Abstract
Increasing public interest in direct-to-consumer (DTC) genetic ancestry testing has been accompanied by growing concern about issues ranging from the personal and societal implications of the testing to the scientific validity of ancestry inference. The very concept of “ancestry” is subject to misunderstanding in both the general and scientific communities. What do we mean by ancestry? How exactly is ancestry measured? How far back can such ancestry be defined and by which genetic tools? How do we validate inferences about ancestry in genetic research? What are the data that demonstrate our ability to do this correctly? What can we say and what can we not say from our research findings and the test results that we generate? This white paper from the American Society of Human Genetics (ASHG) Ancestry and Ancestry Testing Task Force builds upon the 2008 ASHG Ancestry Testing Summary Statement in providing a more in-depth analysis of key scientific and non-scientific aspects of genetic ancestry inference in academia and industry. It culminates with recommendations for advancing the current debate and facilitating the development of scientifically based, ethically sound, and socially attentive guidelines concerning the use of these continually evolving technologies.
Main Text
Introduction
In recent years, advances in genetics and genomics have brought new dimensions to the commercial genetics enterprise in the form of DTC genetic testing. With the click of a mouse, the public now has direct access to personal genetic and genomic information related to health, ancestry, nutrition, physical traits, athletic ability, dating compatibility, and a seemingly infinite list of other attributes. Although health-related DTC genetic testing appropriately continues to receive a substantial amount of attention,1,2 discourse regarding other DTC applications of genetics, in particular the echoing of concern about genetic ancestry testing, is increasing.3–6 There are approximately 40 companies, based in various countries, that currently provide genetic ancestry testing to the public (Table 1). The companies differ in both the range of genetic testing services and the types of ancestry tests that they offer.
Table 1.
Companies Providing Direct-to-Consumer Genetic Ancestry Testing
| Companya | URL | Start Date, Locationb | Genetic Testing Services | Ancestry Tests Offered |
|---|---|---|---|---|
| 1. African Ancestry | http://www.africanancestry.com/ | 2002 | Ancestry | mtDNA, Y chromosome |
| 2. African DNA | http://www.africandna.com/ | 2007 | Ancestry | mtDNA, Y |
| 3. Ancestry.com | http://ancestry.com | 2007 | Ancestry | mtDNA, Y |
| 4. Ancestry by DNA | http://ancestrybydna.com | 2009 | Ancestry | mtDNA, Y, admixture |
| 5. ARGUS Biosciences | http://www.argusbio.com/ | 2003 | Ancestry, cancer tissue screening, personal DNA sequencing | mtDNA-HV, mtDNA-FL, Y |
| 6. Cambridge DNA Services | http://www.cambridgedna.com/ | 2007 UK |
Ancestry | mtDNA, Y, admixture |
| 7. deCODEme | http://www.decodeme.com/ | 2007 Iceland |
R&D, complete scan, cancer scan, cardio scan | mtDNA, Y, map of kinship, genetic atlas |
| 8. Determigene | http://www.determigene.com/ | 2002 | Paternity, immigration DNA testing, infidelity testing, twin zygosity, ancestry, DNA safeguarding | Ancestral origins DNA ancestry map (population matches, native region matches, strength indicators) |
| 9. DNA Ancestry | http://www.easternbiotech.com | 2006 Dubai, UAE |
Ancestry | mtDNA, Y |
| 10. DNA Direct | http://www.dnadirect.com/web/ | 2003 | Screening tests, genetic disorders, drug response, DNA storage, paternity and family tests | Y |
| 11. DNA Identity Testing Center | http://www.800dnaexam.com/ | 2006 | Paternity, family relationship DNA tests, immigration, adoption, forensic, ancestry, identity | mtDNA, Y |
| 12. DNA Heritage | http://www.dnaheritage.com/ | 2003 UK |
Ancestry | mtDNA, Y, surname projects |
| 13. DNA Reference Laboratory | http://www.dnareferencelab.com | 2006 | Paternity, immigration paternity testing, ancestry, forensic, infectious-disease testing, R&D | mtDNA, Y, ethnicity DNA makeup, European ancestry DNA test, Native American ethnicity DNA test |
| 14. DNA Solutions | http://www.dnasolutions.us/ | 2000 | R&D, paternity, sibling DNA, grandparent, twins test, ancestry, bird sexing | mtDNA, Y |
| 15. DNA Testing Systems | http://dnaconsultants.com/ | 2003 | Ancestry, paternity, linkage disequilibrium | mtDNA, Y, admixture, Native American, African, Melungeon test, Hindu single & double for males |
| 16. DNA Tribes | http://www.dnatribes.com/ | 2006 | Ancestry | Autosomal analysis |
| 17. DNA Worldwide | http://www.dna-worldwide.com/ | 2005 UK |
Paternity, relationship, immigration, ancestry, forensic, DNA storage, pet DNA | mtDNA, Y, world DNA match |
| 18. easyDNA | http://www.easy-dna.com/ | 2006 | Paternity, legal DNA test, relationship, DNA profiles, twin zygosity, forensic, ancestry, immigration, maternity | Ancestral origins DNA ancestry map (population matches, native region matches, strength indicators), mtDNA, Y |
| 19. Ethno Ancestry | http://www.ethnoancestry.com/ | 2004 Scotland & Ireland |
Ancestry | mtDNA, Y |
| 20. Family Builder | https://dna.familybuilder.com | 2007 | Ancestry | mtDNA, Y |
| 21. Family Genetics | http://www.familygenetics.co.uk | 2005 UK |
Ancestry | mtDNA, Y |
| 22. Family Tree DNA | http://www.familytreedna.com | 2000 | Ancestry | mtDNA, Y, autosomal, X-STR |
| 23. Genebase | http://www.genebase.com/ | 2005 Canada |
Ancestry | mtDNA, Y, autosomal DNA STR |
| 24. Genelex | http://www.healthanddna.com | 2000 | Paternity, drug sensitivity, ancestry, predictive genetics | mtDNA, Y, Native American DNA, Jewish DNA, African ancestry DNA testing |
| 25. Genetic Testing Laboratories, Inc. | http://www.gtldna.com/ | 2002 | Paternity, ancestry, infidelity, DNA maternity, siblingship, twin zygosity, grandparentage, missing parent, immigration, prenatal | mtDNA, Y, ancestral origins DNA ancestry map (population matches, native region matches, strength indicators) |
| 26. Genetree | http://www.genetree.com/ | 2007 | Ancestry | mtDNA, Y |
| 27. homeDNAdirect | http://www.homednadirect.com/ | 2006 | Paternity, legal DNA testing, relationship, forensic, ancestry | mtDNA, Y, ancestral origins DNA ancestry map (population matches, native region matches, strength indicators) |
| 28. International Biosciences | http://www.ibdna.com | 2007 UK |
Paternity, ancestry, siblingship | mtDNA, Y, ancestral origins DNA ancestry map (population matches, native region matches, strength indicators) |
| 29. Metaphase Paternity Test | http://www.metaphasegenetics.com/ | 2002 | Paternity, siblingship, grandparentage, twins, prenatal, forensic, ancestry | Y |
| 30. Oxford Ancestors | http://www.oxfordancestors.com | 2000 UK |
Ancestry | mtDNA, Y |
| 31. Paternity Experts | http://www.paternityexperts.com | 2004 | Paternity, sibling, ancestry, forensic paternity | Admixture |
| 32. Pathway Genomics | http://www.pathway.com/ | 2009 | Health conditions, ancestry, carrier status, personal traits, monogenic dominants, drug responses | mtDNA, Y |
| 33. Roots for Real | http://www.rootsforreal.com/ | 2002 UK |
Ancestry | mtDNA, Y, admixture |
| 34. Test Country | http://www.testcountry.com | 2001 | Health conditions, substance abuse, health & wellness, pregnancy/fertility, early disease detection, ancestry | Ancestral origins DNA ancestry map (population matches, native region matches, strength indicators) |
| 35. The Genographic Project | https://genographic.nationalgeographic.com/ | 2005 | Research, ancestry | mtDNA, Y |
| 36. Universal Genetics; DNA Testing Laboratory | http://www.dnatestingforpaternity.com/ | 2009 | Paternity, forensic, ancestry | mtDNA, Y |
| 37. Warrior Roots | http://www.warriorroots.com/ | 2009 | Ancestry, linking ancestry to ancient warrior groups, athletic profile | mtDNA, Y |
| 38. 23andMe | https://www.23andme.com/ | 2006 | Complete scan | mtDNA, Y, global similarity |
Last updated February 23, 2010.
This list does not include companies or sites that only promote genetic ancestry testing services. On November 17, 2009 deCODE genetics (owner of deCODEme) announced that it has filed a voluntary Chapter 11 bankruptcy petition but will continue to offer services during its restructuring process. On January 21, 2010, deCODE genetics was purchased by Saga Investments LLC, a consortium that includes Polaris Ventures and ARCH Venture Partners. Since the initial preparation of this table, two companies have been removed from the list – DNA Ancestors has gone out of business, and DNA Diagnostics Center has become a promoter site.
Start date for offering genetic ancestry testing. Some companies were in existence prior to this date. Locations are listed for non-U.S. locations only.
The marketing of genetic tests directly to consumers is a priority area for the ASHG, as demonstrated by its statements on DTC health-related testing7 and ancestry testing.8 This white paper, commissioned by the ASHG, expands discussion of the issues highlighted in the Society's 2008 Ancestry Testing Summary Statement8 and introduces additional pertinent issues. The purpose of our report is twofold: (1) to enlighten and engage ASHG members and the broader scientific and general communities and (2) to assist in determining the appropriate course of action for the Society in responding to the critical concerns.
It is important to note that the genetic tools employed by ancestry testing companies, as well as many of the scientists involved in DTC ancestry testing, have their roots in, and are still a part of, academia. Therefore, in this report, as in the summary statement, we evaluate application of ancestry estimation technologies in both environments because the many overlapping scientific and nonscientific issues in academic research have not, to date, been adequately addressed.
Definition(s) of Ancestry
Our common origin as a species implies that we, as individuals, are all related to one another by varying degrees,9 so it is important to be clear about what frame of reference is being used in discussions of “ancestry” and relationship (Figure 1). For example, because of recombination, each segment of the genome has its own ancestral history, and various segments of an individual's genome may have ancestral histories that trace to different populations.
Figure 1.
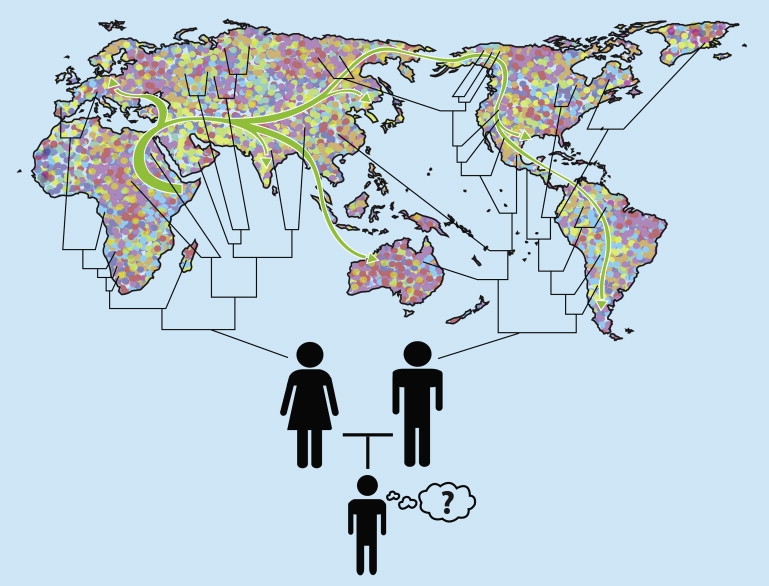
Global Ancestry
The arrows symbolize migration of early human ancestors out of Africa. The color mosaic denotes global population diversity resulting from various subsequent inter- and intra-continental and regional migrations. The pedigree represents the complex network of intermediate and recent ancestors that is the subject of individual genetic genealogy testing.
One commonly employed concept of ancestry is continental ancestry, which assumes the existence of four or five major “parental” populations that gave rise within the last 100,000 years to existing populations.10 This conception of ancestry is frequently equated with that of “race,” and the terms are often used interchangeably; however, this is problematic because in the history of science there have been many “racial” taxonomic schemes.11,12 A related view of ancestry is biogeographic ancestry, in which a person's origin is associated with the geographic location(s) of presumed ancestors inferred by comparison with contemporary populations living in these locations.13,14 There is also lineage or family history, which typically represents a generational narrative about one's relatives through his or her maternal and paternal lines of descent.15 It is this notion of ancestry that people often focus on when considering their genealogy. However, we note that genealogy does not necessarily confer genetic similarity in all biological systems; for example, even one's offspring may not be the ideal match for a kidney transplant or blood transfusion.
In addition to the concepts described above, there are sociopolitical rules about ancestry that guide membership in certain groups. In the United States (US) these include the legal and historic utility of hypodescent (“one-drop” rule) for African Americans and blood quantum laws for Native Americans.16–18 In reality, however, neither are these rules absolute nor have they been consistently applied.18 This further illustrates that ancestry-related social identity—how a society may see or define a person or group in relationship to real or putative ancestry—is to be distinguished from personal or group interpretations of such identity or actual knowledge of genealogy.
Interest in Ancestry Estimation
Consumers and researchers are interested in using genetic information to infer ancestry for a variety of reasons, including genealogical, anthropological, and epidemiological. Most consumers are interested in using ancestry testing to gain, confirm, or extend knowledge about their recent family genealogy.3,19 To permit inferences about shared recent ancestry, commercial testers of genetic ancestry employ a variety of genetic-marker systems to make comparisons between a customer's DNA and genetic databases of individuals sampled from diverse populations and geographic regions. However, although the concept of “ancestry” is least ambiguous when it refers to either very close ancestors (i.e., parents or grandparents) or our most distant ancestors (i.e., the earliest hominids), genetic ancestry tests typically address more intermediate levels of ancestry that are imprecisely defined and identified10,20 (Figure 1). Given this intrinsic imprecision, the power of commercial genetic tests to address specific genealogical questions is contingent on several factors that we will discuss later in this paper.
Population geneticists and anthropologists use genetic markers and comparative datasets similar to those used in commercial ancestry testing to make inferences about population histories and relationships. Ancestry estimation has enormous value in this regard because it has the potential to illuminate patterns of past human migration and provide background information about human genetic variation that is essential for distinguishing the impact of demographic processes from the effects of natural selection.21–23 Unlike commercial ancestry testing, such inferences are nearly always made at the level of populations or groups, rather than at the individual level. As a consequence of this plural focus, these ancestry inferences are more robust with respect to their fundamentally probabilistic nature, and the limitations of ancestry estimation for individuals are comparatively less apparent.
Genetic epidemiologists with an interest in identifying genetic associations with disease employ methods of ancestry inference for specific analytical reasons: either to control for statistical biases related to population stratification among cases and controls24–26 or as a strategy to map susceptibility variants that might be differentially distributed with respect to ancestry within groups whose histories more clearly demonstrate the “mixing” of two or more peoples. Recently admixed groups (such as African Americans or Hispanic Americans) provide the opportunity to perform mapping by admixture linkage disequilibrium, commonly referred to as admixture mapping.27–29 However, epidemiological inferences of genetic ancestry are typically applied to individuals and are nearly always based on the analysis of large collections of single nucleotide polymorphisms (SNPs) or ancestry informative markers (AIMs, described below). Each individual's genome is then mapped as a mosaic of segments inferred to be derived from one or the other ancestral population (or both, in the case where maternal and paternal alleles in the individual are each derived from different ancestral populations).
Human History and Variation
Genetic ancestry estimation is based on an understanding of the distribution of diversity among human populations that reflects the demographic and evolutionary history of our species. Genetic and archaeological evidence indicates that, over the past 100,000 years or so, as the population size of humans increased markedly, humans dispersed from East Africa to populate other parts of the world30,31 (Figure 1). The number of migration events, their magnitude, and the routes that migrants took are still active areas of research. Nevertheless, it is apparent that the dispersal of anatomically modern humans affected the geographic distribution of diversity in at least two important ways. First, founder populations typically carried with them only a subset of the genetic variation found in their most immediate ancestral population while simultaneously developing new mutations and genetic profiles. Second, as founder populations became more widely separated from one another, the probability that two randomly chosen individuals would mate with each other became even lower, and matings were even more likely to occur between people living close to each other, accentuating the divergence between geographically isolated populations.
Over the past two decades, geneticists have characterized the geographic pattern of variation in great detail by using both haploid and diploid genetic markers (described below).32–37 Because different parts of the genome can have different ancestral histories, different marker systems often provide somewhat different information about population history and individual ancestry.
Currently, we only have partial knowledge of how human genetic diversity is distributed across the globe, but initial studies38 are revealing the degree of resolution possible in testing the relationship between genetic ancestry and geographic origins. A number of these studies have used a collection of ∼1100 DNA samples obtained from 51 populations living in different parts of the world; these samples constitute the Human Genetic Diversity Panel (HGDP).39 Analysis of 987 microsatellites typed in the HGDP collection, for example, inferred six population clusters that correspond to continental regions (i.e., Africa, America, Central/South Asia, East Asia, and Oceania).38,40 Analysis of ∼642,000 autosomal SNPs in the HGDP collection enabled clustering of individuals not only to these large geographic regions, but also to specific populations within these regions.38,40 Although the HGDP collection is a useful collection of widely distributed human populations, it is a convenient sample and does not sample densely within any one geographic region; hence, there are limitations to the accuracy of ancestry inference within and among regions. Several studies that sampled populations deeply across Europe have shown that population structure can be inferred even at fine spatial scales (i.e., the scale of several hundreds of kilometers) within Europe.41–45 There is rapidly escalating interest in ancestral histories of other continents and geographic regions, and the deeper timescales of populations from Africa46 and India47 have identified deep splits among geographic regions with complex patterns of past migration and admixture. Further studies of human genetic diversity are clearly needed; these will determine whether such fine-scale geographic patterns can be detected in other parts of the world and assign interpretive values to these patterns.
It is important to note that the diversity of human social structures, intermarriage patterns, and demographic histories makes it likely that the resolution of population structure will be challenging and that the extent of the resolution will vary considerably among populations. The inclusion of individuals with recent migration among ancestors creates the more complex problem of disentangling recently mixed ancestry.
Tools for Inferring Ancestry
Estimates of genetic ancestry are based either on the use of haploid markers (mitochondrial DNA [mtDNA] or Y chromosome haplotypes) or on the use of multiple unlinked autosomal markers that are diploid and sometimes preselected to be “ancestry informative.” As uniparentally inherited haploid markers, mtDNA provides information about the female-to-female transmitted lineage (male children also inherit mtDNA from their mothers but do not transmit it to their offspring), whereas the Y chromosome is informative about male-to-male transmitted lineage. More recently, autosomal markers, which are inherited from both parents, have been used for assessing patterns of genetic variation in worldwide human populations. Commercial genetic ancestry testing primarily utilizes haploid markers to make ancestry inferences (Table 1), whereas estimates of genetic ancestry in epidemiological applications rely almost exclusively on the consideration of allele frequencies of autosomal SNPs. Population geneticists and anthropologists employ both types of markers; which type they use depends on the availability of funds and the questions being addressed.
mtDNA and Y Chromosome Markers
Haploid genetic markers such as mtDNA D-loop region sequences or Y chromosome SNP haplotypes permit direct comparison of the lineages of sampled and reference individuals. As such, and unlike probabilistic estimates of population ancestry, matches among haploid genetic markers are intuitively easy to understand: an exact match of a male's Y chromosome haplotype to a man living in Australia implies that these two men share a common paternal ancestor.
An important issue with regard to lineage-based genetic estimates is that they reflect only a fraction of any person's total genetic ancestry. For example, the Parsis have Y chromosome information that indicates an origin in Iran, consistent with the historical record, whereas the mtDNA originates in Gujarat, a region in northwestern India where the Parsis arrived in approximately 900 AD, before moving eventually to Mumbai, India, and Karachi, Pakistan.48,49 This asymmetry of maternal and paternal ancestry is not a matter of test inconsistency; rather, it reflects the high likelihood that nearly everyone will have ancestors from different geographic locations.
Another problem related to lineage-based comparisons involves the interpretation of exact genetic matches between individuals. Although it is biologically justified to infer that two individuals with the same mtDNA haplotype share a common ancestor, moving from this inference of common ancestry to the conclusion that the match implies something about the biogeographical ancestry of both individuals can be problematic. For example, if someone lives in North America and his or her mtDNA haplotype exactly matches an individual living in Indonesia, the only thing that can be inferred with confidence is that they share a common ancestor. Without more information about family history and/or the geographic distribution of closely related mtDNA haplotypes, it is impossible to say whether this match arises via recent Indonesian ancestors in the North American's family tree, whether both share distant ancestors who lived in an entirely different part of the world, or whether the Indonesian match has recent North American heritage. Similarly, it is difficult to arrive at a robust interpretation of an mtDNA haplotype that exactly matches those sampled from multiple geographic locations, e.g., Indonesia, Thailand, and Papua New Guinea.
Autosomal Variants
In comparison to mtDNA and Y chromosome markers, autosomal markers provide much more comprehensive information on individual ancestry because cumulatively they represent a much greater proportion of genome history (i.e., multiple biparentally inherited loci versus a single locus, as inherited through mtDNA or the Y chromosome). However, because the genome is finite, only a small fraction of ancestors are represented by each given genomic segment in an individual, and every ancestor does not necessarily pass on his or her DNA at any given genomic segment to a descendant, so one can only ever have limited information on the origins of a given individual's ancestors.10
Autosomal variation can be measured by whole-genome sequencing approaches, with genome-wide genotyping panels, or via an assessment of AIMs. Whole-genome sequencing, although ideal and likely to usher in a renaissance in genetic anthropology, is still prohibitively expensive and so is beyond the reach of most academic researchers or commercial testing companies. Genome-wide genotyping arrays, the next-most comprehensive approach, include SNPs that are common in a select subset of populations and lead to ascertainment biases that can impact ancestry estimation.50 AIMs, most often developed to estimate admixture proportions originating from African, European, Asian, and Native American populations,51 offer increased power for ancestry inference in comparison to a random set of autosomal markers. Accordingly, a smaller set of markers can be used, reducing genotyping costs and increasing throughput.
The use of AIMs has facilitated efforts to control for admixture and population stratification in genetic association studies. Specifically, knowing the proportion of an individual's ancestry that originated in different populations and to what degree a group is divided into genetic subpopulations can be useful for both reducing false-positive associations and uncovering true associations. We note, however, that not all people from a given population have the AIM(s) identified with that population, and people from different populations can have the same AIM(s). Gene mapping with AIMs, or admixture mapping, has also been used successfully for identifying genomic regions associated with diseases and health-related traits such as prostate cancer (MIM #176807), hypertension (EHT [MIM #145500]), and white blood cell count.29,52–54 Admixture mapping is most effective for identifying genetic variants associated with health conditions that differ between recently admixed populations (e.g., tropical African and European in the case of most African Americans; and Native American, European, and African populations in the case of Hispanic Americans) and for which this difference has not yet been fully explained by nongenetic factors. Although most DTC tests for ancestry offer lineage testing that uses mtDNA and Y-chromosome markers, DTC testing with autosomal markers, especially with whole-genome SNP chips, is becoming more common. The results reported to the consumer typically estimate admixture proportions from several populations, most often Africans, Europeans, Asians, and Native Americans. However, the interpretation of such estimates by both the scientist and the consumer is unclear.
Accuracy of Ancestry Inferences
Ideally, any quantitative claims about ancestry should have an easily interpreted assessment of confidence or accuracy associated with them. Our interest in accuracy is to assess not only what the accuracy estimate is but also how well we can describe our confidence in the inferences. We also stress the difference between accuracy of a particular individual's ancestry versus the inference of ancestry of a population sample. The former is particularly important in the case of DTC ancestry testing, whereas most scientific research on ancestry inference deals with the latter. The accuracy of ancestry inference methods is a function of (1) how the underlying patterns of human genetic variation are distributed across the geographic range of human habitation, (2) how that diversity is surveyed (i.e., the type and number of genetic markers used) and who was sampled, (3) which populations are used as references, and (4) the statistical methods used for interpreting patterns of variation.
Distribution of Genetic Variation
Accuracy is limited by the fact that every person has hundreds of ancestors going back even a few centuries and thousands of ancestors in just a millennium. There is enormous stochastic variation to the portion of the genome retained in a descendant from a given ancestor, and there is a rough expectation that it halves every generation. Genetic ancestry tests can access only a fraction of these ancestral contributions. Furthermore, the genomic segments contributed by a particular ancestor are far from all being uniquely identifiable, so even if one's genome has those specific contributions, identification of particular ancestry is always uncertain and statistical.
Geneticists also make specific choices about which levels of ancestry to examine. For example, many estimations of genetic ancestry are designed to distinguish contributions from reference populations that live in particular geographic regions (e.g., West Africa, Europe, East Asia, and the Americas) that were prominent in colonial-era population movements. This creates a bias that might lead us to define ancestry in reference to particular sociopolitical groups. Moreover, our knowledge of diversity, and hence the genetic contributions to ancestry, of populations in many other parts of the world (e.g., East Africa, South Asia, Arabian Peninsula, and Southeast Asia) is limited.
Lineage Identification with Uniparental Markers
While it is now possible to identify related groups of Y-chromosome and mtDNA lineages with high accuracy, population-level inferences that have been made from these uniparental systems are substantially less accurate. Two simple examples help to illustrate this point. A large number of single-site changes have served as the basis for breaking Y chromosomes into different “haplogroups,” and it is accurate to say that Y chromosomes within, say, haplogroup C are more closely related to one another than to a Y chromosome from haplogroup J. Thus, if two men both carry haplogroup C Y chromosomes, they are more likely to share a paternal lineage than if they had different haplogroups. Even so, this relationship does not mean that they are more genetically similar overall.
On the other hand, in the scientific literature there has been a connection drawn between one subset of haplogroup C and Ghengis Khan on the basis of the commonness of that branch of the Y chromosome genealogy in parts of the world conquered by Ghengis Khan.55 Although such a connection is by no means impossible, we currently have no way of assessing how much confidence to place in such a connection. We emphasize, however, that whenever formal inferences about population history have been attempted with uniparental systems, the statistical power is generally low. Claims of connections, therefore, between specific uniparental lineages and historical figures or historical migrations of peoples are merely speculative.
Admixture Estimation
For autosomal markers, ancestry inference is most often performed under a discrete-deme admixture model where there is a set of discrete demes (usually or always referred to as “ancestral” or “parental” populations), and each individual inherits proportions of his or her genome from each of these demes. The goal of the method is to estimate this list of admixture proportions for each individual. Strictly speaking, a deme is a breeding population, defined on the basis of population genetic inference of intermixed genetic variation, and it is unlike classical anthropology's “races,” which are defined by morphology.11 Nonetheless, the emphasis of admixture estimation on differences over similarities can be misleading about the overall genetic structure of the human species.
Admixture estimation has greatly advanced the field of ancestry inference; however, there are caveats to the interpretation of its results. First, the “ancestral populations” are not directly observed—although in many applications, samples from related populations are used as a proxy. For example, present-day Yoruba are the most frequently used proxy for inferring African American ancestry, despite the fact that most African Americans derive their ancestry from diverse West African (and other African) populations that existed over a span of several centuries and that might not all be well represented by present-day proxy populations.46,56,57 Second, if some ancestral populations are missing altogether from the analyses, programs such as STRUCTURE58 and FRAPPE59 will force the results into a composite of the reference samples used; therefore, the results will be skewed simply because of how the algorithms work.
If a poor proxy is used for one ancestral population, the method might compensate by adding admixture from other ancestral populations. Consider genetic ancestry testing performed on an individual we will call Joe, whose eight great-grandparents were from southern Europe. The HapMap populations are used as references for testing Joe's genetic ancestry. The HapMap's European samples consist of “northern” Europeans. In regions of Joe's genome that vary between northern and southern Europe (such regions might include the lactase gene, LCT [MIM #603202]), the genetic ancestry test using the HapMap reference populations is likely to incorrectly assign the ancestry of that portion of the genome to a non-European population because that genomic region will appear to be more similar to the HapMap's Yoruba or Han samples than to its (northern) European samples.
Although the discrete-deme admixture method is informative about ancestry in settings where individuals have recent admixture from diverse continental populations, it does not perform well in settings where individuals have more ancestors from across a continuous gradient of genetic diversity. European populations, for example, despite revealing genetic differences, have been shown (as described above) to exhibit mainly continuous spatial patterns of variation. When admixture is estimated for European individuals under the assumption of two ancestral populations,28 the method chooses admixture proportions that make individuals a mixture of “northern” and “southern” ancestral populations even though there is no independent evidence that two such ancestral populations ever existed.
Methods for addressing continuous, spatial population structure are still under development, but principal-components analysis (PCA) has been widely applied in this context.60,61 The expected behavior of PCA on evenly spaced samples from spatially structured data is to return coordinates that are related to the geographic origin of each individual.62 Moreover, there is a clearly established relationship between the genealogical structure of a sample and the principal components, grounding PCA in firm principles of population genetics.63 One caveat of PCA-based approaches is that if individuals are a product of “recent admixture” from disparate origins, it will assign individuals to a single origin that is intermediate between the source populations, which is incorrect (e.g., an individual with an East Asian and European parent will be indistinguishable from an individual from Central Asia). This reinforces the need for models that take these and other limiting factors into account and recognize that in some cases accurate social identifications cannot be made.
Reference Samples
To infer ancestry, researchers rely on comparing any individual's particular genetic profile to that of reference populations. Research geneticists benefit from various publicly available databases such as the HapMap, Human Genome Diversity Panel, Perlegen Human Genome Resources, POPRES project, and Seattle SNPs projects. However, even the databases that researchers consider the most applicable reflect a woefully incomplete sampling of human genetic diversity, and this has important consequences for the accuracy of ancestry inference. One problem is that the “ancestral populations” assumed by some methods are not explicitly represented in databases—and indeed cannot be represented as such because we do not have the ability to sample ancestral populations. A second problem is that populations that are mixtures of the “typical” reference populations (e.g., Africans, Asians, and Europeans) are substantially under-represented in these databases. Recent sampling efforts, such as HapMap Phase III samples, are helping to remedy this problem; however, continued attention to diverse sampling will be an important aspect of any subsequent surveys of human genetic variation.
Some commercial scientists and private groups have their own unpublished databases with the potential to provide more refined information than that available from publicly available resources. In some cases, the commercial interest in ancestry testing is indirectly benefiting public research. For example, the company 23andMe partially funded the genotyping for the Human Genome Diversity Project samples.38 Although such collaborations might be helpful and commendable, vigilance is needed in identifying and addressing potential conflicts of interest. The scientific claims of companies that choose not to disclose the contents of their proprietary databases cannot be assessed; therefore, the reliability of the information they provide to consumers cannot be verified.64
Statistical Methods
Regardless of the methods used or samples referenced, steps should be taken to adequately convey the amount of uncertainty in the inferences about ancestry, whether in the research or commercial setting. Population genetic inference is ultimately a statistical exercise, and rarely can definitive conclusions about ancestry be made beyond the assessment of whether putative close relatives are or are not related. Because ancestry inferences for less simple questions require reliance on complex statistical procedures with inherent uncertainty, both producers and consumers of genetic ancestry estimates need to have a fairly sophisticated understanding of probability.
There are two levels to the inherent uncertainty of these statistical inferences. First, there is uncertainty in parameter estimates (for instance, how large are the confidence intervals of admixture coefficients for an individual?). Second, there is uncertainty in how to interpret these parameters (e.g., what do the admixture coefficients mean—what does an individual's haplogroup say about his or her past?). The context in which ancestry estimation is being used determines the importance of these sources of error. In some research contexts (e.g., when ancestry is used as a covariate in genome-wide association studies), it might be sufficient to have some quantitative variable that represents ancestry. In commercial ancestry-testing applications, however, interpretation often is key because the information that is presented might have direct psychosocial and other personal implications for the individual.
The statistical methods used to perform ancestry inference vary with regard to the assumptions they make, how much of the information available in the genetic data is extracted, and how their statements about inference are summarized for the researcher or the consumer receiving the information. The ease in understanding the statistical confidence in the ancestry inference also varies widely among methods. The most important aspect of reporting confidence in ancestry determinations is to accurately convey the level of uncertainty in the interpretations and to convey the real meaning of that uncertainty.
Ancestry and Health
(Note: unlike in other sections of this report, where we mention “race” to make specific points, in this section we use “race” (and ethnicity) as constructed by the US Office of Management and Budget (OMB) and as used in US social, government, and biomedical research parlance. We realize that there are various connotations and limitations of these terms, but our goal here is only to provide a brief overview of some important issues pertaining to health outcomes and health differences within and among the referent ancestry-linked sociopolitical groups.)
Researchers still poorly understand the relationship of genetic ancestry to individual and population health, but this relationship is a potentially important area for investigation in that it might have social and political consequences.65–68 In the US, it has been commonplace to report disease prevalence for each racial or ethnic group separately, and these prevalence estimates often vary among groups.69 This has led to widespread speculation that racial or ethnic differences in individual or population health are primarily due to genetic factors, including genetic ancestry.68,70 Yet, racial or ethnic identity could be associated with the health of an individual or group in several ways. It might co-vary with different environmental or genetic factors that underlie risk or with different interactions within and between genetic and environmental factors.71–73
There are circumstances in which genetic factors influencing heath-related traits are associated with specific genetic variations that tend to be more prevalent in a particular racial or ethnic group than in the rest of the population. Certain genetic variants associated with hypertension,52 type 2 diabetes mellitus (T2D [MIM #125853]),74 end-stage renal disease (FSGS4 [MIM #612551]),75,76 prostate cancer,53,77 and some treatment responses 78,79 have been shown to differ significantly in frequency among groups. Therefore, disease risk or treatment response is associated with and, in some situations, influenced by genetic factors that vary among racial or ethnic groups. It is not clear how much of this is actually gene expression versus DNA sequence.
Given the complexity and limited understanding of the relationships among genetic variation, ancestry, race, ethnicity, and health and treatment outcomes, the translation of genetic epidemiological research findings to clinical application requires ample consideration of a variety of factors, including personal, social, and other nongenetic factors. This issue might be highlighted in the context of DTC genetic testing, where consumers might share ancestry test results or ancestry-related estimates of disease risk with their healthcare providers and expect that the information be factored into their care.3,68 In view of the ongoing national efforts to increase the public's exploration of family (health) history,80 it is possible that this practice could become widespread as people seek to exhaust the available sources of information about their family history and associated health risks. As such, the healthcare community must be recognized as a key stakeholder in decision making concerning genetic ancestry inference.
Personal and Societal Implications
Ancestry inference—in both its research and commercial applications—prompts a host of psychological, social, legal, political, and ethical concerns from the individual to the global level. These actual or potential consequences have received increasing attention3,6,81–86 and must be considered alongside relevant technical and analytical issues.
Knowledge about genetic ancestry, particularly if undesirable and unexpected, can lead to the reshaping of group, familial, or personal identity.87–91 Anthropological and population-genetics research that postulates or casts doubt on ancestral relationships has historically incited varying degrees of identity-related conflict. Some of the most notable examples include the case of Kennewick Man,92 research linking the Lemba and certain Jews,93,94 and the discovery of family ties between Thomas Jefferson and Sally Hemings.95 The occurrence of, or potential for, emotional distress in people, families, and groups after receipt of conflicting information about their identity through DTC ancestry testing has also been discussed.3,15,87,89–91 Nonetheless, some research focused on consumers of ancestry testing has revealed that although ancestry tests might promote genetic thinking about ancestry and “race,” test takers also were able to construct meaningful narratives of their identity.5 Clearly, additional empirical research will need to adequately explore the relationship between genetic ancestry testing and the identities and overall psychological well-being of test takers, their families, and their communities.
Questions have been raised about privacy and about the security of reference databases that support ancestry-estimation endeavors. For example, for genetic-ancestry-testing companies that are sold or go bankrupt, there are concerns about the future of the privacy policies and other terms under which data were collected.96 Some people also fear that commercial ancestry-testing databases might be more vulnerable than other genetics databases to alternate and inappropriate uses.64,97 The problem of alternate uses of data in the context of ancestry estimation might also be extended to the unauthorized inclusion of population-based genetic research data or samples in ancestry-related studies6,98 or in commercial ancestry-testing databases. These practices bring to the fore consideration about evolving notions of consent, anonymity, respect for communities, group risk-benefit assessment, and benefit sharing, and these issues must also be addressed within the current broader discourse on the sharing and secondary use of genetic and genomic data and samples.99,100
A common concern about scientific efforts to explain origins is the alleged diminished regard for important cultural, religious, social, historical, and political processes that inform origin as well as group membership, identity, and rights.3,16,101 Reports of the use (or intended use) of ancestry test results to make claims for benefits through affirmative action or for rights perceived to be associated with their new-found Native American status have increased unease over the loss or gain of certain rights or entitlements.91,102,103 Entitlement could also be viewed in terms of interest among some DTC-ancestry-test takers in seeking dual citizenship in countries identified as their ancestral homelands.104 This trend is similar to that discussed in relation to some population-genetics research connecting the Lemba and certain Jews.87,105 It remains to be seen what tangible effects (if any) genetic ancestry inference will have on these pre-existing entitlement issues.
Genetic ancestry inference (in particular, the use of AIMs and admixture mapping techniques) could reveal the nuances of ancestry and dispel the notion of race in humans and/or the practice of equating race with ancestry. Paradoxically, it is equally capable of giving credence to the idea that humans subdivide into distinct biological races and implying that there are the clear-cut connections between DNA and specific geographic regions or ethnic groups. There has been substantial anxiety and discussion about the potentially reifying effects of current ancestry-estimation practices.3,16,86,106 Beyond ancestry estimation itself, the routine treatment, in science, of ancestral, ethnic, and so-called racial groups as bounded biological entities perpetuates an inaccurate concept of human variation and increases the possibility of stigmatization and discrimination of the groups and the people within them on the basis of traits, behaviors, diseases, and other attributes.64,70,107 Scientists and the scientific establishment as a whole must attend to this longstanding and pervasive problem of conveying conflicting messages pertaining to human variation.
Although genetic ancestry inference in research and the marketplace is the focus of this report, we are well aware that the technologies are being employed in other arenas. For example, since 2003, the forensic use of DNA to make determinations about ancestry in criminal cases has become more widespread.107–110 More recently, the “Human Provenance pilot project” proposed using DNA ancestry testing to identify the nationalities of people seeking asylum in the UK.111,112 These and other such applications of genetic ancestry estimation also merit scrutiny because they have the same technical problems discussed above and may pose palpable threats to human welfare.
Conclusions and Recommendations
Concerns about analytical procedures, interpretation, and the personal and social implications of genetic ancestry inference make it clear that enormous care is required in the application of ancestry estimation in both research and commercial settings. A major issue regarding commercial ancestry testing is that there is no quality assurance guarantee. This gives rise to the question of whether there is a need for lab certification or accreditation. We tend to lean against anything so formal because it would provide a stamp of approval by any designated accrediting body. It is one thing to certify accuracy of the genotyping procedures, but it might not be very useful to do that and also claim that the inferences from the data are not validated or certified in any way. Determination of feasibility or of mechanisms for certification and validation, as well as specific approaches for enhancing consumer understanding of the scientific and nonscientific issues, will require thoughtful deliberation beyond the scope of work of this task force.
The academic research community cannot afford to be exempt from similar efforts to increase scientific rigor and overall accountability in genetic ancestry estimation. Indeed, the peer-review processes for funding and journal publications are designed to assist in such efforts, but their effectiveness is compromised by the inadequacy and inconsistent application of existing guidelines in this area. Because of the intrinsic uncertainties of the science and the potential societal ramifications, the field of population genetics as a whole could benefit from improved and enforced standards with respect to terminology and methodologies, as well as interpretation and communication of research findings.
Recently, Lee and colleagues6 called for federal regulation of genetic ancestry testing. At this juncture, we offer an alternate approach, one that might itself lead to federal oversight, if subsequently deemed appropriate, necessary, or practical. We believe that effective decision making regarding genetic ancestry inference, in particular DTC genetic ancestry testing, will be best initiated through cooperative interaction among a variety of stakeholders, including suitable federal agencies. Considering that such collective engagement has not yet occurred, it is premature to assume reticence or resistance on the part of any of the players or that federal regulation is the only recourse.
On the basis of our review of the state of the science and the personal, societal, and health-related implications of genetic ancestry inference in academia and industry, we make the following recommendations:
-
(1)
Leadership of the human-genetics community, diverse in its interests and its own identities, should develop mechanisms for promoting thoughtful and rigorous use of genetic ancestry estimation in academic research. This might be implemented through workshops or similar activities to (a) identify criteria for appropriate selection of ancestry-estimation methods and genetic markers, (b) establish standards for the representation of the statistical confidence in ancestry inference results, (c) create guidelines for terminology and the assessment of methodology in peer-reviewed research proposals and publications, and (d) devise strategies for the effective translation of ancestry-mediated research findings to the general public.
-
(2)
Interested scientific and scholarly societies should collaborate to convene a national roundtable discussion of DTC genetic ancestry testing. The goal of this face-to-face conversation among ancestry-testing companies and promoters, consumers, community leaders, advocacy and interest groups, geneticists, social and behavioral scientists, humanists, healthcare providers, legal professionals, federal agencies, media, and other key stakeholders should be to identify major issues of concern and brainstorm practical solutions. Points for consideration must encompass accuracy and the reporting of statistical confidence, proprietary databases, additional research, communication of limitations and potential consequences, public and personal education, interdisciplinary collaboration, and mechanisms for accountability. Findings from this meeting will inform decisions about the next steps.
These interconnected recommendations are intended to foster direct and productive dialog surrounding genetic ancestry inference in academia and industry and move us closer to achieving the most beneficial outcomes for both science and society. In light of the issues at stake and the enduring fruitless attempts at effecting meaningful change, the time is now for no-holds-barred discussions among the players, particularly among scientists who must more purposefully and constructively critique one another's premises, methodologies, findings, and interpretations of findings.
Although there might be general agreement that genetic variation provides a window into human origins, history, interrelation, and identity, differences of opinion about genetic ancestry inference will probably persist. Our desire is that, ongoing conversation and collaboration across disciplines and sectors will address principal concerns and reduce the intensity of the debate. An ever-present challenge and responsibility for scientists engaged in this work is to understand the inherent uncertainties, socio-historical contexts, and potential ramifications of the science and to effectively incorporate this knowledge into efforts to refine their methodologies and improve human well-being.
Acknowledgments
We thank the ASHG leadership and staff, specifically Aravinda Chakravarti, Edward McCabe, Joann Boughman, and Phyllis Edelman, for their substantial support of our deliberations that led to this report. We are grateful to Jill Cooper and Switzon Wigfall, III for providing research and technical assistance. We also appreciate the comments from Karla Holloway, Charles Jonassaint, Shomarka Keita, Jennifer Wagner, Kenneth Weiss, Huntington Willard, members of the ASHG Social Issues Committee, and our anonymous reviewers on earlier drafts of this manuscript.
Web Resources
The URL for data presented herein is as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim/
References
- 1.Evans J.P., Green R.C. Genet. Med. 2009;11:568–569. doi: 10.1097/GIM.0b013e3181afbaed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGuire A.L., Diaz C.M., Wang T., Hilsenbeck S.G. Am. J. Bioeth. 2009;9:3–10. doi: 10.1080/15265160902928209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolnick D.A., Fullwiley D., Duster T., Cooper R.S., Fujimura J.H., Kahn J., Kaufman J.S., Marks J., Morning A., Nelson A. Science. 2007;318:399–400. doi: 10.1126/science.1150098. [DOI] [PubMed] [Google Scholar]
- 4.Greely H.T. Genetic genealogy: Genetics meets the marketplace. In: Koenig B.A., Lee S.S., Richardson S.S., editors. Revisiting Race in a Genomic Age. Rutgers University Press; New Brunswick: 2008. pp. 215–234. [Google Scholar]
- 5.Nelson A. Soc. Stud. Sci. 2008;38:759–783. doi: 10.1177/0306312708091929. [DOI] [PubMed] [Google Scholar]
- 6.Lee S.S., Bolnick D.A., Duster T., Ossorio P., TallBear K. Science. 2009;325:38–39. doi: 10.1126/science.1173038. [DOI] [PubMed] [Google Scholar]
- 7.Hudson K., Javitt G., Burke W., Byers P. Obstet. Gynecol. 2007;110:1392–1395. doi: 10.1097/01.AOG.0000292086.98514.8b. [DOI] [PubMed] [Google Scholar]
- 8.ASHG. (2008). (http://www.ashg.org/pdf/ASHGAncestryTestingStatement_FINAL.pdf)
- 9.Chakravarti A. Nature. 2009;457:380–381. doi: 10.1038/457380a. [DOI] [PubMed] [Google Scholar]
- 10.Weiss K.M., Long J.C. Genome Res. 2009;19:703–710. doi: 10.1101/gr.076539.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keita S.O., Kittles R.A., Royal C.D., Bonney G.E., Furbert-Harris P., Dunston G.M., Rotimi C.N. Nat. Genet. 2004;36:S17–S20. doi: 10.1038/ng1455. [DOI] [PubMed] [Google Scholar]
- 12.Caspari R. Am. J. Phys. Anthropol. 2009;139:5–15. doi: 10.1002/ajpa.20975. [DOI] [PubMed] [Google Scholar]
- 13.Shriver M.D., Parra E.J., Dios S., Bonilla C., Norton H., Jovel C., Pfaff C., Jones C., Massac A., Cameron N. Hum. Genet. 2003;112:387–399. doi: 10.1007/s00439-002-0896-y. [DOI] [PubMed] [Google Scholar]
- 14.Bamshad M., Wooding S., Salisbury B.A., Stephens J.C. Nat. Rev. Genet. 2004;5:598–609. doi: 10.1038/nrg1401. [DOI] [PubMed] [Google Scholar]
- 15.Baylis F. Developing World Bioethics. 2003;3:142–150. doi: 10.1046/j.1471-8731.2003.00070.x. [DOI] [PubMed] [Google Scholar]
- 16.TallBear K. Wicazo Sa Review. 2003;18 88–93 and 98–99. [Google Scholar]
- 17.Sweet F.W. Backintyme; Palm Coast, FL: 2005. Legal History of the Color Line: Rise and Triumph of the One-drop Rule. pp. 3–4. [Google Scholar]
- 18.Haney Lopez I. New York University Press; New York: 2006. White by Law: The Legal Construction of Race. [Google Scholar]
- 19.Wolinsky H. EMBO Rep. 2006;7:1072–1074. doi: 10.1038/sj.embor.7400843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shriver M.D., Kittles R.A. Nat. Rev. Genet. 2004;5:611–618. doi: 10.1038/nrg1405. [DOI] [PubMed] [Google Scholar]
- 21.Akey J.M., Eberle M.A., Rieder M.J., Carlson C.S., Shriver M.D., Nickerson D.A., Kruglyak L. PLoS Biol. 2004;2:e286. doi: 10.1371/journal.pbio.0020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabeti P.C., Schaffner S.F., Fry B., Lohmueller J., Varilly P., Shamovsky O., Palma A., Mikkelsen T.S., Altshuler D., Lander E.S. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen R., Hubisz M.J., Hellmann I., Torgerson D., Andres A.M., Albrechtsen A., Gutenkunst R., Adams M.D., Cargill M., Boyko A. Genome Res. 2009;19:838–849. doi: 10.1101/gr.088336.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reich D., Patterson N., De Jager P.L., McDonald G.J., Waliszewska A., Tandon A., Lincoln R.R., DeLoa C., Fruhan S.A., Cabre P. Nat. Genet. 2005;37:1113–1118. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- 25.Enoch M.A., Shen P.H., Xu K., Hodgkinson C., Goldman D. J. Psychopharmacol. 2006;20:19–26. doi: 10.1177/1359786806066041. [DOI] [PubMed] [Google Scholar]
- 26.Barnholtz-Sloan J.S., McEvoy B., Shriver M.D., Rebbeck T.R. Cancer Epidemiol. Biomarkers Prev. 2008;17:471–477. doi: 10.1158/1055-9965.EPI-07-0491. [DOI] [PubMed] [Google Scholar]
- 27.Smith M.W., O'Brien S.J. Nat. Rev. Genet. 2005;6:623–632. doi: 10.1038/nrg1657. [DOI] [PubMed] [Google Scholar]
- 28.Seldin M.F. Curr. Opin. Genet. Dev. 2007;17:177–181. doi: 10.1016/j.gde.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basu A., Tang H., Arnett D., Gu C.C., Mosley T., Kardia S., Luke A., Tayo B., Cooper R., Zhu X. Obesity (Silver Spring) 2009;17:1226–1231. doi: 10.1038/oby.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jobling M., Hurles M., Tyler-Smith C. Garland; New York: 2004. Human Evolutionary Genetics: Origins, Peoples & Diseases. [Google Scholar]
- 31.Cavalli-Sforza L.L. Annu. Rev. Genomics Hum. Genet. 2007;8:1–15. doi: 10.1146/annurev.genom.8.080706.092403. [DOI] [PubMed] [Google Scholar]
- 32.Jorde L.B., Watkins W.S., Bamshad M.J., Dixon M.E., Ricker C.E., Seielstad M.T., Batzer M.A. Am. J. Hum. Genet. 2000;66:979–988. doi: 10.1086/302825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mesa N.R., Mondragon M.C., Soto I.D., Parra M.V., Duque C., Ortiz-Barrientos D., Garcia L.F., Velez I.D., Bravo M.L., Munera J.G. Am. J. Hum. Genet. 2000;67:1277–1286. doi: 10.1016/s0002-9297(07)62955-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jobling M.A., Tyler-Smith C. Nat. Rev. Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- 35.Malhi R.S., Breece K.E., Shook B.A., Kaestle F.A., Chatters J.C., Hackenberger S., Smith D.G. Hum. Biol. 2004;76:33–54. doi: 10.1353/hub.2004.0023. [DOI] [PubMed] [Google Scholar]
- 36.Fagundes N.J., Kanitz R., Eckert R., Valls A.C., Bogo M.R., Salzano F.M., Smith D.G., Silva W.A., Jr., Zago M.A., Ribeiro-dos-Santos A.K. Am. J. Hum. Genet. 2008;82:583–592. doi: 10.1016/j.ajhg.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auton A., Bryc K., Boyko A.R., Lohmueller K.E., Novembre J., Reynolds A., Indap A., Wright M.H., Degenhardt J.D., Gutenkunst R.N. Genome Res. 2009;19:795–803. doi: 10.1101/gr.088898.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 39.Cann H.M., de Toma C., Cazes L., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 40.Jakobsson M., Scholz S.W., Scheet P., Gibbs J.R., VanLiere J.M., Fung H.C., Szpiech Z.A., Degnan J.H., Wang K., Guerreiro R. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 41.Heath S.C., Gut I.G., Brennan P., McKay J.D., Bencko V., Fabianova E., Foretova L., Georges M., Janout V., Kabesch M. Eur. J. Hum. Genet. 2008;16:1413–1429. doi: 10.1038/ejhg.2008.210. [DOI] [PubMed] [Google Scholar]
- 42.Lao O., Lu T.T., Nothnagel M., Junge O., Freitag-Wolf S., Caliebe A., Balascakova M., Bertranpetit J., Bindoff L.A., Comas D. Curr. Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 43.Novembre J., Johnson T., Bryc K., Kutalik Z., Boyko A.R., Auton A., Indap A., King K.S., Bergmann S., Nelson M.R. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabatti C., Lange K. J. Am. Stat. Assoc. 2008;103:89–100. doi: 10.1198/016214507000000338.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McEvoy B.P., Montgomery G.W., McRae A.F., Ripatti S., Perola M., Spector T.D., Cherkas L., Ahmadi K.R., Boomsma D., Willemsen G. Genome Res. 2009;19:804–814. doi: 10.1101/gr.083394.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tishkoff S.A., Reed F.A., Friedlaender F.R., Ehret C., Ranciaro A., Froment A., Hirbo J.B., Awomoyi A.A., Bodo J.M., Doumbo O. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reich D., Thangaraj K., Patterson N., Price A.L., Singh L. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quintana-Murci L., Chaix R., Wells R.S., Behar D.M., Sayar H., Scozzari R., Rengo C., Al-Zahery N., Semino O., Santachiara-Benerecetti A.S. Am. J. Hum. Genet. 2004;74:827–845. doi: 10.1086/383236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McElreavey K., Quintana-Murci L. Ann. Hum. Biol. 2005;32:154–162. doi: 10.1080/03014460500076223. [DOI] [PubMed] [Google Scholar]
- 50.Clark A.G., Hubisz M.J., Bustamante C.D., Williamson S.H., Nielsen R. Genome Res. 2005;15:1496–1502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halder I., Shriver M., Thomas M., Fernandez J.R., Frudakis T. Hum. Mutat. 2008;29:648–658. doi: 10.1002/humu.20695. [DOI] [PubMed] [Google Scholar]
- 52.Zhu X., Luke A., Cooper R.S., Quertermous T., Hanis C., Mosley T., Gu C.C., Tang H., Rao D.C., Risch N. Nat. Genet. 2005;37:177–181. doi: 10.1038/ng1510. [DOI] [PubMed] [Google Scholar]
- 53.Freedman M.L., Haiman C.A., Patterson N., McDonald G.J., Tandon A., Waliszewska A., Penney K., Steen R.G., Ardlie K., John E.M. Proc. Natl. Acad. Sci. USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nalls M.A., Wilson J.G., Patterson N.J., Tandon A., Zmuda J.M., Huntsman S., Garcia M., Hu D., Li R., Beamer B.A. Am. J. Hum. Genet. 2008;82:81–87. doi: 10.1016/j.ajhg.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zerjal T., Xue Y., Bertorelle G., Wells R.S., Bao W., Zhu S., Qamar R., Ayub Q., Mohyuddin A., Fu S. Am. J. Hum. Genet. 2003;72:717–721. doi: 10.1086/367774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salas A., Carracedo A., Richards M., Macaulay V. Am. J. Hum. Genet. 2005;77:676–680. doi: 10.1086/491675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ely B., Wilson J.L., Jackson F., Jackson B.A. BMC Biol. 2006;4:34. doi: 10.1186/1741-7007-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pritchard J.K., Stephens M., Donnelly P. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexander D.H., Novembre J., Lange K. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patterson N., Price A.L., Reich D. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 62.Novembre J., Stephens M. Nat. Genet. 2008;40:646–649. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McVean G. PLoS Genet. 2009;5:e1000686. doi: 10.1371/journal.pgen.1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sarata, A.K. (2008). CRS Report for Congress: Genetic ancestry testing. (http://assets.opencrs.com/rpts/RS22830_20080312.pdf).
- 65.Bamshad M. JAMA. 2005;294:937–946. doi: 10.1001/jama.294.8.937. [DOI] [PubMed] [Google Scholar]
- 66.Gravlee C.C., Non A.L., Mulligan C.J. PLoS ONE. 2009;4:e6821. doi: 10.1371/journal.pone.0006821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee C. Soc. Sci. Med. 2009;68:1183–1190. doi: 10.1016/j.socscimed.2008.12.036. [DOI] [PubMed] [Google Scholar]
- 68.Via M., Ziv E., Burchard E.G. Clin. Genet. 2009;76:225–235. doi: 10.1111/j.1399-0004.2009.01263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu J., Kochanek K.D., Tejada-Vera B. Natl. Vital Stat. Rep. 2009;58:1–51. [PubMed] [Google Scholar]
- 70.Caulfield T., Fullerton S.M., Ali-Khan S.E., Arbour L., Burchard E.G., Cooper R.S., Hardy B.J., Harry S., Hyde-Lay R., Kahn J. Genome Med. 2009;1:8. doi: 10.1186/gm8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cooper R.S., Kaufman J.S. Hypertension. 1998;32:813–816. doi: 10.1161/01.hyp.32.5.813. [DOI] [PubMed] [Google Scholar]
- 72.Ramos E., Rotimi C. BMC Med. Genomics. 2009;2:29. doi: 10.1186/1755-8794-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gravlee C.C. Am. J. Phys. Anthropol. 2009;139:47–57. doi: 10.1002/ajpa.20983. [DOI] [PubMed] [Google Scholar]
- 74.Steinthorsdottir V., Thorleifsson G., Reynisdottir I., Benediktsson R., Jonsdottir T., Walters G.B., Styrkarsdottir U., Gretarsdottir S., Emilsson V., Ghosh S. Nat. Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 75.Freedman B.I., Hicks P.J., Bostrom M.A., Cunningham M.E., Liu Y., Divers J., Kopp J.B., Winkler C.A., Nelson G.W., Langefeld C.D. Kidney Int. 2009;75:736–745. doi: 10.1038/ki.2008.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kopp J.B., Smith M.W., Nelson G.W., Johnson R.C., Freedman B.I., Bowden D.W., Oleksyk T., McKenzie L.M., Kajiyama H., Ahuja T.S. Nat. Genet. 2008;40:1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robbins C., Torres J.B., Hooker S., Bonilla C., Hernandez W., Candreva A., Ahaghotu C., Kittles R., Carpten J. Genome Res. 2007;17:1717–1722. doi: 10.1101/gr.6782707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chung W.H., Hung S.I., Hong H.S., Hsih M.S., Yang L.C., Ho H.C., Wu J.Y., Chen Y.T. Nature. 2004;428:486. doi: 10.1038/428486a. [DOI] [PubMed] [Google Scholar]
- 79.Ge D., Fellay J., Thompson A.J., Simon J.S., Shianna K.V., Urban T.J., Heinzen E.L., Qiu P., Bertelsen A.H., Muir A.J. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 80.U.S. Department of Health & Human Services. Surgeon General's Family Health History Initiative. (http://www.hhs.gov/familyhistory/).
- 81.Brodwin P. Anthropol. Q. 2002;75:323–330. [Google Scholar]
- 82.Nordgren A., Juengst E. N. Genet. Soc. 2009;28:157–172. [Google Scholar]
- 83.Zoloth L. Developing World Bioethics. 2003;3:128–132. doi: 10.1046/j.1471-8731.2003.00068.x. [DOI] [PubMed] [Google Scholar]
- 84.Palsson G., Helgason A. Developing World Bioethics. 2003;3:159–169. doi: 10.1046/j.1471-8731.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- 85.Tutton R. New Genet. Soc. 2004;23:105–120. doi: 10.1080/1463677042000189606. [DOI] [PubMed] [Google Scholar]
- 86.Duster T. Genewatch. 2009;22:16–17. [Google Scholar]
- 87.Elliott C., Brodwin P. BMJ. 2002;325:1469–1471. doi: 10.1136/bmj.325.7378.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Palsson G. Med. Anthropol. 2002;21:337–367. doi: 10.1080/01459740214078. [DOI] [PubMed] [Google Scholar]
- 89.Kaplan, E. (2003). Black Like I Thought I Was. In LA Weekly. (http://www.laweekly.com/2003-10-09/calendar/black-like-i-thought-i-was).
- 90.Winston C., Kittles R.A. Inferring African Ancestry of African Americans. In: Turner T., editor. Biological Anthropology and Ethics. SUNY Press; New York: 2005. [Google Scholar]
- 91.Koerner, B. (2005). Blood Feud. Wired Magazine Online. (www.wired.com/wired/archive/13.09/seminoles.html>).
- 92.Custred G. Academic Questions. 2000;13:12–30. [Google Scholar]
- 93.Richards M., Macaulay V., Hickey E., Vega E., Sykes B., Guida V., Rengo C., Sellitto D., Cruciani F., Kivisild T. Am. J. Hum. Genet. 2000;67:1251–1276. [PMC free article] [PubMed] [Google Scholar]
- 94.Thomas M.G., Parfitt T., Weiss D.A., Skorecki K., Wilson J.F., le Roux M., Bradman N., Goldstein D.B. Am. J. Hum. Genet. 2000;66:674–686. doi: 10.1086/302749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Foster E.A., Jobling M.A., Taylor P.G., Donnelly P., de Knijff P., Mieremet R., Zerjal T., Tyler-Smith C. Nature. 1998;396:27–28. doi: 10.1038/23835. [DOI] [PubMed] [Google Scholar]
- 96.Moore, L., and Sherlock, E. (2009). Federal Privacy Regulation and the Financially Troubled DTC Genomics Company. Genomics Law Report. (http://www.genomicslawreport.com/index.php/2009/10/27/federal-privacy-regulation-and-the-financially-troubled-dtc-genomics-company/).
- 97.Holloway K. Lit. Med. 2007;26:269–276. [Google Scholar]
- 98.Dalton R. Nature. 2004;430:500–502. doi: 10.1038/430500a. [DOI] [PubMed] [Google Scholar]
- 99.Malin B., Karp D., Scheuermann R.H. J. Investig. Med. 2010;58:11–18. doi: 10.231/JIM.0b013e3181c9b2ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bathe O.F., McGuire A.L. Genet. Med. 2009;11:712–715. doi: 10.1097/GIM.0b013e3181b2e168. [DOI] [PubMed] [Google Scholar]
- 101.Rotimi C.N. Developing World Bioethics. 2003;3:151–158. doi: 10.1046/j.1471-8731.2003.00071.x. [DOI] [PubMed] [Google Scholar]
- 102.Harmon, A. (2006). Seeking Ancestry in DNA Ties Uncovered by Tests. New York Times, April 12, 2006, p A1. [PubMed]
- 103.TallBear K. J. Law Med. Ethics. 2007;35:412–424. doi: 10.1111/j.1748-720X.2007.00164.x. [DOI] [PubMed] [Google Scholar]
- 104.Watanabe, T. (2009). Called back to Africa by DNA. Los Angeles Times (http://articles.latimes.com/2009/feb/18/local/me-africa18).
- 105.Azoulay K.G. Developing World Bioethics. 2003;3:119–126. doi: 10.1046/j.1471-8731.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- 106.Braun L. Int. J. Health Serv. 2006;36:557–573. doi: 10.2190/8JAF-D8ED-8WPD-J9WH. [DOI] [PubMed] [Google Scholar]
- 107.Duster T. Patterns Prejudice. 2006;40:427–441. [Google Scholar]
- 108.Wade, N. (2003). Unusual use of DNA aided in serial killer search. New York Times, June 3, 2003. p. A28.
- 109.Cho M.K., Sankar P. Nat. Genet. 2004;36:S8–S12. doi: 10.1038/ng1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fullwiley D. Genewatch. 2008;21:12–14. [Google Scholar]
- 111.Anonymous. Nature. 2009;461:697. [Google Scholar]
- 112.Travis J. Science. 2009;326:30–31. doi: 10.1126/science.326_30. [DOI] [PubMed] [Google Scholar]


