Abstract
The phenolic pKa of fluorescein varies depending on its environment. The fluorescence of the dye varies likewise. Accordingly, a change in fluorescence can report on the association of a fluorescein-conjugate to another molecule. Here, we demonstrate how to optimize this process with chemical synthesis. The fluorescence of fluorescein-labeled model protein, bovine pancreatic ribonuclease (RNase A), decreases upon binding to its cognate inhibitor protein (RI). Free and RI-bound fluorescein–RNase A have pKa values of 6.35 and 6.70, respectively, leaving the fluorescein moiety largely unprotonated at physiological pH and thus limiting the sensitivity of the assay. To increase the fluorescein pKa and, hence, the assay sensitivity, we installed an electron-donating alkyl group ortho to each phenol group. 2′,7′-Diethylfluorescein (DEF) has spectral properties similar to those of fluorescein but a higher phenolic pKa. Most importantly, free and RI-bound DEF–RNase A have pKa values of 6.68 and 7.29, respectively, resulting in a substantial increase in the sensitivity of the assay. Using DEF–RNase A rather than fluorescein–RNase A in a microplate assay at pH 7.12 increased the Z′-factor from −0.17 to 0.69. We propose that synthetic “tuning” of the pKa of fluorescein and other pH-sensitive fluorophores provides a general means to optimize binding assays.
INTRODUCTION
The xanthene dye fluorescein (1) was first synthesized by Baeyer in 1871.1 Despite its antiquity, fluorescein remains one of the most widely utilized fluorophores in modern biochemical, biological, and medicinal research. This persistence can be attributed to the excellent spectral properties of fluorescein and the established synthetic chemistry of the dye. Fluorescein serves as a modular scaffold that is well-suited for elaboration to create various molecular tools, including ion indicators, fluorogenic enzyme substrates, and fluorescent labels for biomolecules.2,3
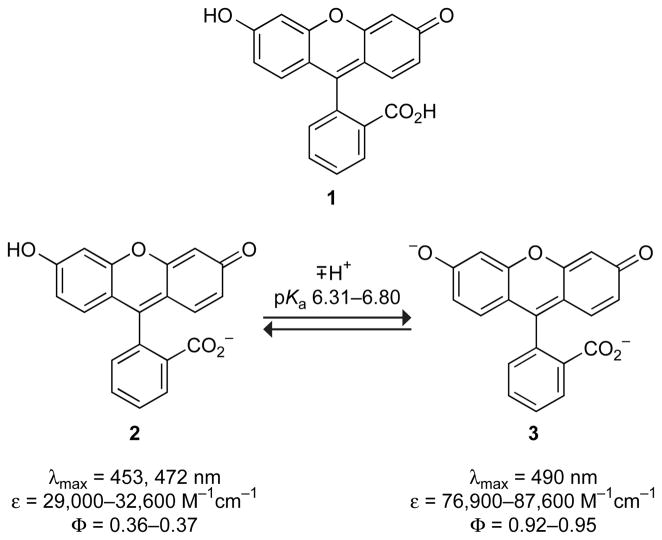
An important (and underappreciated) property of fluorescein is its complex acid–base equilibria in aqueous solution. Fluorescein can exist in seven prototropic forms. The determination of the distinct properties of each of these molecular forms has been the subject of numerous studies.4–16 The monoanion 2 and dianion 3 are the principal ground-state species under biologically-relevant conditions. Scheme 1 shows these two forms of fluorescein with the ranges of reported phenolic pKa values and other spectral properties in aqueous solution. The dianionic form 3 is responsible for the characteristic strong visible absorption band and potent fluorescence emission. The absorbance of the monoionic form 2 is less intense, and the maxima are blue-shifted relative to 3. The quantum yield of the monoanion is also significantly lower than that of the dianion. Because of the dissimilar optical properties of 2 and 3 and the proximity of the pKa value to physiological pH, special care must be taken when using fluorescein and fluorescein-labeled conjugates in biological experiments. To circumvent this pH-sensitivity problem, fluorescein derivatives have been developed that employ the electron-withdrawing nature of chlorine17,18 or fluorine19 substituents to shift this phenolic pKa to a lower value, thereby suppressing the heterogeneity of the protonation state of this phenolic group at physiologically relevant pH values.
Scheme 1.
The sensitivity of the fluorescence of fluorescein to its chemical environment has been exploited in assays of biological processes. Because of the proximity of the phenolic pKa to biologically relevant pH values, small-molecule fluorescein derivatives have been employed as pH sensors.20–26 In addition, the pKa value itself is sensitive to the electrostatic environment around the fluorescein molecule.27–31 Thus, the phenolic pKa of a fluorescein label attached to a protein, for example, can be perturbed depending on the status of the biomolecule. The difference in pKa value translates to an alteration in fluorescent intensity at constant pH. This environmentally-sensitive fluorescence variation has been used as an index to monitor changes in proteins32–37 and nucleic acids.38–41 Studies of protein–protein interactions, in particular, can benefit from assays relying on the pKa shift of a fluorescein label. Such assays circumvent the double-label requirement of Förster resonance energy transfer (FRET),42 and the size limitations of fluorescence polarization (FP).43
Although binding assays relying on the pKa shift of fluorescein are prevalent, the development of such assays is largely empirical. The effects of minor modifications to the fluorescein label remain unexplored. We were especially interested in discerning whether altering the pKa of the dye would have a significant effect on assay performance. Our interest stems from a fluorescence-based assay developed in our laboratory44,45 for the determination of equilibrium dissociation constants for variants of bovine pancreatic ribonuclease (RNase A46) and its homologues in complex with the ribonuclease inhibitor protein (RI47). The ability of these pancreatic-type ribonucleases to evade RI is a prerequisite for their toxic activity toward cancer cells.45 Our assay is based on the decrease in fluorescence (~10–20%) of a fluorescein-labeled ribonuclease variant once bound by RI.48 Competitive binding of RI by unlabeled ribonuclease variants restores fluorescence intensity, allowing accurate determination of equilibrium dissociation constants. Although the assay has proven to be useful, the low dynamic range requires a laborious, cuvette-based assay using sensitive instrumentation and large assay volumes. Expansion of the utility of this assay to a miniaturized, high-throughput system (e.g., microplate format) requires an increase in the fluorescence change without severe modification of other assay parameters.
Here, we describe a general means to improve binding assays through the synthesis and use of a “tuned” fluorescein derivative. As a model system, we use the RI–RNase A interaction, which allows for assays to be performed over a wide range of pH. First, we determine the pKa values of the original fluorescein-labeled RNase A variant in the absence and presence of excess RI. From this analysis, we confirm a shift in the pKa value of the fluorescein label upon complex formation, and surmise that tuning the pKa to a higher value could lead to an improved dynamic range in our assay. Then, we synthesize and evaluate 2′,7′-diethylfluorescein (DEF), which contains an electron-donating ethyl group proximal to each hydroxyl group. We find that DEF displays a higher phenolic pKa than does fluorescein, making it a more useful probe. Next, we synthesize a novel thiol-reactive derivative of DEF for bioconjugation, 2′,7′-diethylfluorescein-5-iodoacetamide (DEFIA), and determine pKa values of a DEF-labeled RNase A in the absence and presence of RI. We observe a substantial increase in assay dynamic range near neutral pH, and show that the diethylfluorescein derivative is a near-optimal probe for this binding assay at our target pH. Lastly, we adapt this improved assay to a microplate format and use this simple, robust system to measure the dissociation constants of several complexes containing RI and variants of RNase A. These findings herald a new and comprehensive strategy for facilitating the analysis of biomolecular interactions.
MATERIALS AND METHODS
General
Fluorescein (reference standard grade) and 5-iodoacetamidofluorescein (5-IAF) were from Molecular Probes (Eugene, OR). Dulbecco’s phosphate-buffered saline49 (DPBS, ΦGibco) was from Invitrogen (Carlsbad, CA). Dithiothreitol (DTT) and tris-(hydroxymethyl)-aminomethane (TRIS) were from Research Products International (Mount Prospect, IL). All other chemicals were from Fisher Scientific (Hanover Park, IL) or Sigma–Aldrich (Milwaukee, WI). Bovine serum albumin (BSA) was obtained as a 20 mg/mL solution (Sigma; Product B8667). 2-(N-Morpholino)-ethanesulfonic acid (MES) was purified as described previously50 to eliminate oligo(vinylsulfonic acid) contamination. MALDI–TOF mass spectra were obtained with a Perkin–Elmer Voyager mass spectrometer in the Biophysics Instrumentation Facility (BIF) at the University of Wisconsin–Madison.
Preparation of Ribonuclease Inhibitor and Fluorophore-labeled Ribonucleases
Human ribonuclease inhibitor (RI), K7A/G88R RNase A, D38R/R39D/N67R/G88R RNase A, and K7A/D38R/R39D/G88R RNase A were prepared as described previously.45 The G88R and A19C/G88R variants of RNase A and the fluorescein-labeled conjugate (fluorescein–RNase A) were prepared as described previously44 with the following change: the proteins were refolded for ≥ 4 days under an inert atmosphere of N2(g). The DEFIA–protein conjugate (DEF–RNase A) was prepared by reaction of A19C/G88R RNase A with ten-fold excess of DEFIA (14) for 2.5 h at ambient temperature and then 16 h at 4 °C. Purification using a HiTrap SP HP cation-exchange column (GE Healthcare, Uppsala, Sweden) afforded the desired conjugate; MS (MALDI): m/z 14,261.00 (expected = 14,258.46). Protein concentration was determined by UV absorption or a bicinchoninic acid (BCA) assay kit from Pierce (Rockford, IL) using wild-type RNase A as a standard.
UV–Visible and Fluorescence Spectroscopy
Absorption spectra were recorded in methacrylate cuvettes having 1-cm path length and 1.5-mL volume from Fisher Scientific on a Cary Model 50 spectrometer from Varian (Sugar Land, TX). Fluorometric measurements were made by using 4.5-mL methacrylate cuvettes from VWR or 4.5-mL glass cuvettes from Starna Cells (Atascadero, CA) and a QuantaMaster1 photon-counting spectrofluorometer from Photon Technology International (South Brunswick, NJ) equipped with sample stirring. All measurements were recorded at ambient temperature (23 ± 2 °C) and buffers were not degassed prior to measurements. Compounds were prepared as stock solutions in DMSO and diluted such that the DMSO concentration did not exceed 1% v/v. The pH was measured with a Beckmann glass electrode that was calibrated prior to each use. Microplate-based experiments were performed in Costar 96-well NBS microplates (Product #3650) from Corning Life Sciences (Acton, MA). The fluorescence intensity was recorded using a Perkin–Elmer EnVison 2100 Plate Reader and an FITC filter set (excitation at 485 nm with 14-nm bandwidth; emission at 535 nm with 25-nm bandwidth; dichroic mirror cutoff at 505 nm) in the Keck Center for Chemical Genomics at the University of Wisconsin–Madison. Graphs were manipulated and parameters were calculated with the programs Microsoft Excel 2003 and GraphPad Prism 4.
Determination of pKa Values
The fluorescein acid–base equilibrium between phenol 2 and phenolate 3 is shown in eq 1:
| (1) |
The observed fluorescence intensity (I) at a given excitation and emission wavelength is given by eq 2:
| (2) |
where f2 and f3 are the fractions of the phenol and phenolate form of fluorescein, respectively, and I2 and I3 are the phenol and phenolate fluorescence intensities.28 The pKa of this acid–base equilibrium can be estimated by measuring the fluorescence intensity as a function of pH and fitting the data to eq 3:
| (3) |
which is derived from eq 2 and the Henderson–Hasselbalch equation.
Buffers contained NaCl (138 mM), DTT (1 mM), and NaOAc, MES, NaH2PO4, TRIS, and NaHCO3 (10 mM each). The pH of the buffered solutions was adjusted with 1.0 M NaOH or 1.0 M HCl. All experiments were performed in cuvettes using λex = 493 nm and λem = 515 nm. The pKa values of the free dyes were determined at a final dye concentration of 50 nM. For determination of the pKa of the protein labels, fluorescein–RNase A or DEF–RNase A was added to the buffer solution at a final concentration of 50 nM, and the initial fluorescence was measured. RI was then added to a final concentration of 350 nM, and the resulting fluorescence intensity was measured. The average pKa values were determined from triplicate experiments involving separate buffer preparations.
Spectral Properties
The extinction coefficient (ε) of fluorescein and 2′,7′-diethylfluorescein was determined with solutions in 0.1 M NaOH (A < 1.0). The absorbance of a series of fluorescein and DEF concentrations were plotted against concentration, and the extinction coefficient was calculated by linear regression using Beer’s Law. The quantum yield of DEF was determined by using dilute samples (A < 0.1) in 0.1 M NaOH. These values were obtained by the comparison of the integrated area of the emission spectrum of the samples with that of fluorescein in 0.1 M NaOH, which has a quantum efficiency of 0.95 ± 0.03.13 The concentration of the fluorescein reference was adjusted to match the absorbance of the test sample at the excitation wavelength. Under these conditions, the quantum yield (Φ) was calculated with eq 4.
| (4) |
Assay Comparison and Z′-factor Determination
Comparison of the gross dynamic range of the two protein conjugates was made using a cuvette format in DPBS containing BSA (2 μg/mL). The fluorescent protein conjugate (fluorescein–RNase A or DEF–RNase A) was added to a final concentration of 50 nM. RI was then added to a final concentration of 350 nM, and the resulting fluorescence change recorded. The Z′-factor was determined by preparing 96-well plates with 50 μL per well of DPBS containing fluorescein–RNase A or DEF–RNase A (100 nM; 2×) and BSA (0.1 mg/mL). The positive control plates also contained RNase A (10 μM; 2×). To these plates were added 50 μL per well of DPBS containing BSA (0.1 mg/mL), DTT (10 mM; 2×), and RI (100 nM; 2×). The fluorescence of each well was quantified after incubation at ambient temperature for 30 min. The Z′-factor was determined with eq 5 where σ+ and σ− are the standard deviations of the positive and negative controls, respectively, and μ+ and μ− are the means of the positive and negative controls.51
| (5) |
Determination of Kd Values
A serial dilution (2×) of the ribonuclease A variant in DPBS with BSA (0.1 mg/mL) was prepared in Eppendorf Protein LoBind Tubes (Fisher Scientific). An aliquot (50 μL) of these serial dilutions was added to the wells of a 96-well plate. A solution (50 μL) of DEF–RNase A (100 nM; 2×) and RI (100 nM; 2×) in DPBS containing DTT (10 mM; 2×) and BSA (0.1 mg/mL) was then added to each well. The negative control contained no ribonuclease and the positive control contained excess RNase A (5 μM). The plate was incubated for 30 min at ambient temperature after which the fluorescence intensity was measured. The observed fluorescence intensity (I) is described by eq 6:
| (6) |
where fF and fB are the fractions of the free and RI-bound form of the fluorescent conjugate, respectively, and IF and IB are the fluorescence intensities of the free and RI-bound states, respectively. The value of IB was determined via linear regression using the intensities of the positive and negative controls, which represent 0% and 84.6% bound respectively based on a Kd value of 1.4 nM for the fluorophore-labeled G88R variant of RNase A.45 The fraction bound (fB) was then calculated using eq 7:
| (7) |
The value of Kd was calculated by plotting fB against the concentration of competing ribonuclease and fitting the data to the mathematical expression for complete competitive binding of two different ligands.52,53
RESULTS AND DISCUSSION
pKa Values of Bound and Free Fluorescein-Labeled RNase A
Our binding assay utilizes the A19C/G88R variant of RNase A. Ala19 resides in a solvent-exposed loop that is not within the interface of the RI·RNase A complex.54 Introduction of a thiol group at this position allows site-specific labeling that does not perturb other properties of the protein, such as enzymatic activity or binding to RI.44 The fluorescein label is attached covalently by reaction of the free thiol-containing protein with 5-iodoacetamidofluorescein (5-IAF) to give fluorescein-labeled RNase A. The RI·RNase A complex exhibits extremely tight binding (Kd = 44 fM, ref 55). Substitution at position 88 attenuates the binding constant of the complex by more than four orders of magnitude (Kd = 1.4 nM, ref 45) and thus allows for effective competition by other ribonucleases with Kd values at or above the nanomolar range. Our assay is typically performed in commercial Dulbecco’s phosphate-buffered saline49 (DPBS) at pH 7.12, which defines the target pH for assay optimization.
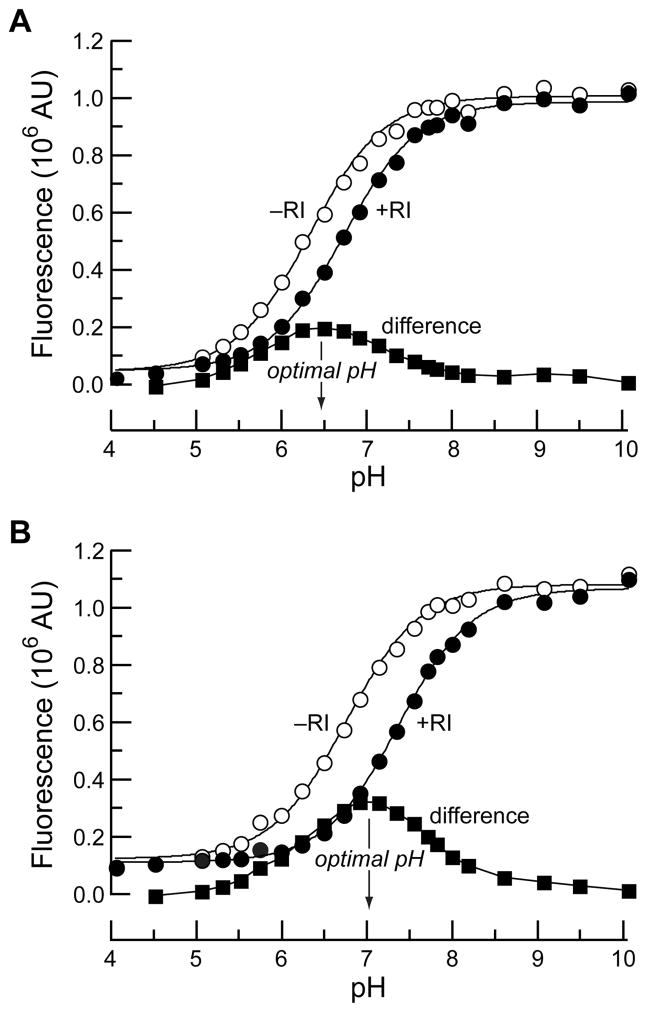
In the original report44 of this assay system, we noted that the cause of the fluorescence change of the fluorescein conjugate of A19C/G88R RNase A (herein, fluorescein–RNase A) upon binding to RI was unclear, but we hypothesized that it arose from a shift in the pKa of the fluorescein label. To test this premise, and gain insight for assay optimization, we measured the pKa values of both bound and free fluorescein–RNase A. A series of buffers were prepared from pH 4 to 10, and the fluorescence intensity of the conjugate in the absence and presence of a 7-fold molar excess of RI was measured and plotted against pH. This surfeit of RI is sufficient to bind >99.5% of the labeled ribonuclease based on the Kd value of 1.4 nM.45 The difference between the curves was also calculated, and the resulting data are shown in Figure 1A.
Figure 1.
Effect of pH on the fluorescence (λex = 493 nm, λem = 515 nm) of fluorophore-labeled RNase A (50 nM) in the absence (○) or presence (●) of excess RI (350 nM), and the difference (■). (A) Fluorescein–RNase A. (B) DEF–RNase A.
The data in Figure 1A conform to a model for pKa shifts of fluorescein protein conjugates described by Garel,32 including the bell-shaped difference trace. We note that at high pH values, complex formation causes only a minor decrease (<5%) in fluorescence intensity. The inhibition of the catalytic activity of RNase A by RI is known to be similar in solutions of different pH,55 indicating that complex formation is unaffected by pH. Thus, the traces in Figure 1A suggest that the pKa shift of fluorescein upon RI binding is the primary cause for the fluorescence modulation. The phenolic pKa value of the free fluorescein–RNase A is 6.35 ± 0.03, and the pKa value for bound fluorescein–RNase A is 6.70 ± 0.02. The maximal difference is at pH 6.5, making that the solution pH at which our assay is most sensitive. Interestingly, the pKa values for the free and RI-bound conjugates are both higher than our measured pKa value for free fluorescein of 6.30 ± 0.03 (vide infra).
The phenolic pKa value of fluorescein can change dramatically upon conjugation to a biomolecule,56 and it remains difficult to predict how the local environment around the label will affect the pKa of the dye. Nonetheless, the shift in pKa value upon binding of fluorescein–RNase A by RI is intuitive, based on the net macromolecular charge of the two proteins. RNase A is a cationic protein (pI = 9.33, ref 57), creating an electropositive field in which the ionization of fluorescein is relatively favorable. Binding of RNase A to the comparatively anionic RI (pI = 4.7, ref 58) can neutralize much of this field,55 leading to an increased phenolic pKa value for the fluorescein label.
The data in Figure 1A suggested to us an optimization strategy that involved “tuning” the phenolic pKa value of the label through chemical synthesis. Changing the pKa of fluorescein would, in effect, shift the pKa curves of the free and RI-bound conjugate either higher or lower along the abscissa, causing the maximal difference between these two curves to align more closely with the desired assay pH. Such a shift could lead to a higher assay dynamic range at the preferred pH. Prior attempts to adjust the pKa of fluorescein labels have focused on decreasing this value as much as possible to abolish pH sensitivity.59 Conversely, our goal was to increase the pKa so as to maximize the fluorescence change at the assay pH.
Design and Synthesis of 2′,7′-Diethylfluorescein
The proximity of the pKa value of fluorescein to biologically relevant pH values allows its use as fluorescent pH indicator in certain experiments.20,22,25 Alkyl substitution on the xanthenyl portion of fluorescein can increase the phenolic pKa of fluorescein, making it more sensitive to changes in pH near neutrality. Such substitution was exploited by Tsien and coworkers to prepare the widely used pH indicator 2′,7′-bis-carboxyethyl-5(6)-carboxyfluorescein (BCECF) that possesses a pKa near 7.0.1,22 Hexyl-substituted fluoresceins also show an increase in the phenolic pKa making them useful fluorescent pH sensors.26 Although these compounds show the desired increase in pKa value, the relatively large appendages could cause unnecessary disruption of the protein if incorporated into a fluorescent label. Substitution with smaller, ethyl groups at the 2′ and 7′ positions can also increase the pKa of fluorescein.60 We reasoned that the negligible perturbation of the fluorescein structure from such ethyl substituents would preserve its utility as a label.
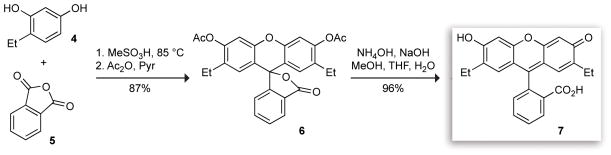
To evaluate the effect of ethyl-group substitution, we synthesized 2′,7′-diethylfluorescein (DEF) by the route shown in Scheme S-1 (see: Supporting Information). This compound was reported in the 1930’s by Novelli in research directed toward the development of fluorescein-based antiseptics.61,62 We used a more contemporary route, taking advantage of the commercial availability of 4-ethylresorcinol 4. This compound was condensed with phthalic anhydride 5 in methanesulfonic acid19 to afford crude DEF. This material was acetylated with acetic anhydride in pyridine, and the resulting diacetate 6 was purified via crystallization. We found that suspension of the crude 6 in cold EtOH prior to crystallization removed a significant amount of polar impurities, thereby improving the yield. Hydrolysis of the ester groups with base followed by acidification gave DEF (7).
Spectral Properties of 2′,7′-Diethylfluorescein
Both fluorescein and 2′,7′-diethylfluorescein were evaluated to confirm the anticipated effect of alkylation on the pKa value and to compare other spectral properties of these dyes in aqueous solution. The measured parameters are listed in Table 1. The pKa values for both fluorescein and DEF were determined in the same buffer system used for the fluorescein–RNase A conjugate (see: Figure S-1 in the Supporting information). The extinction coefficients and quantum yields of fluorescein and DEF were determined in 0.1 M NaOH to isolate the properties of the dianionic dye form.
Table 1.
Spectroscopic Parameters of Fluorescein and 2′,7′-Diethylfluorescein (DEF)
| Parameter | Fluorescein | DEF |
|---|---|---|
| pKa | 6.30 ± 0.02 | 6.61 ± 0.03 |
| λmax (nm) | 491 | 501 |
| ε(mM−1cm−1) | 89.8 ± 0.4 | 98.5 ± 0.4 |
| λem (nm) | 510 | 520 |
| Φ | 0.95 ± 0.03 13 | 0.89 ± 0.03 |
The spectral values determined for the diethyl variant of fluorescein are close to those reported for other dialkyl derivatives.26 The electron-donating character of the ethyl substituents increases the pKa from 6.30 to 6.61. Our pKa value for free fluorescein is slightly lower than the reported range of values8–12,15 because of the relatively high ionic strength of our buffer system.11 The alkyl substitution also elicits bathochromic shifts of 10 nm for both absorption and emission maxima relative to fluorescein (see: Figure S-2 in the Supporting information). The extinction coefficient at maximal absorption of DEF under basic conditions (0.1 M NaOH) is about 10% higher than the absorptivity of fluorescein. The quantum yield of diethylfluorescein is slightly lower than that of fluorescein, again in agreement with other reported dialkylfluorescein derivatives.26 Overall, the ethyl group substitution confers the desired increase in pKa value while causing only minor differences in absorption and fluorescence properties. This similarity of optical characteristics of the two dyes allows for the use of standard fluorescein excitation and emission wavelengths for both dyes and their conjugates.
Synthesis of 2′,7′-Diethylfluorescein-5-iodoacetamide (DEFIA)
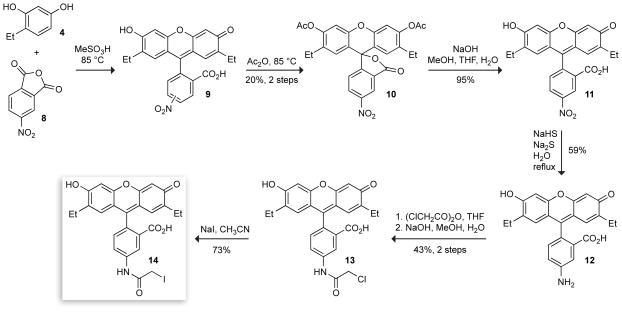
Having confirmed the elevated pKa values of DEF, we next sought to prepare a thiol-reactive derivative for bioconjugation. To ensure an accurate comparison between fluorescein-labeled and DEF-labeled proteins, we designed an analogue of 5-IAF in which introduction of the ethyl groups was the only structural perturbation, thereby eliminating effects due to different linker lengths or conjugation chemistries. The synthesis of 5-aminofluorescein is well documented because of its intermediacy in the preparation of the widely-used fluorescent label: fluorescein-isothiocyanate (FITC).63–65 We were pleased to discover that this established chemistry is sufficiently pliable to allow for introduction of the desired ethyl substituents without dramatic changes in yield. To our knowledge, this is the first example of a fluorescein-derived label in which the phenolic pKa is tuned to a higher rather than a lower value. In addition, this synthesis adds to the sparse reports in the primary scientific literature describing iodoacetamide derivatives of xanthene dyes.66,67
The scheme for the synthesis of 2′,7′-diethylfluorescein-5-iodoacetamide (DEFIA) is shown in Scheme 2. Condensation of 4-ethylresorcinol 4 with commercially available 4-nitrophthalic anhydride (8) gave fluorescein 9 as a mixture of 5- and 6-nitro isomers. This material could be acetylated with acetic anhydride to yield a diacetate. Separation of the two isomers via recrystallization63 afforded the single 5-nitro isomer 10. Hydrolysis of this diacetate with base gave free 5-nitro-2′,7′-diethylfluorescein 11. Reduction of the nitro group with sulfide65 followed by crystallization from aqueous HCl gave the 5-aminofluorescein as the HCl salt. Dissolving this intermediate HCl salt in basic solution followed by precipitation of the free amine with acetic acid64 afforded the 5-amino-2′,7′-diethylfluorescein 12. This material was treated with chloroacetic anhydride to give an intermediate 5-amidofluorescein diester. Hydrolysis of the ester groups using NaOH in the same pot gave chloroacetamide 13, which was taken on to DEFIA (14) via reaction with NaI in acetone.
Scheme 2.
pKa Values of Free and RI-Bound DEF-labeled A19C/G88R RNase A
Thiol-containing A19C/G88R RNase A variant was reacted with DEFIA, and the resulting conjugate was purified with cation-exchange chromatography. The fluorescence intensity of the resulting 2′,7′-diethylfluorescein conjugate of A19C/G88R RNase A (herein, DEF–RNase A) was measured in the absence and presence of RI in buffered aqueous solution over a pH range of 4–10, as with the fluorescein–RNase A conjugate. The fluorescence values and fluorescence difference were plotted against pH as shown in Figure 1B. The pKa values of the free and RI-bound DEF–RNase A were calculated to be 6.68 ± 0.03 and 7.29 ± 0.03, respectively. As with the fluorescein system, these values are higher than those measured for unliganded DEF (pKa = 6.61 ± 0.03). This shift in pKa values increases the maximum of the fluorescence difference to above 7.0.
The bound and free DEFIA conjugates exhibit a larger ΔpKa value than did the fluorescein-labeled protein. This larger difference in pKa likely arises from structural rather then electronic consequences of the two ethyl groups. For example, the DEF label could exist in a different orientation than the fluorescein label such that its pKa value is affected to a greater degree upon binding to RI.
Assay Comparison
A major goal of this work was to improve the assay performance and then investigate the utility of the enhanced system in a microplate format. To quantify the assay improvement, we first determined the maximum dynamic range by measuring the change in fluorescence upon RI binding under typical assay conditions. Addition of excess RI to fluorescein–RNase A elicits a 15% decrease in fluorescence, whereas the change in fluorescence intensity of the DEF–RNase A conjugate is a significantly larger 38% (see: Figure S-3 in the Supporting information).
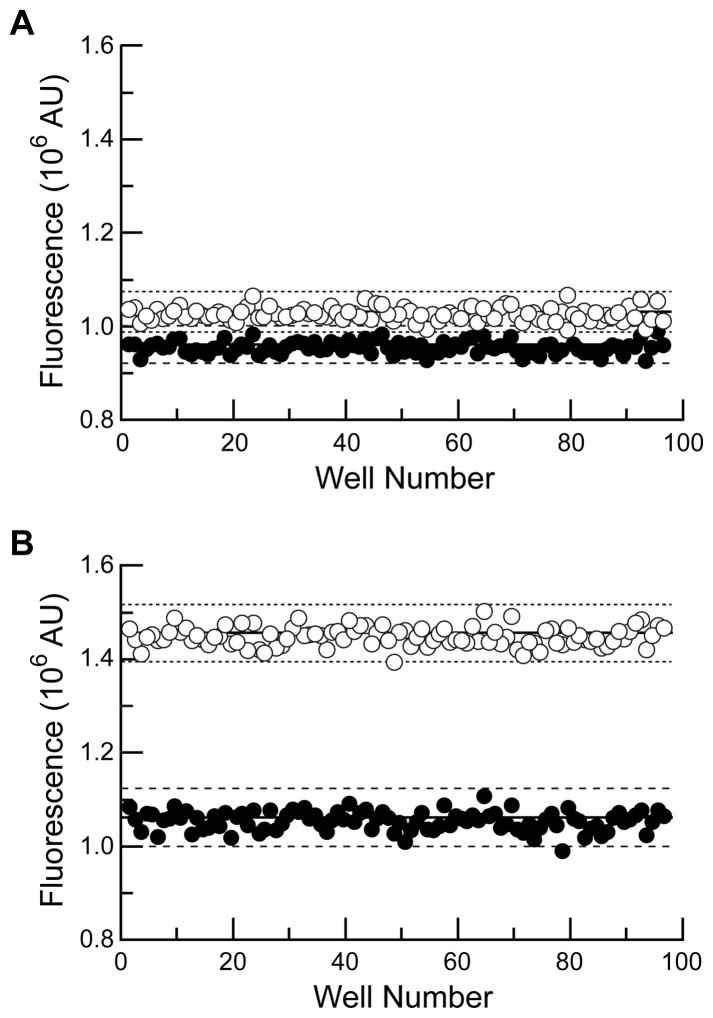
Having confirmed the enhancement of the assay, we then evaluated the utility of this assay in a microplate format. A common quantification of plate-based assay performance is the Z′-factor which uses the standard deviation of controls and the dynamic range to assign a numerical value to assay utility.51 We measured the Z′-factor using microplates containing the labeled conjugate and RI. The positive control also contained excess RNase A to liberate fully the fluorophore-labeled protein. As shown in Figure 2, the fluorescein system had Z′-factor = −0.17, signifying an overlap of the μ ± 3σ levels for the positive and negative controls. In stark contrast, the diethylfluorescein system had a Z′-factor of 0.69. An assay system with a Z′-factor >0.5 is considered to be “excellent”51 and therefore highly useful in microplate systems, including high-thoughput screening (HTS) assays. Thus, in addition to improving assay performance, this new diethylfluorescein label could allow HTS to identify compounds that disrupt the RI–ribonuclease interaction.
Figure 2.
Fluorescence in microplate wells containing a fluorophore-labeled ribonuclease (50 nM) and RI (50 nM) in the absence (○) or presence (●) of excess RNase A (5 μM) at pH 7.12. Horizontal solid lines indicate the mean value; horizontal dashed lines indicate the range (±3σ) of values. (A) Fluorescein–RNase A (Z′-factor −0.17). (B) DEF–RNase A (Z′-factor 0.69).
Microplate-Based Determination of Kd Values
The increase in assay dynamic range and superb Z′-factor prompted us to validate this microplate system to measure RI·ribonuclease dissociation constants. Pancreatic-type ribonucleases are cationic proteins that can enter cells via endocytosis.68 The ubiquitous ribonuclease inhibitor has a high cytosolic concentration (4 μM) and can therefore protect cellular RNA from degradation by invading ribonucleases.69 Amino acid substitutions that disrupt the RI–ribonuclease interface can endow an otherwise benign ribonuclease with cytotoxic activity.45 Thus, the Kd value of a RI·ribonuclease complex is helpful in predicting the cytotoxicity of a novel ribonuclease variant.
We first determined the affinity of DEF–RNase A for RI by direct titration44 and found that the ethyl groups did not cause any significant change in binding constant between the labeled protein and RI (data not shown). We then determined the Kd values for other variants of RNase A using DEF–RNase A in microplates (see: Figure S-4 in the Supporting Information). These values (±SE), which are listed in Table 2, are in gratifying agreement with those determined previously using the cuvette assay.45 The new system proved extremely facile and economical, requiring 5% of the protein and significantly less time compared to the original assay format.
Table 2.
Values of Kd for RI·ERNase A Complexes
| RNase A Variant | Kda (nM) |
|---|---|
| G88R | 1.3 ± 0.2 |
| K7A/G88R | 80 ± 5 |
| D38R/R39D/N67R/G88R | 730 ± 40 |
| K7A/D38R/R39D/G88R | 3100 ± 200 |
Values of Kd (±SE) were determined in microplate assays with DEF–RNase A at pH 7.12.
Conclusion
The ubiquity of fluorescein in biochemical, biological, and medicinal research demands a detailed grasp of the acid–base equilibria of this dye in aqueous solution. Our results show that a thorough understanding and application of the nuances of fluorescein can be useful in the optimization of assays. Shifts in the phenolic pKa are a significant causal force behind many of the fluorescence modulations observed with fluorescein conjugates. Tuning this pKa through chemical synthesis can have dramatic effects on assay dynamic range, leading to significant improvement in throughput and requisite sample volume.
This work could inspire the expansion of the current spectrum of fluorescent labels. A palette of reactive dyes with tuned pKa values could prove useful in the development of new binding assays. In addition to fluorescein, other classes of fluorescent dyes are sensitive to pH and thus could be evaluated and optimized in a similar manner. Of particular interest are dye systems that exhibit a large spectral shift upon protonation.70,71 Use of these fluorophores could permit ratiometric measurement of binding events. Overall, properly tuned fluorescent labels hold the potential to improve existing systems and aid in the development of new assays for characterizing protein–protein interactions and other important biomolecular processes.
Supplementary Material
Scheme 3.
Acknowledgments
We are grateful to C. Schilling for initial synthetic studies, K. J. Kolonko for assistance with NMR spectroscopy, and Z.J. Diwu, B.D. Smith, R.J. Johnson, G.A. Ellis, and S.M. Fuchs for contributive discussions. L.D.L was supported by Biotechnology Training Grant 08349 (NIH) and an ACS Division of Organic Chemistry Graduate Fellowship sponsored by the Genentech Foundation. T.J.R was supported by Biotechnology Training Grant 08349 (NIH) and a William R. & Dorothy E. Sullivan Wisconsin Distinguished Graduate Fellowship. This work was supported by grant CA73808 (NIH). Biophysics Instrumentation Facility was established with grants BIR-9512577 (NSF) and S10 RR13790 (NIH). The Keck Center for Chemical Genomics was established with a grant from the W.M. Keck Foundation. NMRFAM was supported by grant P41 RR02301 (NIH). The MRF was supported by grants CHE-9709065 (NSF), CHE-9208463 (NSF), and S10 RR08389 (NIH).
Footnotes
Supporting Information Available: Additional characterization of the spectral properties of fluorescein (1) and 2′,7′-diethylfluorescein (7, DEF), and of the biochemical properties of fluorescein–RNase A and DEF–RNase A, along with synthetic methods and spectral data for compounds 6, 7, and 10–14 (PDF). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Baeyer A. Ber Dtsch Chem Ges. 1871;4:555–558. [Google Scholar]
- 2.Urano Y, Kamiya M, Kanda K, Ueno T, Hirose K, Nagano T. J Am Chem Soc. 2005;127:4888–4894. doi: 10.1021/ja043919h. and references therein. [DOI] [PubMed] [Google Scholar]
- 3.Haugland RP, Spence MTZ, Johnson ID, Basey A. The Handbook: A Guide to Fluorescent Probes and Labeling Technologies. 10. Molecular Probes; Eugene, OR: 2005. [Google Scholar]
- 4.Zanker V, Peter W. Ber Dtsch Chem Ges. 1958;91:572–580. [Google Scholar]
- 5.Lindqvist L. Ark Kemi. 1960;16:79–138. [Google Scholar]
- 6.Martin MM, Lindqvist L. J Lumin. 1975;10:381–390. [Google Scholar]
- 7.Chen SC, Nakamura H, Tamura Z. Chem Pharm Bull. 1979;27:475–479. [Google Scholar]
- 8.Mchedlov-Petrosyan NO. Zh Anal Khim. 1979;34:1055–1059. [Google Scholar]
- 9.Diehl H. Talanta. 1989;36:413–415. doi: 10.1016/0039-9140(89)80212-7. [DOI] [PubMed] [Google Scholar]
- 10.Diehl H, Markuszewski R. Talanta. 1989;36:416–418. doi: 10.1016/0039-9140(89)80213-9. [DOI] [PubMed] [Google Scholar]
- 11.Sjoback R, Nygren J, Kubista M. Spectrochim Acta, Part A. 1995;51:L7–L21. [Google Scholar]
- 12.Klonis N, Sawyer WH. J Fluoresc. 1996;6:147–157. doi: 10.1007/BF00732054. [DOI] [PubMed] [Google Scholar]
- 13.Lakowicz JR. Principles of Fluorescence Spectroscopy. 2. Kluwer Academic/Plenum; New York: 1999. [Google Scholar]
- 14.Magde D, Wong R, Seybold PG. Photochem Photobiol. 2002;75:327–334. doi: 10.1562/0031-8655(2002)075<0327:fqyatr>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 15.Smith SA, Pretorius WA. Water SA. 2002;28:395–402. [Google Scholar]
- 16.Krol M, Wrona M, Page CS, Bates PA. J Chem Theory Comput. 2006;2:1520–1529. doi: 10.1021/ct600235y. [DOI] [PubMed] [Google Scholar]
- 17.Mchedlov-Petrossyan NO, Rubtsov MI, Lukatskaya LL. Dyes Pigm. 1992;18:179–198. [Google Scholar]
- 18.Sparano BA, Shahi SP, Koide K. Org Lett. 2004;6:1947–1949. doi: 10.1021/ol049537y. and references therein. [DOI] [PubMed] [Google Scholar]
- 19.Sun WC, Gee KR, Klaubert DH, Haugland RP. J Org Chem. 1997;62:6469–6475. [Google Scholar]
- 20.Thomas JA, Buchsbaum RN, Zimniak A, Racker E. Biochemistry. 1979;18:2210–2218. doi: 10.1021/bi00578a012. [DOI] [PubMed] [Google Scholar]
- 21.Paradiso AM, Tsien RY, Machen TE. Proc Natl Acad Sci USA. 1984;81:7436–7440. doi: 10.1073/pnas.81.23.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graber ML, Dilillo DC, Friedman BL, Pastorizamunoz E. Anal Biochem. 1986;156:202–212. doi: 10.1016/0003-2697(86)90174-0. [DOI] [PubMed] [Google Scholar]
- 23.Nedergaard M, Desai S, Pulsinelli W. Anal Biochem. 1990;187:109–114. doi: 10.1016/0003-2697(90)90425-9. [DOI] [PubMed] [Google Scholar]
- 24.Liu JX, Diwu ZJ, Klaubert DH. Bioorg Med Chem Lett. 1997;7:3069–3072. [Google Scholar]
- 25.Wu MM, Llopis J, Adams S, McCaffery JM, Kulomaa MS, Machen TE, Moore HP, Tsien RY. Chem Biol. 2000;7:197–209. doi: 10.1016/s1074-5521(00)00088-0. [DOI] [PubMed] [Google Scholar]
- 26.Schroder CR, Weidgans BM, Klimant I. Analyst. 2005;130:907–916. doi: 10.1039/b501306b. [DOI] [PubMed] [Google Scholar]
- 27.Thelen M, Petrone G, Oshea PS, Azzi A. Biochim Biophys Acta. 1984;766:161–168. doi: 10.1016/0005-2728(84)90228-7. [DOI] [PubMed] [Google Scholar]
- 28.Stanton SG, Kantor AB, Petrossian A, Owicki JC. Biochim Biophys Acta. 1984;776:228–236. doi: 10.1016/0005-2736(84)90212-8. [DOI] [PubMed] [Google Scholar]
- 29.Omelyanenko VG, Jiskoot W, Herron JN. Biochemistry. 1993;32:10423–10429. doi: 10.1021/bi00090a018. [DOI] [PubMed] [Google Scholar]
- 30.Griep MA, Mesman TN. Bioconjugate Chem. 1995;6:673–682. doi: 10.1021/bc00036a003. [DOI] [PubMed] [Google Scholar]
- 31.Agi Y, Walt DR. J Polym Sci, Part A: Polym Chem. 1997;35:2105–2110. [Google Scholar]
- 32.Garel JR. Eur J Biochem. 1976;70:179–189. doi: 10.1111/j.1432-1033.1976.tb10968.x. [DOI] [PubMed] [Google Scholar]
- 33.Labhardt AM, Ridge JA, Lindquist RN, Baldwin RL. Biochemistry. 1983;22:321–327. doi: 10.1021/bi00271a014. [DOI] [PubMed] [Google Scholar]
- 34.Griep MA, Mchenry CS. J Biol Chem. 1990;265:20356–20363. [PubMed] [Google Scholar]
- 35.Goldberg JM, Baldwin RL. Biochemistry. 1998;37:2546–2555. doi: 10.1021/bi972402y. [DOI] [PubMed] [Google Scholar]
- 36.Goldberg JM, Baldwin RL. Biochemistry. 1998;37:2556–2563. doi: 10.1021/bi972403q. [DOI] [PubMed] [Google Scholar]
- 37.Goldberg JM, Baldwin RL. Proc Natl Acad Sci USA. 1999;96:2019–2024. doi: 10.1073/pnas.96.5.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedrich K, Woolley P. Eur J Biochem. 1988;173:227–231. doi: 10.1111/j.1432-1033.1988.tb13988.x. [DOI] [PubMed] [Google Scholar]
- 39.Friedrich K, Woolley P, Steinhauser KG. Eur J Biochem. 1988;173:233–239. doi: 10.1111/j.1432-1033.1988.tb13989.x. [DOI] [PubMed] [Google Scholar]
- 40.Sjoback R, Nygren J, Kubista M. Biopolymers. 1998;46:445–453. doi: 10.1002/(SICI)1097-0282(199812)46:7<445::AID-BIP2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 41.A change in the fluorescence intensity of a fluorescein conjugate is often attributed (imprecisely) to a general alteration in chemical environment, rather than a pKa shift. For a recent example, see: Liu J, Qian N, Morrical SW. J Biol Chem. 2006;281:26308–26319. doi: 10.1074/jbc.M604349200.
- 42.Sapsford KE, Berti L, Medintz IL. Angew Chem Int Ed. 2006;45:4562–4589. doi: 10.1002/anie.200503873. [DOI] [PubMed] [Google Scholar]
- 43.Owicki JC. J Biomol Screen. 2000;5:297–306. doi: 10.1177/108705710000500501. [DOI] [PubMed] [Google Scholar]
- 44.Abel RL, Haigis MC, Park C, Raines RT. Anal Biochem. 2002;306:100–107. doi: 10.1006/abio.2002.5678. [DOI] [PubMed] [Google Scholar]
- 45.Rutkoski TJ, Kurten EL, Mitchell JC, Raines RT. J Mol Biol. 2005;354:41–54. doi: 10.1016/j.jmb.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 46.Raines RT. Chem Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 47.Dickson KA, Haigis MC, Raines RT. Prog Nucleic Acid Res Mol Biol. 2005;80:349–374. doi: 10.1016/S0079-6603(05)80009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.This decrease in total fluorescence and the relatively large size of RNase A confounded attempts to develop a binding assay based on fluorescence polarization (Fuch, S. M; Raines, R. T. unpublished results).
- 49.Dulbecco R, Vogt M. J Exp Med. 1954;99:167–182. doi: 10.1084/jem.99.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith BD, Soellner MB, Raines RT. J Biol Chem. 2003;278:20934–20938. doi: 10.1074/jbc.M301852200. [DOI] [PubMed] [Google Scholar]
- 51.Zhang JH, Chung TD, Oldenburg KR. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 52.Wang ZX. FEBS Lett. 1995;360:111–114. doi: 10.1016/0014-5793(95)00062-e. [DOI] [PubMed] [Google Scholar]
- 53.Roehrl MH, Wang JY, Wagner G. Biochemistry. 2004;43:16056–16066. doi: 10.1021/bi048233g. [DOI] [PubMed] [Google Scholar]
- 54.Kobe B, Deisenhofer J. J Mol Biol. 1996;264:1028–1043. doi: 10.1006/jmbi.1996.0694. [DOI] [PubMed] [Google Scholar]
- 55.Lee FS, Shapiro R, Vallee BL. Biochemistry. 1989;28:225–230. doi: 10.1021/bi00427a031. [DOI] [PubMed] [Google Scholar]
- 56.Klonis N, Clayton AHA, Voss EW, Jr, Sawyer WH. Photochem Photobiol. 1998;67:500–510. [PubMed] [Google Scholar]
- 57.Ui N. Biochim Biophys Acta. 1971;229:567–581. [PubMed] [Google Scholar]
- 58.Blackburn P, Wilson G, Moore S. J Biol Chem. 1977;252:5904–5910. [PubMed] [Google Scholar]
- 59.Most prominant are derivatives of 2′,7′-dichlorofluorescein (pKa ~5.2 17) and 2′,7′-difluorofluorescein (“Oregon Green”; pKa ~4.8 19. Notably, the fluorescein-substitute “Alexa Fluor 488” is not based on fluorescein but is a sulfonated rhodamine with invariant fluorescence at pH 4–10.3
- 60.Diwu Z, Twu JJ, Yi G, Lavis LD, Chen Y-W, Cassutt K. J. Fluorescent pH indicators for intracellular assays. 6,800,765. US Patent. 2004 October 5;
- 61.Novelli A. Anales Farm Bioquim. 1932;3:112–120. [Google Scholar]
- 62.Novelli A. Anales Farm Bioquim. 1933;4:29–35. [Google Scholar]
- 63.Coons AH, Kaplan MH. J Exp Med. 1950;91:1–13. doi: 10.1084/jem.91.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McKinney RM, Spillane JT, Pearce GW. J Org Chem. 1962;27:3986–3988. [Google Scholar]
- 65.Steinbach G. Acta Histochem. 1974;50:19–34. [PubMed] [Google Scholar]
- 66.Krafft GA, Sutton WR, Cummings RT. J Am Chem Soc. 1988;110:301–303. [Google Scholar]
- 67.Corrie JET, Craik JS. J Chem Soc, Perkin Trans 1. 1994:2967–2973. [Google Scholar]
- 68.Haigis MC, Raines RT. J Cell Sci. 2003;116:313–324. doi: 10.1242/jcs.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haigis MC, Kurten EL, Raines RT. Nucleic Acids Res. 2003;31:1024–1032. doi: 10.1093/nar/gkg163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whitaker JE, Haugland RP, Prendergast FG. Anal Biochem. 1991;194:330–344. doi: 10.1016/0003-2697(91)90237-n. [DOI] [PubMed] [Google Scholar]
- 71.Liu J, Diwu Z, Leung WY. Bioorg Med Chem Lett. 2001;11:2903–2905. doi: 10.1016/s0960-894x(01)00595-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.