Abstract
NUP98 gene rearrangements occur in acute myeloid leukemia and result in the expression of fusion proteins. One of the most frequent is NUP98-DDX10 that fuses a portion of NUP98 to a portion of DDX10, a putative DEAD-box RNA helicase. Here we show that NUP98-DDX10 dramatically increases proliferation and self-renewal of primary human CD34+ cells, and disrupts their erythroid and myeloid differentiation. It localizes to their nuclei and extensively deregulates gene expression. Comparison to another leukemogenic NUP98 fusion, NUP98-HOXA9, reveals a number of genes deregulated by both oncoproteins, including HOX genes, COX-2, MYCN, ANGPT1, REN, HEY1, SOX4, and others. These genes may account for the similar leukemogenic properties of NUP98 fusion oncogenes. The YIHRAGRTAR sequence in the DDX10 portion of NUP98-DDX10 represents a major motif shared by DEAD-box RNA helicases that is required for ATP binding, RNA-binding, and helicase functions. Mutating this motif diminished the in vitro transforming ability of NUP98-DDX10, indicating that it plays a role in leukemogenesis. These data demonstrate for the first time the in vitro transforming ability of NUP98-DDX10 and show that it is partially dependent on one of the consensus helicase motifs of DDX10. They also point to common pathways that may underlie leukemogenesis by different NUP98 fusions.
Introduction
At least 21 rearrangements of the NUP98 gene have been described in myeloid malignancies including acute myeloid leukemia (AML), myelodysplastic syndrome (MDS), and blast crisis of chronic myelogenous leukemia (CML) (1, 2). These rearrangements result in fusion of an N-terminal segment of NUP98 to a C-terminal segment of a fusion partner. The inv(11)(p15q22) rearrangement that has been reported in AML, MDS, and CML blast crisis results in the expression of NUP98-DDX10, one of the most common NUP98 fusions. It shows a particularly strong association with therapy-related AML and MDS, especially in patients who had been treated with topoisomerase II inhibitors (1).
About half of the reported NUP98 fusion partners are homeodomain-containing transcription factors. The resulting fusions are believed to act as aberrant transcription factors (3-6). The remaining NUP98 fusion partners are a heterogeneous group and their role in leukemogenesis is not well understood. In particular, the effects of NUP98-DDX10 have not been reported in either mice or in human primary cells. DDX10 is a putative RNA helicase of unknown function that contains 8 highly conserved motifs typical of DEAD box RNA helicases (7, 8). It is thought to be involved in ribosome assembly (8). NUP98-DDX10 consists of the entire FG repeat region of NUP98 fused to a C-terminal portion of DDX10 containing 2 of the 8 conserved helicase motifs.
Here we report on the in vitro transforming effects of NUP98-DDX10 in primary human CD34+ hematopoietic stem/progenitor cells and the underlying changes in global gene expression, and provide evidence that a conserved helicase motif plays a role in these effects. This is the first demonstration of the effects of NUP98-DDX10 and the first report of the effects of a non-homeodomain NUP98 fusion in primary human cells.
Materials and Methods
Plasmid construction
The HA-tagged NUP98-DDX10 cDNA was generated by PCR and subcloned into EcoRI/XbaI sites of pTracer-CMV/Bsd. The NUP98-DDX10/3Q mutant was created by replacing an EagI/HindIIII fragment with a synthetic fragment containing 3 arginine to glutamine mutations in the YIHRAGRTAR motif. HA-tagged NUP98-DDX10 and NUP98-DDX10/3Q were subcloned upstream of IRES into the HpaI site of MSCV-IRES-GFP. PCR products and mutations were confirmed by sequencing. The KBTBD10 and PLN luciferase constructs were previously described (9).
Retrovirus production
GP293 cells were transiently transfected with 4.4 μg of retroviral vector and 1.1 μg of pVSV-G expression vector using Lipofectamine Plus reagent (Invitrogen, Carlsbad, CA, USA). After 48 h, the culture supernatant, containing VSV-G-pseudotyped retrovirus, was collected and used for transduction of PG13 packaging cells by spinoculation in the presence of 8 μg/mL polybrene (Hexadimethrine Bromide; Sigma-Aldrich Corp., St. Louis, MO, USA). The PG13 culture supernatant containing GaLV-pseudotyped retrovirus was used for transduction of CD34+ primary cells.
Immunofluorescence microscopy
Cells were centrifuged onto a slide using a Cytospin centrifuge (Thermo Fisher Scientific, Waltham, MA, USA), fixed with 4% paraformaldehyde in D-PBS for 20 min and permeabilized with 0.1% Triton X-100 for 20 min at room temperature. Two % normal donkey serum in D-PBS with 0.1% Tween 20 was used for blocking and washing. Anti-HA antibody (12CA5; Roche Applied Science, Indianapolis, IN, USA) and Alexa Fluor 647-conjugated anti-mouse IgG (Invitrogen) were used for staining. DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride; Invitrogen) at 300 nM was used for nuclear counterstain. Images were captured with an Eclipse 80i fluorescent microscope (Nikon, Melville, NY, USA) using MetaMorph 6.3r2 software (MDS Analytical Technologies, Downington, PA, USA) and pseudo-colored. Brightness and contrast were adjusted and images were superimposed using Adobe Photoshop CS4, version 11.0 for Mac OS (Adobe Systems, San Jose, CA, USA).
Retroviral transduction and analysis of primary human CD34+ cells
Frozen human CD34+ cells purified from mobilized peripheral blood of two healthy volunteers were purchased from the Fred Hutchinson Cancer Research Center (Seattle, WA, USA). Cells were prestimulated and transduced with retrovirus as described (10). After 46 h, GFP-positive cells were isolated using a FACSVantage SE (BD Biosciences, San Jose, CA, USA) and expression of the transfected gene was confirmed by immunoblotting with anti-HA antibody. Long-term liquid culture of primary cells was performed in the presence of a cytokine cocktail as previously described (10). Colony-forming cell (CFC) and long-term culture-initiating cell (LTC-IC) assays were performed as previously described (10). Cytospin preparations of cells harvested from the CFC plates were stained with Giemsa and a 500-cell differential count was performed using an Olympus BX51 microscope (Olympus America, Center Valley, PA, USA). Photomicrographs were taken with an Olympus DP71 camera with a 60X oil objective.
Flow cytometry
Flow cytometry was performed on a FACScan flow cytometer upgraded to 5 colors and two lasers (BD Biosciences) and a FACSVantage SE flow sorter with 3 lasers and 8 detectors (BD Biosciences), and analyzed using FlowJO v7.2.4 software (Tree Star, Inc., Ashland, OR, USA). The antibodies used for these studies were CD11b (phycoerythrin-conjugated clone ICRF44) from eBioscience (San Diego, CA, USA); CD235a (allophycocyanin-conjugated clone GA-R2) from BD (Franklin Lakes, NJ, USA); and CD33 (allophycocyanin-conjugated clone D3HL60.251) and CD45 (phycoerythrin-Cy7-conjugated clone J.33) from Beckman Coulter (Miami, FL, USA).
Microarray analysis of primary human CD34+ cells
For the 6-hour time point, cells were subjected to nucleofection with either control pTracer-CMV/Bsd plasmid or with plasmid expressing NUP98-DDX10 or NUP98-HOXA9 as previously described (10). For the 3-day and 8-day time points, cells were retrovirally transduced as described above. GFP-sorted cells were snap-frozen and submitted to the Siteman Cancer Center Laboratory for Clinical Genomics where total RNA was isolated and target preparation and microarray hybridization were performed. Labeled targets were hybridized to Affymetrix HG-U133 Plus 2.0 GeneChip microarrays. Data were merged with updated gene annotation data using Spotfire DecisionSite 9.1.1 for Functional Genomics (Spotfire, Somerville, MA). Probe sets were filtered according to present/absent calls and the sets that were absent across all chips were filtered from the analysis. The fold change of each probe set was calculated by dividing the absolute signal intensity from NUP98-HOXA9 or NUP98-DDX10 samples by that of the control sample (vector only). Probes scored as increasing and absent in the numerator, and those scored as decreasing and absent in the denominator were also filtered out. Probe sets induced or repressed by 1.74 fold or greater in two independent experiments were considered differentially expressed. Fold changes shown in Table 1 represent the average of all probesets recognizing each gene in both experiments.
Table 1.
Genes deregulated by both NUP98-DDX10 and NUP98-HOXA9 with possible roles in cell transformation.
| Fold Change (NUP98-DDX10 | |||
|---|---|---|---|
| 6 h | 3 d | 8 d | |
| Upregulated Genes: | |||
| HOXA3 | 3.6 | 9.6 | |
| HOXA5 | 3.7 | 10.2 | |
| HOXA6 | 3.7 | 4.1 | |
| HOXA7 | 2.9 | 3.9 | |
| HOXA9 | 2.1 | 2.5 | 2.3 |
| HOXB3 | 3.8 | 14.6 | |
| HOXB5 | 6.1 | 3.0 | |
| HOXB6 | 5.9 | 13.6 | |
| HOXB7 | 3.6 | 4.5 | |
| MEIS1 | 2.6 | 14.9 | |
| PBX3 | 1.8 | ||
| EGR1 | 3.1 | 3.0 | |
| SOX4 | 2.1 | ||
| MYCN | 3.1 | ||
| ZNF521 | 2.3 | 5.5 | |
| HEY1 | 2.7 | ||
| PTGS2 | 3.0 | ||
| REN | 94.8 | 50.6 | |
| ANGPT1 | 3.9 | ||
| RYK | 2.1 | ||
| STYK1 | 2.4 | ||
| Downregulated genes: | |||
| RAP1A | −21.9 | −20.7 | |
| FCGR2C | −2.5 | ||
| CLEC7A | −3.1 | ||
| ALOX5 | −2.2 | ||
| FPR3 | −3.0 | ||
| A2M | −2.6 | ||
| ELA2 | −2.2 | ||
| GADD45B | −2.2 | ||
| MS4A3 | −2.6 | ||
| TRIM35 | −2.4 | ||
| ZFHX3 | −7.6 | ||
| NDRG2 | −1.9 | ||
| RAD18 | −3.7 | ||
| EED | −2.3 | ||
Luciferase assay
Cells (107) were transfected by electroporation using a Bio-Rad GenePulser (Bio-Rad, Hercules, CA, USA) with 10 μg pGL4.11 vector or pGL4.11 driven by the indicated promoters and 20 μg of either empty pTracer-CMV/Bsd vector, vector expressing HA-tagged NUP98-DDX10 or HA-tagged NUP98-DDX/3Q. To control for efficiency of transfection, 0.5 μg of pRL-TK (Promega), which expresses Renilla luciferase, was included. Luciferase activity was measured 48 h after electroporation using the Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA) and the results were normalized to Renilla luciferase. Relative luciferase activity was obtained by dividing the readings for each promoter construct by the corresponding readings for empty pGL4.11 vector under the same conditions.
Results and Discussion
NUP98-DDX10 localizes to the nuclei of primary human CD34+ cells in a punctate pattern
Wild-type NUP98 is a nucleoporin that localizes primarily to the nuclear pore complex at the nuclear rim with some intranuclear presence (11-13). The intracellular distribution of wild-type DDX10 is not known with certainty. Based on its sequence, it is thought to be involved in ribosome biogenesis (8), and its yeast counterpart, Dbp4p (HCA4), is a putative RNA helicase that functions in ribosome biogenesis (14, 15). Proteomic analysis showed that DDX10 is present in the human nucleolus (16, 17).
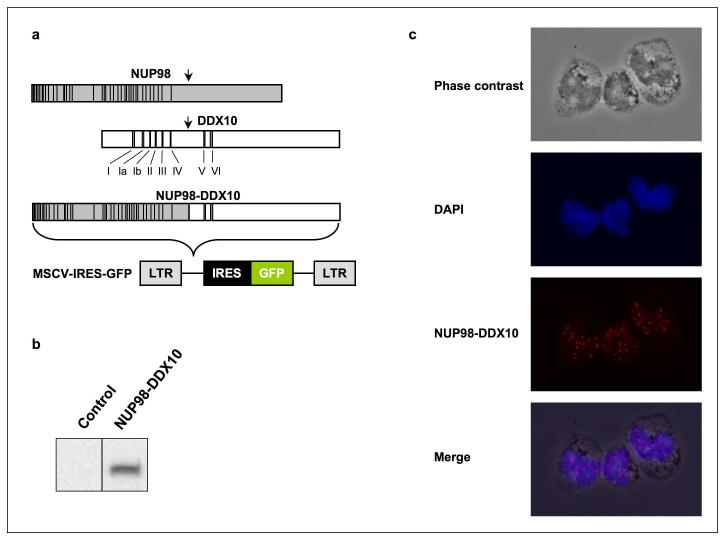
To determine the subcellular localization of NUP98-DDX10, the HA-tagged NUP98-DDX10 open reading frame was subcloned into the MSCV-IRES-GFP retroviral vector (Figure 1a) and introduced into primary human CD34+ cells obtained from peripheral blood. GFP+ cells were sorted and expression of the NUP98-DDX10 protein was confirmed by immunoblotting (Figure 1b). Immunofluorescence showed that NUP98-DDX10 localizes predominantly to distinct punctate structures within the nucleus (Figure 1c). In contrast to wild-type NUP98 (6, 18), there was no detectable localization of NUP98-DDX10 to the nuclear rim. In primary leukemic cells with the NUP98-HOXA9 fusion oncogene, NUP98 was localized within similar intranuclear aggregates (19). In cell lines, NUP98-HOXA9 and other leukemogenic NUP98 fusions have been shown to be predominantly intranuclear, but with a more finely granular or diffuse distribution (3, 6, 20, 21).
Figure 1.
Nuclear expression of NUP98-DDX10 in primary human CD34+ cells. (a) Schematic representation of NUP98, DDX10 and NUP98-DDX10. Vertical lines in the NUP98 portion indicate the locations of FG repeats. The locations of conserved helicase motifs in DDX10 are indicated with Roman numerals. (b) Immunoblotting with anti HA antibody shows NUP98-DDX10 expression in retrovirally transduced primary human CD34+ cells. (c) CD34+ cells retrovirally transduced with HA-NUP98-DDX10 were immunostained with anti-HA antibody and Alexa Fluor 647-conjugated anti-mouse IgG secondary antibody. The corresponding phase contrast image, nuclear counterstain with DAPI, and the merged images are shown. Images were viewed using a Nikon Eclipse 80i microscope with a Nikon 40X, 0.75 numerical aperture CFI Plan Fluor DLL objective and were acquired with a Nikon Coolsnap ES camera using MetaMorph 6.3r2 software with pseudo-coloring. The merged image was obtained by superimposing the 3 images using Photoshop CS4 software.
NUP98-DDX10 increases long-term proliferation and self-renewal in primary human CD34+ cells
Primary human CD34+ cells were transduced with either control retrovirus or retrovirus expressing NUP98-DDX10. After sorting for GFP expression, cells were cultured in liquid media with cytokines with periodic feeding and counting. As shown in Figure 2a, cells expressing NUP98-DDX10 multiplied much faster than cells transduced with empty control retrovirus. The marked long-term proliferation induced by NUP98-DDX10 suggested that it promotes the proliferation of primitive self-renewing cells. To assay for such cells, the LTC-IC assay (22) was used. As shown in Figure 2b, NUP98-DDX10 caused a dramatic increase in the numbers of LTC-ICs. These results indicate that NUP98-DDX10 induces proliferation of human primary hematopoietic cells by increasing the numbers of primitive self-renewing cells. The increases in proliferation and self-renewal are much higher with NUP98-DDX10 than was previously described for another leukemogenic NUP98 fusion, NUP98-HOXA9 (9, 10).
Figure 2.
NUP98-DDX10 induces proliferation and self-renewal in primary human CD34+ cells. Human CD34+ cells were retrovirally transduced with either control MSCV-IRES-GFP vector or vector expressing NUP98-DDX10, and cells were sorted for GFP positivity. (a) The cumulative fold increase in cell numbers compared to day 0 is plotted on a logarithmic scale against time. The experiment was repeated 3 times; results from a representative experiment are shown. (b) LTC-IC assay showing the average fold change compared to control from 3 independent experiments. The error bars represent standard deviations. Pictures of representative LTC-IC plates are shown.
NUP98-DDX10 inhibits the differentiation of primary human CD34+ cells
In order to assess the effects of NUP98-DDX10 on hematopoietic differentiation, primary human CD34+ cells, retrovirally transduced with NUP98-DDX10 or control vectors as described above, were subjected to colony-forming cell (CFC) assays. GFP-sorted cells were seeded in methylcellulose media supplemented with cytokines for 14 days. As shown in Figure 3a, NUP98-DDX10-expressing cells formed many large, prominent erythroid colonies that were readily distinguishable from those in the control samples without magnification. Under low magnification, many of the erythroid colonies in the NUP98-DDX10 sample show a complex branching pattern. The increased numbers of such colonies in cells expressing NUP98-DDX10 suggest that this oncogene increases the numbers of less mature erythroid precursors that have a higher proliferative potential. As shown below, NUP98-DDX10 did indeed cause erythroid hyperplasia and shift to immaturity in primary human cells (Fig. 3b and 3d).
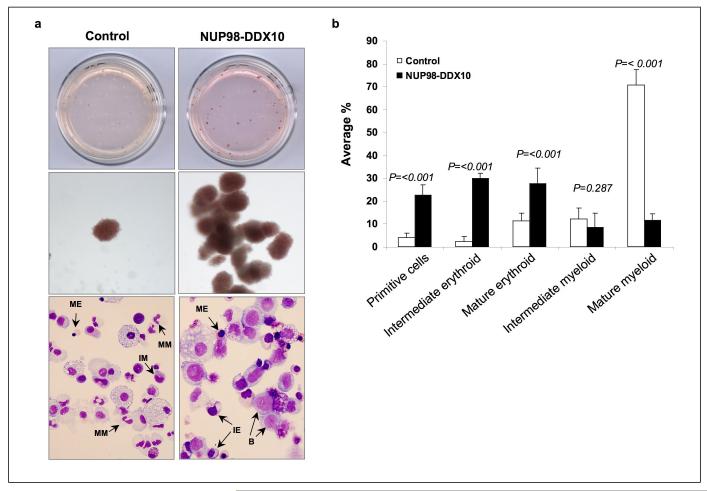
Figure 3.
NUP98-DDX10 disrupts myeloid and erythroid differentiation of primary human CD34+ cells. Human CD34+ cells were retrovirally transduced with either control MSCV-IRES-GFP vector or vector expressing NUP98-DDX10, and cells were sorted for GFP positivity. (a) One thousand cells were seeded into each of two duplicate plates for CFC assay and the experiment was repeated 6 independent times. Representative plates without magnification (upper panels) and low power photomicrographs of representative erythroid colonies (middle panels) are shown. Cytospin smears were prepared from the CFC plates and stained with Giemsa (lower panels). Photomicrographs were taken from representative fields with a 60X oil objective. Representative cells are indicated by arrows. B: blast; MM: mature myeloid; IM: intermediate myeloid; ME: mature erythroid; IE: intermediate erythroid. (b) 500 cell differential counts were performed to distinguish 5 groups of cells. Cells with blast and promyelocyte morphology were counted as primitive; those with myelocyte/metamyelocyte morphology as intermediate myeloid; those with band, segmented neutrophil, monocyte, and macrophage morphology as mature myeloid; those with intermediate hemoglobinization as intermediate erythroid; and those with full hemoglobinization as mature erythroid. Averages from 6 independent experiments are shown; error bars represent standard deviations. The P value was obtained by comparing to control using a two-sample, equal variance, two-tailed distribution t-test. (c) Flow cytometry for myeloid differentiation: Cells from the CFC plates were harvested and stained with CD45, CD33 and CD11b; the CD33+ gate was plotted on a histogram to show CD11b expression compared to control (lower panel). (d) Flow cytometry for erythroid differentiation: Cells from the CFC plates were harvested and stained with antibodies to CD45 and CD235a. The CD235a+ gate was plotted on a histogram (lower panel) to show the expression of CD235a relative to control cells. The percentages of cells falling within each gate are shown.
To further assess differentiation, cells were harvested from the CFC plates and Cytospin smears were prepared and stained with Giemsa (Figure 3a, lower panels). A manual differential count was performed to distinguish 5 groups of cells (Figure 3b). The NUP98-DDX10 samples showed an increase in the number of primitive cells and a shift to immaturity in both the myeloid and erythroid series.
Cells harvested from the CFC plates were also analyzed by flow cytometry to assess erythroid and myeloid differentiation. To assess myeloid differentiation, CD33+ myeloid cells were gated and the level of expression of CD11b was plotted on a histogram (Figure 3c). The intensity of CD11b expression was decreased in cells expressing NUP98-DDX10 indicating a disruption of myeloid differentiation. For erythroid differentiation, CD235a+ erythroid cells were gated and the expression level of CD235a in the gated cells was plotted on a histogram (Figure 3d). In the NUP98-DDX10 samples, the intensity of CD235a+ staining was decreased, consistent with disrupted erythroid differentiation. In addition the flow cytometry data show that NUP98-DDX10 causes a marked decrease in the percentage of CD33+ myeloid cells with a corresponding increase in the percentage of CD235a+ erythroid cells.
In summary, the results show that NUP98-DDX10 disrupts both erythroid and myeloid differentiation in primary human CD34+ cells. The effects are similar to those previously described for the leukemogenic NUP98-HOXA9 fusion.
NUP98-DDX10 and NUP98-HOXA9 deregulate a common set of genes with possible function in leukemogenesis
As described above, the effects of NUP98-DDX10 on the differentiation and proliferation of primary human CD34+ cells are remarkably similar to those previously described for NUP98-HOXA9 under the same conditions (9, 10). It was therefore of interest to determine whether NUP98-DDX10 and NUP98-HOXA9 share a common set of deregulated genes in primary human CD34+ cells. Such genes may offer clues to the shared capacity of NUP98 fusions to induce AML. To identify these genes, expression microarray analysis was carried out at 6 hours, 3 days, and 8 days after introduction of NUP98-DDX10 into primary human CD34+ cells. Each experiment was carried out in duplicate and only genes that were up- or down-regulated in both duplicates were considered significantly deregulated by NUP98-DDX10 (Supplementary Tables S1 – S3). The lists of deregulated genes were then compared to lists obtained from an identical analysis of NUP98-HOXA9. The list of genes deregulated by NUP98-HOXA9 at the 3-day time point has been published (9). The 6-hour and 8-day lists for NUP98-HOXA9 are shown in Supplementary Tables S4 and S5. At each time point, the list of genes deregulated by NUP98-DDX10 was compared to the list of genes deregulated by NUP98-HOXA9 and the intersections are shown as Venn diagrams in Figure 4a. The lists of deregulated genes common to both NUP98-DDX10 and NUP98-HOXA9 are shown in Supplementary Tables S6 – S8. A review of these lists reveals many genes with functions that may explain the leukemogenic effects shared by NUP98 fusions. These genes are listed in Table 1, and some of them are briefly discussed below; “NUP98 fusions” refers to NUP98-DDX10 and NUP98-HOXA9 in this discussion. A more comprehensive list with detailed description and a full list of references is shown in Supplementary Table S9.
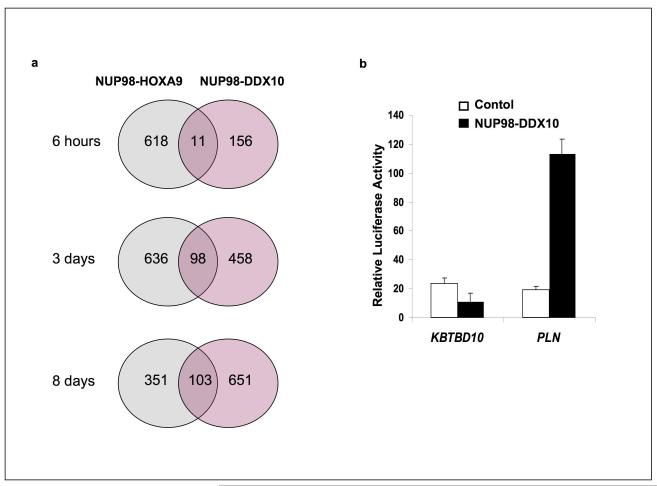
Figure 4.
Deregulation of gene expression by NUP98-DDX10. (a) For the 6 h time point, human CD34+ cells were nucleofected with either control pTracer vector, vector expressing NUP98-DDX10 or vector expressing NUP98-HOXA9. For the 3-day and 8-day time points, human CD34+ cells were retrovirally transduced with either control MSCV-IRES-GFP vector, vector expressing NUP98-DDX10, or vector expressing NUP98-HOXA9. Cells were sorted for GFP positivity and total RNA was subjected to microarray analysis. The experiment was performed two independent times and only genes that showed up- or down-regulation compared to control in both experiments were considered deregulated. Venn diagrams show the intersections of genes deregulated by NUP98-HOXA9 and NUP98-DDX10. (b) K562 cells were transfected by electroporation with a pGL4.11 luciferase construct driven by the KBTBD10 promoter or PLN promoter and either empty pTracer/CMV-Bsd vector (Control) or vector expressing NUP98-DDX10. Firefly luciferase activity was measured 48 h after transfection and normalized to a Renilla luciferase internal control and to empty pGL4.11 vector control. Error bars represent standard deviation.
1. Upregulated genes
Homeobox genes
Several genes that encode homeodomain-containing transcription factors are upregulated by NUP98 fusions. These include the clustered (type I) HOX genes HOXA3, HOXA5, HOXA6, HOXA7, HOXA9, HOXB3, HOXB5, HOXB6, and HOXB7 as well as the non-clustered (type II) homeobox genes, MEIS1 and PBX3. Homeobox genes play important roles in normal hematopoiesis and in leukemogenesis (23, 24). Several are expressed early in hematopoiesis and downregulated with differentiation (25). Overexpression of HOXA9 in mouse bone marrow causes AML that is accelerated by coexpression of MEIS1 (26). MEIS1 also accelerates the induction of AML by NUP98-HOXA9 in mice (26). Several homeobox genes including HOXA9, HOXA7, and MEIS1 are frequently overexpressed in human AML and are associated with worse prognosis (27-31). The induction of several homeobox genes by both NUP98-HOXA9 and NUP98-DDX10 suggests that homeobox genes play an important role in leukemic transformation by NUP98 fusions. This notion is supported by the recent observation that homeobox genes are upregulated in murine bone marrow cells expressing other leukemogenic NUP98 fusions such as NUP98-HOXD13, NUP98-PMX1, NUP98-NSD1, and NUP98-HHEX (20, 32-35).
Other oncogenic transcription factors
EGR1
This zinc finger transcription factor is upregulated in human AML with monocytic differentiation (36) and in a mouse model of acute promyelocytic leukemia (37). There is evidence that EGR1 mediates the proliferative effects of erythropoietin on mouse erythroblasts (38), suggesting that its upregulation of by NUP98 fusions may contribute to the erythroid hyperplasia we observed in vitro (Figure 3d).
SOX4
This transcription factor can cooperate with either MEF2C or EVI1 to induce AML in mice (39, 40). It is induced by the AML-associated AML1-ETO fusion oncoprotein in primary human hematopoietic cells (41).
MYCN
This basic helix-loop-helix/leucine zipper transcription factor is well known for its amplification and overexpression in neuroblastoma, other pediatric tumors, and lymphomas (42-44). More recently, it was reported that MYCN is overexpressed in many cases of pediatric AML and that its overexpression in mice leads to the development of AML (45).
HEY1
This b-HLH protein acts as a transcriptional repressor and a mediator in the Notch signaling pathway (46). Notch signaling is believed to be important in the self-renewal of hematopoietic stem cells and in the pathogenesis of some hematologic malignancies (47). HEY1 is also associated with increased numbers of erythroid cells (48) and a block in their differentiation (49), suggesting that it may mediate these effects of NUP98 fusions.
PTGS2 (COX-2)
Cyclooxygenase-2 (COX-2) is overexpressed in many tumors and its inhibition with drugs such as aspirin can be used in the prevention and treatment of cancer (50). A number of hematopoietic malignancies overexpress COX-2, which is associated with a worse prognosis (51). There is evidence that the use of aspirin and other non-steroidal anti-inflammatory drugs (NSAIDs) is associated with a lower incidence of lymphomas and acute leukemia (52). Specifically, NSAID use can reduce the risk of AML, particularly the FAB M2 subtype (AML with maturation) (53), which is the most common subtype in cases with NUP98 gene rearrangements (1). The upregulation of PTGS2 by NUP98 fusions may contribute to the worse prognosis of AML associated with NUP98 gene rearrangements, and raises the possibility that COX-2 blockade could help in the treatment of these leukemias.
Growth factors
REN
Previous microarray studies, confirmed by q-PCR, have shown that renin (REN) is upregulated by NUP98-HOXA9 (10). In the current study, microarray analysis showed upregulation of renin by NUP98-DDX10 (Table 1 and Supplementary Tables S2 and S3). This was confirmed by q-PCR, which showed an average upregulation of 547-fold at 3 days and an average upregulation of 2527-fold at 8 days. Renin is a constituent of the renin-angiotensin system (RAS) (54). In addition to its role as a regulator of blood pressure and electrolyte balance, the RAS plays a role in hematopoiesis and AML (55). Angiotensin II increases the proliferation of both primitive and committed hematopoietic progenitors (56, 57). Renin, angiotensinogen and angiotensin converting enzyme (ACE) are expressed in bone marrow from AML patients (58-61). Interestingly, drugs that inhibit the RAS, including ACE inhibitors and losartan inhibit growth and induce apoptosis in AML cell lines in vitro (62). These data raise the possibility that such drugs may be useful in the treatment of AML associated with NUP98 gene rearrangements.
ANGPT1
Angiopoietin-1 is an angiogenic growth factor that is co-expressed with its receptor, Tie2 (TEK) in AML cells and is thought to act as an autocrine and paracrine factor that enhances angiogenesis and the growth of AML cells (63-66).
2. Downregulated genes
GADD45B
This is one of a closely related family of genes associated with terminal myeloid differentiation and involved in the induction of cell cycle arrest and apoptosis (67). Its early downregulation by NUP98 fusions may contribute to blocked differentiation and increased proliferation.
MS4A3
This is one of a family of membrane proteins expressed predominantly in hematopoietic cells. It is involved in the regulation of the cell cycle in hematopoietic cells where its overexpression leads to cell cycle arrest at the G0/G1 phase (68). Its downregulation is thought to mediate the oncogenic effects of another AML fusion oncogene, AML1-ETO (69).
TRIM35
This gene is thought to act as a tumor suppressor that induces apoptosis and is associated with erythroid-to-myeloid lineage switch (70-72). Its repression by NUP98 fusions may explain the skewing of differentiation towards the erythroid lineage; it also raises the possibility that the increased numbers of cells in long-term culture may be due in part to decreased apoptosis.
NUP98-DDX10 deregulates gene expression at the transcriptional level
Several leukemogenic NUP98 fusions, including NUP98-HOXA9, NUP98-PMX1, and NUP98-NSD1, have been shown to deregulate transcription (3-6, 34, 35, 73). However, in these cases the fusion partners are proteins with well-known functions in transcriptional regulation: HOXA9 and PMX1 are homeobox transcription factors, and NSD1 is a histone methyltransferase. On the other hand, DDX10 is a putative DEAD-box RNA helicase whose function is not clear. Interestingly, recent data have implicated some RNA helicases in transcriptional regulation (74, 75).
To determine whether NUP98-DDX10 can deregulate gene transcription, luciferase reporter assays were carried out in the K562 myeloid cell line. We have previously identified a set of genes deregulated by the leukemogenic NUP98-HOXA9 fusion in K562 cells (5). This set is different from the set of genes deregulated by NUP98-HOXA9 in primary human CD34+ cells (10), and includes 2 genes that are transactivated by NUP98-HOXA9 in K562 cells by two distinct mechanisms: transactivation of the KBTBD10 gene requires direct DNA binding of NUP98-HOXA9 through the homeodomain whereas transactivation of the PLN gene does not (9). We tested the effect of NUP98-DDX10 on the KBTBD10 and PLN promoters in K562 cells. NUP98-DDX10 transactivated the PLN promoter but not the KBTBD10 promoter (Figure 4b). These data suggest that at least some of the effects of NUP98-DDX10 on gene expression are mediated at the transcriptional level, and that NUP98-DDX10 shares one of the transactivation mechanisms with NUP98-HOXA9, namely, the one that does not involve homeodomain-DNA binding. The exact mechanism by which NUP98-DDX10 regulates transcription remains to be determined, but the data described below suggest that a conserved motif within the DDX10 moiety may play a role.
A conserved helicase motif plays a role in in vitro transformation and transcriptional deregulation by NUP98-DDX10
Two types of NUP98-DDX10 fusions have been reported, the vast majority of which are so-called type II fusions that join exon 14 of NUP98 to exon 7 of DDX10 (7, 76, 77). Of the eight highly conserved motifs of the DEAD-box helicase family, only motifs V and VI are consistently present in these fusions. Motif VI contains 3 invariant arginine residues; its sequence in DDX10 is YIHRAGRTAR (8). Structural data show that motif VI is involved in ATP and RNA binding (78-81). Deletion of this region or mutation of the invariant arginine residues in RNA helicases results in loss of RNA binding, ATP hydrolysis, and helicase activity (80, 82, 83).
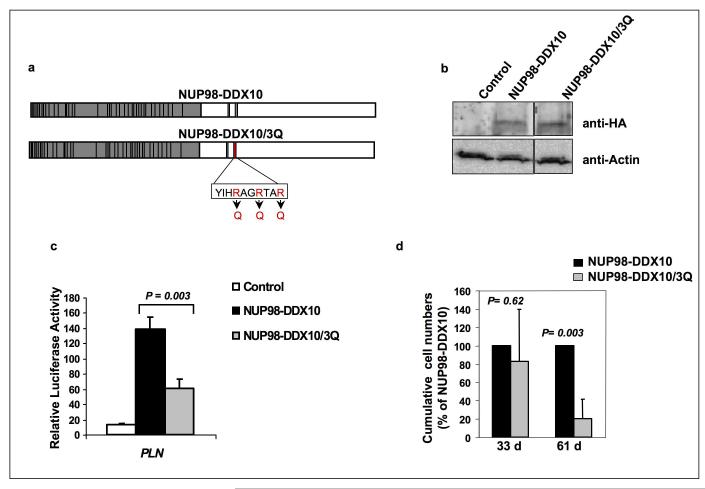
Recent data indicate that some of the DEAD-box family of proteins, including DDX5 (p68) and DDX17 (p72), can participate in transcriptional regulation by interacting with the transcription machinery (74, 75). Interestingly, deletion of motif VI from DDX17 results in loss of its ability to modulate transcription (83). We therefore sought to determine whether this motif plays a role in the ability of NUP98-DDX10 to regulate transcription and in its effects on primary human CD34+ cells. Mutating the 3 invariant arginine residues to glutamines has been previously shown to abolish the functions of motif VI in a DEAD-box RNA helicase (82). These mutations were therefore introduced into NUP98-DDX10 (Figure 5a). The resulting construct, NUP98-DDX10/3Q, was expressed at a level comparable to that of NUP98-DDX10 (Figure 5b).
Figure 5.
Mutation of the YIHRAGTAR motif inhibits the ability of NUP98-DDX10 to induce transcription and cell proliferation. (a) Schematic representation of the NUP98-DDX10/3Q mutant produced by replacing the three arginine residues in the YIHRAGTAR motif with glutamines. (b) Immunoblotting with anti-HA antibody shows equivalent expression of NUP98-DDX10 and NUP98-DDX10/3Q in retrovirally transduced human CD34+ cells (upper panel). Anti-actin control is shown in the lower panel. (c) K562 cells were transfected by electroporation with a luciferase construct driven by the PLN promoter and either empty pTracer/CMV-Bsd vector (Control) or vector expressing NUP98-DDX10 or NUP98-DDX10/3Q. Firefly luciferase activity was measured 48 h after transfection and normalized to a Renilla luciferase internal control and to empty pGL4.11 vector control. The numbers represent averages of 3 independent experiments; error bars represent standard deviations. (d) Long-term liquid culture of primary human CD34+ cells transduced with NUP98-DDX10 or NUP98-DDX10/3Q. Cumulative cell numbers are expressed as percentages compared to unmutated NUP98-DDX10, which is taken as 100%. The numbers represent averages of 3 independent experiments; error bars represent standard deviation. The P value was obtained by comparing NUP98-DDX10 to NUP98-DDX10/3Q using a two-sample, equal variance, two-tailed distribution t-test.
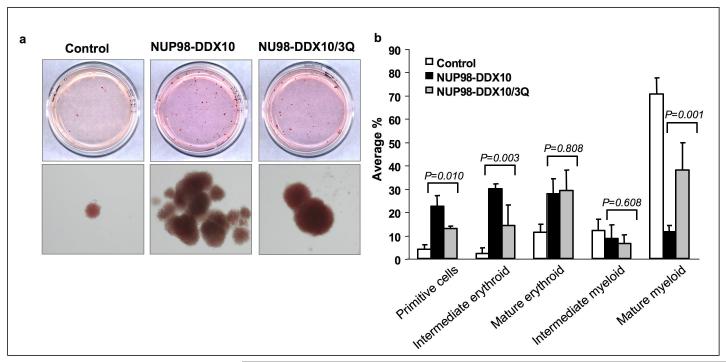
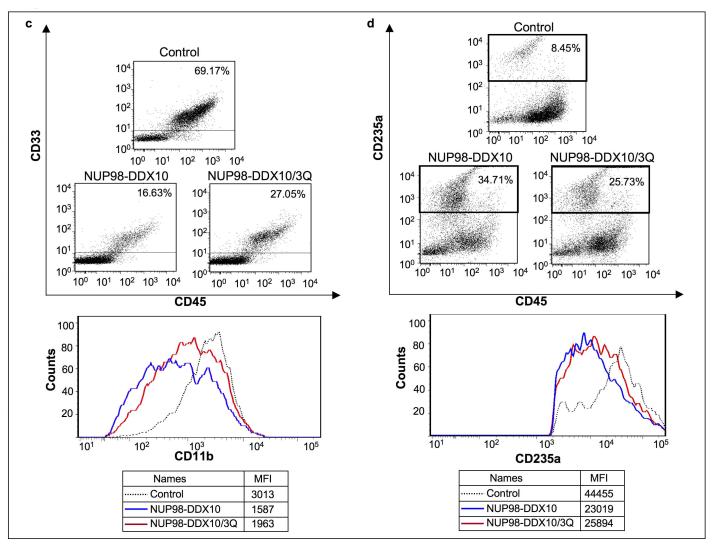
Luciferase assays showed that the motif VI mutation caused a significant reduction in the ability of NUP98-DDX10 to regulate transcription from the PLN promoter (Figure 5c). In addition, after 2 months in long-term liquid culture, primary human CD34+ cells transduced with the NUP98-DDX10/3Q mutant showed significantly fewer cells than those transduced with NUP98-DDX10 (Figure 5d). CFC assays were carried out to compare the effects of NUP98-DDX10 to those of NUP98-DDX10/3Q on erythroid and myeloid differentiation of primary human CD34+ cells. NUP98-DDX10/3Q samples showed less prominent erythroid colonies than samples expressing NUP98-DDX10 (Figure 6a). Differential counts revealed a significant decrease in the numbers of primitive cells and intermediate erythroid cells, and a significant increase in the numbers of mature myeloid cells in samples expressing NUP98-DDX10/3Q, indicating that the motif VI mutation resulted in a partial reversal of the effects of NUP98-DDX10 on erythroid and myeloid differentiation (Figure 6b). In addition, flow cytometry showed a mild shift to erythroid and myeloid maturity in samples expressing NUP98-DDX10/3Q (Figure 6c and 6d).
Figure 6.
Mutation of the YIHRAGTAR motif diminishes the ability of NUP98-DDX10 to disrupt the differentiation of primary human CD34+ cells. Primary human CD34+ cells were retrovirally transduced with either control MSCV-IRES-GFP vector or vector expressing NUP98-DDX10 or vector expressing NUP98-DDX10/3Q, and cells were sorted for GFP positivity. (a) One thousand cells were seeded into each of two duplicate plates for CFC assay and the experiment was repeated 3 independent times. Representative plates without magnification (upper panel) and low power photomicrographs of representative erythroid colonies (lower panel). (b) 500 cell differential counts were performed to distinguish 5 groups of cells. Averages from 3 – 6 independent experiments are shown; error bars represent standard deviations. The P value was obtained by comparing the samples using a two-sample, equal variance, two-tailed distribution t-test. (c) Flow cytometry for myeloid differentiation: Cells from the CFC plates were harvested and stained with CD45, CD33 and CD11b; the CD33+ gate was plotted on a histogram to show CD11b expression (lower panel). (d) Flow cytometry for erythroid differentiation: Cells from the CFC plates were harvested and stained with antibodies to CD45 and CD235a. The CD235a+ gate was plotted on a histogram (lower panel) to show the expression of CD235a. The percentages of cells falling within each gate are shown. The mean fluorescence intensity (MFI) is the arithmetic mean of the linear scaled fluorescence intensity.
Taken together, these data show that mutating motif VI in NUP98-DDX10 resulted in significant decrease in its ability to deregulate transcription, as well as a decrease in its ability to induce proliferation and to block differentiation in primary human CD34+ cells. Several studies have been performed on other leukemogenic NUP98 fusions to determine whether point mutations that target the function of the fusion partner have an effect on the transforming ability of the oncogene. Some mutations have drastic effects on the ability of the fusions to transform cells and deregulate transcription. For example, in NUP98-homeodomain fusions such as NUP98-HOXA9, NUP98-HOXD13 and NUP98-PMX1, point mutations that disrupt the ability of the homeodomain to bind DNA severely curtail their proliferative, leukemogenic, and transcriptional effects (6, 9, 32-34). Similarly, point mutations that abolish the histone methyl transferase activity of NSD1 drastically curtail the ability of NUP98-NSD1 to immortalize mouse myeloid progenitors and to deregulate transcription (35). In contrast, mutating the topoisomerase I catalytic site of NUP98-TOP1 had no discernible effect on its ability to induce proliferation of mouse bone marrow cells in vitro and its leukemogenic potential in vivo (21). In comparison, the role of motif VI in the transcriptional and transforming properties of NUP98-DDX10 falls in between these two extremes: its mutation clearly affects the in vitro transforming and transcriptional activities of the oncogene, but does not entirely abolish them.
Conclusion
The findings demonstrate that NUP98-DDX10 induces dramatic increases in the proliferation and self-renewal of primary human CD34+ cells and blocks their differentiation. These effects are accompanied by extensive changes in gene expression that are mediated at least in part by transcriptional deregulation. A number of the genes deregulated by NUP98-DDX10 are also deregulated by NUP98-HOXA9, suggesting that they may be important in mediating leukemogenesis by NUP98 fusion oncogenes. Questions to be addressed in future studies include the mechanisms underlying transcriptional deregulation by NUP98-DDX10 and the contribution of motif VI to this transcriptional deregulation. It also remains to be determined which of the deregulated genes discussed above play the most important roles in mediating leukemic transformation by NUP98 fusions.
Supplementary Material
Acknowledgements
We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, MO, for the use of the High Speed Cell Sorter Core, which provided flow cytometry analysis and sorting services and for the use of the Multiplexed Gene Analysis Core, which provided Affymetrix GeneChip microarray services. This work was supported by National Institutes of Health grants R01 HL082549 and K02 HL084179 (N.R.Y.), and by T32 CA009547 (A.M.A.). The Siteman Cancer Center is supported in part by an NCI Cancer Center Support Grant #P30 CA91842.
Footnotes
Supplementary information is available at the Leukemia web site (http://www.nature.com/leu).
References
- 1.Romana SP, Radford-Weiss I, Ben Abdelali R, Schluth C, Petit A, Dastugue N, et al. NUP98 rearrangements in hematopoietic malignancies: a study of the Groupe Francophone de Cytogenetique Hematologique. Leukemia. 2006 Apr;20(4):696–706. doi: 10.1038/sj.leu.2404130. [DOI] [PubMed] [Google Scholar]
- 2.Slape C, Aplan PD. The role of NUP98 gene fusions in hematologic malignancy. Leuk Lymphoma. 2004 Jul;45(7):1341–1350. doi: 10.1080/10428190310001659325. [DOI] [PubMed] [Google Scholar]
- 3.Bai XT, Gu BW, Yin T, Niu C, Xi XD, Zhang J, et al. Trans-repressive effect of NUP98-PMX1 on PMX1-regulated c-FOS gene through recruitment of histone deacetylase 1 by FG repeats. Cancer Res. 2006 May 1;66(9):4584–4590. doi: 10.1158/0008-5472.CAN-05-3101. [DOI] [PubMed] [Google Scholar]
- 4.Bei L, Lu Y, Eklund EA. HOXA9 activates transcription of the gene encoding gp91Phox during myeloid differentiation. J Biol Chem. 2005 Apr 1;280(13):12359–12370. doi: 10.1074/jbc.M408138200. [DOI] [PubMed] [Google Scholar]
- 5.Ghannam G, Takeda A, Camarata T, Moore MA, Viale A, Yaseen NR. The oncogene Nup98-HOXA9 induces gene transcription in myeloid cells. J Biol Chem. 2004 Jan 9;279(2):866–875. doi: 10.1074/jbc.M307280200. [DOI] [PubMed] [Google Scholar]
- 6.Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19(1):764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arai Y, Hosoda F, Kobayashi H, Arai K, Hayashi Y, Kamada N, et al. The inv(11)(p15q22) chromosome translocation of de novo and therapy- related myeloid malignancies results in fusion of the nucleoporin gene, NUP98, with the putative RNA helicase gene, DDX10. Blood. 1997;89(11):3936–3944. [PubMed] [Google Scholar]
- 8.Savitsky K, Ziv Y, Bar-Shira A, Gilad S, Tagle DA, Smith S, et al. A human gene (DDX10) encoding a putative DEAD-box RNA helicase at 11q22-q23. Genomics. 1996 Apr 15;33(2):199–206. doi: 10.1006/geno.1996.0184. [DOI] [PubMed] [Google Scholar]
- 9.Yassin ER, Sarma NJ, Abdul-Nabi AM, Dombrowski J, Han Y, Takeda A, et al. Dissection of the transformation of primary human hematopoietic cells by the oncogene NUP98-HOXA9. PLoS One. 2009;4(8):e6719. doi: 10.1371/journal.pone.0006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeda A, Goolsby C, Yaseen NR. NUP98-HOXA9 induces long-term proliferation and blocks differentiation of primary human CD34+ hematopoietic cells. Cancer Res. 2006 Jul 12;66(13):6628–6637. doi: 10.1158/0008-5472.CAN-06-0458. [DOI] [PubMed] [Google Scholar]
- 11.Powers MA, Macaulay C, Masiarz FR, Forbes DJ. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J Cell Biol. 1995 Mar;128(5):721–736. doi: 10.1083/jcb.128.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fontoura BM, Dales S, Blobel G, Zhong H. The nucleoporin Nup98 associates with the intranuclear filamentous protein network of TPR. Proc Natl Acad Sci U S A. 2001 Mar 13;98(6):3208–3213. doi: 10.1073/pnas.061014698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz TU. Modularity within the architecture of the nuclear pore complex. Curr Opin Struct Biol. 2005 Apr;15(2):221–226. doi: 10.1016/j.sbi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Kos M, Tollervey D. The Putative RNA Helicase Dbp4p Is Required for Release of the U14 snoRNA from Preribosomes in Saccharomyces cerevisiae. Mol Cell. 2005 Oct 7;20(1):53–64. doi: 10.1016/j.molcel.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 15.Liang WQ, Clark JA, Fournier MJ. The rRNA-processing function of the yeast U14 small nucleolar RNA can be rescued by a conserved RNA helicase-like protein. Mol Cell Biol. 1997 Jul;17(7):4124–4132. doi: 10.1128/mcb.17.7.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, et al. Nucleolar proteome dynamics. Nature. 2005 Jan 6;433(7021):77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 17.Scherl A, Coute Y, Deon C, Calle A, Kindbeiter K, Sanchez JC, et al. Functional proteomic analysis of human nucleolus. Mol Biol Cell. 2002 Nov;13(11):4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81(2):215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 19.Moore MA, Chung KY, Plasilova M, Schuringa JJ, Shieh JH, Zhou P, et al. NUP98 dysregulation in myeloid leukemogenesis. Ann N Y Acad Sci. 2007 Jun;1106:114–142. doi: 10.1196/annals.1392.019. [DOI] [PubMed] [Google Scholar]
- 20.Jankovic D, Gorello P, Liu T, Ehret S, La Starza R, Desjobert C, et al. Leukemogenic mechanisms and targets of a NUP98/HHEX fusion in acute myeloid leukemia. Blood. 2008 Jun 15;111(12):5672–5682. doi: 10.1182/blood-2007-09-108175. [DOI] [PubMed] [Google Scholar]
- 21.Gurevich RM, Aplan PD, Humphries RK. NUP98-topoisomerase I acute myeloid leukemia-associated fusion gene has potent leukemogenic activities independent of an engineered catalytic site mutation. Blood. 2004 Aug 15;104(4):1127–1136. doi: 10.1182/blood-2003-10-3550. [DOI] [PubMed] [Google Scholar]
- 22.Sutherland HJ, Lansdorp PM, Henkelman DH, Eaves AC, Eaves CJ. Functional characterization of individual human hematopoietic stem cells cultured at limiting dilution on supportive marrow stromal layers. Proc Natl Acad Sci U S A. 1990 May;87(9):3584–3588. doi: 10.1073/pnas.87.9.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007 Oct 15;26(47):6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 24.Sitwala KV, Dandekar MN, Hess JL. HOX Proteins and Leukemia. Int J Clin Exp Pathol. 2008;1(6):461–474. [PMC free article] [PubMed] [Google Scholar]
- 25.Pineault N, Helgason CD, Lawrence HJ, Humphries RK. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp Hematol. 2002 Jan;30(1):49–57. doi: 10.1016/s0301-472x(01)00757-3. [DOI] [PubMed] [Google Scholar]
- 26.Kroon E, Thorsteinsdottir U, Mayotte N, Nakamura T, Sauvageau G. NUP98-HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. Embo J. 2001;20(3):350–361. doi: 10.1093/emboj/20.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawagoe H, Humphries RK, Blair A, Sutherland HJ, Hogge DE. Expression of HOX genes, HOX cofactors, and MLL in phenotypically and functionally defined subpopulations of leukemic and normal human hematopoietic cells. Leukemia. 1999 May;13(5):687–698. doi: 10.1038/sj.leu.2401410. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence HJ, Rozenfeld S, Cruz C, Matsukuma K, Kwong A, Komuves L, et al. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia. 1999;13(12):1993–1999. doi: 10.1038/sj.leu.2401578. [DOI] [PubMed] [Google Scholar]
- 29.Afonja O, Smith JE, Jr., Cheng DM, Goldenberg AS, Amorosi E, Shimamoto T, et al. MEIS1 and HOXA7 genes in human acute myeloid leukemia. Leuk Res. 2000;24(10):849–855. doi: 10.1016/s0145-2126(00)00059-x. [DOI] [PubMed] [Google Scholar]
- 30.Drabkin HA, Parsy C, Ferguson K, Guilhot F, Lacotte L, Roy L, et al. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia. 2002;16(2):186–195. doi: 10.1038/sj.leu.2402354. [DOI] [PubMed] [Google Scholar]
- 31.Bullinger L, Dohner K, Bair E, Frohling S, Schlenk RF, Tibshirani R, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004 Apr 15;350(16):1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 32.Palmqvist L, Pineault N, Wasslavik C, Humphries RK. Candidate genes for expansion and transformation of hematopoietic stem cells by NUP98-HOX fusion genes. PLoS ONE. 2007;2(1):e768. doi: 10.1371/journal.pone.0000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pineault N, Buske C, Feuring-Buske M, Abramovich C, Rosten P, Hogge DE, et al. Induction of acute myeloid leukemia in mice by the human leukemia-specific fusion gene NUP98-HOXD13 in concert with Meis1. Blood. 2003 Jun 1;101(11):4529–4538. doi: 10.1182/blood-2002-08-2484. [DOI] [PubMed] [Google Scholar]
- 34.Hirose K, Abramovich C, Argiropoulos B, Humphries RK. Leukemogenic properties of NUP98-PMX1 are linked to NUP98 and homeodomain sequence functions but not to binding properties of PMX1 to serum response factor. Oncogene. 2008 Oct 9;27(46):6056–6067. doi: 10.1038/onc.2008.210. [DOI] [PubMed] [Google Scholar]
- 35.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007 Jul;9(7):804–812. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 36.Tokura Y, Shikami M, Miwa H, Watarai M, Sugamura K, Wakabayashi M, et al. Augmented expression of P-gp/multi-drug resistance gene by all-trans retinoic acid in monocytic leukemic cells. Leuk Res. 2002 Jan;26(1):29–36. doi: 10.1016/s0145-2126(01)00094-7. [DOI] [PubMed] [Google Scholar]
- 37.Yuan W, Payton JE, Holt MS, Link DC, Watson MA, DiPersio JF, et al. Commonly dysregulated genes in murine APL cells. Blood. 2007 Feb 1;109(3):961–970. doi: 10.1182/blood-2006-07-036640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang J, Menon M, Kapelle W, Bogacheva O, Bogachev O, Houde E, et al. EPO modulation of cell-cycle regulatory genes, and cell division, in primary bone marrow erythroblasts. Blood. 2007 Oct 1;110(7):2361–2370. doi: 10.1182/blood-2006-12-063503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du Y, Spence SE, Jenkins NA, Copeland NG. Cooperating cancer-gene identification through oncogenic-retrovirus-induced insertional mutagenesis. Blood. 2005 Oct 1;106(7):2498–2505. doi: 10.1182/blood-2004-12-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyd KE, Xiao YY, Fan K, Poholek A, Copeland NG, Jenkins NA, et al. Sox4 cooperates with Evi1 in AKXD-23 myeloid tumors via transactivation of proviral LTR. Blood. 2005 Oct 4; doi: 10.1182/blood-2003-05-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tonks A, Pearn L, Musson M, Gilkes A, Mills KI, Burnett AK, et al. Transcriptional dysregulation mediated by RUNX1-RUNX1T1 in normal human progenitor cells and in acute myeloid leukaemia. Leukemia. 2007 Dec;21(12):2495–2505. doi: 10.1038/sj.leu.2404961. [DOI] [PubMed] [Google Scholar]
- 42.Pession A, Tonelli R. The MYCN oncogene as a specific and selective drug target for peripheral and central nervous system tumors. Curr Cancer Drug Targets. 2005 Jun;5(4):273–283. doi: 10.2174/1568009054064606. [DOI] [PubMed] [Google Scholar]
- 43.Mao X, Onadim Z, Price EA, Child F, Lillington DM, Russell-Jones R, et al. Genomic alterations in blastic natural killer/extranodal natural killer-like T cell lymphoma with cutaneous involvement. J Invest Dermatol. 2003 Sep;121(3):618–627. doi: 10.1046/j.1523-1747.2003.12406.x. [DOI] [PubMed] [Google Scholar]
- 44.Schwaenen C, Nessling M, Wessendorf S, Salvi T, Wrobel G, Radlwimmer B, et al. Automated array-based genomic profiling in chronic lymphocytic leukemia: development of a clinical tool and discovery of recurrent genomic alterations. Proc Natl Acad Sci U S A. 2004 Jan 27;101(4):1039–1044. doi: 10.1073/pnas.0304717101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawagoe H, Kandilci A, Kranenburg TA, Grosveld GC. Overexpression of N-Myc rapidly causes acute myeloid leukemia in mice. Cancer Res. 2007 Nov 15;67(22):10677–10685. doi: 10.1158/0008-5472.CAN-07-1118. [DOI] [PubMed] [Google Scholar]
- 46.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003 Mar;194(3):237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 47.Nefedova Y, Gabrilovich D. Mechanisms and clinical prospects of Notch inhibitors in the therapy of hematological malignancies. Drug Resist Updat. 2008 Dec;11(6):210–218. doi: 10.1016/j.drup.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henning K, Schroeder T, Schwanbeck R, Rieber N, Bresnick EH, Just U. mNotch1 signaling and erythropoietin cooperate in erythroid differentiation of multipotent progenitor cells and upregulate beta-globin. Exp Hematol. 2007 Sep;35(9):1321–1332. doi: 10.1016/j.exphem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 49.Elagib KE, Xiao M, Hussaini IM, Delehanty LL, Palmer LA, Racke FK, et al. Jun blockade of erythropoiesis: role for repression of GATA-1 by HERP2. Mol Cell Biol. 2004 Sep;24(17):7779–7794. doi: 10.1128/MCB.24.17.7779-7794.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris RE. Cyclooxygenase-2 (cox-2) and the inflammogenesis of cancer. Subcell Biochem. 2007;42:93–126. doi: 10.1007/1-4020-5688-5_4. [DOI] [PubMed] [Google Scholar]
- 51.Bernard MP, Bancos S, Sime PJ, Phipps RP. Targeting cyclooxygenase-2 in hematological malignancies: rationale and promise. Curr Pharm Des. 2008;14(21):2051–2060. doi: 10.2174/138161208785294654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robak P, Smolewski P, Robak T. The role of non-steroidal anti-inflammatory drugs in the risk of development and treatment of hematologic malignancies. Leuk Lymphoma. 2008 Aug;49(8):1452–1462. doi: 10.1080/10428190802108854. [DOI] [PubMed] [Google Scholar]
- 53.Pogoda JM, Katz J, McKean-Cowdin R, Nichols PW, Ross RK, Preston-Martin S. Prescription drug use and risk of acute myeloid leukemia by French-American-British subtype: results from a Los Angeles County case-control study. Int J Cancer. 2005 Apr 20;114(4):634–638. doi: 10.1002/ijc.20788. [DOI] [PubMed] [Google Scholar]
- 54.Dzau VJ. Circulating versus local renin-angiotensin system in cardiovascular homeostasis. Circulation. 1988 Jun;77(6 Pt 2):I4–13. [PubMed] [Google Scholar]
- 55.Haznedaroglu IC, Ozturk MA. Towards the understanding of the local hematopoietic bone marrow renin-angiotensin system. Int J Biochem Cell Biol. 2003 Jun;35(6):867–880. doi: 10.1016/s1357-2725(02)00278-9. [DOI] [PubMed] [Google Scholar]
- 56.Mrug M, Stopka T, Julian BA, Prchal JF, Prchal JT. Angiotensin II stimulates proliferation of normal early erythroid progenitors. J Clin Invest. 1997 Nov 1;100(9):2310–2314. doi: 10.1172/JCI119769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodgers KE, Xiong S, Steer R, diZerega GS. Effect of angiotensin II on hematopoietic progenitor cell proliferation. Stem Cells. 2000;18(4):287–294. doi: 10.1634/stemcells.18-4-287. [DOI] [PubMed] [Google Scholar]
- 58.Wulf GG, Jahns-Streubel G, Nobiling R, Strutz F, Hemmerlein B, Hiddemann W, et al. Renin in acute myeloid leukaemia blasts. Br J Haematol. 1998;100(2):335–337. doi: 10.1046/j.1365-2141.1998.00565.x. [DOI] [PubMed] [Google Scholar]
- 59.Sde L Inigo, Casares MT, Jorge CE, Leiza SM, Santana GS, de Laguna SJ Bravo, et al. Relevance of renin expression by real-time PCR in acute myeloid leukemia. Leuk Lymphoma. 2006 Mar;47(3):409–416. doi: 10.1080/10428190500305927. [DOI] [PubMed] [Google Scholar]
- 60.Casares M Teresa Gomez, de la Iglesia S, Perera M, Lemes A, Campo C, Miguel JD Gonzalez San, et al. Renin expression in hematological malignancies and its role in the regulation of hematopoiesis. Leuk Lymphoma. 2002 Dec;43(12):2377–2381. doi: 10.1080/1042819021000040080. [DOI] [PubMed] [Google Scholar]
- 61.Beyazit Y, Aksu S, Haznedaroglu IC, Kekilli M, Misirlioglu M, Tuncer S, et al. Overexpression of the local bone marrow renin-angiotensin system in acute myeloid leukemia. J Natl Med Assoc. 2007 Jan;99(1):57–63. [PMC free article] [PubMed] [Google Scholar]
- 62.De la Iglesia Inigo S, Lopez-Jorge CE, Gomez-Casares MT, Castellano A Lemes, Cabrera P Martin, Brito J Lopez, et al. Induction of apoptosis in leukemic cell lines treated with captopril, trandolapril and losartan: A new role in the treatment of leukaemia for these agents. Leuk Res. 2008 Nov 14; doi: 10.1016/j.leukres.2008.09.029. [DOI] [PubMed] [Google Scholar]
- 63.Hatfield KJ, Hovland R, Oyan AM, Kalland KH, Ryningen A, Gjertsen BT, et al. Release of angiopoietin-1 by primary human acute myelogenous leukemia cells is associated with mutations of nucleophosmin, increased by bone marrow stromal cells and possibly antagonized by high systemic angiopoietin-2 levels. Leukemia. 2008 Feb;22(2):287–293. doi: 10.1038/sj.leu.2404985. [DOI] [PubMed] [Google Scholar]
- 64.Hatfield K, Oyan AM, Ersvaer E, Kalland KH, Lassalle P, Gjertsen BT, et al. Primary human acute myeloid leukaemia cells increase the proliferation of microvascular endothelial cells through the release of soluble mediators. Br J Haematol. 2009 Jan;144(1):53–68. doi: 10.1111/j.1365-2141.2008.07411.x. [DOI] [PubMed] [Google Scholar]
- 65.Albitar M. Angiogenesis in acute myeloid leukemia and myelodysplastic syndrome. Acta Haematol. 2001;106(4):170–176. doi: 10.1159/000046613. [DOI] [PubMed] [Google Scholar]
- 66.Muller A, Lange K, Gaiser T, Hofmann M, Bartels H, Feller AC, et al. Expression of angiopoietin-1 and its receptor TEK in hematopoietic cells from patients with myeloid leukemia. Leuk Res. 2002 Feb;26(2):163–168. doi: 10.1016/s0145-2126(01)00110-2. [DOI] [PubMed] [Google Scholar]
- 67.Liebermann DA, Hoffman B. Myeloid differentiation (MyD) primary response genes in hematopoiesis. Blood Cells Mol Dis. 2003 Sep-Oct;31(2):213–228. doi: 10.1016/s1079-9796(03)00160-8. [DOI] [PubMed] [Google Scholar]
- 68.Donato JL, Ko J, Kutok JL, Cheng T, Shirakawa T, Mao XQ, et al. Human HTm4 is a hematopoietic cell cycle regulator. J Clin Invest. 2002 Jan;109(1):51–58. doi: 10.1172/JCI14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dunne J, Cullmann C, Ritter M, Soria NM, Drescher B, Debernardi S, et al. siRNA-mediated AML1/MTG8 depletion affects differentiation and proliferation-associated gene expression in t(8;21)-positive cell lines and primary AML blasts. Oncogene. 2006 Oct 5;25(45):6067–6078. doi: 10.1038/sj.onc.1209638. [DOI] [PubMed] [Google Scholar]
- 70.Endersby R, Majewski IJ, Winteringham L, Beaumont JG, Samuels A, Scaife R, et al. Hls5 regulated erythroid differentiation by modulating GATA-1 activity. Blood. 2008 Feb 15;111(4):1946–1950. doi: 10.1182/blood-2007-04-085746. [DOI] [PubMed] [Google Scholar]
- 71.Kimura F, Suzu S, Nakamura Y, Nakata Y, Yamada M, Kuwada N, et al. Cloning and characterization of a novel RING-B-box-coiled-coil protein with apoptotic function. J Biol Chem. 2003 Jul 4;278(27):25046–25054. doi: 10.1074/jbc.M303438200. [DOI] [PubMed] [Google Scholar]
- 72.Lalonde JP, Lim R, Ingley E, Tilbrook PA, Thompson MJ, McCulloch R, et al. HLS5, a novel RBCC (ring finger, B box, coiled-coil) family member isolated from a hemopoietic lineage switch, is a candidate tumor suppressor. J Biol Chem. 2004 Feb 27;279(9):8181–8189. doi: 10.1074/jbc.M306751200. [DOI] [PubMed] [Google Scholar]
- 73.Nakamura T, Yamazaki Y, Hatano Y, Miura I. NUP98 is fused to PMX1 homeobox gene in human acute myelogenous leukemia with chromosome translocation t(1;11)(q23;p15) Blood. 1999;94(2):741–747. [PubMed] [Google Scholar]
- 74.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34(15):4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fuller-Pace FV, Ali S. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem Soc Trans. 2008 Aug;36(Pt 4):609–612. doi: 10.1042/BST0360609. [DOI] [PubMed] [Google Scholar]
- 76.Ahuja HG, Felix CA, Aplan PD. Potential role for DNA topoisomerase II poisons in the generation of t(11;20)(p15;q11) translocations. Genes Chromosomes Cancer. 2000;29(2):96–105. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1013>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 77.Morerio C, Acquila M, Rapella A, Tassano E, Rosanda C, Panarello C. Inversion (11)(p15q22) with NUP98-DDX10 fusion gene in pediatric acute myeloid leukemia. Cancer Genet Cytogenet. 2006 Dec;171(2):122–125. doi: 10.1016/j.cancergencyto.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 78.Caruthers JM, Johnson ER, McKay DB. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci U S A. 2000 Nov 21;97(24):13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006 Apr 21;125(2):287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 80.Cheng Z, Coller J, Parker R, Song H. Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA. 2005 Aug;11(8):1258–1270. doi: 10.1261/rna.2920905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Story RM, Li H, Abelson JN. Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proc Natl Acad Sci U S A. 2001 Feb 13;98(4):1465–1470. doi: 10.1073/pnas.98.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pause A, Methot N, Sonenberg N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol Cell Biol. 1993 Nov;13(11):6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Watanabe M, Yanagisawa J, Kitagawa H, Takeyama K, Ogawa S, Arao Y, et al. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J. 2001 Mar 15;20(6):1341–1352. doi: 10.1093/emboj/20.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.