Abstract
The t(8;21)(q22;q22) translocation, present in ~5% of adult acute myeloid leukemia (AML) cases, produces the AML1/ETO fusion protein. Dysregulation of the POU domain-containing transcription factor POU4F1 is a recurring abnormality in t(8;21) AML. Here, we show that POU4F1 over-expression is highly correlated with, but not caused by AML1/ETO. AML1/ETO markedly increases the self-renewal capacity of myeloid progenitors from murine bone marrow or fetal liver and drives expansion of these cells in liquid culture. POU4F1 is neither necessary nor sufficient for these AML1/ETO-dependent properties, suggesting that it contributes to leukemia through novel mechanisms. To identify targets of POU4F1, we performed gene expression profiling in primary mouse cells with genetically defined levels of POU4F1 and identified 140 differentially expressed genes. This expression signature was significantly enriched in human t(8;21) AML samples and was sufficient to cluster t(8;21) AML samples in an unsupervised hierarchical analysis. Among the most highly differentially expressed genes, half are known AML1/ETO targets, implying that the unique transcriptional signature of t(8;21) AML is, in part, attributable to POU4F1 and not AML1/ETO itself. These genes provide novel candidates for understanding the biology and developing therapeutic approaches for t(8;21) AML.
Keywords: POU4F1, AML1/ETO, acute myeloid leukemia, gene expression profiling
introduction
The t(8;21)(q22;q22) translocation produces an in-frame fusion of the first five exons of AML1 (RUNX1) to all but the first exon of ETO (MTG8) (1). This translocation is detectable in ~10% of de novo acute myeloid leukemia (AML) cases of the French-American-British M2 subtype and ~5% of all AML cases (2). AML1 is the DNA binding subunit of core binding factor (CBF), a multimeric transcription factor complex that includes CBFβ and additional transcriptional cofactors. The chimeric AML1/ETO protein has dominant negative effects on genes typically regulated by CBF (3). ETO, also a transcription factor, contains four nervy homology regions that contribute directly to the negative regulation of CBF-responsive genes (4). Despite these effects on gene regulation, AML1/ETO is not sufficient to cause AML (5-8), implying that additional genetic events are required.
Genome-wide expression profiling of primary human AML samples, performed by several groups, has identified a robust gene expression profile that distinguishes t(8;21) from other AML subtypes (9, 10). ETO is part of the t(8;21) expression signature. This is not unexpected, since most of the ETO coding sequence is contained within the AML1/ETO fusion transcript. These studies have also demonstrated that the POU4F1 gene is consistently dysregulated in t(8;21) human patient samples (9, 10).
POU4F1 is a transcription factor, originally identified in rat brain (11). The mouse and human orthologs are highly homologous (95% nucleic acid identity, 99% amino acid identity). POU4F1 contains a homeodomain and a POU-specific domain, both of which are required for DNA binding (11). Pou4f1 is important for embryonic brain development and is expressed beginning at E11.0 in mice (12), but has no reported role in normal or leukemic hematopoiesis. Pou4f1 null mice die postnatally with developmental anomalies in both the central and peripheral nervous system (13, 14).
The striking correlation between AML1/ETO and POU4F1 expression in human AML led us to hypothesize that POU4F1 might be a transcriptional target of AML1/ETO. Surprisingly, we found that POU4F1 dysregulation is not caused by AML1/ETO and that POU4F1 is dispensable for AML1/ETO function in vitro, but it is an important driver of the unique transcriptional profile associated with t(8;21) AML.
MATERIALS AND METHODS
Plasmids
Recombinant murine stem cell proviral plasmids MSCV2.2-ires-GFP (MIG) and MSCV2.2-AML1/ETO-ires-GFP (MAIG) were provided by Michael Tomasson (Washington University, St. Louis, MO). MSCV2.2-ires-YFP (MIY) was created by removing the GFP cDNA from MIG and replacing it with YFP from pEYFP-N1 (Clontech, Mountain View, CA). MSCV 2.2-Pou4f1-ires-YFP (MPIY) and MSCV2.2-Pou4f1-ires-GFP (MPIG) were generated by subcloning the mouse Pou4f1 cDNA (provided by Eric Turner, University of CA, San Diego) into MIY or MIG, respectively.
Mice
Pou4f1(Brn3a) mutant mice were provided by Eric Turner (University of CA, San Diego) (15). Embryos from timed matings were genotyped by PCR (primer sequences provided in Supplementary Table 1). Sca+/GFP and Sca+/AE mice were generated, as previously described (8, 16). All mice were backcrossed at least 10 generations to a C57BL/6J background.
RT-PCR analysis
RNA was made in Trizol LS (Invitrogen, Carlsbad, CA). All samples, excluding RNA from human AML samples, were treated with DNAse (Roche, Palo Alto, CA). cDNA was made from RNA using M-MLV reverse transcriptase (Promega, Madison, Wisconsin). qRT-PCR was performed using TaqMan Universal PCR Master Mix (Roche) (primer and probe sequences provided in Supplementary Table 1). All samples were run in triplicate on a 7300 Real Time PCR system (Applied Biosystems, Foster City, CA) and analyzed using the standard curve method.
Retroviral transduction
Retroviral supernatants were generated by transient transfection of 293T cells with Ecopac (Cell Genesys, Forster City, CA) and the MSCV-based retroviral constructs. 48 hrs after transfection, retroviral supernatants were harvested and titered on 3T3 cells using flow cytometry. Bone marrow cells were cultured in complete media (RPMI containing 1% L-glutamine, 20% fetal bovine serum) supplemented with recombinant hematopoietic cytokines (100 ng/mL stem cell factor, 6 ng/mL interleukin-3, 50 ng/mL Fms-related tyrosine kinase 3 ligand, and 10 ng/mL thrombopoietin; all from Peprotech, Rocky Hill, NJ) for 48 hours. 3.0-4.5 × 106 cells were infected on two consecutive days by centrifugation at 2500g for 90 minutes in the presence of 10 ug/ml polybrene (Sigma; St. Louis, MO) and 33 uM HEPES with retroviral supernatants using multiplicities of infection (MOIs) ranging from 1.5-3. Two days post-infection, the cells were seeded in cytokine-supplemented media at 105cells/ml in triplicate. In some experiments, transduced cells were selected by sorting on GFP or YFP (MoFlo, Beckman Coulter, Fullerton, CA) prior to initiating the cultures. Cells were counted twice-weekly and replated in fresh media, maintaining a concentration of 105 cell/mL. The proportion of transduced cells was monitored weekly by flow cytometry (FACScan, Becton Dickinson).
Hematopoietic progenitor analysis
Bone marrow cells from adult mice or 14.5-16.5 dpc fetal liver cells were transduced, as above. 48 hrs after the first round of infection, the cells were washed three times with RPMI, plated (6.7 × 103 cells/mL for bone marrow or 3.3 ×104 cells/mL for fetal liver) in cytokine-supplemented methylcellulose media (MethoCult M3434,Stem Cell Technologies; Vancouver, BC, Canada), and incubated at 37°C in 5% CO2. Seven days after plating, the colonies were examined under a fluorescence microscope. GFP+ or YFP+ colonies were individually selected, disrupted, and replated at one colony-equivalent per well in 24-well plates containing fresh methylcellulose media. Colonies were scored for survival and serially replated weekly.
Gene expression profiling
Murine 14.5-16.5dpc fetal liver cells were sorted for GFP or YFP expression 48 hours after retroviral transduction. Three experimental groups were generated: “Pou4f1 null” (Pou4f1−/− cells transduced with MIY), “Pou4f1 wildtype” (Pou4f1+/+ cells transduced with MIY), and “Pou4f1 high” (Pou4f1+/+ cells transduced with MPIY). Three independent samples were generated for each group (n=9 total samples). RNA was purified using Trizol LS, quantified by UV spectroscopy (Nanodrop Technologies), and qualitatively assessed using a BioAnalyzer 2100 and the RNA NanoChip assay (Agilent Technologies, Palo Alto CA). All samples were linear amplified, labeled, and hybridized to Affymetrix MOE430v2.0 GeneChip microarrays (Affymetrix, Santa Clara, CA) using standard protocols from the Siteman Cancer Center Multiplexed Gene Analysis Core Facility (for protocols, see http://Pathimm.wustl.edu/~mgacore/index.htm). Data are available from the Gene Expression Omnibus (GSE19997).
Probe raw signal intensities were summarized into probeset values using RMA (17, 18), and samples were quantile normalized. Only probesets called ‘Present’ by MAS5 software in all triplicates of at least one experimental group were retained for cluster and differential gene expression analysis (n=17,568 probesets). The overall expression profiles were highly similar for all samples (average Pearson correlation = 0.959) except for one from the wildtype group (average Pearson coefficient = 0.923 compared to the remaining 8 samples). This sample was retained, since the results of downstream analysis were not affected. We performed one-way analysis of variance (ANOVA) to identify probesets exhibiting differential expression between experimental groups. We performed 17,568 statistical tests, and applied q-value to the resulting p-values to estimate the genome-wide false discovery rate (19). Gene Ontology enrichment analysis was performed using DAVID (20).
Total RNA from 111 de novo M0-M7 human AML samples was profiled on Affymetrix U133+2 arrays, as previously described (21). Data are available from the Gene Expression Omnibus (GSE10358). Human orthologs of the dysregulated murine genes were identified (n=285 probesets) using BioMart (22). Testing for the enrichment of the Pou4f1 gene set in human AML samples was performed using Gene Set Enrichment Analysis (23, 24). Samples with or without the t(8;21) were compared, and the genes ranked based on the correlation between their expression and the class distinction using both signal2noise and ratio-of-classes gene ranking metrics (24). Wards hierarchical clustering was performed using Spotfire DecisionSite 8.2 (TIBCO Software Inc, Somerville, Mass). The P-value of the t(8;21) clustering was assessed by determining the number of times that a random selection of 285 probesets would result in the t(8;21) samples being nearest neighbors (distance metric = 1-Pearson correlation), divided by the number of random samplings (n=10,000). The POU4F1-independent gene expression profile was identified by removing the probesets for POU4F1 and its targets from the human AML data. Probesets with fewer than 25% present calls or a CV less than 0.5 were also removed. The remaining 13,700 probesets were used to cluster the AML samples with or without t(8;21). Significant differences in expression were identified by SAM using an FDR threshold <0.05 (25).
results
POU4F1 is associated with t(8;21) AML
We and others have noted that POU4F1 expression is dysregulated in t(8;21) AML (9, 26-30). We performed gene expression profiling using primary human samples and found that POU4F1 is not expressed in normal human CD34+ bone marrow cells or in AML samples from most FAB subtypes (Figure 1). High POU4F1 expression is restricted to M2 samples, with the highest levels noted in t(8;21) positive AML. To confirm these results, we performed quantitative RT-PCR using three samples with t(8;21) and nine randomly selected M2 samples without t(8;21). The t(8;21) samples tested had significantly higher levels of POU4F1 expression compared to M2 samples lacking t(8;21) (P<0.001) (Figure 1).
Figure 1. POU4F1 is upregulated in t(8;21) AML.
(a) Normal human CD34+ bone marrow cells (n=5) and FAB M0-M7 AML patient samples (n=111) were analyzed on the Affymetrix U133 Plus 2.0 array. High POU4F1 expression is restricted to the M2 samples and is highest in the subset with t(8;21) (filled triangles). The mean across all arrays (dotted line) and 2 standard deviations above (dashed line) for POU4F1 (probeset 211341_at) are shown. (b) qRT-PCR validation demonstrates significantly higher POU4F1 RNA expression levels in t(8;21) (●) vs. non t(8;21) (■) M2 samples (P<0.0001). The mean expression in each group (dotted line) is shown in comparison to the t(8;21) positive Kasumi cell line.
POU4F1 over-expression is AML1/ETO independent
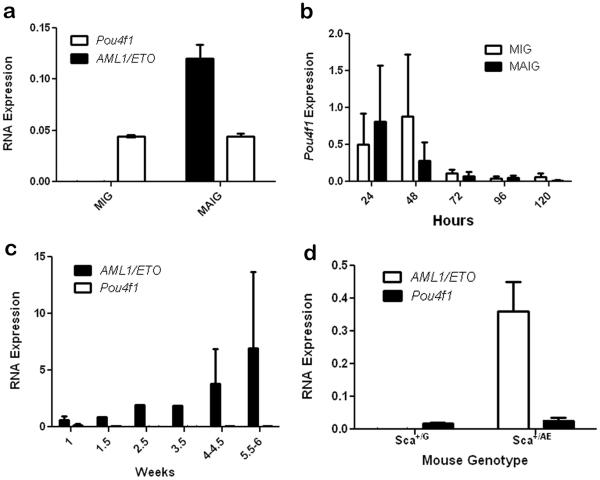
We hypothesized that high POU4F1 expression in t(8;21) AML is caused by altered transcriptional activity mediated by AML1/ETO (AE). Transient transfection of K562 cells with a construct containing AE (MAIG) or a control (MIG) plasmid did not increase POU4F1 mRNA above baseline levels (Figure 2). Next, we infected primary mouse bone marrow cells with the MAIG or MIG retroviruses. The cells were placed in culture and serial samples were taken for qRT-PCR analysis to measure AE and Pou4f1 expression. AE mRNA was readily detectable in cells infected with MAIG, but Pou4f1 levels were not increased in cells expressing AE (Figure 2). Similar results were obtained when bone marrow cells were first enriched for stem/progenitors by cell sorting (lineage-Kit+Sca+) prior to retroviral transduction (not shown). Finally, Pou4f1 levels were similar in bone marrow cells from mice heterozygous for an AE allele targeted to the Sca1 locus, compared to control Sca+/GFP mice (Figure 2). Taken together, these data suggest that high POU4F1 expression, though correlated with t(8;21), is not caused by AE.
Figure 2. AML1/ETO does not activate Pou4f1 expression.
(a) K562 cells were transfected with AML1/ETO (MAIG) or a control vector (MIG) and analyzed by qRT-PCR. AML1/ETO is present in MAIG transfected cells, as expected. Pou4f1 levels are unchanged. (b) Mouse bone marrow cells were infected with MAIG vs. MIG and analyzed by qRT-PCR. Mean transduction efficiencies were 19.4% for MIG and 14.9% for MAIG. There is a transient non-specific increase in Pou4f1 levels after infection, but no significant difference in cells expressing AML1/ETO vs. control. (c) AML1/ETO expression increases over 5 weeks in MAIG-infected bone marrow cells with no detectable expression of Pou4f1. (d) Pou4f1 is expressed at similar levels in bone marrow cells from mice heterozygous for either GFP or AML1/ETO targeted to the Sca1 locus.
Role of POU4F1 in AE-dependent hematopoietic cell growth
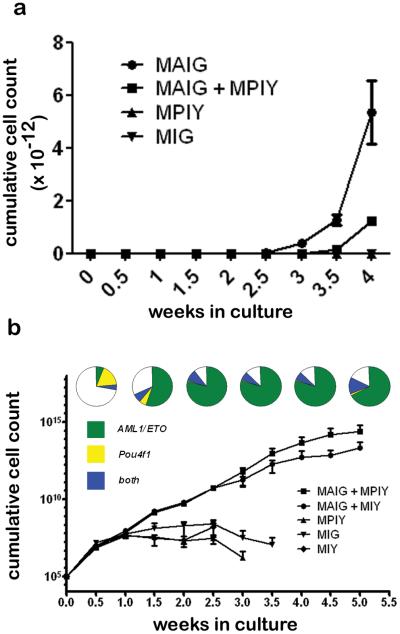
To test the impact of AE and Pou4f1 on the survival and proliferation of hematopoietic cells in vitro, we performed retroviral transduction of primary murine bone marrow cells. Transduced cells were enriched by flow sorting for GFP and/or YFP expression, and cultured in cytokine-supplemented media. Cells transduced with Pou4f1 (MPIY) or empty vector (MIY) died within two weeks, whereas cells transduced with AE (with or without Pou4f1) expanded rapidly beyond three weeks (Figure 3). Coexpression of Pou4f1 with AE provided no consistent growth advantage compared to AE alone.
Figure 3. Pou4f1 does not affect the growth of bone marrow cells.
(a) Mouse bone marrow cells were transduced with AML1/ETO (MAIG), POU4F1 (MPIY), or a control vector (MIG), positively selected by flow sorting, and cultured for up to 5 weeks, with cell counts and replating performed twice weekly. Cell growth was significantly enhanced by AML1/ETO, but not by Pou4f1. (b) Bone marrow cells were transduced as in (a) and cultured directly without sorting. The proportion of cells expressing AML1/ETO (GFP+), Pou4f1 (YFP+), or both (GFP+YFP+) are shown at weekly intervals for cells transduced with both AML1/ETO (MAIG) and Pou4f1 (MPIY). The relative expansion of AML1/ETO single positive cells (green) is greater than the expansion of AML1/ETO/Pou4f1 double positive cells (blue).
Next, we performed long term liquid culture of unsorted, retrovirally transduced cells and monitored the proportion of cells expressing GFP (AE reporter) and/or YFP (Pou4f1 reporter). Once again, cells infected with Pou4f1 alone were rapidly lost, whereas cells transduced with AE (with or without Pou4f1) had rapid and indistinguishable growth kinetics (Figure 3). In three independent experiments, the percentage of cells expressing AE only (GFP+YFP−) expanded 8.1-fold (range 5.4-9.6) over 6 weeks, compared to a 0.6-fold increase (range 0.14-6) for AE/Pou4f1 double positive cells. Although the growth of AE and AE/Pou4f1 positive cells are similar, the growth is attributable to an increasing population of AE single positive cells. The slight decrease in growth seen in the AE/Pou4f1 population therefore, suggests that Pou4f1 may have a modest inhibitory effect on cell growth stimulated by the presence of AE, although this difference was not statistically significant (p= .22, two-tailed paired t-test).
Pou4f1 is not required for AE-induced myeloid progenitor self-renewal
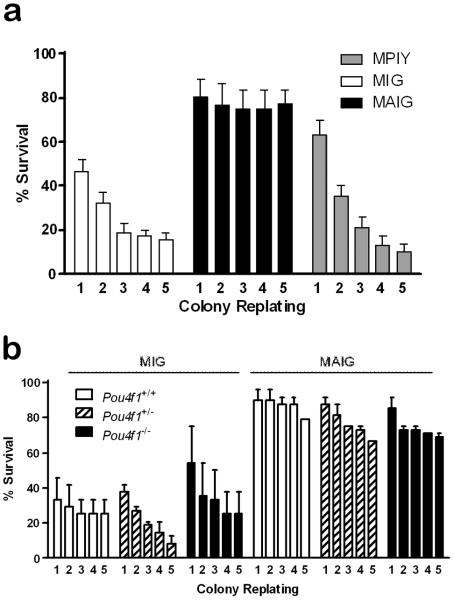
To test the importance of these genes for self-renewal of bone marrow myeloid progenitors, we performed serial replating of colonies retrovirally transduced with AE or Pou4f1 into cytokine-supplemented methocellulose media. In three independent experiments, colonies infected with the AE virus could be replated for at least five weeks (mean =17; range =15-20 per 24 colonies plated), which was significantly longer compared to colonies infected with the Pou4f1 virus (mean = 2; range = 1-4 per 24 colonies plated; p<0.001) or the control virus (mean = 3; range = 0-5;p<0.001), indicating that AE, but not Pou4f1, is sufficient to enhance progenitor self-renewal (Figure 4). To determine whether cells required Pou4f1 to self-renew, we serially replated colonies from Pou4f1−/−, Pou4f1+/−, or Pou4f1+/+ fetal liver cells infected with AE or the control virus. Significantly more colonies infected with the AE virus could be replated for 5 weeks (mean = 19; range = 16-24 per 24 colonies), compared to cells infected with the control virus (mean = 4; range = 1-9 per 24 colonies). Similar results were obtained regardless of Pou4f1 genotype (Figure 4). Therefore, AE promotes self-renewal of myeloid progenitors independent of Pou4f1.
Figure 4. Effect of AML1/ETO and POU4F1 on self-renewal of hematopoietic progenitors.
(a) Bone marrow cells infected with AML1/ETO (MAIG), Pou4f1 (MPIY), or control (MIG) virus were serially replated in methylcellulose. Colonies expressing AML1/ETO have increased self-renewal. (b) Fetal liver cells from Pou4f1−/−, Pou4f1+/− or Pou4f1+/+ mice were transduced with AML1/ETO (MAIG) or control (MIG) virus and serially plated in methylcellulose. Pou4f1 deficiency does not impair colony self-renewal induced by AML1/ETO.
Pou4f1-dependent transcriptional profile
Since POU4F1 dysregulation is not caused by AE, we reasoned that POU4F1 might regulate factors that can cooperate with AE in leukemogenesis. Therefore, we utilized microarray analysis to determine the gene expression profile induced by POU4F1. RNA was obtained from flow sorted murine fetal liver cells expressing high, wildtype, or null levels of Pou4f1. The global gene expression profiles of all samples were highly similar (>95% correlated); suggesting that over-expression of Pou4f1 did not dramatically alter the global transcriptional state of murine fetal liver cells, compared to wild-type or Pou4f1−/−cells. We identified 167 probesets (140 unique genes) that were differentially expressed between at least two experimental groups (Supplementary Table 2). The probeset comparisons were significant between cells expressing high levels of Pou4f1 compared to cells expressing endogenous levels or no Pou4f1. The genes are enriched in several biological pathways relevant for cancer (Supplementary Table 3). Pou4f1 itself is not part of the gene set, since the Pou4f1 probesets on the microarray are all 3′ of sequences contained within the transduced Pou4f1 cDNA.
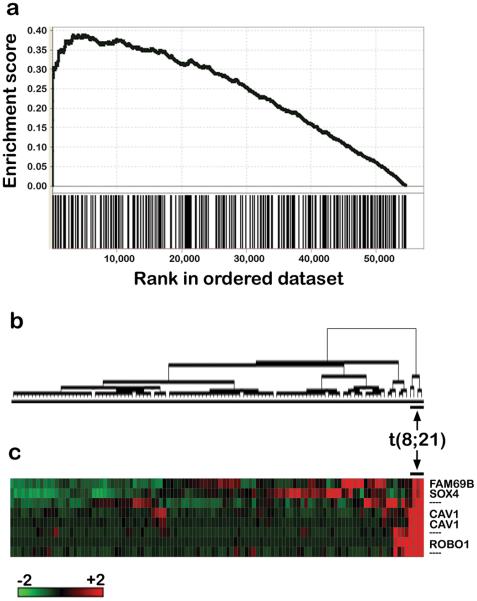
We next sought to determine whether the set of 140 Pou4f1-regulated genes discovered in mouse cells was relevant for human AML. Because POU4F1 is highly expressed in t(8;21) human AML samples, we hypothesized that the human orthologs of the mouse Pou4f1 gene set would be enriched in a comparison of AML samples with or without t(8;21). To test this hypothesis, we applied Gene Set Enrichment Analysis (GSEA) to the expression profiles of 111 human AML samples using ratio-of-classes and signal2noise (similar results obtained with both metrics). Significant enrichment of the Pou4f1 gene set was demonstrated in the t(8;21) positive samples (FDR q-value < 0.001), suggesting that the Pou4f1 expression signature is robust across species (Figure 5).
Figure 5. Contribution of Pou4f1 target genes to the transcriptional profile of t(8;21) AML.
(a) Gene Set Enrichment Analysis was performed on de novo AML samples (n=111) using the human orthologs of the 140 genes differentially regulated by POU4F1 expression. The POU4F1 gene set is significantly enriched in human t(8;21) positive AML samples (FDR<0.001). (b) Dendrogram of AML samples after unsupervised hierarchal clustering using the Pou4f1 gene set demonstrates segregation of the t(8;21) patients. (c) Four annotated genes (and 3 unannotated probesets) are consistently and significantly (P < 0.01) differentially expressed between AML samples with or without t(8;21).
Next, we performed unsupervised hierarchical clustering of the human AML samples using the 140 gene (n=285 probeset) POU4F1 expression signature and found that the t(8;21) samples segregated into a distinct cluster (Figure 5). Because t(8;21) AML samples have a strong, reproducible expression signature (9, 10), these samples might be predicted to cluster using many combinations of 285 probesets. We tested this hypothesis using a random sampling strategy and found that the t(8;21) samples cluster together rarely by chance when gene sets of identical size were used (P < 0.05).
Limiting our comparison to the M2 AML patients (n=25), we found that the POU4F1 gene set again segregated the t(8;21) samples (not shown). When each of the signature genes was tested for differences in expression between samples with or without t(8;21), eight annotated genes were identified (Table 1). Four of these genes show marked (> 2-fold), consistent upregulation in the t(8;21) samples, compared to other AML subtypes (Figure 5).
Table 1.
Gene expression differences in FAB M2 AML samples with or without t(8;21).
| Gene | Fold Change* |
|---|---|
| CAV1 | 34.48 |
| CAV1 | 11.78 |
| ROBO1 | 26.01 |
| FAM69B | 2.54 |
| SOX4 | 1.97 |
| PELI2 | 1.69 |
| PPAPDC1B | 1.56 |
| H2AFV | 1.49 |
| PLXDC2 | 0.12 |
Fold change = mean expression in t(8;21) samples divided by mean expression in AML samples without t(8;21). All values are significant by unpaired two-tailed t-test (P < 0.01).
To ask which genes are have significant differential expression independent of POU4F1 in t(8;21) AML samples, we used the Significance Analysis of Microarray algorithm after removing the POU4F1 gene set. 115 annotated genes (183 probesets) remained significant in this comparison (Supplementary Table 4).
discussion
Previous work has demonstrated that expression of the AE fusion gene is not sufficient to induce AML. AE transcripts remain detectable in bone marrow cells from patients with t(8;21) AML in durable remission (5, 31). Expression of an AE cDNA in transgenic mice (6-8) or by retroviral transduction/transplantation (32, 33) does not cause AML, unless additional mutations are induced (6, 7, 32). Gene expression profiling experiments have implicated POU4F1 as a candidate cooperating factor in t(8;21) AML (9, 26-30). Here, we confirm the observation that POU4F1 dysregulation is highly correlated with t(8;21) AML and explore the biological consequences of POU4F1 expression in hematopoietic cells.
Several lines of evidence indicate that POU4F1 dysregulation is not caused by AE. First, we ectopically expressed the AE cDNA in mouse and human cells by transfection, retroviral transduction, or transgenesis and could not detect differences in POU4F1 expression under any of these conditions. These results are consistent with previous experiments in cell lines and transgenic zebrafish (34-37). Next, high POU4F1 expression is detectable in a minority of t(8;21) negative AML cases, suggesting AE is not required for its transcriptional activation. Finally, Pou4f1 is not expressed in normal hematopoietic or lymphoid cells (38) and is not required for hematopoiesis (11, 13, 14). Taken together, this suggests that AE does not cause POU4F1 upregulation, nor does it cause POU4F1 levels to increase by expanding a cellular population that normally expresses POU4F1. POU4F1 and AE, therefore, are concordantly over-expressed in t(8;21) AML, but through independent mechanisms, suggesting that they provide non-redundant signals important for leukemogenesis.
These results support a model in which POU4F1 upregulation precedes acquisition of the t(8;21). This raises many questions for future investigation, including: what genetic and/or epigenetic mechanism(s) activate POU4F1? Do POU4F1-dependent signals facilitate acquisition of the t(8;21) or provide a selective advantage for cells that undergo this translocation? Do POU4F1 and AE cooperate during induction of leukemia in vivo?
POU4F1 is a member of the highly conserved family of POU (Pit/Oct/Unc) domain-containing transcription factors. Although we have shown that Pou4f1 is dispensable for self-renewal of hematopoietic progenitors in vitro, transplantation experiments will be required to address the importance of this factor for pluripotency and self-renewal of hematopoietic stem cells in vivo. The closely related POU family member, POU5F1 (encoding OCT4), is required to maintain pluripotency in embryonic stem cells (39) and elevated levels drive their differentiation along mesodermal and endodermal lineages (40). POU5F1 has also been implicated in carcinogenesis. It is over-expressed in breast and germ cell tumors (41, 42). Inducible expression in mice causes dysplasia in the skin and GI tract (43).
Previous studies of POU4F1 have been restricted to neuronal cells, where it has been shown to promote cell survival and inhibit apoptosis, in part by antagonizing p53 and p73 to increase Bcl2 and Bax expression (44, 45). In hematopoietic cells, we observed different effects of Pou4f1 on transcription and cell growth (in fact, Pou4f1 appears to restrain cell growth that was stimulated by AE). These apparently contradictory results may reflect differences in experimental design or cellular context.
We took an unbiased approach to identify transcriptional targets of Pou4f1 in primary murine hematopoietic cells and then cross-validated this expression signature in primary human AML samples. The Pou4f1 expression signature is sufficient to cluster t(8;21) AML samples in an unsupervised analysis and the Pou4f1 gene set is enriched in t(8;21) AML, suggesting that expression of these target genes discovered in mouse is relevant in human cells and preserved in fully transformed leukemias. Four of these genes (SOX, CAV1, ROBO1, PPADC1B) are also regulated independently by AE (35-37). The nerve growth factor receptor TrkA has also been shown to be a target of both Pou4f1 and AE (46, 47). The other four genes in the POU4F1 signature (PELI2, FAM69B, H2AFV, PLXDC2) are POU4F1-specific (i.e., they are over-expressed in t(8;21) AML samples, but not activated by AE). These results imply that the gene expression profile of t(8;21) AML is, in part, attributable to POU4F1 and not solely AE itself.
Several members of the Pou4f1 gene set have been previously implicated in cancer. SOX4 is a transcription factor important for regulation of embryonic development and determination of cell fate (48-50). Depending on context, SOX4 has both oncogenic (51-53) and tumor suppressor properties (54). SOX4 is activated by retroviral integration in mice, and cooperates with Evi1 in the induction of AML (55). CAV1 is a lipid raft protein that binds to the G-protein coupled receptor, GLP-1, and is involved in subcellular localization, trafficking, and signaling (56). In AML samples, CAV1 and MDR1 colocalize and their expression is highly correlated (57). ROBO1 is a receptor for SLIT and interacts with Rho GTPase activating proteins important for cell motility and angiogenesis (58, 59). Downregulation of ROBO1 in breast cancer may initiate a metastatic phenotype (60). PPADC1B encodes a transmembrane protein phosphatase that can induce anchorage-independent growth in 3T3 fibroblasts (61).
Although additional work is needed to elucidate the mechanism by which POU4F1 contributes to t(8;21) AML, it is clear that this transcription factor is a critical driver of disordered gene expression in this disease. Further research on factors both upstream and downstream of POU4F1 should shed new light on the pathogenesis of t(8;21) positive AML.
Supplementary Material
Acknowledgments
Supporting by funding from the NIH (P01 CA101937) and the G&P Foundation. Cell sorting and gene expression profiling were performed in Siteman Cancer Center core facilities that are supported by the NCI (P30 CA91842). We thank Eric Turner and Michael Tomasson for providing plasmids and Mieke Hoock for expert mouse colony management.
Footnotes
conflict of interest
The authors declare no conflicts of interest.
Supplementary Information
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu).
REFERENCES
- 1.Zhang Y, Strissel P, Strick R, Chen J, Nucifora G, Le Beau MM, et al. Genomic DNA breakpoints in AML1/RUNX1 and ETO cluster with topoisomerase II DNA cleavage and DNase I hypersensitive sites in t(8;21) leukemia. Proc Natl Acad Sci U S A. 2002 Mar 5;99(5):3070–3075. doi: 10.1073/pnas.042702899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC; Lyon: 2008. [Google Scholar]
- 3.Meyers S, Lenny N, Hiebert SW. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol Cell Biol. 1995 Apr;15(4):1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci U S A. 1998 Sep 1;95(18):10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nucifora G, Larson RA, Rowley JD. Persistence of the 8;21 translocation in patients with acute myeloid leukemia type M2 in long-term remission. Blood. 1993 Aug 1;82(3):712–715. [PubMed] [Google Scholar]
- 6.Higuchi M, O'Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002 Feb;1(1):63–74. doi: 10.1016/s1535-6108(02)00016-8. [DOI] [PubMed] [Google Scholar]
- 7.Yuan Y, Zhou L, Miyamoto T, Iwasaki H, Harakawa N, Hetherington CJ, et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc Natl Acad Sci U S A. 2001 Aug 28;98(18):10398–10403. doi: 10.1073/pnas.171321298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fenske TS, Pengue G, Mathews V, Hanson PT, Hamm SE, Riaz N, et al. Stem cell expression of the AML1/ETO fusion protein induces a myeloproliferative disorder in mice. Proc Natl Acad Sci U S A. 2004 Oct 19;101(42):15184–15189. doi: 10.1073/pnas.0400751101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004 Apr 15;350(16):1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 10.Bullinger L, Dohner K, Bair E, Frohling S, Schlenk RF, Tibshirani R, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004 Apr 15;350(16):1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 11.He X, Treacy MN, Simmons DM, Ingraham HA, Swanson LW, Rosenfeld MG. Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature. 1989 Jul 6;340(6228):35–41. doi: 10.1038/340035a0. [DOI] [PubMed] [Google Scholar]
- 12.Theil T, McLean-Hunter S, Zornig M, Moroy T. Mouse Brn-3 family of POU transcription factors: a new aminoterminal domain is crucial for the oncogenic activity of Brn-3a. Nucleic Acids Res. 1993 Dec 25;21(25):5921–5929. doi: 10.1093/nar/21.25.5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McEvilly RJ, Erkman L, Luo L, Sawchenko PE, Ryan AF, Rosenfeld MG. Requirement for Brn-3.0 in differentiation and survival of sensory and motor neurons. Nature. 1996 Dec 12;384(6609):574–577. doi: 10.1038/384574a0. [DOI] [PubMed] [Google Scholar]
- 14.Xiang M, Gan L, Zhou L, Klein WH, Nathans J. Targeted deletion of the mouse POU domain gene Brn-3a causes selective loss of neurons in the brainstem and trigeminal ganglion, uncoordinated limb movement, and impaired suckling. Proc Natl Acad Sci U S A. 1996 Oct 15;93(21):11950–11955. doi: 10.1073/pnas.93.21.11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quina LA, Pak W, Lanier J, Banwait P, Gratwick K, Liu Y, et al. Brn3a-expressing retinal ganglion cells project specifically to thalamocortical and collicular visual pathways. J Neurosci. 2005 Dec 14;25(50):11595–11604. doi: 10.1523/JNEUROSCI.2837-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson P, Mathews V, Marrus SH, Graubert TA. Enhanced green fluorescent protein targeted to the Sca-1 (Ly-6A) locus in transgenic mice results in efficient marking of hematopoietic stem cells in vivo. Exp Hematol. 2003 Feb;31(2):159–167. doi: 10.1016/s0301-472x(02)01021-4. [DOI] [PubMed] [Google Scholar]
- 17.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003 Feb 15;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolstad B, Irizarry R, Astrand M, Speed T. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003 Jan 22;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. 2003. [DOI] [PubMed] [Google Scholar]
- 19.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003 Aug 5;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Tomasson MH, Xiang Z, Walgren R, Zhao Y, Kasai Y, Miner T, et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood. 2008 May 1;111(9):4797–4808. doi: 10.1182/blood-2007-09-113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, et al. BioMart--biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003 Jul;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001 Apr 24;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bullinger L, Rucker FG, Kurz S, Du J, Scholl C, Sander S, et al. Gene-expression profiling identifies distinct subclasses of core binding factor acute myeloid leukemia. Blood. 2007 Aug 15;110(4):1291–1300. doi: 10.1182/blood-2006-10-049783. [DOI] [PubMed] [Google Scholar]
- 27.Ichikawa H, Tanabe K, Mizushima H, Hayashi Y, Mizutani S, Ishii E, et al. Common gene expression signatures in t(8;21)- and inv(16)-acute myeloid leukaemia. Br J Haematol. 2006 Nov;135(3):336–347. doi: 10.1111/j.1365-2141.2006.06310.x. [DOI] [PubMed] [Google Scholar]
- 28.Schoch C, Kohlmann A, Schnittger S, Brors B, Dugas M, Mergenthaler S, et al. Acute myeloid leukemias with reciprocal rearrangements can be distinguished by specific gene expression profiles. Proc Natl Acad Sci U S A. 2002 Jul 23;99(15):10008–10013. doi: 10.1073/pnas.142103599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Debernardi S, Lillington DM, Chaplin T, Tomlinson S, Amess J, Rohatiner A, et al. Genome-wide analysis of acute myeloid leukemia with normal karyotype reveals a unique pattern of homeobox gene expression distinct from those with translocation-mediated fusion events. Genes Chromosomes Cancer. 2003 Jun;37(2):149–158. doi: 10.1002/gcc.10198. [DOI] [PubMed] [Google Scholar]
- 30.Ross ME, Mahfouz R, Onciu M, Liu HC, Zhou X, Song G, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004 Dec 1;104(12):3679–3687. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- 31.Miyamoto T, Weissman IL, Akashi K. AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc Natl Acad Sci U S A. 2000 Jun 20;97(13):7521–7526. doi: 10.1073/pnas.97.13.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grisolano JL, O'Neal J, Cain J, Tomasson MH. An activated receptor tyrosine kinase, TEL/PDGFbetaR, cooperates with AML1/ETO to induce acute myeloid leukemia in mice. Proc Natl Acad Sci U S A. 2003 Aug 5;100(16):9506–9511. doi: 10.1073/pnas.1531730100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Guzman CG, Warren AJ, Zhang Z, Gartland L, Erickson P, Drabkin H, et al. Hematopoietic stem cell expansion and distinct myeloid developmental abnormalities in a murine model of the AML1-ETO translocation. Mol Cell Biol. 2002 Aug;22(15):5506–5517. doi: 10.1128/MCB.22.15.5506-5517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimada H, Ichikawa H, Ohki M. Potential involvement of the AML1-MTG8 fusion protein in the granulocytic maturation characteristic of the t(8;21) acute myelogenous leukemia revealed by microarray analysis. Leukemia. 2002 May;16(5):874–885. doi: 10.1038/sj.leu.2402465. [DOI] [PubMed] [Google Scholar]
- 35.Tonks A, Pearn L, Musson M, Gilkes A, Mills KI, Burnett AK, et al. Transcriptional dysregulation mediated by RUNX1-RUNX1T1 in normal human progenitor cells and in acute myeloid leukaemia. Leukemia. 2007 Dec;21(12):2495–2505. doi: 10.1038/sj.leu.2404961. [DOI] [PubMed] [Google Scholar]
- 36.Gardini A, Cesaroni M, Luzi L, Okumura AJ, Biggs JR, Minardi SP, et al. AML1/ETO oncoprotein is directed to AML1 binding regions and co-localizes with AML1 and HEB on its targets. PLoS Genet. 2008 Nov;4(11):e1000275. doi: 10.1371/journal.pgen.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeh JR, Munson KM, Chao YL, Peterson QP, Macrae CA, Peterson RT. AML1-ETO reprograms hematopoietic cell fate by downregulating scl expression. Development. 2008 Jan;135(2):401–410. doi: 10.1242/dev.008904. [DOI] [PubMed] [Google Scholar]
- 38.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004 Apr 20;101(16):6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998 Oct 30;95(3):379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 40.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000 Apr;24(4):372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 41.Jin T, Branch DR, Zhang X, Qi S, Youngson B, Goss PE. Examination of POU homeobox gene expression in human breast cancer cells. Int J Cancer. 1999 Mar 31;81(1):104–112. doi: 10.1002/(sici)1097-0215(19990331)81:1<104::aid-ijc18>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 42.Cheng L. Establishing a germ cell origin for metastatic tumors using OCT4 immunohistochemistry. Cancer. 2004 Nov 1;101(9):2006–2010. doi: 10.1002/cncr.20566. [DOI] [PubMed] [Google Scholar]
- 43.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005 May 6;121(3):465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 44.Budram-Mahadeo V, Morris PJ, Latchman DS. The Brn-3a transcription factor inhibits the pro-apoptotic effect of p53 and enhances cell cycle arrest by differentially regulating the activity of the p53 target genes encoding Bax and p21(CIP1/Waf1) Oncogene. 2002 Sep 5;21(39):6123–6131. doi: 10.1038/sj.onc.1205842. [DOI] [PubMed] [Google Scholar]
- 45.Hudson CD, Sayan AE, Melino G, Knight RA, Latchman DS, Budhram-Mahadeo V. Brn-3a/POU4F1 interacts with and differentially affects p73-mediated transcription. Cell Death Differ. 2008 Aug;15(8):1266–1278. doi: 10.1038/cdd.2008.45. [DOI] [PubMed] [Google Scholar]
- 46.Ma L, Lei L, Eng SR, Turner E, Parada LF. Brn3a regulation of TrkA/NGF receptor expression in developing sensory neurons. Development. 2003 Aug;130(15):3525–3534. doi: 10.1242/dev.00582. [DOI] [PubMed] [Google Scholar]
- 47.Mulloy JC, Jankovic V, Wunderlich M, Delwel R, Cammenga J, Krejci O, et al. AML1-ETO fusion protein up-regulates TRKA mRNA expression in human CD34+ cells, allowing nerve growth factor-induced expansion. Proc Natl Acad Sci U S A. 2005 Mar 15;102(11):4016–4021. doi: 10.1073/pnas.0404701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M, et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996 Apr 25;380(6576):711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 49.Cheung M, Abu-Elmagd M, Clevers H, Scotting PJ. Roles of Sox4 in central nervous system development. Brain Res Mol Brain Res. 2000 Jun 23;79(1-2):180–191. doi: 10.1016/s0169-328x(00)00109-1. [DOI] [PubMed] [Google Scholar]
- 50.Schilham MW, Moerer P, Cumano A, Clevers HC. Sox-4 facilitates thymocyte differentiation. Eur J Immunol. 1997 May;27(5):1292–1295. doi: 10.1002/eji.1830270534. [DOI] [PubMed] [Google Scholar]
- 51.Graham JD, Hunt SM, Tran N, Clarke CL. Regulation of the expression and activity by progestins of a member of the SOX gene family of transcriptional modulators. J Mol Endocrinol. 1999 Jun;22(3):295–304. doi: 10.1677/jme.0.0220295. [DOI] [PubMed] [Google Scholar]
- 52.Bangur CS, Switzer A, Fan L, Marton MJ, Meyer MR, Wang T. Identification of genes over-expressed in small cell lung carcinoma using suppression subtractive hybridization and cDNA microarray expression analysis. Oncogene. 2002 May 23;21(23):3814–3825. doi: 10.1038/sj.onc.1205480. [DOI] [PubMed] [Google Scholar]
- 53.Ahn SG, Cho GH, Jeong SY, Rhim H, Choi JY, Kim IK. Identification of cDNAs for Sox-4, an HMG-Box protein, and a novel human homolog of yeast splicing factor SSF-1 differentially regulated during apoptosis induced by prostaglandin A2/delta12-PGJ2 in Hep3B cells. Biochem Biophys Res Commun. 1999 Jun 24;260(1):216–221. doi: 10.1006/bbrc.1999.0856. [DOI] [PubMed] [Google Scholar]
- 54.Pan X, Zhao J, Zhang WN, Li HY, Mu R, Zhou T, et al. Induction of SOX4 by DNA damage is critical for p53 stabilization and function. Proc Natl Acad Sci U S A. 2009 Mar 10;106(10):3788–3793. doi: 10.1073/pnas.0810147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boyd KE, Xiao YY, Fan K, Poholek A, Copeland NG, Jenkins NA, et al. Sox4 cooperates with Evi1 in AKXD-23 myeloid tumors via transactivation of proviral LTR. Blood. 2006 Jan 15;107(2):733–741. doi: 10.1182/blood-2003-05-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Syme CA, Zhang L, Bisello A. Caveolin-1 regulates cellular trafficking and function of the glucagon-like Peptide 1 receptor. Mol Endocrinol. 2006 Dec;20(12):3400–3411. doi: 10.1210/me.2006-0178. [DOI] [PubMed] [Google Scholar]
- 57.Pang A, Au WY, Kwong YL. Caveolin-1 gene is coordinately regulated with the multidrug resistance 1 gene in normal and leukemic bone marrow. Leuk Res. 2004 Sep;28(9):973–977. doi: 10.1016/j.leukres.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 58.Wong K, Ren XR, Huang YZ, Xie Y, Liu G, Saito H, et al. Signal transduction in neuronal migration: roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit-Robo pathway. Cell. 2001 Oct 19;107(2):209–221. doi: 10.1016/s0092-8674(01)00530-x. [DOI] [PubMed] [Google Scholar]
- 59.Wang B, Xiao Y, Ding BB, Zhang N, Yuan X, Gui L, et al. Induction of tumor angiogenesis by Slit-Robo signaling and inhibition of cancer growth by blocking Robo activity. Cancer Cell. 2003 Jul;4(1):19–29. doi: 10.1016/s1535-6108(03)00164-8. [DOI] [PubMed] [Google Scholar]
- 60.Marlow R, Strickland P, Lee JS, Wu X, Pebenito M, Binnewies M, et al. SLITs suppress tumor growth in vivo by silencing Sdf1/Cxcr4 within breast epithelium. Cancer Res. 2008 Oct 1;68(19):7819–7827. doi: 10.1158/0008-5472.CAN-08-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernard-Pierrot I, Gruel N, Stransky N, Vincent-Salomon A, Reyal F, Raynal V, et al. Characterization of the recurrent 8p11-12 amplicon identifies PPAPDC1B, a phosphatase protein, as a new therapeutic target in breast cancer. Cancer Res. 2008 Sep 1;68(17):7165–7175. doi: 10.1158/0008-5472.CAN-08-1360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.