells | Decreased apoptosis in CD8+ T cells |
| Reduced SLAMF-dependent NK cell killing | Reduced SLAMF-dependent NK cell killing |
| Absence of NKT cells | Absence of NKT cells |
| B cell lymphomas | |
| Dys-gammaglobulinemia | Impaired primary and secondary antibody responses |
| Progressive loss of IgG levels and specific antibody responses | Defective Germinal Center formation due to dysfunctional CD4+ ⇔ B cell interactions |
Mutations in the gene encoding the SAP gene cause XLP
Ten years ago, the SH2D1A gene, which encodes SAP, was found to be defective in XLP patients with macro/microdeletions, splice-site mutations, nonsense mutations, or missense mutations [21–23]. Whereas most XLP patients have mutations in the SH2D1A (SAP) gene, more than 20% express intact SAP but have a mutation in the X-linked inhibitor of apoptosis (XIAP) gene [24–26]. Interestingly, these patients lack NKT cells, a phenotype which was not found in the XIAP-deficient mouse [27].
Missense mutations that disrupt the binding of SAP to SLAM family receptors interfere (a) with the interactions involving the N-terminal amino acids in the ITSM motif, e.g., T53I, (b) binding in the pY pocket, e.g., R32T, C42W, or (c) interactions with the Y+3 amino acid in the hydrophobic cleft, E67D, T68. Interestingly, the mutant T53I binds to the phosphorylated SLAM but fails to bind to SLAMF3 and SLAMF4 [28, 29]. A number of the missense mutations that decrease the half-life of SAP are conserved amino acids in SAP isolated from human, mouse, and other species, e.g., Y7C, S28R, Q99P, P101L, V102G, Y54C, F87S, and I84T [28, 30]. Surprisingly, mutations Y54C and F87S also affect SAP binding to some, but not all the SLAM family receptors, suggesting that the requirement of SAP binding might differ between receptors.
SAP, a single SH2-domain protein, binds with high affinity to SLAM family receptors and is an adapter, which recruits Fyn
Although SH2 domains are highly conserved noncatalytic modules, they exhibit considerable flexibility in the way they bind to their respective pY ligands. SH2 domains associate with the pY motif in a “two-pronged fashion”: a pY-binding pocket and a “specificity determining” region that interacts with an amino acid C-terminal of the pY By contrast, the SH2 domain of SAP interacts with the ITSM motif in a “three-pronged binding mode”, which results in a high affinity binding to the phosphorylated ITSM-based peptide SLAMF1 Y281 (Fig. 2) [21]. This SH2 domain is also unique in that it also binds to the nonphosphorylated ITSM-based peptide SLAMF1 Y281 [21, 31–33]. This “three-pronged binding mode” is caused by the additional interactions involving three residues N-terminal to the SLAMF1 Y281 residue [21, 33].
Fig. 2.
Cytoplasmic tails of human SLAM receptors and the functional ITSM motifs and other docking sites. Cytoplasmic tails of human full-length SLAMF1 and SLAMF3-7 contain several ITSM motifs (tyrosine (Y) in red) which are docking sites for the SAP and EAT-2 adapters. Additional tyrosines are in the cytoplasmic tail of the SLAMF molecules (Y in black) are involved in the association with other effector molecules, e.g., SLAMF3 binds to the adaptor molecules AP-2 and Grb-2. Numbers indicate the tyrosine position relative to the molecule's N-terminal amino acid accession numbers between brackets correspond to Ensembl ID
Because of its high affinity for phosphorylated ITSM motifs, SAP should outcompete SH2 domain proteins that bind to the same phosphotyrosine motif with lesser affinity. Although support for the concept that SAP is an inhibitor of the recruitment of the other SH2 proteins to SLAMF receptors exists [31, 34] (Fig. 4), further experiments are required. The second SLAMF adapter EAT-2 binds to the ITSM motifs using a similar three pronged interaction [35]. However, in cells that express both adapters, SAP seems to outcompete EAT-2.
Fig. 4.
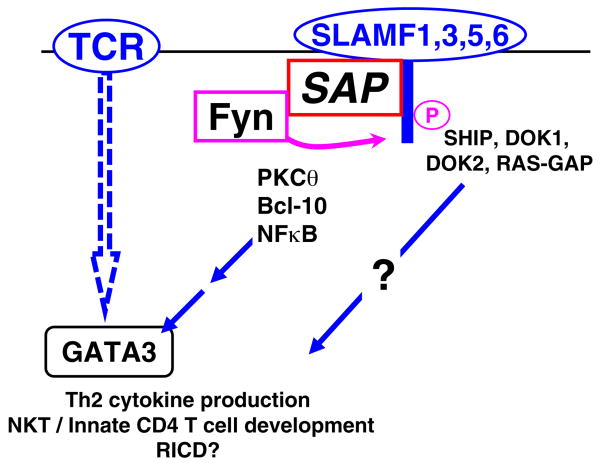
Signal transduction by SLAM family surface receptors in CD4+ T cells. During formation of the immune synapse, clustering of SLAMF receptors brings SAP to its cytoplasmic tails and mediates recruitment and activation of Fyn. Thus, SLAMF receptors modulate TCR signaling by inducing a sustained recruitment of PKCθ and Bcl-10, which in turn leads to the activation of NF-κB and consequently to the production of Th2 cytokines and participation in NKT/innate CD4 T cell development. Phosphorylated tyrosines in the SLAMF cytoplasmic tails triggers the association with SHIP, docking protein 1 (DOK1), DOK2, and RAS-GAP, which may participate in these processes
The second unique and somewhat surprising property of the single SH2 domain protein SAP is that it is an adapter, which couples the protein tyrosine kinase Fyn to the SLAMF receptors. Biochemical studies and structural analyses unveiled the remarkable mechanism of interaction between the SH2 domain of SAP and the SH3 domain of Fyn that directly couples Fyn to SLAM receptor [36, 37]. The structure of the SAP/FynSH3 complex reveals that SAP binding to Fyn domain does not involve canonical SH3 interactions. Instead at the point of interaction, the positively charged SAP surface interacts with negatively charged Fyn SH3 surface at a dissociation constant of 3.45 μM for the entire complex. On the SAP SH2 domain, R78 appears to be particularly critical for the interaction, as it forms dual salt-bridge hydrogen bonds with D100 in Fyn, and its guanidinium moiety stacks with the side chain of W119 in the Fyn SH3 domain. Indeed the R78 mutation of SAP almost abolishes the interaction with Fyn, and conversely, mutations of key residues of Fyn reduce binding to SAP [36, 38]. The site of the SAP SH2 domain, which interacts with Fyn, does not overlap with the SLAMF binding site, and, in fact, SAP mutants that are unable to bind to the SLAMF cytoplasmic tail still bind to Fyn. Consequently, the SH2 domain of SAP is able to bind simultaneously to SLAM and Fyn [36, 38]. Importantly, structural considerations predict that the SAP/FynSH3 complex cannot be formed by the auto-inhibitory form of Fyn. In vitro experiments indicate that SAP can activate the inactive form of Fyn [39, 40]. Thus, the interaction of SAP SH2 with its ligands may control two key events: the tyrosine site location of signaling molecules within the cell and the switch to the inactive/active configuration of Src kinases. These two events allow the activation of Src-like kinases at the right time and in the correct location. It is conceivable that SAP bound to SLAM could bind to a Fyn molecule that has been activated by other mechanisms. The association of the negatively charged surface of Fyn with the positively charged surface of SAP is unique because most of the amino acids involved in the SH3 binding surface of Fyn are not conserved in other Src kinases. As the EAT-2 SH2 domain does not contain the positively charged residues that are requisite for the interaction with the Fyn SH3 domain, the role of EAT-2 in enhancing the phosphorylation of SLAMF receptors [35] requires further clarification.
SAP may also bind directly to Lck, as suggested by the results from in vitro studies and yeast two-hybrid analyses [39]. Although the putative binding of SAP to the kinase domains of Fyn and Lck remains ill-defined, Lck phosphorylates SLAMF1 and SLAMF3-5. The fact though that SLAMF1 phosphorylation is reduced, but not eliminated, in Fyn−/− T cells supports the notion that other Src kinases are involved in SLAMF1 phosphorylation [39]. Comparative studies with SAP-deficient and Fyn-deficient mice clearly indicate that the two proteins control distinct, yet overlapping mechanisms [41–44].
SAP binds to proteins other than SLAMF receptors
Whereas the “three-pronged binding mechanism” increases substrate specificity and the affinity of the SAP SH2 domain for its phosphorylated ITSM ligand, it is otherwise a conventional SH2 domain characterized by a central β-sheet flanked by two alpha helices (αA and αB) and a small β-sheet. Consequently, the SAP SH2 domain could in principle bind to other pY containing peptides. The association of SAP with the SH3 domain of the PAK-interacting protein (β-PIX), a guanine nucleotide exchange factor specific for Rac/Cdc42 GTPase, was recently demonstrated. Similar to the SAP/Fyn interaction, the binding of SAP to β-PIX is mediated by the C-terminal region of the SAP SH2 domain and the β-PIX SH3 domain [45]. The mutant SAPR78A, but not SAPR32Q, fails to bind β-PIX, suggesting that β-PIX and Fyn might compete with each other for SAP binding [45]. SAP recruits β-PIX to SLAMF4, suggesting that β-PIX may mediate some biological functions of the SLAMF receptors [45].
In spite of major progress in our understanding of the functions of SAP and of SLAMF receptors in adaptive immunity, the mechanisms involved in the pathogenesis of XLP need to be better understood. We have only begun to understand the role of SAP in signaling networks, which prevent the extreme proliferation of CD8 cells in response to viruses (“Aberrant CD8+ responses in the absence of SAP” section), the role of SAP in CD4 T cells, which partake in the germinal center (GC) reaction (“Defective humoral response in SAP−/− mice” section), NKT cell development and function (“The role of SLAMF and SAP in the development and function of T cells selected on thymocytes, e.g., NKT cells and innate CD8+ T cells” section) and NK cell killing. Furthermore, as no genotype/phenotype correlation has been observed in XLP patients, modifying genes and environmental factors could very well play a significant role in the diverse disease manifestations, which are caused by mutations in the SH2D1A gene.
Aberrant CD8+ responses in the absence of SAP
Responses of SAP−/− mice to viral infections
CD8+ T cell responses are aberrant in XLP patients, in particular during the fatal infectious mononucleosis, in which EBV-specific CD8 cells proliferate without a contraction of the response. Although EBV, of course, does not infect mouse cells, the study of the response to lymphocytic choriomeningitis virus (LCMV), murine gammaherpesvirus (MHV)-68, and influenza virus in SAP−/− mice has helped us to understand cell-specific defects and in elucidating the molecular mechanisms that are altered in the absence of the adapter. Increased numbers of antigen-specific CD8+ T cells, IFN-γ secreting CD8+ T cells, and elevated production of IFN-γ per cell occur upon infection with LCMV. This response subsides after 2 weeks, as it does in control mice. However, whereas SAP−/− mice were able to survive acute infection, they failed to resolve chronic infection and died most likely because of a lack of antibody responses. [46, 47].
Upon infection with MHV-68, SAP−/− mice also respond by increasing the number of virus-specific CTL to resolve the acute infection as effectively as wt mice. By contrast to an LCMV infection, the chronic infection with MHV-68 is more efficiently controlled by SAP−/− CTL than by wt CTL, as judged by the increased number of virus-specific CD8+ T cells [48, 49]. Strikingly, mouse SAP−/− CD8+ T cells display greater cytotoxicity toward MHV68-infected cells than their wt counterparts. The observations made in SAP−/− mice are in contrast to some analyses of CD8+ T cells from XLP patients, which exhibit defective lysis of EBV-infected B cells and a decrease in numbers of IFN-γ-producing cells [6]. However, as shown in the next section, the general principle that CD8+ T cells proliferate more due to the absence of SAP has been found in human cells as well.
Naïve human and mouse CD8+ T cells express several SLAMF receptors, i.e., SLAMF2, 3, 5, and 6, and upon activation, human and mouse SLAMF1 and SLAMF4 are upregulated. Because CD8+ T cells co-express these receptors, it was not surprising that the functions of CD8 cells isolated from individual SLAMF receptor knockout mice were not different from those of wt mice [50–52]. As discussed in “The role of SLAMF and SAP in the development and function of T cells selected on thymocytes, e.g., NKT cells and innate CD8+ T cells” section, downregulation of both SLAMF1 and SLAMF6 affected NKT cell development, while only minor defects were detected in single knockout mice.
The role of SAP in CD8+ T cell apoptosis
Recent experiments provide direct evidence that the hyperproliferation of SAP-deficient CD8+ T cells is related to impaired apoptosis and RICD.
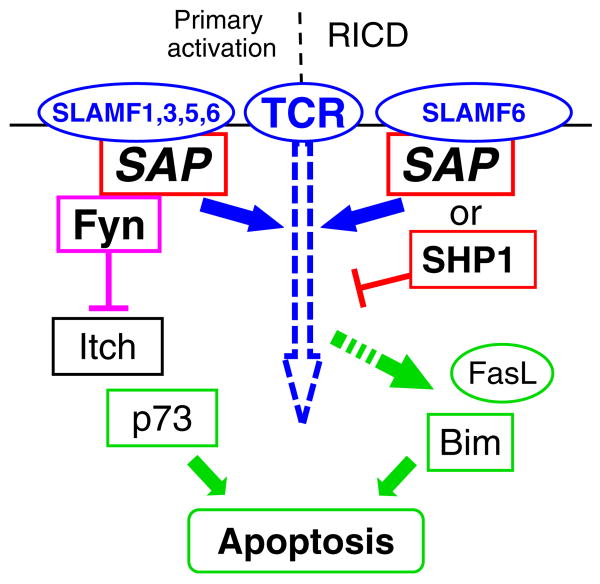
Chen et al. found that ovalbumin-peptide-activated SAP−/− OT-I CD8+ T cells have lower AICD compared to their wt counterparts [53]. Furthermore, the induction of p73, a key mediator of TCR-induced apoptosis through the mitochondrial apoptotic pathway, was significantly reduced at both the mRNA and protein levels in the activated SAP−/− cells. Meanwhile, a reduced level of active caspase 9, the initiator caspase of the mitochondrial apoptotic pathway, was observed in the mutant cells. These observations led to the conclusion that impaired p73 induction in the mitochondrial apoptotic pathway is mainly responsible for the reduction of AICD in activated SAP−/−CD8+ T cells, which may explain the hyperresponsiveness of CD8+ T cells in SAP−/− mice. Although the precise underlying mechanism is still unclear, one can hypothesize that defective Fyn activation in the absence of SAP results in decreased activity of Itch, a ubiquitin ligase shown to be essential in the regulation of p73 protein stability (Fig. 3).
Fig. 3.
A model depicting how apoptosis in CD8+ T cells is controlled by SAP in a Fyn-dependent and Fyn-independent manner. Signaling of SLAMF receptors through SAP binding and subsequent recruitment of Fyn fine-tunes the strength of the TCR signals and contributes in the p73-mediated way of apoptosis during primary T cell responses. In this model, activated Fyn could block the ubiquitin ligase Itch and therefore inhibit the degradation of p73. During the re-activation phase of T cells, SLAMF receptors, e.g., SLAMF6, might promote apoptosis by a SAP-dependent but Fyn-independent mechanism. In this model, SAP dislodges the phosphatase SHP1 from the cytoplasmic tail of SLAM molecules, which facilitates a stronger level of TCR signals sufficient to induce the transcription of RICD mediators such as FasL and Bim. Rectangles: intracellular signaling molecules, ellipses: membrane receptors or their secreted forms (FasL)
SAP also plays a “pro-apoptotic” role in human T and B lymphocyte-derived lines. In clones of a T-ALL tumor cell line, high SAP levels correlate with succumbed AICD. Importantly, retroviral expression of SAP into lymphoblastoid cell lines established from XLP patients leads to elevated apoptotic response to DNA damage. Surprisingly, similar results were obtained upon ectopic expression of SAP in an osteosarcoma line. Because SAP binds to the anti-apoptotic protein valosin-containing protein, a SLAMF receptor-independent mechanism could be involved [54].
A more detailed analysis by Snow et al. showed that the lack of susceptibility of pre-activated T cells to RICD after re-encountering the antigen is a substantial problem in XLP derived CD8+ T cells [55]. In this process, SAP is suggested to deliver SLAMF6 signaling in a Fyn-independent manner, facilitating TCR signal strength for reaching the required threshold for the induction of apoptosis in normal T cells. Indeed, downmodulation of SAP and SLAMF6 expression were found to block TCR-induced upregulation of RICD execution molecules FasL and Bim. In contrast, in XLP-derived T cells where SAP-mediated dissociation of SHP-1 from SLAM receptors does not occur SLAMF6 and SHP-1 contribute to RICD resistance [55].
Taken together, the SAP mutation of XLP patients or mice may abolish multiple pathways of the homeostatic regulation of T cells at different stages of the immune response. As EAT-2 has been detected in a subset of CD8 cells, its role in viral infections must be examined as well.
Defective humoral response in SAP−/− mice
Defects in primary and secondary humoral responses and an aberrant germinal center formation were observed in SAP−/− mice upon infections with LCMV, influenza virus, MHV68, or after immunization with proteins or haptens [56–61]. As GCs are specific sites for the development of long-term humoral immunity, their absence could account for the significant decrease in circulating memory B cells and long-lived plasma cells resulting in a reduced levels of serum IgG. The absence of SAP-dependent signaling networks impairs T helper functions and consequently germinal center formation, as shown by a number of adoptive transfer experiments [56–58].
The role of SAP in T cell function is not yet fully understood. While the strongly reduced IL-4 secretion has an impact, the contribution of dysregulation of other cytokines, e.g., IL-6 and IL-21 or CD40L, are less clear [46, 56, 57, 62, 63]. The notion that SAP is dispensable for the priming of naïve CD4+ T cells into a subset of effector cells programmed to help B cells, but critical for the communication with B lymphocytes is supported by different experiments. As judged by intravital fluorescence microscopy, SAP−/− T cells form conjugates with antigen pulsed DCs as efficiently as their wt counterparts [64]. Further evidence that SAP is not requisite for primary activation comes from experiments in which T cells were retrovirally transfected with SAP 5 days after their APC-mediated priming. These cells become potent B cell activators [65]. Qi et al. provided elegant evidence for the concept that the duration of the interaction between B cells and SAP−/− T cells is rather short-lived, which in turn results in inefficient B cell proliferation and germinal center development [64, 66].
In the last decade, a class of CD4+ T cells, i.e., T follicular helper cells (Tfh), localized in B cell follicles through expression of the chemokine receptor CXCR5 has attracted attention since they appeared to be particularly potent B cell helpers [67–69]. Further investigation revealed that their gene expression profile is distinct from of conventional Th1 and Th2 cells, as they express transcription factor BCL-6, high levels of IL-21, and surface receptors ICOS and PD-1.
In agreement with the SAP-independent nature of normal T-DC interactions, primed wt and SAP−/− CD4+ T cells were found to upregulate CXCR5 and ICOS in a comparable fashion [64, 65]. Curiously, in some studies, attenuated ICOS expression and abnormal CD40L expression have been observed on SAP−/− T cells after in vitro or in vivo stimulation [58, 62].
According to a current study by Eddahri et al., IL-6/STAT3 promoted IL-21 secretion correlates with the acquisition of B cell help without significantly affecting expression of characteristic surface markers of Tfh cells [70]. Consistent with this model, the prolonged CD40L expression of SAP−/− CD4+ T cells is uncoupled from their diminished capacity to help B cells in influenza-infected SAP−/− mice [65].
Cytokine responses, the other major factors by which T cells facilitate and shape humoral defense, exhibit a dramatic bias in SAP−/− CD4+ T cells, as they display an impaired ability to differentiate into Th2 cells but produce elevated levels of IFN-γ upon TCR stimulation in vitro and in vivo [46, 71]. Although there were no differences observed in the IL-4 or IFN-γ production, IL-10 secretion of CD4+ T cells from XLP patients was also found to be severely impaired [62]. Further biochemical analyses revealed an important role of SAP in the recruitment of PKCθ to the immune synapse followed by NF-κB activation and induction of GATA-3, the master transcription factor of IL-4 [43] (Fig. 4). However, while these signal transduction pathways are dependent on the Fyn recruiting ability of SAP [72], defective induction of germinal centers could be rescued by retroviral reconstitution of T cells with SAP-R78A, a Fyn binding mutant form of SAP [58].
Since T–B conjugates are bidirectional stimulating units; not only B cells require sustained interaction for their comprehensive activation and subsequent germinal center formation but differentiation of Tfh cells also appears to depend on a prolonged, cognate engagement [64]. Nevertheless, it still remains to be determined whether the development of incompetent Tfh cells participates in defective germinal reactions of SAP-deficient mice. Furthermore, the SAP dependence of suitable contact formation between GC B cells and GC Tfh cells in the second round of T–B interaction also needs further clarification.
Another remaining question is the contribution of the different SLAM family receptors in T cell-mediated B cells responses. Mice carrying single deletions of SLAMF1, SLAMF3, or SLAMF6 did not recapitulate the humoral defect of SAP−/− mice ([ [73], [50]] and unpublished observations), supporting the notion that multiple SLAM receptors are implicated.
The role of SLAMF and SAP in the development and function of T cells selected on thymocytes, e.g., NKT cells and innate CD8+ T cells
NKT cell development in the thymus
NKT cells form a separate lineage of unconventional T lymphocytes operating on the border of innate and adaptive immune systems. Unlike conventional T cells, they co-express NK cell-associated receptors and a limited repertoire of TCRs specialized for glycolipid antigen recognition presented in complex with the major histocompatibility complex (MHC)-like molecule CD1d [74–76]. Accordingly to their memory-like phenotype, they mount a rapid cytokine response to TCR stimulation secreting Th1 and Th2 cytokines as well as IL-17 [75, 77, 78].
Recent studies have established a crucial role for SAP in NKT cell ontogeny as they were absent in XLP patients and SAP-deficient mice [79–81]. NKT cells arise from common CD4+CD8+ thymic T cell precursors with conventional T cells and the fact that only the development of NKT cells is affected suggested that SAP is crucial for a specific step in their selection process. A partial block of NKT cell maturation was also detected in Fyn−/− mice [44, 82, 83]. As SAP has been shown to couple SLAM family receptors to Fyn in the periphery, SAP–Fyn interaction is a plausible component of signaling pathways involved in NKT cell development [6, 36, 39]. However, the recruitment of Fyn to SAP was indeed found to be involved in NKT cell maturation; it is not the exclusive mechanism by which SAP controls the development of NKT cells. Fyn binding mutant of SAP (R78A) can restore the generation of NKT cells, albeit less efficiently than wt SAP, in a OP9-DL1-based in vitro system as well as in SAP-R78A knock-in mice and SAP-R78A competitive bone marrow chimeras [84].
Because the positive selection of NKT cells depends on thymocyte–thymocyte interactions, instead of being selected on cortical epithelial cells [85, 86], SLAMF receptors represent obvious candidates to support NKT development because of their SAP-dependent signal transduction and their homotypic cellular interactions [44]. SLAMF1 and SLAMF6 are not only highly expressed on DP thymocytes and immature NKT cells, SLAMF1−/− as well as SLAMF6−/− mice develop slightly reduced NKT cell compartment. In addition, overlapping functions of these two receptors were observed [51, 52]. To investigate the conceivable redundant functions of SLAMF1 and SLAMF6 in NKT cell maturation, pseudodouble knockouts were generated in bone marrow chimeras [42]. As predicted, NKT cell development was severely impaired in SLAMF1/SLAMF6 functional double mutant chimeras. Thus, SLAMF1 and SLAMF6 complement each other in the positive selection of NKT cells, mediated through their homophilic interactions between DP thymocytes.
It is important to note that although the reduction of NKT cells is dramatic in SAP-deficient mice, the reduction of NKT cells in SLAMF1/SLAMF6 double mutant chimeras as in Fyn-deficient mice is not complete, suggesting a possible role of other protein kinases (e.g., Lck or SLAMF members such as SLAMF3 and SLAMF5) [42]. Although the precise signaling mechanism by which SLAMF1/SLAMF6-SAP– Fyn control NKT cell development remains to be elucidated, PKCθ and possibly NF-κB might be involved [44, 87, 88].
It is conceivable that XLP disease manifestations can be explained, in part, by an absence of NKT cells. For example, they have been described to promote antiviral responses by stimulating NK cells and CD8+ T lymphocytes through robust cytokine production or indirectly via enhanced antigen presentation and pDC activation [89, 90]. Additionally, CD1d−/− mice, which lack NKT cells, recapitulate the same CD8+ T cell hyperproliferation observed in SAP−/− mice after viral infection [90]. By controlling cytotoxic responses, NKT cells have been also implicated in tumor surveillance in human and mouse [91–93]. Furthermore, several recent studies suggest that activated NKT cells enhance multiple levels of antibody responses by cognate and noncognate interactions with CD1d-expressing B cells as well as through dendritic cells and helper T cells [94–96]. Because SAP R78A knock-in mice develop a significant number of NKT cells, they have been exploited to investigate the importance of SAP–Fyn interaction in response of NKT cells following in vivo TCR stimulation. Interestingly, contrary to regular CD4+ T cells, Fyn binding of SAP is required neither for normal IL-4 and IFN-γ production nor the activation of bystander lymphocytes [84]. In another study, the role of SAP in NKT function was examined by generating SAP-deficient NKT cells using SAP−/− mice expressing the invariant TCR (Vα14Jα18) encoding transgene [97]. Although these SAP−/− NKT cells appeared to have an immature phenotype, the defective cytokine response and corresponding absence of key transcription factors GATA-3 and T-bet suggest a complex function of SAP in NKT cell development, homeostasis, and function [97, 98].
Innate CD8+ T cells
The selection and differentiation of innate-like CD8+ T cells differs from those required for conventional T cell populations [99, 100]. They are selected by nonclassical MHC class I molecules expressed on hematopoietic cells rather than on thymic epithelial cells. Itk and Rlk, members of the Tec family tyrosine kinases, are necessary for the development of conventional T lymphocytes but are dispensable for the selection of innate-like CD8+ T cells [101–103]. Consequently, the majority of CD8+ T cells that develop in Itk- and Itk/Rlk-deficient thymuses exhibit the characteristic phenotype of innate-like T cells, providing an opportunity to study the specifications of their development as well as their function [102, 104, 105]. According to the observation by Horai et al., the thymic selection of this group of innate-like CD8+ T cells also requires SAP-dependent signal transduction [106]; however till date, we still do not have information which SLAM receptors participate.
SAP governs development of T-CD4 cells and a subset of innate CD4+ T cells
MHC class II-expressing double-positive thymocytes, which are generated from specific class II transgenic mice, induce progression of conventional CD4+ T cell development as efficiently as cortical thymic epithelial cells [107, 108]. These thymocyte-selected CD4+ T cells, or T-CD4 cells, require the same signaling components as NKT cells, namely SAP, Fyn, and PKCθ. Furthermore, SAP is critical for IL-4 production by T-CD4 cells, and T-bet is necessary to produce the maximum amount of IFN-γ for CD4+ T cells regardless of the selection pathway. Thus, in contrast to epithelial cell-selected CD4+ T cells, the two distinct lineages of T cells selected by thymocytes, i.e., T-CD4 and NKT cells, both utilize the SAP–Fyn–PKCθ pathway for their development and function. Likewise in the case of NKT cells, the lack of SLAMF6 resulted only a slight decrease of T-CD4 cells, suggesting that other SLAM receptors are also involved in their maturation [41]. Although these cells do not occur naturally in the mouse, they exist in humans and they might be absent in XLP patients.
A unique subset of innate-like T cells was identified recently among the thymocytes of Itk- and Itk/Rlk-deficient mice [109]. Interestingly, these innate CD4+ T cells, unlike innate CD8+ T cells described in the same mice, are not selected by hematopoietic cells, but they require MHC class II expression on the thymic epithelium for their development [100]. Surprisingly, they still appear to be dependent on the interactions with the SLAM family receptors present on hematopoietic cells during thymic maturation [100].
According to an elegant interpretation of Berg and colleagues, the balance of TCR and SLAM family signaling pathways determines conventional versus innate T cell lineage fate: Strong TCR signaling restricts the effect of SLAM signals promoting the development of conventional αβ CD4+ and CD8+ T cells, while NKT cells emerge upon strong signaling of both TCR and SLAM family receptors, whereas weak TCR signaling combined with intact SLAM signals facilitate innate-like CD4+ and CD8+ T cell maturation [100].
Do SLAMF polymorphisms play a role in the pathogenesis of systemic lupus erythematosus?
SLAMF polymorphisms and isoforms
SLAMF receptors on chromosome 1 differ in coding and noncoding sequences in inbred mouse strains. Two major haplotypes exist in the region containing the seven SLAMF genes: One is represented by C57BL/6 and the second by 129Sv, NZW, or BALB/c. The ectodomains of SLAMF3 and SLAMF4 differ the most. Less variation is found in the ectodomains of SLAMF2, SLAMF8, and SLAMF9 and none in the other SLAMF ectodomains. Ectodomain amino acid sequence variation has not yet been found in humans, but increasing numbers of cDNA's lacking one of the extracellular domains, part of the cytoplasmic tail, or the transmembrane segment of a SLAM family member have been reported for both mouse and human receptors. These variations are caused by alternative splicing events. The loss of one of the ectodomains will, expectedly, affect the receptor/ligand interactions in the restricted space requirements of the immune synapse [8, 110]. Variants of the cytoplasmic tail will affect signal transduction because one of the ITSM motifs involved in binding of SAP and/or EAT-2 will be lacking. Several of the isoforms are expressed as proteins, but the complexity of these SLAMF isoforms is not yet understood.
SLAMF gene polymorphisms are likely to be susceptibility factors that contribute to the pathogenesis of SLE
Systemic lupus erythematosus (SLE) is a multisystem autoimmune disease, marked by a range of autoantibodies with a long prodromal phase of autoantibody development and epitope spreading. Although the pathogenesis of this disease and primary mechanisms underlying the loss of tolerance remains largely unknown, it would appear that a multitude of genes are involved, as is the case for other autoimmune diseases [111–113]. A number of studies clearly demonstrate that disease manifestations involve dysregulation of T/B cell interactions, as well as aberrant DC and macrophage functions [114].
Genome-wide linkage scans of families with multiple members affected with SLE have consistently demonstrated the presence of a susceptibility locus on chromosome 1q23, which includes the SLAMF genes [115–121]. In addition, it established a first-generation haplotype map, using 63 single nucleotide polymorphisms (SNPs) across the SLAM region in a set of 254 SLE trios and 160 discordant sibling pairs [122]. These preliminary results suggest that there is variation within the human SLAM region that is associated with SLE.
More recently, a specific association with SLE disease susceptibility was found using the nonsynonymous SNP rs509749 SNP, which is located in exon 8 of SLAMF3 [123]. This SNP encodes a Val/Met change in amino acid 602 in the cytoplasmic domain of the SLAMF3 protein (Table 1; Fig. 2). Only two of the eight tyrosine's in its cytoplasmic tail are embedded in ITSM motifs and have been shown to associate with SAP and/or EAT-2 [35, 124]. The C-terminal tyrosine binds Grb2 via its Sh2 domain [13]. Whether the V602M replacement in the −1 position of the first ITSM affects function requires further investigation, especially because the M602 allele is present in a substantial percentage of the human population (NCBI, dbSNP).
SLAMF gene polymorphisms as susceptibility factors in murine SLE
In mice, the region syntenic to human 1q23 has been implicated in three different models of spontaneous lupus: the (NZB × NZW) F2 intercross, the NZM/Aeg2410 New Zealand mouse, and the BXSB mouse [125–127]. The disease phenotype of these mice is similar to that of SLE patients, both in terms of production of autoantibodies as well as severe nephritis and multi-organ involvement. A strong, spontaneous, antinuclear antibody response occurred in congenic mice (B6.Sle1) derived from crosses between the lupus-prone mouse strain NZM2410 (NZB × NZW/F1) and C57BL/6. The presence of a small NZW-derived chromosome #1 segment (Sle1) on a C57BL/6 background was sufficient to generate these autoantibody responses [128, 129]. Fine mapping of the Sle1 locus determined that three loci within this congenic interval, termed Sle1a, Sle1b, and Sle1c, are implicated cause a loss of tolerance to chromatin [128–132].
The Sle1b locus, which mediates a breach in tolerance to nuclear antigens [128, 129], is an approximately 0.9-Mb segment that includes the SLAMF locus [129]. Although this locus contains 24 expressed genes, the NZW-based SLAMF variants embedded in the C57BL/6 background are thought to be the major contributors to the pathogenesis of SLE. This notion is supported by studies with congenic strains derived from crosses between 129 and C57BL/6 mice, which develop spontaneous autoimmunity [133, 134]. One congenic strain, B6.129chr1b, carrying a 129 interval contains the SLAMF locus. Thus, the SLAMF-haplotype II, e.g., derived from NZW or 129, segments Sle1b and 129chr1b in the C57BL/6 background induce loss of tolerance to nuclear antigens. We do, however, not know which C57BL/6 genes are involved in the putative epistatic pathogenic networks between SLAMF genes and other genes [135].
As judged by sequence and single nucleotide polymorphism analyses, the NZW-derived or 129-derived SLAMF locus embedded in the C57BL/6 genome of B6.Sle1b or B6.129Chr1b is identical to the SLAMF locus of the 129 mouse strain [129]. However, although B6.129Chr1b carries SLAMF haplotype II, the 129 mouse itself does not develop autoimmunity. Thus, an interplay between SLAMF haplotype II variants and the background must be maintained by either “suppressive” 129 genes or by epistatic activating genes in the C57BL/6 genome. While there are marked differences in a variety of immune responses between the C57BL/6 and 129 mouse [136–140], surprisingly, 129 mice are more susceptible to certain autoimmune stimuli than C57BL/6 [141, 142]. This might indicate that a specific epistatic interaction exist in C57BL/6 ×129 congenic mice rather than just a generally suppressed autoimmunity in 129. This notion would be consistent with the development of autoimmunity in the other SLAMF-haplotype II mouse, namely B6.Sle1b.
While Sle1b subcongenic line shows a pronounced B cell phenotype (similar to the one of the whole Sle1 locus), the effect of congenic region on T cell compartment seems to be more elusive. A broad range of autoimmune manifestations have been observed in the congenic B6. Sle1 and partially in aged B6.129Chr1b T cells (e.g., CD4+ T cell activation, decreased number of T regulatory cells, increased proliferation and cytokine production, presence of histone-specific T cells) [131, 134, 143]. In contrast, the subcongenic Sle1b T cell phenotype is restricted to an increased percentage of activated cells and an elevated calcium flux in response to receptor cross-linking [129, 131]. Interestingly, this activation phenotype of T cells could be amplified in an epistatic interaction between Sle1b and Yaa or Faslpr [ [132], [144]]. This interplay resulted a hyperactivation of PI3K/AKT/mTOR signaling pathway in Sle1bx Faslpr.
Mohan, Wakeland, and colleagues postulate that among the SLAM family members, Slamf6 in the context of the C57BL/6 background may be the strongest candidate to be causally correlated with the B6.Sle1b phenotype. They find that the long isoform of murine Ly108, i.e., Ly108-1, is upregulated, while the short isoform (Ly108-2) is down-regulated in T and B cells of B6.Sle1b mice compared to C57BL/6 [129]. Supporting this hypothesis, transfection of Ly108-2 was able to sensitize the immature B cell line WEHI-231 to undergo apoptosis, suggesting that Ly108 could regulate the stringency with which self-reactive B cells are censored during early development [145].
The role of SAP in murine SLE
Aberrant T cell functions caused by the SAP mutation have been studied in a few lupus-prone mouse strains. For instance, SAP−/− animals are resistant to pristane-induced lupus [59] or crossing SAP−/− mice with B6.Sle1b resulted in a decrease in penetrance of autoantibody production upon aging [146] and the SLE phenotype of MRL-Faslpr is greatly reduced by a spontaneous SAP mutation [147]. Vinuesa and colleagues have shown recently that SAP null mutation also abrogates the lupus-like phenotype of Roquinsan/san mice [148]. This finding provides more insight into the role of SAP in T cell-mediated humoral autoimmunity since their studies support the theory that autoimmune phenotype of Roquinsan/san is mediated by Tfh cells. The authors demonstrated that aberrant expansion of Tfh cells and induction of humoral autoimmunity is T cell intrinsic in Roquinsan/san mice and SAP deficiency effectively corrected these phenotypes along with the observed spontaneous GC formation. However, the connection between SAP signaling and lupus is likely to be indirect because of the lack of germinal center formation caused by the absence of SAP affects autoantibody production.
Taken together, an increasing body of evidence shows that SAP and SLAM family members are involved in controlling humoral autoimmunity in several murine lupus models by T lymphocytes, in particular CD4+ Th cells. Studies of SAP-deficient mice point to a possible role of Tfh population and their role in mediating negative and positive selection of GC B cells. Furthermore, murine SLAM family receptors are distinguished among candidate genes of chromosome 1 lupus congenic models due to their highly polymorphic nature and their generic expression on lymphoid lineage. They have the potential to regulate T cell function in the level of thymic development and cytokine production in periphery. Because of their functional diversity and overlapping signaling, one can assume that their control over self-tolerance is not confined to only one SLAMF receptor.
Acknowledgments
We are grateful for support from the NIH (AI-15066, AI-076210, AI-065687 to CT) and the CCFA (to XR).
Contributor Information
Cynthia Detre, Email: cdetre@bidmc.harvard.edu, BIDMC Division of Immunology, Harvard Center for Life Sciences, Rm. CLS 938, 3 Blackfan Circle, Boston MA 02115 USA.
Marton Keszei, BIDMC Division of Immunology, Harvard Center for Life Sciences, Rm. CLS 938, 3 Blackfan Circle, Boston MA 02115 USA.
Xavier Romero, BIDMC Division of Immunology, Harvard Center for Life Sciences, Rm. CLS 938, 3 Blackfan Circle, Boston MA 02115 USA.
George C. Tsokos, BIDMC Division of Rheumatology, Harvard Center for Life Sciences, Rm. CLS 937 3 Blackfan Circle, Boston, MA 02115, USA
Cox Terhorst, Email: cterhors@bidmc.harvard.edu, BIDMC Division of Immunology, Harvard Center for Life Sciences, Rm. CLS 938, 3 Blackfan Circle, Boston MA 02115 USA, Harvard Medical School, Boston MA USA.
References
- 1.Wang N, Morra M, Wu C, Gullo C, Howie D, Coyle T, Engel P, Terhorst C. CD150 is a member of a family of genes that encode glycoproteins on the surface of hematopoietic cells. Immunogenetics. 2001;53:382–394. doi: 10.1007/s002510100337. [DOI] [PubMed] [Google Scholar]
- 2.Tovar V, del Valle J, Zapater N, Martin M, Romero X, Pizcueta P, Bosch J, Terhorst C, Engel P. Mouse novel Ly9: a new member of the expanding CD150 (SLAM) family of leukocyte cell-surface receptors. Immunogenetics. 2002;54:394–402. doi: 10.1007/s00251-002-0483-3. [DOI] [PubMed] [Google Scholar]
- 3.Fraser CC, Howie D, Morra M, Qiu Y, Murphy C, Shen Q, Gutierrez-Ramos JC, Coyle A, Kingsbury GA, Terhorst C. Identification and characterization of SF2000 and SF2001, two new members of the immune receptor SLAM/CD2 family. Immunogenetics. 2002;53:843–850. doi: 10.1007/s00251-001-0415-7. [DOI] [PubMed] [Google Scholar]
- 4.Staunton DE, Thorley-Lawson DA. Molecular cloning of the lymphocyte activation marker Blast-1. EMBO J. 1987;6:3695–3701. doi: 10.1002/j.1460-2075.1987.tb02703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yokoyama S, Staunton D, Fisher R, Amiot M, Fortin JJ, Thorley-Lawson DA. Expression of the Blast-1 activation/adhesion molecule and its identification as CD48. J Immunol. 1991;146:2192–2200. [PubMed] [Google Scholar]
- 6.Calpe S, Wang N, Romero X, Berger SB, Lanyi A, Engel P, Terhorst C. The SLAM and SAP gene families control innate and adaptive immune responses. Adv Immunol. 2008;97:177–250. doi: 10.1016/S0065-2776(08)00004-7. [DOI] [PubMed] [Google Scholar]
- 7.Engel P, Eck MJ, Terhorst C. The SAP and SLAM families in immune responses and X-linked lymphoproliferative disease. Nat Rev Immunol. 2003;3:813–821. doi: 10.1038/nri1202. [DOI] [PubMed] [Google Scholar]
- 8.Cao E, Ramagopal UA, Fedorov A, Fedorov E, Yan Q, Lary JW, Cole JL, Nathenson SG, Almo SC. NTB-A receptor crystal structure: insights into homophilic interactions in the signaling lymphocytic activation molecule receptor family. Immunity. 2006;25:559–570. doi: 10.1016/j.immuni.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 9.Yan Q, Malashkevich VN, Fedorov A, Fedorov E, Cao E, Lary JW, Cole JL, Nathenson SG, Almo SC. Structure of CD84 provides insight into SLAM family function. Proc Natl Acad Sci. 2007;104:10583–10588. doi: 10.1073/pnas.0703893104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roda-Navarro P, Mittelbrunn M, Ortega M, Howie D, Terhorst C, Sanchez-Madrid F, Fernandez-Ruiz E. Dynamic redistribution of the activating 2B4/SAP complex at the cytotoxic NK cell immune synapse. J Immunol. 2004;173:3640–3646. doi: 10.4049/jimmunol.173.6.3640. [DOI] [PubMed] [Google Scholar]
- 11.Howie D, Okamoto S, Rietdijk S, Clarke K, Wang N, Gullo C, Bruggeman JP, Manning S, Coyle AJ, Greenfield E, Kuchroo V, Terhorst C. The role of SAP in murine CD150 (SLAM)-mediated T-cell proliferation and interferon gamma production. Blood. 2002;100:2899–2907. doi: 10.1182/blood-2002-02-0445. [DOI] [PubMed] [Google Scholar]
- 12.Rethi B, Gogolak P, Szatmari I, Veres A, Erdos E, Nagy L, Rajnavolgyi E, Terhorst C, Lanyi A. SLAM/SLAM interactions inhibit CD40-induced production of inflammatory cytokines in monocyte-derived dendritic cells. Blood. 2006;107:2821–2829. doi: 10.1182/blood-2005-06-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin M, Del Valle JM, Saborit I, Engel P. Identification of Grb2 as a novel binding partner of the signaling lymphocytic activation molecule-associated protein binding receptor CD229. J Immunol. 2005;174:5977–5986. doi: 10.4049/jimmunol.174.10.5977. [DOI] [PubMed] [Google Scholar]
- 14.Del Valle JM, Engel P, Martin M. The cell surface expression of SAP-binding receptor CD229 is regulated via its interaction with clathrin-associated adaptor complex 2 (AP-2) J Biol Chem. 2003;278:17430–17437. doi: 10.1074/jbc.M301569200. [DOI] [PubMed] [Google Scholar]
- 15.Purtilo DT, Cassel CK, Yang JP, Harper R. X-linked recessive progressive combined variable immunodeficiency (Duncan's disease) Lancet. 1975;1:935–940. doi: 10.1016/s0140-6736(75)92004-8. [DOI] [PubMed] [Google Scholar]
- 16.Harrington DS, Weisenburger DD, Purtilo DT. Malignant lymphoma in the X-linked lymphoproliferative syndrome. Cancer. 1987;59:1419–1429. doi: 10.1002/1097-0142(19870415)59:8<1419::aid-cncr2820590807>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 17.Egeler RM, de Kraker J, Slater R, Purtilo DT. Documentation of Burkitt lymphoma with t(8;14) (q24;q32) in X-linked lymphoproliferative disease. Cancer. 1992;70:683–687. doi: 10.1002/1097-0142(19920801)70:3<683::aid-cncr2820700324>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 18.Schuster V, Terhorst C. Primary immunodeficiency diseases: a molecular and cellular approach. 2nd. Oxford University Press; Oxford: 2006. pp. 470–484. [Google Scholar]
- 19.Morra M, Howie D, Grande MS, Sayos J, Wang N, Wu C, Engel P, Terhorst C. X-linked lymphoproliferative disease: a progressive immunodeficiency. Annu Rev Immunol. 2001;19:657–682. doi: 10.1146/annurev.immunol.19.1.657. [DOI] [PubMed] [Google Scholar]
- 20.Sumegi J, Huang D, Lanyi A, Davis JD, Seemayer TA, Maeda A, Klein G, Seri M, Wakiguchi H, Purtilo DT, Gross TG. Correlation of mutations of the SH2D1A gene and Epstein–Barr virus infection with clinical phenotype and outcome in X-linked lymphoproliferative disease. Blood. 2000;96:3118–3125. [PubMed] [Google Scholar]
- 21.Poy F, Yaffe MB, Sayos J, Saxena K, Morra M, Sumegi J, Cantley LC, Terhorst C, Eck MJ. Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition. Mol Cell. 1999;4:555–561. doi: 10.1016/s1097-2765(00)80206-3. [DOI] [PubMed] [Google Scholar]
- 22.Morra M, Silander O, Calpe S, Choi M, Oettgen H, Myers L, Etzioni A, Buckley R, Terhorst C. Alterations of the X-linked lymphoproliferative disease gene SH2D1A in common variable immunodeficiency syndrome. Blood. 2001;98:1321–1325. doi: 10.1182/blood.v98.5.1321. [DOI] [PubMed] [Google Scholar]
- 23.Parolini O, Kagerbauer B, Simonitsch-Klupp I, Ambros P, Jaeger U, Mann G, Haas OA, Morra M, Gadner H, Terhorst C, Knapp W, Holter W. Analysis of SH2D1A mutations in patients with severe Epstein–Barr virus infections, Burkitt's lymphoma, and Hodgkin's lymphoma. Ann Hematol. 2002;81:441–447. doi: 10.1007/s00277-002-0490-3. [DOI] [PubMed] [Google Scholar]
- 24.Rigaud S, Fondaneche MC, Lambert N, Pasquier B, Mateo V, Soulas P, Galicier L, Le Deist F, Rieux-Laucat F, Revy P, Fischer A, de Saint BG, Latour S. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. 2006;444:110–114. doi: 10.1038/nature05257. [DOI] [PubMed] [Google Scholar]
- 25.Latour S. Natural killer T cells and X-linked lymphopro-liferative syndrome. Curr Opin Allergy Clin Immunol. 2007;7:510–514. doi: 10.1097/ACI.0b013e3282f1bad6. [DOI] [PubMed] [Google Scholar]
- 26.Marodi L, Notarangelo LD. Immunological and genetic bases of new primary immunodeficiencies. Nat Rev Immunol. 2007;7:851–861. doi: 10.1038/nri2195. [DOI] [PubMed] [Google Scholar]
- 27.Rumble JM, Oetjen KA, Stein PL, Schwartzberg PL, Moore BB, Duckett CS. Phenotypic differences between mice deficient in XIAP and SAP, two factors targeted in X-linked lymphoproliferative syndrome (XLP) Cell Immunol. 2009;259:82–89. doi: 10.1016/j.cellimm.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morra M, Simarro-Grande M, Martin M, Chen AS, Lanyi A, Silander O, Calpe S, Davis J, Pawson T, Eck MJ, Sumegi J, Engel P, Li SC, Terhorst C. Characterization of SH2D1A missense mutations identified in X-linked lymphoproliferative disease patients. J Biol Chem. 2001;276:36809–36816. doi: 10.1074/jbc.M101305200. [DOI] [PubMed] [Google Scholar]
- 29.Wu C, Sayos J, Wang N, Howie D, Coyle A, Terhorst C. Genomic organization and characterization of mouse SAP, the gene that is altered in X-linked lymphoproliferative disease. Immunogenetics. 2000;51:805–815. doi: 10.1007/s002510000215. [DOI] [PubMed] [Google Scholar]
- 30.Hare NJ, Ma CS, Alvaro F, Nichols KE, Tangye SG. Missense mutations in SH2D1A identified in patients with X-linked lymphoproliferative disease differentially affect the expression and function of SAP. Int Immunol. 2006;18:1055–1065. doi: 10.1093/intimm/dxl039. [DOI] [PubMed] [Google Scholar]
- 31.Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D, van Schaik S, Notarangelo L, Geha R, Roncarolo MG, Oettgen H, De Vries JE, Aversa G, Terhorst C. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature. 1998;395:462–469. doi: 10.1038/26683. [DOI] [PubMed] [Google Scholar]
- 32.Hwang PM, Li C, Morra M, Lillywhite J, Muhandiram DR, Gertler F, Terhorst C, Kay LE, Pawson T, Forman-Kay JD, Li SC. A “three-pronged” binding mechanism for the SAP/SH2D1A SH2 domain: structural basis and relevance to the XLP syndrome. EMBO J. 2002;21:314–323. doi: 10.1093/emboj/21.3.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finerty PJ, Muhandiram R, Forman-Kay JD. Side-chain dynamics of the SAP SH2 domain correlate with a binding hot spot and a region with conformational plasticity. J Mol Biol. 2002;322:605–620. doi: 10.1016/s0022-2836(02)00803-3. [DOI] [PubMed] [Google Scholar]
- 34.Eissmann P, Beauchamp L, Wooters J, Tilton JC, Long EO, Watzl C. Molecular basis for positive and negative signaling by the natural killer cell receptor 2B4 (CD244) Blood. 2005;105:4722–4729. doi: 10.1182/blood-2004-09-3796. [DOI] [PubMed] [Google Scholar]
- 35.Morra M, Lu J, Poy F, Martin M, Sayos J, Calpe S, Gullo C, Howie D, Rietdijk S, Thompson A, Coyle AJ, Denny C, Yaffe MB, Engel P, Eck MJ, Terhorst C. Structural basis for the interaction of the free SH2 domain EAT-2 with SLAM receptors in hematopoietic cells. EMBO J. 2001;20:5840–5852. doi: 10.1093/emboj/20.21.5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan B, Lanyi A, Song HK, Griesbach J, Simarro-Grande M, Poy F, Howie D, Sumegi J, Terhorst C, Eck MJ. SAP couples Fyn to SLAM immune receptors. Nat Cell Biol. 2003;5:155–160. doi: 10.1038/ncb920. [DOI] [PubMed] [Google Scholar]
- 37.Seemayer TA, Gross TG, Egeler RM, Pirruccello SJ, Davis JR, Kelly CM, Okano M, Lanyi A, Sumegi J. X-linked lymphoproliferative disease: twenty-five years after the discovery. Pediatr Res. 1995;38:471–478. doi: 10.1203/00006450-199510000-00001. [DOI] [PubMed] [Google Scholar]
- 38.Latour S, Roncagalli R, Chen R, Bakinowski M, Shi X, Schwartzberg PL, Davidson D, Veillette A. Binding of SAP SH2 domain to FynT SH3 domain reveals a novel mechanism of receptor signalling in immune regulation. Nat Cell Biol. 2003;5:149–154. doi: 10.1038/ncb919. [DOI] [PubMed] [Google Scholar]
- 39.Simarro M, Lanyi A, Howie D, Poy F, Bruggeman J, Choi M, Sumegi J, Eck MJ, Terhorst C. SAP increases FynT kinase activity and is required for phosphorylation of SLAM and Ly9. Int Immunol. 2004;16:727–736. doi: 10.1093/intimm/dxh074. [DOI] [PubMed] [Google Scholar]
- 40.Chen R, Latour S, Shi X, Veillette A. Association between SAP and FynT: inducible SH3 domain-mediated interaction controlled by engagement of the SLAM receptor. Mol Cell Biol. 2006;26:5559–5568. doi: 10.1128/MCB.00357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, Sofi MH, Rietdijk S, Wang N, Terhorst C, Chang CH. The SLAM-associated protein signaling pathway is required for development of CD4+ T cells selected by homotypic thymocyte interaction. Immunity. 2007;27:763–774. doi: 10.1016/j.immuni.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, Mamchak AA, Terhorst C, Bendelac A. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 2007;27:751–762. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cannons JL, Yu LJ, Hill B, Mijares LA, Dombroski D, Nichols KE, Antonellis A, Koretzky GA, Gardner K, Schwartzberg PL. SAP regulates T(H)2 differentiation and PKC-theta-mediated activation of NF-kappaB1. Immunity. 2004;21:693–706. doi: 10.1016/j.immuni.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 44.Borowski C, Bendelac A. Signaling for NKT cell development: the SAP–FynT connection. J Exp Med. 2005;201:833–836. doi: 10.1084/jem.20050339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu C, Tangye SG, Sun X, Luo Y, Lin Z, Wu J. The X-linked lymphoproliferative disease gene product SAP associates with PAK-interacting exchange factor and participates in T cell activation. Proc Natl Acad Sci. 2006;103:14447–14452. doi: 10.1073/pnas.0606624103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Czar MJ, Kersh EN, Mijares LA, Lanier G, Lewis J, Yap G, Chen A, Sher A, Duckett CS, Ahmed R, Schwartzberg PL. Altered lymphocyte responses and cytokine production in mice deficient in the X-linked lymphoproliferative disease gene SH2D1A/DSHP/SAP. Proc Natl Acad Sci USA. 2001;98:7449–7454. doi: 10.1073/pnas.131193098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crotty S, McCausland MM, Aubert RD, Wherry EJ, Ahmed R. Hypogammaglobulinemia and exacerbated CD8 T-cell-mediated immunopathology in SAP-deficient mice with chronic LCMV infection mimics human XLP disease. Blood. 2006;108:3085–3093. doi: 10.1182/blood-2006-04-018929. [DOI] [PubMed] [Google Scholar]
- 48.Chen G, Tai AK, Lin M, Chang F, Terhorst C, Huber BT. Signaling lymphocyte activation molecule-associated protein is a negative regulator of the CD8 T cell response in mice. J Immunol. 2005;175:2212–2218. doi: 10.4049/jimmunol.175.4.2212. [DOI] [PubMed] [Google Scholar]
- 49.Yin L, Al Alem U, Liang J, Tong WM, Li C, Badiali M, Medard JJ, Sumegi J, Wang ZQ, Romeo G. Mice deficient in the X-linked lymphoproliferative disease gene sap exhibit increased susceptibility to murine gammaherpesvirus-68 and hypogammaglobulinemia. J Med Virol. 2003;71:446–455. doi: 10.1002/jmv.10504. [DOI] [PubMed] [Google Scholar]
- 50.Graham DB, Bell MP, McCausland MM, Huntoon CJ, van Deursen J, Faubion WA, Crotty S, McKean DJ. Ly9 (CD229)-deficient mice exhibit T cell defects yet do not share several phenotypic characteristics associated with. J Immunol. 2006;176:291–300. doi: 10.4049/jimmunol.176.1.291. [DOI] [PubMed] [Google Scholar]
- 51.Howie D, Laroux FS, Morra M, Satoskar AR, Rosas LE, Faubion WA, Julien A, Rietdijk S, Coyle AJ, Fraser C, Terhorst C. Cutting edge: the SLAM family receptor Ly108 controls T cell and neutrophil functions. J Immunol. 2005;174:5931–5935. doi: 10.4049/jimmunol.174.10.5931. [DOI] [PubMed] [Google Scholar]
- 52.Wang N, Satoskar A, Faubion W, Howie D, Okamoto S, Feske S, Gullo C, Clarke K, Sosa MR, Sharpe AH, Terhorst C. The cell surface receptor SLAM controls T cell and macrophage functions. J Exp Med. 2004;199:1255–1264. doi: 10.1084/jem.20031835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen G, Tai AK, Lin M, Chang F, Terhorst C, Huber BT. Increased proliferation of CD8+ T cells in SAP-deficient mice is associated with impaired activation-induced cell death. Eur J Immunol. 2007;37:663–674. doi: 10.1002/eji.200636417. [DOI] [PubMed] [Google Scholar]
- 54.Nagy N, Matskova L, Kis LL, Hellman U, Klein G, Klein E. The proapoptotic function of SAP provides a clue to the clinical picture of X-linked lymphoproliferative disease. Proc Natl Acad Sci USA. 2009;106:11966–11971. doi: 10.1073/pnas.0905691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snow AL, Marsh RA, Krummey SM, Roehrs P, Young LR, Zhang K, van Hoff J, Dhar D, Nichols KE, Filipovich AH, Su HC, Bleesing JJ, Lenardo MJ. Restimulation-induced apoptosis of T cells is impaired in patients with X-linked lymphoproliferative disease caused by SAP deficiency. J Clin Invest. 2009;119:2976–2989. doi: 10.1172/JCI39518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crotty S, Kersh EN, Cannons J, Schwartzberg PL, Ahmed R. SAP is required for generating long-term humoral immunity. Nature. 2003;421:282–287. doi: 10.1038/nature01318. [DOI] [PubMed] [Google Scholar]
- 57.Morra M, Barrington RA, Abadia-Molina AC, Okamoto S, Julien A, Gullo C, Kalsy A, Edwards MJ, Chen G, Spolski R, Leonard WJ, Huber BT, Borrow P, Biron CA, Satoskar AR, Carroll MC, Terhorst C. Defective B cell responses in the absence of SH2D1A. Proc Natl Acad Sci USA. 2005;102:4819–4823. doi: 10.1073/pnas.0408681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cannons JL, Yu LJ, Jankovic D, Crotty S, Horai R, Kirby M, Anderson S, Cheever AW, Sher A, Schwartzberg PL. SAP regulates T cell-mediated help for humoral immunity by a mechanism distinct from cytokine regulation. J Exp Med. 2006;203:1551–1565. doi: 10.1084/jem.20052097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hron JD, Caplan L, Gerth AJ, Schwartzberg PL, Peng SL. SH2D1A regulates T-dependent humoral autoimmunity. J Exp Med. 2004;200:261–266. doi: 10.1084/jem.20040526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma CS, Pittaluga S, Avery DT, Hare NJ, Maric I, Klion AD, Nichols KE, Tangye SG. Selective generation of functional somatically mutated IgM+CD27+, but not Ig isotype-switched, memory B cells in X-linked lymphoproliferative disease. J Clin Invest. 2006;116:322–333. doi: 10.1172/JCI25720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kamperschroer C, Dibble JP, Meents DL, Schwartzberg PL, Swain SL. SAP is required for Th cell function and for immunity to influenza. J Immunol. 2006;177:5317–5327. doi: 10.4049/jimmunol.177.8.5317. [DOI] [PubMed] [Google Scholar]
- 62.Ma CS, Hare NJ, Nichols KE, Dupre L, Andolfi G, Roncarolo MG, Adelstein S, Hodgkin PD, Tangye SG. Impaired humoral immunity in X-linked lymphoproliferative disease is associated with defective IL-10 production by CD4+ T cells. J Clin Invest. 2005;115:1049–1059. doi: 10.1172/JCI23139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malbran A, Belmonte L, Ruibal-Ares B, Bare P, Massud I, Parodi C, Felippo M, Hodinka R, Haines K, Nichols KE, de Bracco MM. Loss of circulating CD27+ memory B cells and CCR4+ T cells occurring in association with elevated EBV loads in XLP patients surviving primary EBV infection. Blood. 2004;103:1625–1631. doi: 10.1182/blood-2003-07-2525. [DOI] [PubMed] [Google Scholar]
- 64.Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. SAP-controlled T–B cell interactions underlie germinal centre formation. Nature. 2008;455:764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamperschroer C, Roberts DM, Zhang Y, Weng NP, Swain SL. SAP enables T cells to help B cells by a mechanism distinct from Th cell programming or CD40 ligand regulation. J Immunol. 2008;181:3994–4003. doi: 10.4049/jimmunol.181.6.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwartzberg PL, Mueller KL, Qi H, Cannons JL. SLAM receptors and SAP influence lymphocyte interactions, development and function. Nat Rev Immunol. 2009;9:39–46. doi: 10.1038/nri2456. [DOI] [PubMed] [Google Scholar]
- 67.McHeyzer-Williams LJ, Pelletier N, Mark L, Fazilleau N, McHeyzer-Williams MG. Follicular helper T cells as cognate regulators of B cell immunity. Curr Opin Immunol. 2009;21:266–273. doi: 10.1016/j.coi.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.King C, Tangye SG, Mackay CR. T follicular helper (TFH) cells in normal and dysregulated immune responses. Annu Rev Immunol. 2008;26:741–766. doi: 10.1146/annurev.immunol.26.021607.090344. [DOI] [PubMed] [Google Scholar]
- 69.Fazilleau N, Mark L, McHeyzer-Williams LJ, McHeyzer-Williams MG. Follicular helper T cells: lineage and location. Immunity. 2009;30:324–335. doi: 10.1016/j.immuni.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eddahri F, Denanglaire S, Bureau F, Spolski R, Leonard WJ, Leo O, Andris F. Interleukin-6/STAT3 signaling regulates the ability of naive T cells to acquire B-cell help capacities. Blood. 2009;113:2426–2433. doi: 10.1182/blood-2008-04-154682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu C, Nguyen KB, Pien GC, Wang N, Gullo C, Howie D, Sosa MR, Edwards MJ, Borrow P, Satoskar AR, Sharpe AH, Biron CA, Terhorst C. SAP controls T cell responses to virus and terminal differentiation of TH2 cells. Nat Immunol. 2001;2:410–414. doi: 10.1038/87713. [DOI] [PubMed] [Google Scholar]
- 72.Davidson D, Shi X, Zhang S, Wang H, Nemer M, Ono N, Ohno S, Yanagi Y, Veillette A. Genetic evidence linking SAP, the X-linked lymphoproliferative gene product, to Src-related kinase FynT in T(H)2 cytokine regulation. Immunity. 2004;21:707–717. doi: 10.1016/j.immuni.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 73.McCausland MM, Yusuf I, Tran H, Ono N, Yanagi Y, Crotty S. SAP regulation of follicular helper CD4 T cell development and humoral immunity is independent of SLAM and Fyn kinase. J Immunol. 2007;178:817–828. doi: 10.4049/jimmunol.178.2.817. [DOI] [PubMed] [Google Scholar]
- 74.Kronenberg M, Kinjo Y. Innate-like recognition of microbes by invariant natural killer T cells. Curr Opin Immunol. 2009;21:391–396. doi: 10.1016/j.coi.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 76.Godfrey DI, McCluskey J, Rossjohn J. CD1d antigen presentation: treats for NKT cells. Nat Immunol. 2005;6:754–756. doi: 10.1038/ni0805-754. [DOI] [PubMed] [Google Scholar]
- 77.Godfrey DI, Kronenberg M. Going both ways: immune regulation via CD1d-dependent NKT cells. J Clin Invest. 2004;114:1379–1388. doi: 10.1172/JCI23594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pichavant M, Goya S, Meyer EH, Johnston RA, Kim HY, Matangkasombut P, Zhu M, Iwakura Y, Savage PB, Dekruyff RH, Shore SA, Umetsu DT. Ozone exposure in a mouse model induces airway hyperreactivity that requires the presence of natural killer T cells and IL-17. J Exp Med. 2008;205:385–393. doi: 10.1084/jem.20071507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chung B, Aoukaty A, Dutz J, Terhorst C, Tan R. Cutting edge: signaling lymphocytic activation molecule-associated protein controls NKT cell functions. J Immunol. 2005;174:3153–3157. doi: 10.4049/jimmunol.174.6.3153. [DOI] [PubMed] [Google Scholar]
- 80.Nichols KE, Hom J, Gong SY, Ganguly A, Ma CS, Cannons JL, Tangye SG, Schwartzberg PL, Koretzky GA, Stein PL. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Med. 2005;11:340–345. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pasquier B, Yin L, Fondaneche MC, Relouzat F, Bloch-Queyrat C, Lambert N, Fischer A, Saint-Basile G, Latour S. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J Exp Med. 2005;201:695–701. doi: 10.1084/jem.20042432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eberl G, Lowin-Kropf B, MacDonald HR. Cutting edge: NKT cell development is selectively impaired in Fyn-deficient mice. J Immunol. 1999;163:4091–4094. [PubMed] [Google Scholar]
- 83.Gadue P, Morton N, Stein PL. The Src family tyrosine kinase Fyn regulates natural killer T cell development. J Exp Med. 1999;190:1189–1196. doi: 10.1084/jem.190.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nunez-Cruz S, Yeo WC, Rothman J, Ojha P, Bassiri H, Juntilla M, Davidson D, Veillette A, Koretzky GA, Nichols KE. Differential requirement for the SAP–Fyn interaction during NK T cell development and function. J Immunol. 2008;181:2311–2320. doi: 10.4049/jimmunol.181.4.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CDId. Nature Immunol. 2001;2:971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- 86.Zhou D, Mattner J, Cantu C, III, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, Teneberg S, Wang D, Proia RL, Levery SB, Savage PB, Teyton L, Bendelac A. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 87.Schmidt-Supprian M, Tian J, Grant EP, Pasparakis M, Maehr R, Ovaa H, Ploegh HL, Coyle AJ, Rajewsky K. Differential dependence of CD4+CD25+ regulatory and natural killer-like T cells on signals leading to NF-kappaB activation. Proc Natl Acad Sci USA. 2004;101:4566–4571. doi: 10.1073/pnas.0400885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sivakumar V, Hammond KJ, Howells N, Pfeffer K, Weih F. Differential requirement for Rel/nuclear factor [kappa]B family members in natural killer T cell development. J Exp Med. 2003;197:1613–1621. doi: 10.1084/jem.20022234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Diana J, Griseri T, Lagaye S, Beaudoin L, Autrusseau E, Gautron AS, Tomkiewicz C, Herbelin A, Barouki R, von Herrath M, Dalod M, Lehuen A. NKT cell-plasmacytoid dendritic cell cooperation via OX40 controls viral infection in a tissue-specific manner. Immunity. 2009;30:289–299. doi: 10.1016/j.immuni.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 90.Roberts TJ, Lin Y, Spence PM, Van Kaer L, Brutkiewicz RR. CD1d1-dependent control of the magnitude of an acute antiviral immune response. J Immunol. 2004;172:3454–3461. doi: 10.4049/jimmunol.172.6.3454. [DOI] [PubMed] [Google Scholar]
- 91.Yuling H, Ruijing X, Li L, Xiang J, Rui Z, Yujuan W, Lijun Z, Chunxian D, Xinti T, Wei X, Lang C, Yanping J, Tao X, Mengjun W, Jie X, Youxin J, Jinquan T. EBV-induced human CD8+ NKT cells suppress tumorigenesis by EBV-associated malignancies. Cancer Res. 2009;69:7935. doi: 10.1158/0008-5472.CAN-09-0828. [DOI] [PubMed] [Google Scholar]
- 92.Terabe M, Berzofsky JA. The role of NKT cells in tumor immunity. Adv Cancer Res. 2008;101:277–348. doi: 10.1016/S0065-230X(08)00408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Terabe M, Berzofsky JA. NKT cells in immunoregulation of tumor immunity: a new immunoregulatory axis. Trends Immunol. 2007;28:491–496. doi: 10.1016/j.it.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 94.Galli G, Pittoni P, Tonti E, Malzone C, Uematsu Y, Tortoli M, Maione D, Volpini G, Finco O, Nuti S, Tavarini S, Dellabona P, Rappuoli R, Casorati G, Abrignani S. Invariant NKT cells sustain specific B cell responses and memory. Proc Natl Acad Sci USA. 2007;104:3984–3989. doi: 10.1073/pnas.0700191104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lang GA, Devera TS, Lang ML. Requirement for CD1d expression by B cells to stimulate NKT cell-enhanced antibody production. Blood. 2008;111:2158–2162. doi: 10.1182/blood-2007-10-117309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tonti E, Galli G, Malzone C, Abrignani S, Casorati G, Dellabona P. NKT-cell help to B lymphocytes can occur independently of cognate interaction. Blood. 2009;113:370–376. doi: 10.1182/blood-2008-06-166249. [DOI] [PubMed] [Google Scholar]
- 97.Cen O, Ueda A, Guzman L, Jain J, Bassiri H, Nichols KE, Stein PL. The adaptor molecule signaling lymphocytic activation molecule-associated protein (SAP) regulates IFN-gamma and IL-4 production in V alpha 14 transgenic NKT cells via effects on GATA-3 and T-bet expression. J Immunol. 2009;182:1370–1378. doi: 10.4049/jimmunol.182.3.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bassiri H, Janice Yeo WC, Rothman J, Koretzky GA, Nichols KE. X-linked lymphoproliferative disease (XLP): a model of impaired anti-viral, anti-tumor and humoral immune responses. Immunol Res. 2008;42:145–159. doi: 10.1007/s12026-008-8048-7. [DOI] [PubMed] [Google Scholar]
- 99.Berg LJ. Signalling through TEC kinases regulates conventional versus innate CD8+ T-cell development. Nat Rev Immunol. 2007;7:479–485. doi: 10.1038/nri2091. [DOI] [PubMed] [Google Scholar]
- 100.Prince AL, Yin CC, Enos ME, Felices M, Berg LJ. The Tec kinases Itk and Rlk regulate conventional versus innate T-cell development. Immunol Rev. 2009;228:115–131. doi: 10.1111/j.1600-065X.2008.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lucas JA, Atherly LO, Berg LJ. The absence of Itk inhibits positive selection without changing lineage commitment. J Immunol. 2002;168:6142–6151. doi: 10.4049/jimmunol.168.12.6142. [DOI] [PubMed] [Google Scholar]
- 102.Atherly LO, Lucas JA, Felices M, Yin CC, Reiner SL, Berg LJ. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 2006;25:79–91. doi: 10.1016/j.immuni.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 103.Readinger JA, Mueller KL, Venegas AM, Horai R, Schwartzberg PL. Tec kinases regulate T-lymphocyte development and function: new insights into the roles of Itk and Rlk/Txk. Immunol Rev. 2009;228:93–114. doi: 10.1111/j.1600-065X.2008.00757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dubois S, Waldmann TA, Muller JR. ITK and IL-15 support two distinct subsets of CD8+ T cells. Proc Natl Acad Sci. 2006;103:12075–12080. doi: 10.1073/pnas.0605212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Broussard C, Fleischecker C, Horai R, Chetana M, Venegas AM, Sharp LL, Hedrick SM, Fowlkes BJ, Schwartzberg PL. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 2006;25:93–104. doi: 10.1016/j.immuni.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 106.Horai R, Mueller KL, Handon RA, Cannons JL, Anderson SM, Kirby MR, Schwartzberg PL. Requirements for selection of conventional and innate T lymphocyte lineages. Immunity. 2007;27:775–785. doi: 10.1016/j.immuni.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li W, Kim MG, Gourley TS, McCarthy BP, Sant'Angelo DB, Chang CH. An alternate pathway for CD4 T cell development: thymocyte-expressed MHC class II selects a distinct T cell population. Immunity. 2005;23:375–386. doi: 10.1016/j.immuni.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 108.Choi EY, Jung KC, Park HJ, Chung DH, Song JS, Yang SD, Simpson E, Park SH. Thymocyte–thymocyte interaction for efficient positive selection and maturation of CD4 T cells. Immunity. 2005;23:387–396. doi: 10.1016/j.immuni.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 109.Hu J, August A. Naive and innate memory phenotype CD4+ T cells have different requirements for active Itk for their development. J Immunol. 2008;180:6544–6552. doi: 10.4049/jimmunol.180.10.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yan Q, Malashkevich VN, Fedorov A, Fedorov E, Cao E, Lary JW, Cole JL, Nathenson SG, Almo SC. Structure of CD84 provides insight into SLAM family function. Proc Natl Acad Sci USA. 2007;104:10583–10588. doi: 10.1073/pnas.0703893104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N Engl J Med. 2009;360:1646–1654. doi: 10.1056/NEJMra0808284. [DOI] [PubMed] [Google Scholar]
- 112.Coenen MJ, Gregersen PK. Rheumatoid arthritis: a view of the current genetic landscape. Genes Immun. 2009;10:101–111. doi: 10.1038/gene.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zuvich RL, McCauley JL, Pericak-Vance MA, Haines JL. Genetics and pathogenesis of multiple sclerosis. Semin Immunol. 2009;21:328–333. doi: 10.1016/j.smim.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Krishnan S, Chowdhury B, Tsokos GC. Autoimmunity in systemic lupus erythematosus: integrating genes and biology. Semin Immunol. 2006;18:230–243. doi: 10.1016/j.smim.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 115.Moser KL, Gray-McGuire C, Kelly J, Asundi N, Yu H, Bruner GR, Mange M, Hogue R, Neas BR, Harley JB. Confirmation of genetic linkage between human systemic lupus erythematosus and chromosome 1q41. Arthritis Rheum. 1999;42:1902–1907. doi: 10.1002/1529-0131(199909)42:9<1902::AID-ANR16>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 116.Shai R, Quismorio FP, Jr, Li L, Kwon OJ, Morrison J, Wallace DJ, Neuwelt CM, Brautbar C, Gauderman WJ, Jacob CO. Genome-wide screen for systemic lupus erythematosus susceptibility genes in multiplex families. Hum Mol Genet. 1999;8:639–644. doi: 10.1093/hmg/8.4.639. [DOI] [PubMed] [Google Scholar]
- 117.Tsao BP, Cantor RM, Grossman JM, Kim SK, Strong N, Lau CS, Chen CJ, Shen N, Ginzler EM, Goldstein R, Kalunian KC, Arnett FC, Wallace DJ, Hahn BH. Linkage and interaction of loci on 1q23 and 16q12 may contribute to susceptibility to systemic lupus erythematosus. Arthritis Rheum. 2002;46:2928–2936. doi: 10.1002/art.10590. [DOI] [PubMed] [Google Scholar]
- 118.Tsao BP, Cantor RM, Kalunian KC, Chen CJ, Badsha H, Singh R, Wallace DJ, Kitridou RC, Chen SL, Shen N, Song YW, Isenberg DA, Yu CL, Hahn BH, Rotter JI. Evidence for linkage of a candidate chromosome 1 region to human systemic lupus erythematosus. J Clin Invest. 1997;99:725–731. doi: 10.1172/JCI119217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Johanneson B, Lima G, von Salome J, Alarcon-Segovia D, Alarcon-Riquelme ME. A major susceptibility locus for systemic lupus erythematosus maps to chromosome 1q31. Am J Hum Genet. 2002;71:1060–1071. doi: 10.1086/344289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gray-McGuire C, Moser KL, Gaffney PM, Kelly J, Yu H, Olson JM, Jedrey CM, Jacobs KB, Kimberly RP, Neas BR, Rich SS, Behrens TW, Harley JB. Genome scan of human systemic lupus erythematosus by regression modeling: evidence of linkage and epistasis at 4p16–15.2. Am J Hum Genet. 2000;67:1460–1469. doi: 10.1086/316891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wakeland EK, Liu K, Graham RR, Behrens TW. Delineating the genetic basis of systemic lupus erythematosus. Immunity. 2001;15:397–408. doi: 10.1016/s1074-7613(01)00201-1. [DOI] [PubMed] [Google Scholar]
- 122.Vyse TJ, Richardson AM, Walsh E, Farwell L, Daly MJ, Terhorst C, Rioux JD. Mapping autoimmune disease genes in humans: lessons from IBD and SLE. Novartis Found Symp. 2005;267:94–107. [PubMed] [Google Scholar]
- 123.Graham DSC, Vyse TJ, Fortin PR, Montpetit A, Cai Y, Lim S, McKenzie T, Farwell L, Rhodes B, Chad L, Hudson TJ, Sharpe A, Terhorst C, Greenwood CMT, Wither J, Rioux JD. Association of LY9 in UK and Canadian SLE families. Genes Immun. 2008;9:93–102. doi: 10.1038/sj.gene.6364453. [DOI] [PubMed] [Google Scholar]
- 124.Sayos J, Martin M, Chen A, Simarro M, Howie D, Morra M, Engel P, Terhorst C. Cell surface receptors Ly-9 and CD84 recruit the X-linked lymphoproliferative disease gene product SAP. Blood. 2001;97:3867–3874. doi: 10.1182/blood.v97.12.3867. [DOI] [PubMed] [Google Scholar]
- 125.Rozzo S, Vyse T, Drake C, Kotzin B. Effect of genetic background on the contribution of New Zealand Black loci to autoimmune lupus nephritis. Proc Natl Acad Sci. 1996;93:15164–15168. doi: 10.1073/pnas.93.26.15164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kono DH, Burlingame RW, Owens DG, Kuramochi A, Balderas RS, Balomenos D, Theofilopoulos AN. Lupus susceptibility loci in New Zealand mice. Proc Natl Acad Sci. 1994;91:10168–10172. doi: 10.1073/pnas.91.21.10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hogarth MB, Slingsby JH, Allen PJ, Thompson EM, Chandler P, Davies KA, Simpson E, Morley BJ, Walport MJ. Multiple lupus susceptibility loci map to chromosome 1 in BXSB mice. J Immunol. 1998;161:2753–2761. [PubMed] [Google Scholar]
- 128.Morel L, Blenman KR, Croker BP, Wakeland EK. The major murine systemic lupus erythematosus susceptibility locus, Sle1, is a cluster of functionally related genes. Proc Natl Acad Sci. 2001;98:1787–1792. doi: 10.1073/pnas.031336098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wandstrat AE, Nguyen C, Limaye N, Chan AY, Subramanian S, Tian XH, Yim YS, Pertsemlidis A, Garner HR, Jr, Morel L, Wakeland EK. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity. 2004;21:769–780. doi: 10.1016/j.immuni.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 130.Chen YF, Perry D, Boackle SA, Sobel ES, Molina H, Croker BP, Morel L. Several genes contribute to the production of autoreactive B and T cells in the murine lupus susceptibility locus Sle1c. J Immunol. 2005;175:1080–1089. doi: 10.4049/jimmunol.175.2.1080. [DOI] [PubMed] [Google Scholar]
- 131.Chen YF, Cuda C, Morel L. Genetic determination of T cell help in loss of tolerance to nuclear antigens. J Immunol. 2005;174:7692–7702. doi: 10.4049/jimmunol.174.12.7692. [DOI] [PubMed] [Google Scholar]
- 132.Croker BP, Gilkeson G, Morel L. Genetic interactions between susceptibility loci reveal epistatic pathogenic networks in murine lupus. Genes Immun. 2003;4:575–585. doi: 10.1038/sj.gene.6364028. [DOI] [PubMed] [Google Scholar]
- 133.Bygrave AE, Rose KL, Cortes-Hernandez J, Warren J, Rigby RJ, Cook HT, Walport MJ, Vyse TJ, Botto M. Spontaneous autoimmunity in 129 and C57BL/6 mice—implications for autoimmunity described in gene-targeted mice. PLoS Biol. 2004;2:E243. doi: 10.1371/journal.pbio.0020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Carlucci F, Cortes-Hernandez J, Fossati-Jimack L, Bygrave AE, Walport MJ, Vyse TJ, Cook HT, Botto M. Genetic dissection of spontaneous autoimmunity driven by 129-derived chromosome 1 Loci when expressed on C57BL/6 mice. J Immunol. 2007;178:2352–2360. doi: 10.4049/jimmunol.178.4.2352. [DOI] [PubMed] [Google Scholar]
- 135.Chen YF, Morel L. Genetics of T cell defects in lupus. Cell Mol Immunol. 2005;2:403–409. [PubMed] [Google Scholar]
- 136.Kaminski DA, Stavnezer J. Antibody class switching differs among SJL, C57BL/6 and 129 mice. Int Immunol. 2007;19:545–556. doi: 10.1093/intimm/dxm020. [DOI] [PubMed] [Google Scholar]
- 137.Corcoran LM, Metcalf D. IL-5 and Rp105 signaling defects in B cells from commonly used 129 mouse substrains. J Immunol. 1999;163:5836–5842. [PubMed] [Google Scholar]
- 138.McVicar DW, Winkler-Pickett R, Taylor LS, Makrigiannis A, Bennett M, Anderson SK, Ortaldo JR. Aberrant DAP12 signaling in the 129 strain of mice: implications for the analysis of gene-targeted mice. J Immunol. 2002;169:1721–1728. doi: 10.4049/jimmunol.169.4.1721. [DOI] [PubMed] [Google Scholar]
- 139.O'Neill SM, Brady MT, Callanan JJ, Mulcahy G, Joyce P, Mills KH, Dalton JP. Fasciola hepatica infection downregulates Th1 responses in mice. Parasite Immunol. 2000;22:147–155. doi: 10.1046/j.1365-3024.2000.00290.x. [DOI] [PubMed] [Google Scholar]
- 140.Mo XY, Sangster M, Sarawar S, Coleclough C, Doherty PC. Differential antigen burden modulates the gamma interferon but not the immunoglobulin response in mice that vary in susceptibility to Sendai virus pneumonia. J Virol. 1995;69:5592–5598. doi: 10.1128/jvi.69.9.5592-5598.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fritz RB, Zhao ML. Active and passive experimental autoimmune encephalomyelitis in strain 129/J (H-2b) mice. J Neurosci Res. 1996;45:471–474. doi: 10.1002/(SICI)1097-4547(19960815)45:4<471::AID-JNR17>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 142.Mittleman BB, Morse HC, III, Payne SM, Shearer GM, Mozes E. Amelioration of experimental systemic lupus erythematosus (SLE) by retrovirus infection. J Clin Immunol. 1996;16:230–236. doi: 10.1007/BF01541229. [DOI] [PubMed] [Google Scholar]
- 143.Morel L, Blenman KR, Croker BP, Wakeland EK. The major murine systemic lupus erythematosus susceptibility locus, Sle1, is a cluster of functionally related genes. Proc Natl Acad Sci USA. 2001;98:1787–1792. doi: 10.1073/pnas.031336098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xie C, Patel R, Wu T, Zhu J, Henry T, Bhaskarabhatla M, Samudrala R, Tus K, Gong Y, Zhou H, Wakeland EK, Zhou XJ, Mohan C. PI3K/AKT/mTOR hypersignaling in autoimmune lymphoproliferative disease engendered by the epistatic interplay of Sle1b and FASlpr. Int Immunol. 2007;19:509–522. doi: 10.1093/intimm/dxm017. [DOI] [PubMed] [Google Scholar]
- 145.Kumar KR, Li L, Yan M, Bhaskarabhatla M, Mobley AB, Nguyen C, Mooney JM, Schatzle JD, Wakeland EK, Mohan C. Regulation of B cell tolerance by the lupus susceptibility gene Ly108. Science. 2006;312:1665–1669. doi: 10.1126/science.1125893. [DOI] [PubMed] [Google Scholar]
- 146.Jennings P, Chan A, Schwartzberg P, Wakeland EK, Yuan D. Antigen-specific responses and ANA production in B6. Sle1b mice: a role for SAP. J Autoimmun. 2008;31:345–353. doi: 10.1016/j.jaut.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Komori H, Furukawa H, Mori S, Ito MR, Terada M, Zhang MC, Ishii N, Sakuma N, Nose M, Ono M. A signal adaptor SLAM-associated protein regulates spontaneous autoimmunity and Fas-dependent lymphoproliferation in MRL-Faslpr lupus mice. J Immunol. 2006;176:395–400. doi: 10.4049/jimmunol.176.1.395. [DOI] [PubMed] [Google Scholar]
- 148.Linterman MA, Rigby RJ, Wong RK, Yu D, Brink R, Cannons JL, Schwartzberg PL, Cook MC, Walters GD, Vinuesa CG. Follicular helper T cells are required for systemic autoimmunity. J Exp Med. 2009;206:561–576. doi: 10.1084/jem.20081886. [DOI] [PMC free article] [PubMed] [Google Scholar]