Abstract
Background
Immune features of infants with food allergy have not been delineated.
Objectives
To explore basic mechanisms responsible for food allergy and identify biomarkers, e.g. prick skin tests (PST), food-specific IgE, and mononuclear cell responses in a cohort of infants with likely milk/egg allergy at increased risk of developing peanut allergy.
Methods
Infants aged 3–15 months were enrolled with a positive PST to milk or egg and either a corresponding convincing clinical history of allergy to milk or egg, or with moderate to severe atopic dermatitis (AD). Infants with known peanut allergy were excluded.
Results
Overall, 512 infants (67% males) were studied with 308 (60%) having a history of a clinical reaction. Skin tests and/or detectable food-specific IgE revealed sensitization as follows: milk-78%, egg-89% and peanut-69%. PST and food-specific IgE levels were discrepant for peanut: 15% IgE ≥ 0.35 kUA/L/PST- versus 8% PST+/IgE < 0.35, p = 0.001. Mononuclear cell allergen stimulation screening for CD25, CISH, FOXP3, GATA3, IL-10, IL-4, IFN-gamma and TBET expression using casein, egg white and peanut revealed that only allergen-induced IL-4 expression was significantly increased in those with clinical allergy to milk (compared to non-allergic) and in those sensitized to peanut, despite the absence of an increase in GATA-3 mRNA expression.
Conclusions
Infants with likely milk/egg allergy are at considerably high risk of having elevated peanut-specific IgE (potential allergy). Peanut-specific serum IgE was a more sensitive indicator of sensitization than PST. Allergen-specific IL-4 expression may be a marker of allergic risk. Absence of an increase in GATA-3 mRNA expression suggests that allergen-specific IL-4 may not be of T cell origin.
Keywords: food allergy, sensitization, atopy
INTRODUCTION
Food allergy is estimated to affect approximately 4–6% of young children, with egg, milk and peanut allergy being the most common.(1;2) Food allergy appears to be increasing in prevalence in westernized countries, with specific evidence for an increase in peanut allergy in children within the past decade.(3–5) Although allergy to egg and milk typically resolves over time, peanut allergy typically persists,(6) and studies indicate that milk and egg allergies are also becoming more persistent.(7;8) While genetic factors clearly predispose an individual to food allergy,(9) the observed recent increases and persistence of childhood food allergies must be attributable to environmental factors. Food allergy can be life-threatening or fatal(10) and seriously impacts quality of life.(11) Considering the increasing prevalence and seriousness of this disease, studies to determine risk factors, prevention strategies, better diagnostic tests and definitive treatments are needed.
The Consortium of Food Allergy Research (CoFAR), funded by the National Institutes of Allergy and Infectious Diseases, is comprised of 5 clinical sites in the United States with established expertise in food allergic disorders that are undertaking observational and treatment studies of food allergy. To address the immunologic, genetic and environmental factors that impact the course of food allergy, we established a cohort of infants with likely egg and/or milk allergy who are predicted to be at increased risk to have or develop peanut allergy. These children will be followed longitudinally to determine the course of their egg and milk allergies as well as the development or resolution of peanut allergy.
Insights into the basic mechanisms responsible for the development of food allergies are lacking. In the current study, we sought to determine if mononuclear cell expression of key cytokine and regulatory genes were markers of current milk/egg allergy or associated with sensitization to peanut in these infants. In what we believe to be the largest and most comprehensive study to date, we present data confirming the expected importance of IL-4, but do not detect an associated increase in GATA-3 transcripts or a shift in the ratio of GATA-3/TBET, findings that raise questions as to the role of T cells in the early development of food allergy. The infants enrolled into the study have demographic features representing common food allergy presentations; however, we found clinically important observations including an unexpectedly high rate of peanut sensitization, and we characterize discrepancies in common diagnostic tests in this large cohort, observing that serum testing for peanut sensitization is more sensitive that PST.
METHODS
Study rationale, design and enrollment criteria
We aim to study markers of development of peanut allergy and the natural course of egg/milk allergy and therefore we did not intend to enroll children with likely peanut allergy, only those “at risk”. We developed enrollment criteria for this cohort based upon the results of various studies of childhood food allergies and atopic dermatitis (AD),(12–16) estimating that children with a convincing clinical reaction to milk and/or egg with a positive PST to the responsible food and/or children with moderate to severe AD and a positive prick skin test (PST) to milk and/or egg would be likely (~20–30%) to develop a peanut allergy and have a 25–50% chance of resolving their egg or milk allergy by 5–7 years of age. We enrolled children ages 3 to 15 months to allow adequate recall of their feeding history and to reduce the chance that they already would have a peanut allergy. Infants known to have a peanut allergy or a peanut-specific IgE level > 5 kUA/L (representing a more likely current peanut allergy as described in the Online Repository) prior to screening were therefore excluded from enrollment because they already had evidence of peanut allergy/sensitization and therefore could not be evaluated for the basic question to be addressed prospectively. To maintain uniformity and an observational approach, the study design includes evaluations, care for food allergy, and instructions on dietary management that were uniform among the 5 clinical centers, and reflect practice parameters for AD,(17) food allergy,(18) and the American Academy of Pediatrics recommendations for allergy prevention published in 2000, which were current at the initiation of the study.(19)
Enrollment required either 1) a history of a convincing immediate allergic reaction to cow’s milk (and/or egg) as described further in the ONLINE REPOSITORY and a positive prick skin test (≥ ,3 mm larger than the negative control) to cow’s milk (and/or egg, if the clinical reaction was to egg), and/or 2) moderate to severe AD at the time of enrollment (as described in the ONLINE REPOSITORY), and a positive prick skin test to milk and/or egg.
Children were excluded if they had chronic disease (other than asthma, AD, rhinitis) requiring therapy (e.g., heart disease, diabetes), were participating in any interventional study, were unable to discontinue antihistamines for routine tests, had a sibling enrolled in the observational study, or already had confirmed or convincing evidence of peanut allergy (see ONLINE REPOSITORY). Study procedures were reviewed and approved by an NIAID Data Safety Monitoring Board and by local site Institutional Review Boards and written signed consents were obtained.
Definitions of atopic diseases, categorization of food allergy
Atopic diseases (asthma, allergic rhinitis, AD) were diagnosed and graded by severity as described in the ONLINE REPOSITORY. Food allergy was diagnosed based upon clinical criteria (described in the ONLINE REPOSITORY) because as an observational natural history study, repeated scheduled oral food challenges could not be imposed. At enrollment, diagnoses were categorized as confirmed or convincing when there was a clear clinical history and sensitization to the causal food, and in this report these children are categorized as allergic. Those ingesting the food or lacking sensitization are categorized as not allergic. These two clinical categories are the primary clinical endpoints evaluated in the current study. Additional enrollees with potential allergy are described in the ONLINE REPOSITORY. The total study group was also evaluated with regard to their sensitization status as described below.
Prick skin tests (PST)
PSTs were performed using the GreerPick® (Lenoir, NC) with participants avoiding antihistamines for at least 5 half-lives of the specific agent. Tests were performed on the infant’s back and at 15 minutes the wheal was outlined in pen and transferred by tape to paper. The size of the longest diameter and its longest perpendicular were averaged. Additional details are in the ONLINE REPOSITORY.
Plasma food-specific IgE assay
The concentration of specific IgE antibody to egg, milk and peanut were measured from plasma at a single central laboratory using the ImmunoCAP® system (Phadia, Uppsala, Sweden) reported in kUA/L. Those at or above 0.35 kUA/L are described as IgE sensitized.
Mononuclear cell stimulation and PCR analysis
Peripheral blood mononuclear cell isolation was performed by Ficoll-Paque density gradient centrifugation and cultures were performed at each clinical site on fresh venous blood samples. The lab processing protocol was standardized and reproducibility across sites was confirmed in pilot studies. Four million cells per condition were cultured for 48 hours in AIM-V serum-free media (Invitrogen) with aqueous peanut extract (50 μg/ml), tetanus toxoid (5 μg/ml), purified α, β, and κ-caseins (50 μg each/ml), and egg white protein (50 μg/ml). Control stimulations were performed with medium alone and anti-CD3/-CD28 beads (5 μl; DYNAL). At the end of the culture period, cells expressing CD25 were enriched by selection with anti-CD25 coated para-magnetic beads according to the manufacturer’s protocol (Miltenyi). Pilot experiments demonstrated ~10-fold enrichment of CD25+ cells with 70–80% of selected cells co-expressing CD3, CD4 and CD25 as measured by flow cytometry. The entire selected fraction of cells was immediately lysed in RLT buffer (Qiagen) containing ~0.14M β-mercaptoethanol and stored at −80°C until RNA purification. Semi-automated RNA isolation was performed using RNeasy Micro kit on the QIACube robotic centrifuge (both by Qiagen) according to the manufacturer’s protocols adjusted to the smaller cell lysate volume of ~100 μl. A two-step quantitative RT-PCR protocol was implemented. The first strand cDNA was synthesized using the TaqMan kit (Applied Biosystems). The quantitative polymerase chain reaction (qPCR) was carried out according to the in-house established protocol utilizing SYBR Green I fluorescence detection in a 384 well plate on ABI 7900 (Applied Biosystems).
Raw PCR analysis and annotation were performed on coded samples. Threshold cycle number (Ct) was set by software with confirmation and adjustment as necessary to define the threshold of linear amplification. Upon reviewing data and filtering out artifacts (e.g. by inspection of amplification curves and dissociation analysis) data were exported into Excel for data upload to the central database for further analysis. For the gene expression data, ddCt was calculated by subtracting the RPS9 reporter gene Ct and then normalizing by subtracting the standardized medium control response. Negative values indicate relatively higher activity with ‘fold-increase’ relative to the medium control calculated as 2-ddCt. Non-detected genes were arbitrarily assigned a Ct of 40. Due to the significant cohort size, the qPCR was performed in one repetition for each gene per sample. Data were compared for those with confirmed/convincing milk allergy (versus those not-allergic) and for those sensitized (serum IgE > 0.35 kUA/L) or not for all 3 foods.
Statistics
Standard descriptive measures and univariate methods were used including McNemar’s test for paired data. Log transformed IgE values were analyzed with non-detectable values set to 0.001. Relationships among the standardized gene ddCt values were examined with principal component analyses, a multivariate analysis technique that constructs coordinate systems based on linear combinations of the gene expression levels to identify potential important combined influences.(20)
RESULTS
Demographic characteristics
A total of 512 children were enrolled and included in these analyses. However, blood samples were not obtained for technical reasons from 9 participants whose results are therefore excluded in figures and tables concerning IgE results. Specific food skin tests to milk (n=1) and egg (n=1) were not performed in 2 infants due to a recent history of severe anaphylaxis to the trigger food, but these children qualified for enrollment by fulfilling criteria that did not rely on these results and had sensitization to the respective foods by blood tests. There were 287 children screened who were not enrolled because PSTs to milk and egg were negative. There were 104 subjects excluded from enrollment at time of screening because of likely peanut allergy (e.g., peanut-IgE > 5 kUA/L) having been determined prior to the enrollment blood test. Overall, 308 (60.2%) were enrolled with clinical reactions to egg and/or milk.
Table E1 in the ONLINE REPOSITORY shows the demographic features of the study participants. Among 308 infants enrolled with a clinical reaction, 71.4% reacted to cow’s milk, 39.9% to egg, and 11.4% to both foods. Among the 204 enrolled with AD but no acute food-allergic reactions, 116 (56.9%) had a positive PST to cow’s milk, 181 (88.7%) to egg, and 93 (45.6%) had skin test sensitivity to both. Table 1 shows the clinical categories of the enrolled infants and Table E2 shows sensitization rates to milk, egg, and peanut. Although we specifically avoided enrolling subjects with known clinical allergic symptoms when exposed to peanut, as described above, 69% of the study group had evidence of sensitization to peanut. Of note, 26.6% had a significantly elevated peanut-IgE level (> 5 kUA/L) at enrollment.
Table 1.
Food allergy diagnostic categories at time of enrollment (N=503) (see ONLINE REPOSITORY for definitions).
| Milk | Egg | Peanut | ||||
|---|---|---|---|---|---|---|
| N | % | N | % | N | % | |
| Confirmed IgE mediated reaction (approx. 97% certainty) | 88 | 17.5 | 69 | 13.7 | - | - |
| Convincing, but not confirmed IgE mediated reaction (approx. 95% accurate) | 155 | 30.8 | 72 | 14.3 | - | - |
| Serological diagnosis | 24 | 4.8 | 165 | 32.8 | 134 | 26.6 |
| Potential allergy | 116 | 23.1 | 125 | 24.9 | 195 | 38.8 |
| Not allergic-sensitized | 43 | 8.6 | 16 | 3.2 | 17 | 3.4 |
| Not allergic-not sensitized | 63 | 12.5 | 6 | 1.2 | 8 | 1.6 |
| Not sensitized-never ingested | 14 | 2.8 | 50 | 9.9 | 149 | 29.6 |
Sensitization
We performed analyses to compare PST and serum IgE with regard to their relative sensitivity in detecting food-specific IgE (not clinical outcomes). Enrollment criteria required subjects to have a positive PST to milk and/or egg and so analysis of peanut is least affected by this bias. Figure E1a–c in the ONLINE REPOSITORY shows the relationship of skin test size to serum IgE levels for milk, egg and peanut. Of note, there are discordances where one sensitization test is negative while the other is positive. Table 2 summarizes the discrepancies for the entire study group by calculating the percent who are determined to be sensitized by serum IgE among those with a negative PST, (e.g., PSTnegative with IgEpositive/[(PSTnegative with IgEpositive) + (PSTnegative with IgEnegative)]) and the percent determined to be sensitized by PST among all of those with a undetectable IgE. When evaluating only participants with confirmed or convincing milk/egg allergies, test discordance was less pronounced as shown in Figure E1a–b (Milk 5.4% IgE ≥ 0.35 kUA/L/PST- versus 20.2% PST+/IgE < 0.35 kUA/L; Egg 0% IgE ≥ 0.35 kUA/L/PST- versus 9.4% PST+/IgE < 0.35 kUA/L The pattern of discrepancy for peanut indicates that the serum test was more sensitive than the PST (p=0.001, McNemar) in detecting a sensitized participant. Of note, enrollees were required to have a positive skin test to milk/egg and therefore only the analysis of peanut sensitization comparing serum with PST results are unaffected by this type of selection bias. Conversely, despite the exclusion of infants with known peanut-specific IgE > 5 kUA/L prior to enrollment, the serum test was still more likely to detect sensitization than the skin test.
Table 2.
Discrepancies in detection of specific IgE by PST versus serum IgE
| Allergen | Percent with detectable serum IgE among all of those with a negative PST | Percent with a positive PST among all those with undetectable serum IgE |
|---|---|---|
| Milk* | 28% | 42% |
| Egg* | 12% | 56% |
| Peanut | 33% | 21% |
The data show that serum IgE was more sensitive than PST for detecting sensitization to peanut (p = 0.001). Data for milk and egg must be interpreted with caution because enrollment required a positive PST to one or both of these allergens. Calculations are based upon the entire study group; see text for sub-analysis of children with confirmed/convincing milk/egg allergy.
Comparison of clinical allergy: mononuclear cell gene expression
Quality control measures for PCR reactions and inter-site processing are described in the ONLINE REPOSITORY, including Figure E2 and Figure E3 that demonstrate consistency of PCR products for housekeeping genes and results obtained across the 5 sites.
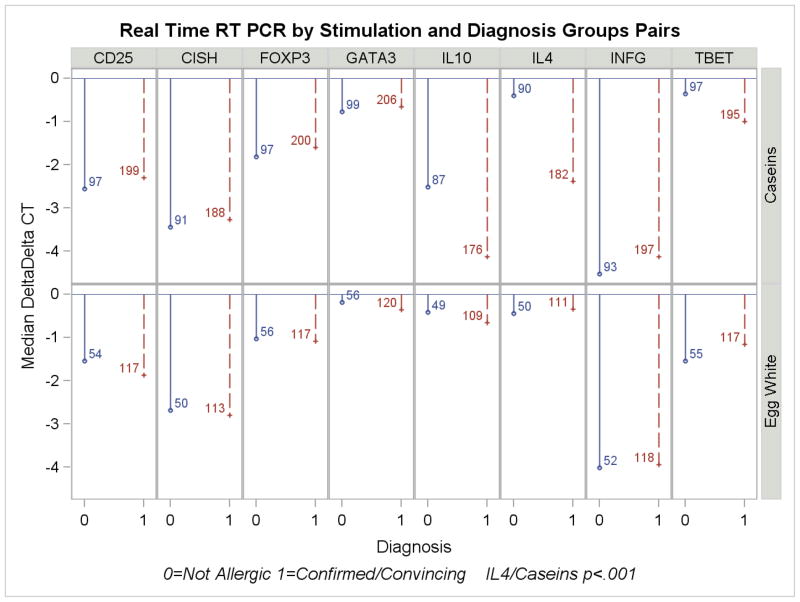
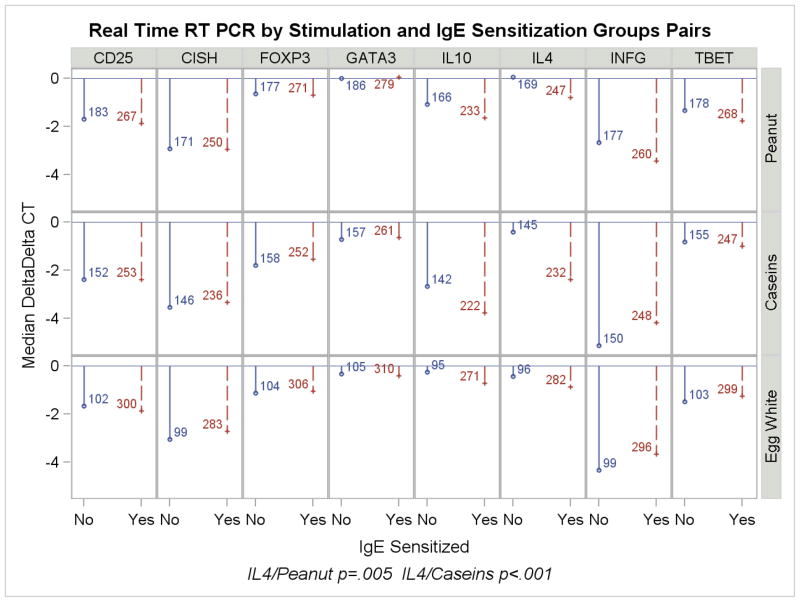
For purposes of analyzing the outcomes related to gene transcription responses, allergic phenotypes were categorized in two ways: 1) allergic (e.g., clinical reaction) versus not allergic (e.g., clinically tolerant or not sensitized); and 2) sensitized (by serum results) versus not-sensitized. The results of stimulation with egg white, caseins and peanut are shown according to calculated delta-delta Ct in Figure 1a,b) and raw data, including controls with tetanus and anti-CD3/28, are presented in Table E3 in the ONLINE REPOSITORY. By design, no enrollees had confirmed clinical peanut allergy and therefore gene expression induced by peanut is categorized by serologic status.
Figure 1.
a,b: Real time RT PCR results by stimulant comparing children with confirmed/convincing milk or egg allergy (1a) or sensitization status to milk, egg or peanut (1b). Values shown are median delta-delta Ct as described in the text. Negative values indicate increasing expression. The number of infants in each category is shown alongside the ddCt value.
Following 48 hours stimulation in culture, cells expressing CD25+ constitutively or as a consequence of activation were selected by bead-coupled monoclonal antibody [CD3+CD4+CD25+ = 70% – 80%], mRNA was isolated and expression later analyzed by RT-PCR. Consistent with other reports of allergen-induced immune responses in food allergic children,(21;22) significantly higher IL-4 expression was associated with both allergy to milk (allergic vs. not-allergic; ratio of medians, 3.9 fold, p < .001) and with sensitization to milk or peanut (3.9 fold [p <.001] and 1.8 fold [p=.005], respectively). There was no evidence of increased regulatory T cell responses in any group as measured by the levels of FoxP3 and IL-10 expression. Despite the association of increased IL-4 with clinical allergy and IgE sensitization, there was no evidence of preferential activation (i.e., CD25 upregulation) of Th2-polarized T cells in allergic or sensitized individuals, as indicated by the absence of significant differences in either GATA-3 expression or the GATA-3/TBET ratio.
Principal component analyses failed to identify other important multivariate relationships that discriminated between the clinical groups. IgE and gene products relationships were examined and only milk and peanut-specific IgE showed weak correlations with induction of food-specific IL-4 (Spearman r=−.25 p<0.001, r=−.14 p=.003, respectively for ddCt).
DISCUSSION
It is estimated that 2.5% of young children have milk allergy, 1–2% have egg allergy and 1% have peanut allergy.(2;5;23–25) and that these allergies are increasing in prevalence and becoming more persistent.(3;7;8;26) However, little is known about the underlying mechanisms resulting in the development of allergy or tolerance to these foods.(2;26) The cohort described herein was established to evaluate and elucidate environmental, genetic and immunologic factors associated with the development or persistence of these common food allergies with the goal of determining better diagnostic and therapeutic modalities. The cohort will be followed longitudinally in this observational study to characterize the outcomes of milk, egg, and peanut allergy, including the conduct of oral food challenges as clinically indicated. The current study focuses upon immune characteristics associated with clinical allergy and sensitization in these infants at enrollment.
We undertook analysis of activated mononuclear cell expression of several key markers of immune regulation and Th1/Th2 bias. We found that allergen-induced IL-4 expression in peripheral mononuclear cells of this high risk cohort was associated with clinical allergy to milk and IgE-sensitization to milk and peanut. This is consistent with other reports of a Th2 bias in food allergy as well as with our current understanding of the role of IL-4 in inducing IgE class switching in activated B cells.(27) Differentiated CD4 Th2 cells have been shown to express high levels of GATA-3 and low levels of TBET -- master transcriptional regulators for many Th2 and Th1 genes, respectively.(28) We hypothesized that allergen-specific T cells activated in vitro would be predominantly Th2 effector memory cells expressing a high GATA-3/TBET ratio and that selection of activated cells on the basis of their upregulation of CD25 would be reflected by an increase in the GATA-3/TBET ratio of transcripts detectable by PCR. Though previous studies have associated Th2 immune responses with food allergy, the immune phenotype of such young children with early evidence of food allergy or sensitization has not been previously studied on this scale. We did not find evidence of an increased GATA-3/TBET ratio with allergen activation. The lack of detectable enrichment for GATA-3 expressing cells was not due to a failure of detecting GATA-3, which was readily detectable and expressed at higher levels on average than IL-4 and several other transcripts (see Table E5, ONLINE REPOSITORY). Activation of cells, reflected by an increase in CD25 expression over medium alone was also detectable (not shown) though it did not differ significantly between clinical groups (Figure 1). This finding suggests that strongly polarized allergen-specific T cell differentiation may not have occurred in these subjects, even though they have produced IgE in vivo and allergen stimulation in vitro induced IL-4. In this study we looked specifically at CD25+ cells isolated following stimulation with antigen in order to capture both potential regulatory (constitutive CD25 expression) and activated (induced CD25 expression) T cell populations, however it is known that CD25 is also induced or constitutively expressed on many non-lymphoid cells, including NK T cells and basophils, potent potential sources of IL-4.
Basophils constitutively express CD25,(29) are enriched with mononuclear cells during density gradient isolation and produce high levels of IL-4 in sensitized subjects.(30) Recently, basophils have been implicated in allergy model systems for playing an important role in priming and enhancing memory Th2 responses.(31–33) Murine basophils have been shown to express IL-4 in the absence of detectable GATA-3 or c-maf, expression, suggesting that IL-4 may be regulated distinctly in these cells.(29) Thus, new paradigms are emerging that basophils play a key role in directing Th2 responses and our data may provide additional support for this observation. Preliminary studies utilizing flow cytometry revealed that basophils (CD3- CD123+ CD203+ HLA-DR dim and IL4+, data not shown) are present in the CD25 preparation. NKT cells can also produce significant IL-4, although the role of these cells in allergic sensitization or disease is controversial.
We cannot rule out the possibility that the frequency of Th2 effector subsets is so low in the peripheral blood that as a fraction of total CD25+ cells enriched by selection, the overall impact on GATA-3 transcript number on the subsequently isolated mRNA is too small to be detected, while the activation-induced change in IL4 in those same few cells is detectable. There were not sufficient cell numbers to both phenotype the CD25+ fraction and perform RT-PCR in our initial protocol, and therefore we cannot draw clear conclusions regarding the potential role of non-T cells. We are currently investigating the potential role for these populations in the observed in vitro IL-4 response. It will also be interesting to observe in this cohort, whether a T cell signature of Th2 polarization develops over time and if the early IL-4 response, potentially from other cells, proves to be important for priming clinical allergy. If so, this would corroborate findings in animal models and suggest new targets for primary prevention of disease.
We observed several differences in gene expression in response to egg stimulation compared to stimulation with milk or peanut. CD25 expression was significantly but only slightly increased for those with egg allergy but IL4 expression was not increased. The observed increase in CD25 expression following egg stimulation in vitro was marginal and did not reach the magnitude of significance observed for IL4 expression following in vitro stimulation with milk or peanut. The explanation for the difference in response to egg compared to milk and peanut (e.g., lack of IL4 increase) remains uncertain, but may be related to particular proteins used in the stimulation experiments. For example, the antigens used for milk stimulation were comprised of caseins, the major milk allergens, whereas the egg stimulation was performed with whole egg protein extracts. In future studies we will explore whether enhancement with specific egg proteins (e.g., Gal d 1) results in different patterns of response.
In addition to the observed immunologic characteristics, several key clinically-relevant observations were made. Although children with known peanut allergy or previously known elevated (> 5 kUA/L ) peanut-specific IgE were not eligible for enrollment, there was a strikingly high rate of peanut sensitization and likely peanut allergy: 69% of the cohort were sensitized and 28% of 503 with IgE quantization had IgE concentrations to peanut over 5 kUA/L, which we estimate is likely associated with clinical peanut allergy (as described in the ONLINE REPOSITORY). Although there are no prior studies with precisely our enrollment criteria, a large international study of infants with active AD where 68% had moderate-severe eczema, 24% were sensitized to peanut.(15) Our observation indicates that infants presenting with likely milk/egg allergy without already known peanut allergy are at high risk of also having peanut sensitization and, therefore, possible peanut allergy. A recent report concerning the young age of presentation of children with peanut allergy supports the concern that young egg or milk allergic children, or eczematous children, are at high risk for exhibiting peanut allergy on their first known ingestion.(34)
We evaluated the relative sensitivity of PST and serum IgE in detecting sensitization (not clinical allergy) in this cohort because there are currently very limited data about these relationships in young potentially food-allergic infants/children. We found a strong relationship between PST wheal size and serum IgE concentration for the 3 foods. As indicated in Table 2 and Figures E1a–c, when tests were discordant for peanut, the serum IgE test was more sensitive than the skin tests in detecting peanut sensitization (i.e., 15% of those tested had detectable peanut-specific serum IgE but negative peanut PST compared to 8% who had undetectable peanut IgE and a positive peanut PST, p < 0.001), which is in contrast to general conceptions that PST are more sensitive than serum tests. The reason for this observation could include our use of a more sensitive serum assay than previous generation tests,(35) reduced skin test reactivity in infants,(36) or other factors such as differences in the proteins being tested in the 2 methods. These findings are important when selecting tests to evaluate infants for possible food hypersensitivity, and substantially dispel the long held notion that skin testing is uniformly more sensitive (more likely to be positive) than serologic testing, especially in younger children.(37) Unlike the test results for peanut, those for milk and egg must be interpreted with caution and may be biased because a positive PST to milk or egg was required for enrollment into the study. Conversely, we did not enroll infants with serum testing strongly positive (>5 kUA/L) to peanut prior to their screening, so the bias would have been against our finding the serum test to be a more sensitive indicator of sensitization. We emphasize that sensitization to peanut is not indicative of peanut allergy.(38) We are therefore not discussing clinical allergy outcomes but detection of IgE by these methods. As this cohort ages, we will determine whether elevated IgE levels in the infants are associated with clinical reactions.
In summary, we have established a cohort of infants with likely egg and/or milk allergy who will be studied longitudinally for the resolution of egg and milk allergies or the development peanut allergy. We have focused upon relationships of concurrent immune determinants with clinical allergy to milk/egg and sensitization to the 3 most common early childhood food allergens, egg, milk and peanut. Data about parallel PST and serum IgE tests reveal that these tests are highly correlated, but there are also potentially significant discrepancies between them. Mononuclear cell stimulation results indicate that IL-4 expression is associated with allergy to milk and sensitization to milk/peanut in the absence of detectable induction of GATA-3 in these cells. Lastly, we documented an alarmingly high rate of peanut sensitization (68.8%), including 26.6% with a significantly elevated peanut-specific IgE level > 5 kUA/L. This observation may have clinical implications for pediatricians and allergists evaluating young children with evidence of milk and/or egg allergy or more than mild AD and evidence of sensitization to milk or egg. The results underscore the need for caution in managing these infants’ early diet. Specifically, children with these clinical features may represent those for whom testing for food allergy is appropriate prior to advancing the diet.
Supplementary Material
Acknowledgments
Supported by NIH-NIAID U19AI066738 and U01AI066560. We also acknowledge the National Center for Research Resources supported Clinical Research Centers: RR-024128 (Duke), RR-00052 (Johns Hopkins University School of Medicine) and the Clinical Translational Scientific Award UL1 RR025780 (National Jewish), UL1 RR 029887 (Mount Sinai).
Additional Site Investigators: D Atkins, TT Perry, AM Scurlock, M Masilamani, B Vickery.
Coordinators and support: D Brown, M Mishoe, S Walsh, L Talarico, S Noone, M Beksinska, J Grabowska, K Mudd, S Driggers, P Steele, J Kamilaris, S Carlisle,T Hubbart, A Hiegel, L Christie, M Groetch, J Slinkard, J Stone, S Leung, K Morgan, and K Brown-Engelhardt. We thank M Plaut, MD, Medical Officer for the program and J Poyser. We thank the families who kindly participated.
We thank Greer (Lenoir, NC) and Phadia (Uppsala, Sweden) for providing reagents.
Abbreviations
- AD
atopic dermatitis
- CoFAR
Consortium of Food Allergy Research
- PST
prick skin test
- CISH
cytokine-inducible SH2-containing protein
Footnotes
Clinical Implication: Infants presenting with an acute allergic reaction to milk or egg, or with moderate to severe atopic dermatitis (and evidence of sensitization to milk/egg), are at increased risk for peanut sensitization (potential allergy).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rona RJ, Keil T, Summers C, Gislason D, Zuidmeer L, Sodergren E, et al. The prevalence of food allergy: a meta-analysis. J Allergy Clin Immunol. 2007;120(3):638–46. doi: 10.1016/j.jaci.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 2.Sicherer SH, Sampson HA. 9. Food allergy. J Allergy Clin Immunol. 2006;117(2 Suppl Mini-Primer):S470–S475. doi: 10.1016/j.jaci.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 3.Branum AM, Lukacs SL. Food Allergy Among Children in the United States. Pediatrics. 2009 doi: 10.1542/peds.2009-1210. [DOI] [PubMed] [Google Scholar]
- 4.Sicherer SH, Munoz-Furlong A, Sampson HA. Prevalence of peanut and tree nut allergy in the United States determined by means of a random digit dial telephone survey: a 5-year follow-up study. J Allergy Clin Immunol. 2003;112(6):1203–7. doi: 10.1016/s0091-6749(03)02026-8. [DOI] [PubMed] [Google Scholar]
- 5.Hourihane JO, Aiken R, Briggs R, Gudgeon LA, Grimshaw KE, Dunngalvin A, et al. The impact of government advice to pregnant mothers regarding peanut avoidance on the prevalence of peanut allergy in United Kingdom children at school entry. J Allergy Clin Immunol. 2007;119(5):1197–202. doi: 10.1016/j.jaci.2006.12.670. [DOI] [PubMed] [Google Scholar]
- 6.Fleischer DM, Conover-Walker MK, Christie L, Burks AW, Wood RA. The natural progression of peanut allergy: Resolution and the possibility of recurrence. J Allergy Clin Immunol. 2003;112(1):183–9. doi: 10.1067/mai.2003.1517. [DOI] [PubMed] [Google Scholar]
- 7.Savage JH, Matsui EC, Skripak JM, Wood RA. The natural history of egg allergy. J Allergy Clin Immunol. 2007;120(6):1413–7. doi: 10.1016/j.jaci.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 8.Skripak JM, Matsui EC, Mudd K, Wood RA. The natural history of IgE-mediated cow’s milk allergy. J Allergy Clin Immunol. 2007;120(5):1172–7. doi: 10.1016/j.jaci.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 9.Sicherer SH, Furlong TJ, Maes HH, Desnick RJ, Sampson HA, Gelb BD. Genetics of peanut allergy: a twin study. J Allergy Clin Immunol. 2000;106(1 Pt 1):53–6. doi: 10.1067/mai.2000.108105. [DOI] [PubMed] [Google Scholar]
- 10.Bock SA, Munoz-Furlong A, Sampson HA. Further fatalities caused by anaphylactic reactions to food, 2001–2006. J Allergy Clin Immunol. 2007;119(4):1016–8. doi: 10.1016/j.jaci.2006.12.622. [DOI] [PubMed] [Google Scholar]
- 11.Cohen BL, Noone S, Munoz-Furlong A, Sicherer SH. Development of a questionnaire to measure quality of life in families with a child with food allergy. J Allergy Clin Immunol. 2004;114(5):1159–63. doi: 10.1016/j.jaci.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Burks AW, James JM, Hiegel A, Wilson G, Wheeler JG, Jones SM, et al. Atopic dermatitis and food hypersensitivity reactions. J Pediatr. 1998;132(1):132–6. doi: 10.1016/s0022-3476(98)70498-6. [DOI] [PubMed] [Google Scholar]
- 13.Sampson HA, Scanlon SM. Natural history of food hypersensitivity in children with atopic dermatitis. J Pediatr. 1989;115:23–7. doi: 10.1016/s0022-3476(89)80323-3. [DOI] [PubMed] [Google Scholar]
- 14.Eigenmann PA, Sicherer SH, Borkowski TA, Cohen BA, Sampson HA. Prevalence of IgE-mediated food allergy among children with atopic dermatitis. Pediatrics. 1998;101(3):E8. doi: 10.1542/peds.101.3.e8. [DOI] [PubMed] [Google Scholar]
- 15.Hill DJ, Hosking CS, de Benedictis FM, Oranje AP, Diepgen TL, Bauchau V. Confirmation of the association between high levels of immunoglobulin E food sensitization and eczema in infancy: an international study. Clin Exp Allergy. 2008;38(1):161–8. doi: 10.1111/j.1365-2222.2007.02861.x. [DOI] [PubMed] [Google Scholar]
- 16.Hill DJ, Hosking CS. The cow milk allergy complex: overlapping disease profiles in infancy. Eur J Clin Nutr. 1995;49 (Suppl 1):S1–12. [PubMed] [Google Scholar]
- 17.Leung DY, Nicklas RA, Li JT, Bernstein IL, Blessing-Moore J, Boguniewicz M, et al. Disease management of atopic dermatitis: an updated practice parameter. Joint Task Force on Practice Parameters. Ann Allergy Asthma Immunol. 2004;93(3 Suppl 2):S1–21. doi: 10.1016/s1081-1206(10)61385-3. [DOI] [PubMed] [Google Scholar]
- 18.Food allergy: a practice parameter. Ann Allergy Asthma Immunol. 2006;96(3 Suppl 2):S1–68. [PubMed] [Google Scholar]
- 19.American Academy of Pediatrics. Committee on Nutrition. Hypoallergenic infant formulas. Pediatrics. 2000;106(2 Pt 1):346–9. [PubMed] [Google Scholar]
- 20.Anderson TW. Introduction to Multivariate Statistical Analysis. 2. John Wiley and Sons; 1984. [Google Scholar]
- 21.Tiemessen MM, Van Ieperen-Van Dijk AG, Bruijnzeel-Koomen CA, Garssen J, Knol EF, Van Hoffen E. Cow’s milk-specific T-cell reactivity of children with and without persistent cow’s milk allergy: key role for IL-10. J Allergy Clin Immunol. 2004;113(5):932–9. doi: 10.1016/j.jaci.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Tsuge I, Kondo Y, Tokuda R, Kakami M, Kawamura M, Nakajima Y, et al. Allergen-specific helper T cell response in patients with cow’s milk allergy: Simultaneous analysis of proliferation and cytokine production by carboxyfluorescein succinimidyl ester dilution assay. Clin Exp Allergy. 2006;36(12):1538–45. doi: 10.1111/j.1365-2222.2006.02600.x. [DOI] [PubMed] [Google Scholar]
- 23.Eggesbo M, Botten G, Halvorsen R, Magnus P. The prevalence of allergy to egg: a population-based study in young children. Allergy. 2001;56(5):403–11. doi: 10.1034/j.1398-9995.2001.056005403.x. [DOI] [PubMed] [Google Scholar]
- 24.Eggesbo M, Botten G, Halvorsen R, Magnus P. The prevalence of CMA/CMPI in young children: the validity of parentally perceived reactions in a population-based study. Allergy. 2001;56(5):393–402. doi: 10.1034/j.1398-9995.2001.056005393.x. [DOI] [PubMed] [Google Scholar]
- 25.Kagan RS, Joseph L, Dufresne C, Gray-Donald K, Turnbull E, Pierre YS, et al. Prevalence of peanut allergy in primary-school children in Montreal, Canada. J Allergy Clin Immunol. 2003;112(6):1223–8. doi: 10.1016/j.jaci.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 26.Sicherer SH, Sampson HA. Peanut allergy: emerging concepts and approaches for an apparent epidemic. J Allergy Clin Immunol. 2007;120(3):491–503. doi: 10.1016/j.jaci.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 27.Lebman DA, Coffman RL. Interleukin 4 causes isotype switching to IgE in T cell-stimulated clonal B cell cultures. J Exp Med. 1988;168(3):853–62. doi: 10.1084/jem.168.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grogan JL, Locksley RM. T helper cell differentiation: on again, off again. Curr Opin Immunol. 2002;14(3):366–72. doi: 10.1016/s0952-7915(02)00340-0. [DOI] [PubMed] [Google Scholar]
- 29.Min B, Prout M, Hu-Li J, Zhu J, Jankovic D, Morgan ES, et al. Basophils produce IL-4 and accumulate in tissues after infection with a Th2-inducing parasite. J Exp Med. 2004;200(4):507–17. doi: 10.1084/jem.20040590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devouassoux G, Foster B, Scott LM, Metcalfe DD, Prussin C. Frequency and characterization of antigen-specific IL-4- and IL-13- producing basophils and T cells in peripheral blood of healthy and asthmatic subjects. J Allergy Clin Immunol. 1999;104(4 Pt 1):811–9. doi: 10.1016/s0091-6749(99)70292-7. [DOI] [PubMed] [Google Scholar]
- 31.Denzel A, Maus UA, Rodriguez GM, Moll C, Niedermeier M, Winter C, et al. Basophils enhance immunological memory responses. Nat Immunol. 2008;9(7):733–42. doi: 10.1038/ni.1621. [DOI] [PubMed] [Google Scholar]
- 32.Mukai K, Matsuoka K, Taya C, Suzuki H, Yokozeki H, Nishioka K, et al. Basophils play a critical role in the development of IgE-mediated chronic allergic inflammation independently of T cells and mast cells. Immunity. 2005;23(2):191–202. doi: 10.1016/j.immuni.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 33.Sokol CL, Barton GM, Farr AG, Medzhitov R. A mechanism for the initiation of allergen-induced T helper type 2 responses. Nat Immunol. 2008;9(3):310–8. doi: 10.1038/ni1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green TD, LaBelle VS, Steele PH, Kim EH, Lee LA, Mankad VS, et al. Clinical characteristics of peanut-allergic children: recent changes. Pediatrics. 2007;120(6):1304–10. doi: 10.1542/peds.2007-0350. [DOI] [PubMed] [Google Scholar]
- 35.Sampson HA, Albergo R. Comparison of results of skin tests, RAST, and double-blind, placebo-controlled food challenges in children with atopic dermatitis. J Allergy Clin Immunol. 1984;74:26–33. doi: 10.1016/0091-6749(84)90083-6. [DOI] [PubMed] [Google Scholar]
- 36.Menardo JL, Bousquet J, Rodiere M, Astruc J, Michel FB. Skin test reactivity in infancy. J Allergy Clin Immunol. 1985;75(6):646–51. doi: 10.1016/0091-6749(85)90088-0. [DOI] [PubMed] [Google Scholar]
- 37.Bernstein IL, Li JT, Bernstein DI, Hamilton R, Spector SL, Tan R, et al. Allergy diagnostic testing: an updated practice parameter. Ann Allergy Asthma Immunol. 2008;100(3 Suppl 3):S1–148. doi: 10.1016/s1081-1206(10)60305-5. [DOI] [PubMed] [Google Scholar]
- 38.Nicolaou N, Poorafshar M, Murray C, Simpson A, Winell H, et al. Allergy or tolerance in children sensitized to peanut: Prevalence and differentiation using component-resolved diagnostics. J Allergy Clin Immunol. 2010;125:191–7. doi: 10.1016/j.jaci.2009.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.