This study utilizes inbred strains of mice to demonstrate that murine central corneal thickness is highly influenced by genetic background. Mice with strain-dependent differences in corneal thickness were used to determine cellular and molecular factors that contribute to these differences.
Abstract
Purpose.
Central corneal thickness (CCT) exhibits broad variability. For unknown reasons, CCT also associates with diseases not typically considered corneal, particularly glaucoma. The purpose of this study was to test the strain dependence of CCT variability among inbred mice and identify cellular and molecular factors associated with differing CCT.
Methods.
Methodology for measuring murine CCT with ultrasound pachymetry was developed and used to measure CCT among 17 strains of mice. Corneas from three strains with nonoverlapping differences in CCT (C57BLKS/J, C57BL/6J, and SJL/J) were compared by histology, transmission electron microscopy, and expression profiling with gene microarrays.
Results.
CCT in mice was highly strain dependent. CCT exhibited continuous variation from 89.2 μm in C57BLKS/J to 123.8 μm in SJL/J. Stromal thickness was the major determinant of the varying murine CCT, with epithelial thickness also contributing. Corneal expression levels of many genes differed between strains with differing CCT, but most of these changes did not correlate with the changes observed in previously studied corneal diseases nor did they correlate with genes encoding major structural proteins of the cornea.
Conclusions.
Murine CCT has been measured with a variety of different techniques, but only among a limited number of different strains. Here, pachymetry was established as an additional tool and used to conduct a broad survey of different strains of inbred mice. These results demonstrated that murine CCT was highly influenced by genetic background and established a baseline for future genetic approaches to further elucidate mechanisms regulating CCT and its disease associations.
In the healthy human eye, central corneal thickness (CCT) exhibits a broad variability between different individuals and ethnicities. The normal human eye is expected to have a CCT of approximately 544 μm,1 but many eyes deviate substantially from this value. Part of this variability is ethnicity based. For example, African-Americans and Japanese tend to have thinner corneas than Caucasians or Hispanics.2 Factors beyond ethnicity presumably also contribute to CCT variability. Within ethnically matched cohorts, a substantial fraction of eyes have CCTs outside the range of 500 to 600 μm. These broad distributions and ethnicity-dependent differences in CCT have been observed repeatedly in several studies.2–7
More than a mere anatomic curiosity, these variations in CCT are thought to reflect minute differences in the function of many important physiologic processes and are also associated with several disease conditions.8 Physiologic processes influencing CCT occur within all layers of the cornea. Disruption of either the epithelial or endothelial layer causes corneal swelling, reflecting their important functions as barriers and regulators of water flux. Equally important is the corneal stroma. Approximately 90% of the total corneal thickness in humans is contributed by the thickness of the stroma, which largely consists of a matrix of collagen fibrils and interfibrillary substance produced by keratocytes.9 Pathologic events occurring within each of these layers have been linked to multiple corneal diseases, but the processes causing individual and ethnic differences in CCT among overtly healthy corneas remain largely unknown.
One approach to the study of the factors influencing CCT is through genetics. Several lines of evidence suggest that CCT is strongly influenced by heredity, including ethnic,2 twin,10 and familial studies.11,12 However, it is likely that CCT represents a polygenic trait and that identification of the individual contributing genes may be difficult in the human population. As with many complex phenotypes, genetic studies with inbred mouse strains offer a synergistic approach that has often been successful. We initiated such a genetic approach for studying CCT in inbred strains of mice. In previous studies murine CCT has been measured with a variety of approaches, but only within a relatively limited number of strains.13–22 The purpose of this study was to conduct a broad strain survey of CCT among inbred strains of mice. To accomplish this, we first established ultrasound pachymetry as a valid tool for measuring murine CCT and then used pachymetry to study 17 different inbred mouse strains. Having identified strains free of overt corneal disease, but with nonoverlapping differences in CCT, we began to assess factors contributing to CCT regulation by comparing corneal histology and identifying gene expression patterns correlating with corneal thickness.
Methods
Mouse Husbandry
All mice were obtained from The Jackson Laboratory. The mice were subsequently housed and bred at the University of Iowa Research Animal Facility, maintained on a 4% fat NIH 31 diet provided ad libitum, and housed in cages containing dry bedding (Cellu-dri; Shepherd Specialty Papers, Kalamazoo, MI). The environment was kept at 21°C in a 12-hour light–12-hour dark cycle. All animals were treated in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. All experimental protocols were approved by the Animal Care and Use Committee of The University of Iowa.
Slit Lamp Examination
Corneas and anterior chambers of conscious mice were examined with a slit lamp (SL-D7; Topcon, Tokyo, Japan) and photodocumented with a digital camera (D100; Nikon, Tokyo, Japan). All photographs were taken with identical camera settings and prepared with identical image software processing.
Ultrasound Pachymetry
Probe movements of the ultrasound pachymeter (Corneo-Gage Plus; Sonogage, Cleveland, OH) were controlled with a micromanipulator under the guidance of a dissecting microscope, to achieve perpendicular alignments between the probe and central cornea (the Corneo-Gage Plus ultrasound pachymeter has a small acceptance angle of 5%, which further helped to promote consistent perpendicular probe placement). Mice were anesthetized with a mixture of ketamine (100 mg/kg) and xylazine (10 mg/kg). Balanced salt solution (BSS; Alcon Laboratories, Inc., Fort Worth, TX) was applied to the eye to maintain a consistent tear film. CCT was measured by aligning the probe beside the pupil and gently moving it forward until the pachymeter began to record values. An average of 20 consecutive CCT measurements was recorded in each eye. Balanced salt solution was reapplied during recovery from anesthesia. Unless otherwise noted, values reported herein represent the mean ± SD. The number of eyes measured is reported for each experiment.
Analysis of Cryosections
Enucleated eyes were immediately embedded in medium (Tissue-Tek OCT Compound; Sakura Finetek USA, Inc., Torrance, CA). Sections (10-μm) were cut and transferred to glass slides using a tape system to avoid potential compression artifacts (CryoJane; Instrumedics, Inc., St. Louis, MO) and allowed to dry at room temperature for 15 minutes. Sections were judged to be central by the presence of the pupil and optic nerve. For initial experiments comparing pachymetry-recorded measurements versus thickness measured from cryosections, 10 consecutive unstained cryosections per eye of the central cornea were analyzed by light microscopy. For later experiments characterizing features of C57BLKS/J, C57BL/6J, and SJL/J corneas, cryosections of central cornea were stained with hematoxylin and eosin (H&E) and measurements were made with NIH ImageJ software (developed by Wayne Rasband, National Institutes of Health, Bethesda, MD; available at http://rsb.info.nih.gov/ij/index.html). Epithelial and stromal thicknesses were measured in five eyes per strain with two to four cryosections per eye. Stromal cell densities were calculated by collecting 400× images of the central corneal in three eyes per strain (two to four cryosections per eye) and counting the number of nuclei in each field.
Corneal Endothelial Cell Density
Enucleated eyes were dissected in phosphate-buffered saline (PBS), and a punch of central cornea was collected with a 2-mm biopsy punch (Miltex, Inc., York, PA). Corneal punches were stained with 1% alizarin red for 2 minutes and rinsed in PBS. Based on analysis of punches from four eyes per strain, endothelial cell density was estimated by measuring the area of 50 contiguous endothelial cells in each punch with NIH ImageJ software.
Plastic-Embedded Histology
The anterior cup of each eye was dissected, fixed overnight at room temperature with 2.5% glutaraldehyde in 0.1 M sodium cacodylate, and post-fixed with 1% osmium for 1 hour. A series of acetone dehydrations were performed followed by an infiltration protocol (Embed-812/DDSA/NMA/DMP-30; EMS, Hatfield, PA) for 24 hours. For light microscopy, 0.5-μm sections were cut with an ultramicrotome (UC6; Leica EM, Wetzler, Germany), and the sections were stained with 1% toluidine blue. For transmission electron microscopy (TEM), the same ultramicrotome was used to cut 100-nm sections. Postsectioning staining with uranyl acetate and Sato's lead stain was performed. A transmission electron microscope (JEM-1230; JEOL, Tokyo, Japan) equipped with CCD camera (USC1000 2Kx2K; Gatan, Pleasanton, CA) and located at the University of Iowa Central Microscopy Core Facility, was used to collect 15 images of each corneal layer from individual eyes of each strain. Collagen diameters were measured in cross section in TEM images from the anterior and posterior third of the stroma; measuring 10 fibers per image from six to nine images per strain. The percentage of overall CCT contributed by Descemet's membrane was calculated using TEM measurements of Descemet's membrane and the average cryosection-measured thickness for each strain.
Gene Expression Analyses
Gene expression profiling was performed on inbred mice from strains with thin (C57BLKS/J), intermediate (C57BL/6J), and thick (SJL/J) corneas. Enucleated eyes were dissected in PBS and a 2-mm punch of central cornea was collected. Two central corneal punches (left and right eyes) were pooled from each mouse to form one sample. Three samples were analyzed per strain. Corneal samples were homogenized (Tissue-Tearor; Biospec Products, Bartlesville, OK), RNA was extracted, treated with DNase I, and purified (Aurum Total RNA Mini Kit; Bio-Rad Laboratories, Hercules, CA). RNA yields were assessed by absorbance at 260 nm (NanoDrop 1000 Spectrophotometer; Thermo Scientific, Wilmington, DE) and the integrity confirmed on a bioanalyzer (Model 2100; Agilent Technologies, Inc., Palo Alto, CA). According to the manufacturer's recommended methodology, the DNA Core Facility of the University of Iowa converted RNA into biotin-labeled cRNA and probed arrays (GeneChip Mouse Genome 430 2.0 Arrays; Affymetrix, Santa Clara, CA).
Raw data obtained were normalized with the GCRMA algorithm.23 Genes that did not appear to be expressed (i.e., probesets that did not yield normalized values >100 in at least two samples) or did not display differences in expression (more than twofold expression difference between the lowest and highest value among all individuals) were removed from further analysis. The remaining values were log2-transformed and evaluated for expression differences using multiclass analysis within the statistical analysis for microarrays software package.24 Δ = 1.78 was chosen, which resulted in a false discovery rate of 0.06% based on 100 permutations. Data were further filtered to identify probesets with at least twofold differences in expression values between C57BLKS/J and SJL/J and intermediate values in C57BL/6J. To avoid exclusion of genes that are expressed at approximately equal levels between C57BL/6J and one of these strains, genes with expression values in C57BL/6J that were within 10% of the expression level of C57BLKS/J or SJL/J were also included. Statistical evaluations of expression changes for individual candidate genes were additionally performed with an unpaired two-tailed Student's t-test. Functional annotation clustering was performed with DAVID, a web-accessible program integrating functional genomic annotations from array data.25 The complete data sets contributing to the array analysis have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus under accession number GSE14270. Transcripts with elevated expression levels in C57BLKS/J and SJL/J were compared with previous microarray studies of laser-captured murine basal epithelial cells (GDS2433)26 and cultured murine corneal keratocytes (GDS857).27
Statistical Analysis
Unless stated otherwise, all P-values from statistical comparisons were calculated with an unpaired two-tailed Student's t-test.
Results
Accuracy and Reproducibility of Measuring Murine CCT with Pachymetry
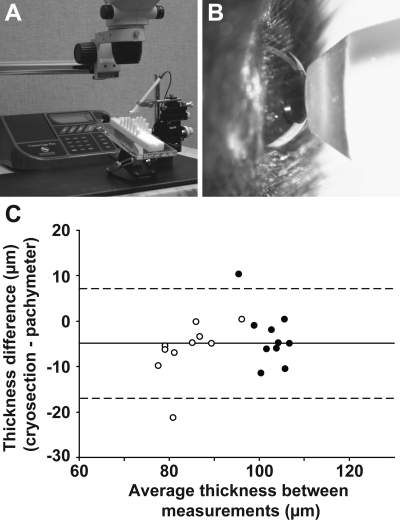
Although not previously used to study murine CCT, ultrasound pachymetry offers several advantages, including noninvasiveness and a comparatively low instrument cost. The primary challenge in using pachymetry in mice is the small size of the mouse eye. Many commercially available instruments are designed for human use and are not able to record values in the thickness range expected of the mouse cornea (∼100 μm).15,20 However, some ultrasound pachymeters can be used intraoperatively to monitor flap thicknesses in LASIK eye surgery and can measure thicknesses as thin as 35 μm.28 Thus, we reasoned that some pachymeters might also be capable of measuring thickness of the relatively thin mouse cornea. To test this, we evaluated the performance of an ultrasound pachymeter (Corneo-Gage Plus; Sonogage) in measuring murine CCT in the widely used C57BL/6J strain.
Using a micromanipulator to promote consistent probe placement (Fig. 1A), we made serial measurements on each eye of anesthetized C57BL/6J mice. Despite the relatively large size of the probe with respect to the size of the mouse eye (Fig. 1B), CCTs near 100 μm were readily attained, which is consistent with previously reported CCTs of 91 to 105 μm for various strains of adult mice measured by optical low coherence reflectometry.16,20 The procedure with pachymetry was rapid, easy to perform, and well tolerated by the mice. Analysis of serial measurements recorded after a single probe placement yielded an average SD of 1.1 μm between measurements (n = 8 eyes, each with 20 serial measurements). Analysis of repeat measurements from individual eyes after repositioning of the mouse and independent placement of the probe yielded an average SD of 0.9 μm (n = 9 eyes of 44-day-old C57BL/6J males; comparing the average of 20 serial measurements in duplicate, means of test/retest measurements were 103.0 and 102.5 μm, respectively). These results indicated that ultrasound pachymetry is capable of measuring CCT in mice and can do so reproducibly.
Figure 1.
Ultrasound pachymetry of mice. (A) Equipment used to measure murine CCT. (B) Pachymeter probe in contact with mouse cornea. (C) Bland-Altman plot indicating that CCT measured by pachymeter or cryosection yields similar values, though cryosection measurements are slightly thinner. Solid line: average difference; dotted lines: 95% limits of agreement (±1.96 SD); (○): 21-day-old mice; (●): 10-month-old mice.
Performance of ultrasound pachymetry was further tested by comparing measurements obtained by pachymetry to unfixed cryosections of the same eyes (Fig. 1C). Two groups of C57BL/6J mice were used; a group of young mice expected to have a relatively thin CCT (n = 10 eyes; 21-day-old mice) and a group of adult mice expected to have a somewhat thicker CCT (n = 10 eyes; 10-month-old mice). CCTs measured with these two methods were very similar, although cryosection measurements yielded consistently thinner values. The average difference of measurements between methods was slightly, but significantly, different from 0 (average cryosection − average pachymeter = −4.9 μm; P = 0.0019, paired two-tailed Student's t-test). To investigate whether the difference between these two methods was dependent on CCT, a linear regression was conducted using this difference as the dependent variable and the sum of the measurements from these two methods as the independent variable. The slope of the regression line was not significantly different from 0 (P = 0.28), indicating that there is no significant change in the difference between ultrasound pachymetry and cryosection measurements over a wide range of values.
Influence of Age, Sex, and Bodyweight to CCT in Adult Mice
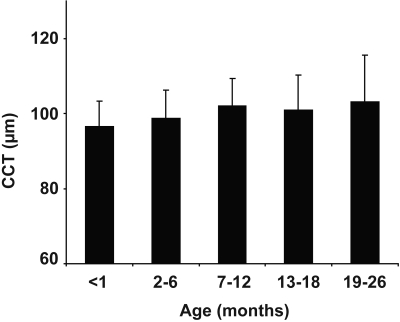
Using a large cohort of C57BL/6J mice (n = 187 eyes; 3 weeks to 26 months old), we next used pachymetry to determine baseline CCT with respect to age (Fig. 2). In agreement with previous studies,29 CCT changed notably during the first month. Values from 3-week-old mice were significantly thinner than those of 4-week-old mice (93.6 and 99.1 μm, respectively; P = 0.0061). CCT measurements from mice 4 to 8 weeks old indicated a trend of subtle growth, although it was not statistically significant (99.1 and 103.2 μm, respectively; P > 0.05). Among all mice in these aging cohorts, no correlations between sex and CCT or bodyweight and CCT were detectable (P > 0.05 in all pair-wise comparisons).
Figure 2.
Influence of age on CCT. Past 1 month of age, CCT remains relatively constant within C57BL/6J mice. There is a slight but statistically nonsignificant trend toward growth (P > 0.05 between all groups). Number of eyes per age group: <1 month (n = 44), 2–6 months (n = 33), 7–12 months (n = 46), 13–18 months (n = 52), and 19–26 months (n = 12). Mean ± SD.
Although ultrasound pachymetry was well tolerated by the mice, the experiments did uncover a minor caveat of the technique. Repeat measurements of an individual cornea sometimes caused a white corneal opacity to appear (after 10 weeks of weekly measurement, 32% of corneas exhibited opacities; n = 11 mice 3–13 weeks of age). By slit lamp examination, the opacities resembled band keratopathy and, in a sampling of eyes, were confirmed to contain calcium with von Kossa stain. The opacities evidently impeded sound waves from the ultrasound pachymeter from passing through the entire length of the cornea, resulting in artifactually thin CCTs (50–60 μm, the approximate depth of the opacity on histologic sections). Opacities were completely avoided by limiting the number of separate measurements per cornea to three measurements within three consecutive weeks. All values reported herein were from mice free of this induced corneal opacity.
CCT Differences between Inbred Strains of Mice
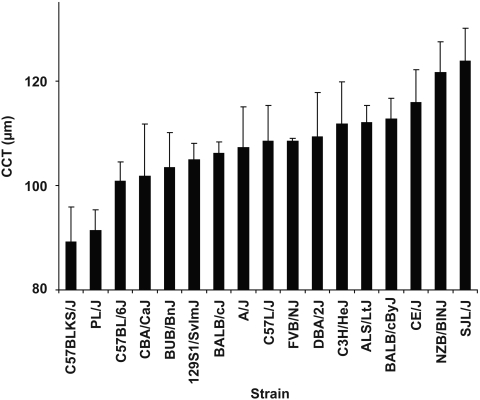
To test the extent to which genetic context influences CCT in mice, we next conducted a strain survey among a variety of different inbred strains of mice (Fig. 3). CCT exhibited a wide distribution among mice of these differing genetic backgrounds. Mean CCT ranged from 89.2 ± 6.6 μm in C57BLKS/J mice to 123.8 ± 6.2 μm in SJL/J mice, with a near continuous variation in other strains between these extremes. C57BL/6J mice had a CCT of 100.8 ± 5.7 μm; the overall average observed across all strains examined was 107.6 ± 9.0 μm. Considering the phylogenetic relationships known among inbred mouse strains,30,31 there was no readily apparent relationship between lineage and CCT. The two strains with thinnest CCTs (C57BLKS/J and PL/J) were not closely related to each other, nor were the strains with thickest CCTs closely related to each other (SJL/J and NZB/BlNJ). Combined, these results indicated that CCT is a genetically dependent trait in mice.
Figure 3.
Strain survey of CCT in inbred mice. Among 17 stains of inbred mice, CCT was thinnest in C57BLKS/J mice (89.2 ± 6.6 μm), was thickest in SJL/J mice (123.8 ± 6.2 μm), and varied continuously between those thicknesses (mean ± SD). All mice were 100 to 120 days of age. Number of male/female eyes per strain: C57BLKS/J, 10/10; PL/J, 8/0; C57BL/6J, 14/6; CBA/CaJ, 8/0; BUB/BnJ, 4/26; 129S1/SvlmJ, 10/10; BALB/cJ, 0/8; A/J, 0/10; C57L/J, 0/10; FVB/NJ, 0/8; DBA/2J, 0/8; C3H/HeJ, 0/10; ALS/LtJ, 0/8; BALB/cByJ, 10/10; CE/J, 10/0; NZB/BINJ, 0/10; SJL/J, 8/12.
Comparative Analysis of Cellular Contributions to Differing CCTs
Having identified inbred strains with differing CCTs, we next selected three strains representing different points of the CCT spectrum for analyzing in more detail. Two strains from opposite ends of the survey (C57BLKS/J, thinnest; SJL/J, thickest), along with an intermediate strain (C57BL/6J), were analyzed to study anatomic features contributing to differing CCTs. The pachymetry measured CCTs of these three strains were significantly different from one other (C57BLKS/J versus C57BL/6J, P = 0.0103; C57BLKS/J versus SJL/J, P = 0.0002; and C57BL/6J versus SJL/J, P = 0.0058). Aside from the albinism associated with SJL/J mice, anterior chambers of all three strains appeared healthy and normal by slit lamp examination (Figs. 4A–C). With only rare exception, corneas of all three strains were clear at all ages examined (n = 14–18 eyes examined per strain, 3–4-month-old mice, a single C57BL/6J eye exhibited a cloudy cornea). Although SJL/J mice are albino, seven other strains included in the survey (PL/J, BUB/BnJ, BALB/cJ, A/J, FVB/NJ, ALS/LtJ, and BALB/cByJ) were also albino and exhibited widely dispersed CCTs. Thus, the albinism of SJL/J is likely to have little influence on CCT.
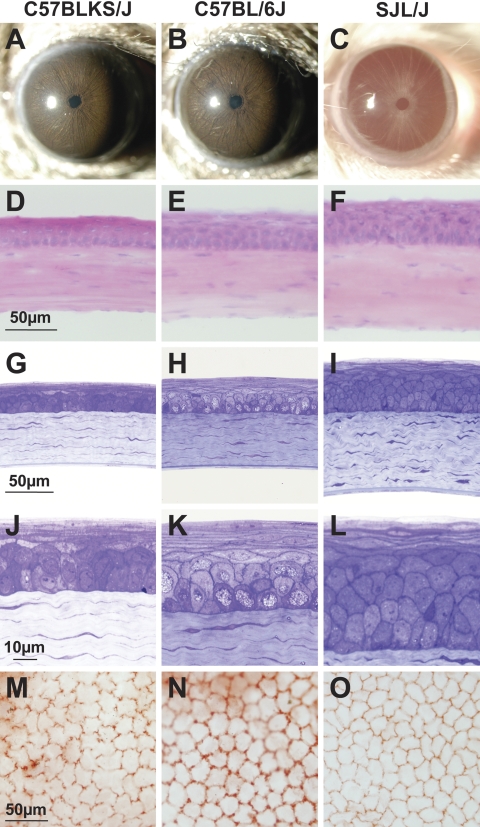
Figure 4.
Comparative corneal anatomy of strains with differing CCTs. Representative images from C57BLKS/J, C57BL/6J, and SJL/J eyes. (A–C) Slit lamp images indicated normal and healthy-appearing anterior chambers. (D–F) Unfixed cryosections stained with H&E showed an absence of overt disease and that the epithelium and stroma both contribute to strain-specific differences in CCT. (G–I) Glutaraldehyde-fixed, plastic-embedded sections stained with toluidine blue, again indicated an absence of corneal disease and contributions to CCT from both the stroma and epithelium. (J–L) At higher magnification, plastic-embedded sections demonstrated differences in the thickness of the epithelium. There was a striking difference in the number of wing cells; C57BLKS/J, C57BL/6J, and SJL/J corneas had on average one, two, and three layers of wing cells, respectively. All three strains had a similar monolayer of columnar basal cells and three to four layers of flattened squamous cells. Brightness and contrast of (J–L) were digitally modified, compared with (G–I), for better visualization of the epithelium. (M–O) Flatmounts stained with alizarin red showing greater cell density of corneal endothelial cells in SJL/J eyes than in C57BLKS/J or C57BL/6J eyes.
An absence of overt disease was further confirmed by an examination of unfixed cryosections stained with H&E (Figs. 4D–F). Both the stroma (C57BLKS/J, 73.9 ± 11.3 μm, n = 5 eyes; C57BL/6J, 74.1 ± 7.9 μm, n = 5; and SJL/J, 91.7 ± 9.2 μm, n = 5) and the epithelium (C57BLKS/J, 24.7 ± 6.3 μm, n = 5 eyes; C57BL/6J, 32.2 ± 5.8 μm, n = 5; and SJL/J, 42.2 ± 4.8 μm, n = 5) contributed to the strain-specific differences in CCT. The epithelial contribution to total CCT was similar in C57BL/6J and SJL/J mice (30.3% and 31.5% of total CCT, respectively), but was decreased in C57BLKS/J (25.1%). Estimates of stromal cell density followed CCT (C57BLKS/J, 1300 ± 540 cells/mm2; C57BL/6J, 1500 ± 70 cells/mm2; and SJL/J, 1900 ± 280 cells/mm2).
Corneas were further evaluated for potential anatomic features associated with differing CCT between strains, by examining glutaraldehyde-fixed, plastic-embedded sections of each strain with light microscopy (Figs. 4G–L). In all samples (n = 5 eyes per strain), differences in stromal and epithelial thickness were again apparent. Differences in epithelial thickness were primarily due to the number of wing cells present. C57BLKS/J, C57BL/6J, and SJL/J corneas had on average one, two, and three layers of wing cells, respectively. All three strains had a monolayer of columnar basal cells and three to four layers of flattened squamous cells.
In all analyses, the corneal endothelium of each strain was continuous throughout the cornea and did not appear to contribute to the strain-specific differences in CCT. However, endothelial cell density did differ between the strains (Figs. 4M–O). C57BLKS/J and C57BL/6J corneas displayed endothelial cell densities that were statistically equal, whereas the SJL/J density was significantly greater than the other strains. (All corneas analyzed were from mice 114 to 167 days of age. C57BLKS/J, 2282.6 ± 235.9 cells/mm2; C57BL/6J, 2296.6 ± 279.2 cells/mm2; and SJL/J, 3060.9 ± 194.1 cells/mm2. C57BLKS/J versus C57BL/6J, P = 0.94; C57BLKS/J versus SJL/J, P = 0.0022; and C57BL/6J versus SJL/J, P = 0.0041).
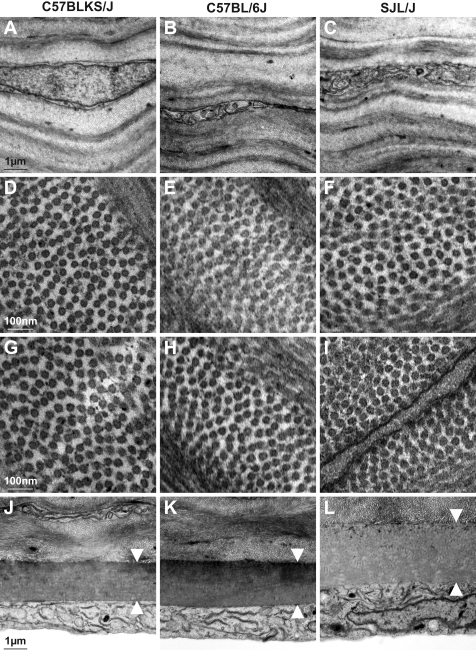
Several additional anatomic observations were made with TEM (Fig. 5). First, differing thickness of the stroma appeared to be due to an increased number of lamellae, and not to the thickness of each individual lamellae (Figs. 5A–C). Second, collagen fiber diameters were well organized and of uniform diameter among all three strains in both the anterior (Figs. 5D–F) and posterior (Figs. 5G–I) corneal stroma. Collagen fiber diameters in the anterior/posterior cornea for each strain were 31.5/32.4, 32.5/31.8, and 31.6/31.8 nm (n = 2, 1, and 2 eyes of mice 100 to 120 days of age; C57BLKS/J, C57BL/6J, and SJL/J, respectively). Finally, the thickness of Descemet's membrane correlated with overall corneal thickness (Figs. 5J–L), contributing 1.7%, 1.7%, and 2.0% to the overall corneal thickness (C57BLKS/J, C57BL/6J, and SJL/J, respectively). In sum, the findings of these comparative analyses demonstrated by multiple methods that the differences in CCT between these strains was not a simple consequence of an underlying corneal disease, but rather, that the differences reflected extremes of the normal physiological range of CCT exhibited by mice.
Figure 5.
TEM of the corneal stroma and endothelium. Representative images from C57BLKS/J, C57BL/6J, and SJL/J eyes. (A–C) There were no notable differences in architecture of the stromal lamellae or keratocytes. Rather, differences in CCT appeared to be due to an increased number of structurally normal-appearing lamellae. (D–I) Collagen fibers in cross section from the anterior (D–F) and posterior (G–I) cornea are well organized and of uniform diameter. (J–L) Thickness of Descemet's membrane (arrowheads) follows overall thickness of the cornea, although it is only responsible for a small portion of the corneal thickness.
Expression Array Profiles Associated with Differing CCT
The study of molecular mechanisms contributing to differences in CCT was begun by gene expression profiling of punches of central cornea from C57BLKS/J, C57BL/6J, and SJL/J mice that were 100 to 120 days of age. A genome-wide analysis of transcription levels was performed with mouse genome expression arrays. To help focus this experiment, we specifically searched for genes with expression levels correlating to CCT between these three strains. For example, because CCT manifests as SJL/J > C57BL/6J > C57BLKS/J, we hypothesized that genes promoting corneal thickness might exhibit expression values that are highest in SJL/J, intermediate in C57BL/6J, and lowest in C57BLKS/J. Conversely, genes limiting corneal thickness (such as matrix-degrading factors) would exhibit an opposite expression pattern (highest in C57BLKS/J, intermediate in C57BL/6J, and lowest in SJL/J).
This analysis resulted in the identification of many genes with expression levels correlating to CCT. Of the 45,037 probesets contained on the array, 17,097 probesets representing 10,395 unique genes were expressed in the cornea of at least two individuals. Of these, 376 unique genes were identified as differentially expressed between the three mouse strains. There were 142 genes with expression levels correlated to CCT; 87 genes with highest expression in SJL/J (Table 1) and 55 genes with highest expression in C57BLKS/J (Table 2). Differentially expressed genes included genes shown by others to be expressed by multiple corneal cell types, including epithelial cells26 and keratocytes.27 Transcripts more prevalent in SJL/J cornea can be assigned to several functional groups, including development, oxidoreductase activity, secreted/ECM, signal transduction, transcriptional regulation, and protein transport. Of interest, all the same functional groups were also present among transcripts more prevalent in C57BLKS/J cornea, but with different gene members. Several transcripts more prevalent in C57BLKS/J cornea were also related to the functional group immune response.
Table 1.
Transcripts Correlating to CCT with Elevated Expression in SJL/J Mice
| Gene Symbol | Gene Names | C57BLKS/J | C57B1/6J | SJL/J | Change (x-fold) | E/K |
|---|---|---|---|---|---|---|
| Development | ||||||
| BC024561 | cDNA sequence BC024561 | 2.0 | 2.0 | 7.2 | 36.1 | |
| Ceacam10 | CEA-related cell adhesion molecule 10 | 2.2 | 3.2 | 11.2 | 486.6 | |
| Clec2g | C-type lectin domain family 2, member g | 7.5 | 8.3 | 10.0 | 5.5 | |
| Klf9 | Kruppel-like factor 9 | 4.9 | 8.6 | 8.7 | 13.7 | |
| Lpin1 | Lipin 1 | 6.3 | 8.2 | 8.3 | 3.9 | |
| Map3k7 | Mitogen activated protein kinase kinase kinase 7 | 8.5 | 9.6 | 9.6 | 2.3 | K |
| Nif3l1 | Nggl interacting factor 3-like 1 | 6.7 | 8.6 | 8.5 | 3.4 | |
| Spon1 | Spondin 1 | 6.4 | 9.5 | 11.5 | 34.1 | |
| Oxidoreductase Activity | ||||||
| 4833423E24Rik | RIKEN cDNA 4833423E24 gene | 2.3 | 2.3 | 7.1 | 28.3 | |
| Cyp17a1 | Cytochrome P450, 17a1 | 2.3 | 2.3 | 7.2 | 28.9 | |
| Cyp24a1 | Cytochrome P450, 24a1 | 7.5 | 9.4 | 11.0 | 11.4 | E |
| Ero1l | ERO1-like | 9.8 | 10.0 | 12.6 | 6.8 | K |
| LOC547349 | Similar to MHC class 1 antigen precursor | 2.6 | 2.6 | 9.1 | 88.3 | |
| Mlstd2 | Male sterility domain containing 2 | 9.3 | 11.2 | 11.6 | 4.8 | |
| Mogat1 | Monoacylglycerol O-acyltransferase 1 | 2.8 | 3.7 | 7.2 | 21.1 | |
| Txnl5 | Thioredoxin-like 5 | 4.9 | 6.9 | 6.9 | 3.9 | |
| Secreted/ECM | ||||||
| Ccl28 | Chemokine (C–C motif) ligand 28 | 2.3 | 2.3 | 10.4 | 283.3 | |
| Csn3 | Casein kappa | 2.7 | 3.0 | 8.8 | 68.7 | |
| D930028F11Rik | RIKEN cDNA D930028F11 gene | 3.2 | 3.8 | 11.7 | 365.3 | |
| LOC547343 | H-2 class 1 histocompatibility antigen, L-D alpha | 7.9 | 8.4 | 10.8 | 7.3 | |
| Sectm1b | Secreted and transmembrane 1B | 2.2 | 2.2 | 7.0 | 27.6 | |
| Tm7sf3 | Transmembrane 7 superfamily member 3 | 5.8 | 7.7 | 7.7 | 3.8 | K |
| Tmem45b | Transmembrane protein 45b | 4.0 | 5.4 | 10.6 | 94.1 | |
| Signal Transduction | ||||||
| 2610305D13Rik | RIKEN cDNA 2610305D13 gene | 2.5 | 2.5 | 8.1 | 48.5 | |
| A630033E08Rik | RIKEN cDNA A630033E08 gene | 2.3 | 8.4 | 9.0 | 107.5 | |
| E2f6 | E2F transcription factor 6 | 4.5 | 7.7 | 8.9 | 21.2 | |
| Pld2 | Phospholipase D2 | 7.8 | 9.0 | 9.0 | 2.4 | E/K |
| Sh3rf1 | SH3 domain containing ring finger 1 | 4.9 | 7.7 | 7.8 | 7.3 | K |
| Snx6 | Sorting nexin 6 | 2.3 | 8.0 | 9.2 | 116.2 | K |
| Stk35 | Serine/threonine kinase 35 | 10.0 | 10.2 | 12.5 | 5.7 | |
| Transcriptional Regulation | ||||||
| Polr3gl | Polymerase (RNA) III polypeptide G | 4.4 | 4.4 | 7.6 | 9.3 | |
| Rbpms2 | RNA binding protein with multiple splicing 2 | 3.7 | 8.1 | 8.5 | 26.9 | |
| Ssu72 | RNA polymerase II CTD phosphatase homolog | 2.3 | 2.3 | 7.2 | 30.1 | K |
| Zfp277 | Zinc finger protein 277 | 7.7 | 9.6 | 10.0 | 5.2 | K |
| Pcm1 | Pericentriolar material 1 | 2.3 | 2.3 | 8.2 | 58.7 | |
| Trove2 | TROVE domain family, member 2 | 3.8 | 4.0 | 7.9 | 17.1 | |
| Protein Transport | ||||||
| Epb4.111 | Erythrocyte protein band 4.1-like 1 | 8.6 | 8.6 | 11.3 | 6.8 | |
| LOC631721 | Similar to vacuolar protein sorting 52 | 6.3 | 7.9 | 8.4 | 4.1 | |
| Pldn | Pallidin | 5.1 | 5.0 | 9.3 | 18.9 | |
| Slc4a7 | Sodium bicarbonate transporter 2 | 7.2 | 9.2 | 9.2 | 4.0 | E/K |
Expression levels are given as log2 values, showing 40 of 87 total. E/K column identifies transcripts expressed in previous microarray studies. E, transcripts expressed in laser captured murine basal epithelial cells (GDS2433); K, transcripts expressed in cultured murine corneal keratocytes (GDS857).
Table 2.
Transcripts Correlating to CCT with Elevated Expression in C57BLKS/J Mice
| Gene Symbol | Gene Name | C57BLKS/J | C57BL/6J | SJL/J | Change (x-fold) | E/K |
|---|---|---|---|---|---|---|
| Development | ||||||
| Apoe | Apolipoprotein E | 12.4 | 11.2 | 8.7 | 12.4 | K |
| Apod | Apolipoprotein D | 9.1 | 8.6 | 2.3 | 114.2 | |
| Ext1 | Exostoses (multiple)1 | 8.0 | 7.8 | 2.0 | 63.9 | K |
| Ngfr | Nerve growth factor receptor | 9.3 | 9.2 | 7.2 | 4.5 | |
| Otx1 | Orthodenticle homolog 1 (Drosophila) | 11.5 | 9.9 | 6.3 | 35.3 | |
| Rhou | Ras homolog gene family, member U | 9.8 | 9.6 | 7.9 | 3.8 | K |
| Scmh1 | Sex comb on midleg homolog 1 | 8.5 | 7.8 | 5.3 | 8.7 | E/K |
| Tmie | Transmembrane inner ear | 10.0 | 9.4 | 6.7 | 9.8 | |
| Ptpn21 | Protein tyrosine phosphatase, non-receptor type 21 | 8.9 | 7.8 | 6.5 | 5.6 | |
| Oxidoreductase Activity | ||||||
| Akr1e1 | Aldo-keto reductase family 1, member E1 | 11.2 | 10.8 | 8.6 | 6.1 | |
| Cyp2a4 | Cytochrome P450, family 2, subfamily a, polypeptide 4 | 9.3 | 7.8 | 3.6 | 54.2 | |
| Efcbp1 | EF hand calcium binding protein 1 | 8.0 | 7.8 | 4.0 | 16.1 | |
| Htatip2 | HIV-1 tat interactive protein 2 | 8.0 | 7.5 | 2.3 | 50.6 | K |
| Mod1 | Malic enzyme, supernatant | 7.5 | 7.1 | 4.9 | 6.1 | |
| Secreted/ECM | ||||||
| Fmod | Fibromodulin | 9.6 | 8.5 | 4.6 | 33.1 | E/K |
| Gpc4 | Glypican 4 | 10.8 | 9.2 | 8.1 | 6.4 | K |
| Signal Transduction | ||||||
| 6430701C03Rik | RIKEN cDNA 6430701C03 gene | 6.8 | 6.5 | 3.7 | 9.0 | |
| Kcnh1 | Potassium voltage-gated channel, H1 | 8.0 | 7.4 | 5.6 | 5.3 | |
| Lrig1 | Leucine-rich repeats and Ig-like domains 1 | 9.1 | 9.0 | 3.4 | 54.2 | |
| Map3k6 | Mitogen-activated protein kinase kinase kinase 6 | 11.8 | 11.7 | 9.6 | 4.5 | |
| Poqr8 | Progestin and adipoQ receptor family member VIII | 6.6 | 6.2 | 3.1 | 11.4 | E |
| Synj2bp | Synaptojanin 2 binding protein | 11.0 | 10.6 | 8.8 | 4.6 | K |
| Transcriptional Regulation | ||||||
| Ccdc122 | Coiled-coil domain containing 122 | 9.5 | 9.4 | 3.6 | 61.0 | |
| Rbm39 | RNA binding motif protein 39 | 9.3 | 8.9 | 4.6 | 26.3 | E/K |
| Ssbp2 | Single-stranded DNA binding protein 2 | 9.5 | 9.1 | 3.7 | 53.2 | K |
| Protein Transport | ||||||
| Abhd10 | Abhydrolase domain containing 10 | 8.6 | 8.1 | 6.8 | 3.5 | |
| Chit1 | Chitinase 1 (chitotriosidase) | 9.8 | 8.4 | 6.4 | 10.1 | |
| Ctsl | Cathepsin L | 10.4 | 10.1 | 2.6 | 216.9 | E/K |
| H2-D1 | Histocompatibility 2, D region locus 1 | 8.9 | 2.5 | 2.6 | 80.7 | K |
| Ptpn21 | Protein tyrosine phosphatase, non-receptor type 21 | 9.0 | 7.8 | 6.5 | 5.6 | K |
| Serping1 | Serine peptidase inhibitor 1, clade G | 11.7 | 11.6 | 10.6 | 2.2 | K |
| Skiv212 | Superkiller viralicidic activity 2-like 2 | 7.8 | 2.4 | 2.5 | 40.3 | |
| Upb1 | Ureidopropionase, beta | 8.7 | 7.1 | 2.5 | 72.6 | |
| Usp48 | Ubiquitin specific peptidase 48 | 8.1 | 7.6 | 3.4 | 25.5 | |
| Immune Response | ||||||
| Creb312 | cAMP responsive element binding protein 3-like 2 | 8.7 | 8.3 | 6.9 | 3.6 | |
| H2-Ea | Histocompatibility 2, class II antigen E alpha | 7.6 | 2.1 | 2.2 | 44.9 | |
| Il1f8 | Interleukin 1 family, member 8 | 6.5 | 4.9 | 2.4 | 17.0 | E |
| Il1rn | Interleukin 1 receptor antagonist | 9.4 | 8.5 | 6.4 | 8.2 | K |
| Pttg1 | Pituitary tumor-transforming 1 | 11.5 | 11.2 | 9.1 | 5.3 | E |
| Serping1 | Serine peptidase inhibitor 1, clade G | 11.7 | 11.6 | 10.6 | 2.2 | |
Expression levels are given as log2 values, showing 40 of 55 total. E/K column identifies transcripts expressed in previous microarray studies. E, transcripts expressed by laser captured murine basal epithelial cells (GDS2433); K, transcripts expressed by cultured murine corneal keratocytes (GDS857).
Because transcripts maintained at relatively high expression levels may be particularly attractive candidates for regulating CCT, array data were also analyzed to identify genes with the highest expression levels (Table 3). Consistent with previous analyses of mouse corneal gene expression,32 our analysis detected high transcript levels of transketolase; aldehyde dehydrogenase family 3, A1; thymosin, beta 4; glutathione S-transferase omega 1; and many other genes. None of the highly expressed transcripts were also significantly correlated with CCT (compare Table 3 with Tables 1 and 2). Genes with highest levels of corneal transcript expression had limited correlation to the currently known most prevalent corneal proteins.33 Genes encoding highly prevalent corneal proteins also tended to not be significantly correlated with differences in CCT (comparing the results of Karring et al.33 to Tables 1 and 2); only two of the genes encoding the 141 proteins of the known corneal proteome also correlated significantly with CCT (apolipoprotein D and apolipoprotein E, both expressed at higher levels in C57BLKS/J than SJL/J).
Table 3.
Transcripts with Highest Measured Expression
| Gene Symbol | Gene Name | Mean | SD |
|---|---|---|---|
| Gsta4 | Glutathione S-transferase, alpha 4 | 13.3 | 0.15 |
| Tmsb4x | Thymosin, beta 4, X chromosome | 13.1 | 0.12 |
| A830036E02Rik | RIKEN cDNA A830036E02 gene | 13.0 | 0.19 |
| Slurp1 | Secreted Ly6/Plaur domain containing 1 | 13.0 | 0.15 |
| Aqp5 | Aquaporin 5 | 12.9 | 0.15 |
| S100all | S100 calcium binding protein All (calizzarin) | 12.9 | 0.14 |
| Eroll | ERO1-like | 12.8 | 0.14 |
| Aldh3a1 | Aldehyde dehydrogenase family 3, A1 | 12.8 | 0.15 |
| Perp | PERP, TP53 apoptosis effector | 12.8 | 0.21 |
| Tkt | Transketolase | 12.8 | 0.15 |
| 4930583H14Rik | RIKEN cDNA 4930583H14 gene | 12.7 | 0.16 |
| Mal | Myelin and lymphocyte protein | 12.7 | 0.20 |
| Txn1 | Thioredoxin 1 | 12.7 | 0.23 |
| Tpt1 | Tumor protein, translationally controlled 1 | 12.7 | 0.14 |
| Anxa2 | Annexin A2 | 12.7 | 0.12 |
| Sdc1 | Syndecan 1 | 12.7 | 0.20 |
| Pebp1 | Phosphatidylethanolamine binding protein 1 | 12.7 | 0.12 |
| Actg1 | Actin, gamma, cytoplasmic 1 | 12.7 | 0.18 |
| 1100001122Rik | RIKEN cDNA 1100001122 gene | 12.7 | 0.14 |
| Fth1 | Ferritin heavy chain 1 | 12.7 | 0.20 |
| Lypd2 | Ly6/Plaur domain containing 2 | 12.6 | 0.16 |
| Gsto1 | Glutathione S-transferase omega 1 | 12.6 | 0.31 |
| Csda | Cold shock domain protein A | 12.6 | 0.17 |
| Hk1 | Hexokinase 1 | 12.6 | 0.13 |
| S100a6 | S100 calcium binding protein A6 (calcyclin) | 12.6 | 0.16 |
| Stfa3 | Stefin A3 | 12.6 | 0.19 |
| Uba52 | Ubiquitin A-52 protein fusion 1 product | 12.6 | 0.15 |
| Krt6a | Keratin 6A | 12.6 | 0.17 |
| Tegt | Testis enhanced gene transcript | 12.6 | 0.15 |
| Krt6b | Keratin 6B | 12.6 | 0.24 |
| Vdac1 | Voltage-dependent anion channel 1 | 12.6 | 0.21 |
| Actb | Actin, beta, cytoplasmic | 12.6 | 0.17 |
| Mrfap1 | Morf4 family associated protein 1 | 12.5 | 0.12 |
| Cd24a | CD24a antigen | 12.5 | 0.16 |
| Eef1b2 | Eukaryotic translation elongation factor 1b2 | 12.5 | 0.20 |
| Hmgn1 | High mobility group nucleosomal binding 1 | 12.5 | 0.18 |
| Dsp | Desmoplakin | 12.5 | 0.16 |
| Eeflal | Eukaryotic translation elongation factor 1a1 | 12.5 | 0.15 |
| LOC622534 | Similar to ribosomal protein L36 | 12.5 | 0.13 |
| Fan | Finkel-Biskis-Reilly murine sarcoma virus | 12.5 | 0.13 |
| Cox7a2 | Cytochrome c oxidase, subunit Vlla 2 | 12.5 | 0.13 |
| Ahnak | AHNAK nucleoprotein (desmoyokin) | 12.5 | 0.18 |
| A2m | alpha-2-macroglobulin | 12.5 | 0.15 |
| Gnb2l1 | guanine nucleotide binding protein b2 like 1 | 12.5 | 0.12 |
| Dynll1 | Dynein light chain LC8-type 1 | 12.5 | 0.15 |
| Eif1 | Eukaryotic translation initiation factor 1 | 12.5 | 0.23 |
| Prdx1 | Peroxiredoxin 1 | 12.5 | 0.18 |
| Arf6 | ADP-ribosylation factor 6 | 12.5 | 0.19 |
| Dcn | Decorin | 12.4 | 0.15 |
| Hspa1b | Heat shock protein 1B | 12.4 | 0.34 |
Expression levels are given as log2 values, showing top 50 transcripts as identified by individual probesets yielding the highest signal averaged across all nine arrays. Genes encoding ribosomal proteins and poorly annotated transcripts have been removed.
In testing for the presence of smaller magnitude, but perhaps still biologically important differences, expression levels of several candidates were also examined individually for differential corneal expression between C57BLKS/J and SJL/J mice. Most candidates examined failed to demonstrate significant differences in expression (all present probesets P > 0.01), including collagen, type I, α1; collagen, type V, α1; collagen, type VI, α1; keratocan; gelsolin; lumican; and transforming growth factor β1. Analysis of candidates potentially influencing the corneal epithelium, such as genes encoding keratins, were also largely negative, with the exception of two transcripts both expressed at higher levels in C57BLKS/J than SJL/J corneas, keratin 4 (2.2-fold, P = 0.007) and keratin 13 (3.2-fold, P = 0.0002). The interesting CCT candidates forkhead box C1 (Foxc1) and paired-like homeodomain transcription factor 2 (Pitx2), which when mutant both contribute to Axenfeld-Rieger malformations and altered corneal thickness,13,34 were also unchanged in this experiment. Combined, these results identified several candidates worthy of further consideration for potentially influencing CCT, but broadly suggested that the genes influencing CCT may not necessarily be those with highly abundant transcript or protein levels, nor were they easily ascribed to known pathways. Rather, the transcriptional events accompanying changes in CCT were complex and involved multiple classes of biological events.
Discussion
In humans, CCT exhibits a broad biological variability. With an estimated heritability of 0.95, CCT in humans is strongly influenced by genetic effects.10 In this study, we demonstrated that the same is also true of mice. Among 17 different inbred strains of mice, CCT ranged from a low of 89.2 μm in C57BLKS/J mice to a high of 123.8 μm in SJL/J mice. Because CCT exhibits a continual variation in different strains of inbred mice between these extremes, these data suggest that CCT is under multigenic influence. Identification of these strains will empower future genetic experiments to identify genes regulating CCT, and as demonstrated in this study, such experiments represent a powerful resource for studying the cellular and molecular changes associated with CCT variability.
We initially focused on studying events in C57BLKS/J, C57BL/6J, and SJL/J corneas. A priori, there are few clues as to why these strains might exhibit such different CCT values. At the cellular level, it is clear that differences in the corneal epithelium and stroma both contribute to differences in CCT between these strains. As in humans, the mouse corneal stroma constitutes the largest portion of the corneal thickness. The stroma largely consists of a highly structured extracellular matrix with collagen fibrils arranged in lamellar sheets extending the entire length of the cornea. Within each lamellae, the collagen fibrils are oriented in the same direction, are of equal diameter, and have uniform spacing. If one of these parameters does not occur, the cornea will typically become cloudy. Based on our histologic analysis, and because the corneas of C57BLKS/J, C57BL/6J, and SJL/J mice are clear, strain-dependent changes in CCT do not appear to involve gross alterations in the architecture or spacing of the collagen fibers. Rather, differing CCTs among these strains appears to result from changes in the number of normal-appearing stromal lamellae. Presumably, a consequence of altered keratocyte function, we speculate that the C57BLKS/J and SJL/J genetic backgrounds harbor alleles influencing the function of these cells. Ongoing experiments are testing this directly. Also of interest, we observed that epithelial thickness contributes significantly to murine CCT. A similar observation has been made in mice with a targeted mutation of Pitx2.13
Expression profiling offers one approach for identifying molecular pathways potentially contributing to these differences in CCT. Among the individual transcripts with altered corneal expression between C57BLKS/J and SJL/J mice, there are several interesting observations. First, among the transcripts correlating to CCT, a few genes have been investigated as potentially influencing corneal structure, including fibromodulin35 and kruppel-like factor 9.36 Additional experiments are needed to determine whether these or any of the other detected changes are causative factors or markers of differing CCT. Second, also present among the CCT-correlated transcripts are several with very large changes in expression. Some of these changes are likely to indicate the presence of strain-specific alleles resulting in message instability (such as premature stop codons), but to our knowledge no such single-nucleotide-polymorphisms (SNPs) have yet been described in these genes. Third, keratin 4 (a type I keratin) and keratin 13 (a type II keratin), encoding proteins that are frequently found together in epithelia,37 are both modestly but significantly expressed at higher levels in C57BLKS/J corneas. Finally, changes in corneal genes potentially important in other corneal diseases, but with no known links to corneal structure, were detected. For example, apolipoprotein E, which is expressed at higher levels in the C57BLKS/J cornea, has been shown to influence ocular herpes pathogenesis.38 Independent of a potential role related to corneal thickness, knowledge of these strain-specific differences may prove useful in the study of additional corneal phenotypes.
Another class of interesting candidate CCT-regulatory genes pertains to electro-osmosis in the cornea. Fluid transport across the corneal endothelium and epithelium is a key mechanism that promotes corneal clarity and CCT constancy. Because of the high water-binding capacity of proteoglycans, the stroma has a natural tendency to imbibe water and swell. Under normal conditions, stroma-to-aqueous fluid transport by the endothelium opposes stromal swelling, promoting a relatively dehydrated stromal matrix of regularly spaced collagen fibers. Although much remains unknown about the details of endothelial transport, several candidates participating in transport have been suggested, including: Na+/K+-ATPase (such as ATPA1A); Na+/HCO3− cotransporter (such as SLC4A4, commonly referred to as NBC1); aquaporins (AQP1, -3, and -5), cystic fibrosis transmembrane conductance regulator (CFTR); and epithelial sodium channels (such as SCNN1A, -B, and -G). In mice, aquaporins have been shown to regulate corneal water movement both in the epithelium (AQP3 and -5) and endothelium (AQP1),39,40 and have been shown to affect corneal thickness. Aqp1-null mice have a cornea approximately 20% thinner than that of wild-types, whereas Aqp5-null mice are approximately 20% thicker.40,41 Although it was not highlighted by our current experiments examining differences in corneal gene expression between mouse strains with differing CCTs, genes influencing fluid transport of the corneal endothelium and epithelium nonetheless remain promising candidates for CCT regulation.
Among the most intriguing facets of CCT variability in humans is the association between CCT and glaucoma. In the Ocular Hypertension Treatment Study, a multicenter randomized study involving 1636 participants with ocular hypertension, participants with a CCT of 555 μm or less had a threefold greater risk of primary open-angle glaucoma compared with participants who had a CCT of more than 588 μm.3,42–44 Additional support of a role for CCT in glaucoma risk has also been found in subsequent studies.45 The reasons that CCT correlates with glaucoma risk in humans remain hotly debated. One hypothesis suggests that differing CCT creates artifacts in intraocular pressure (IOP) measurement (because applanation assumes that corneal thickness and rigidity are constant between people).3 Accordingly, the differing glaucoma risk reflects the possibility that people with thin corneas have had IOP much higher than that actually measured. However, other data suggest that the correlation is more complex. Even after correcting IOP measurements for the true IOP, thinner CCT remains associated with worse glaucoma outcomes.46 Therefore, a second hypothesis worthy of consideration is that there may be biological correlates caused by molecules that influence both CCT and some other tissue more directly involved in glaucomatous disease, such as the trabecular meshwork, retinal nerve fiber layer, or lamina cribrosa.47,48 Perhaps one of the most important avenues that can stem from the work presented herein is the opportunity to identify CCT-regulating genes and experimentally test hypotheses such as these directly.
Before our work, a variety of other approaches have been used to noninvasively measure CCT among a limited number of strains.15–18,20–22 Although successful, each of these past approaches has also required relatively expensive instrumentation that most investigators do not have access to within their animal facilities. One advance from the current work is the demonstration that the ultrasound pachymeter (Corneo-Gage Plus; Sonogage) is also capable of reproducibly measuring CCT in different strains of mice. Because ultrasound pachymeters are comparatively inexpensive, addition of this technique to those already existing for measuring murine CCT is a useful advance. However, pachymetry is not without potential disadvantages. The steep curvature of the mouse cornea renders pachymetry prone to error. Although we have not performed side-by-side comparisons, we suspect that non–contact-based methods such as interferometry may be superior to pachymetry for measuring murine CCT in many instances; our results show only that pachymetry is sufficient for uses such as the strain survey performed here. Also, the mouse and human cornea are similar, but not identical. Species-specific differences in how CCT is maintained may exist and limit the ability to translate insights from mouse studies to humans. These drawbacks aside, continued studies of the cellular and molecular events regulating the strain-specific differences in CCT identified herein are likely to uncover many processes that are evolutionarily conserved in the human cornea and that add new insight into the basic biological events that regulate complex extracellular matrices such as the corneal stroma.
In conclusion, we have developed a methodology for measuring murine CCT with ultrasound pachymetry, performed a strain survey, and characterized the cornea of three strains at differing points of the murine CCT spectrum that are free of overt corneal disease. Because these strains have inherent genetic diversity, they represent an ideal resource for genetic crosses mapping loci that influence CCT. In our ongoing work, we intend to complete these crosses, identify genes regulating CCT, and ultimately study the mechanisms by which these genes influence CCT and glaucoma susceptibility.
Acknowledgments
The authors thank Alan Burns (Baylor College of Medicine) for helpful technical suggestions regarding corneal processing for histology, and John Fingert and Wallace Alward (University of Iowa) for helpful suggestions and advice regarding ultrasound pachymetry. The authors are especially grateful for helpful discussions with the late Norm Hawes of The Jackson Laboratory.
Footnotes
Supported by National Glaucoma Research, a program of the American Health Assistance Foundation, and National Eye Institute Grant EY018825.
Disclosure: G.D. Lively, None; B. Jiang, None; A. Hedberg-Buenz, None; B. Chang, None; G.E. Petersen, None; K. Wang, None; M.H. Kuehn, None; M.G. Anderson, None
References
- 1.Doughty MJ, Zaman ML. Human corneal thickness and its impact on intraocular pressure measures: a review and meta-analysis approach. Surv Ophthalmol 2000; 44: 367–408 [DOI] [PubMed] [Google Scholar]
- 2.Aghaian E, Choe JE, Lin S, Stamper RL. Central corneal thickness of Caucasians, Chinese, Hispanics, Filipinos, African Americans, and Japanese in a glaucoma clinic. Ophthalmology 2004; 111: 2211–2219 [DOI] [PubMed] [Google Scholar]
- 3.Brandt JD. Corneal thickness in glaucoma screening, diagnosis, and management. Curr Opin Ophthalmol 2004; 15: 85–89 [DOI] [PubMed] [Google Scholar]
- 4.Dai E, Gunderson CA. Pediatric central corneal thickness variation among major ethnic populations. J AAPOS 2006; 10: 22–25 [DOI] [PubMed] [Google Scholar]
- 5.Doughty MJ, Laiquzzaman M, Muller A, Oblak E, Button NF. Central corneal thickness in European (white) individuals, especially children and the elderly, and assessment of its possible importance in clinical measures of intra-ocular pressure. Ophthalmic Physiol Opt 2002; 22: 491–504 [DOI] [PubMed] [Google Scholar]
- 6.Hahn S, Azen S, Ying-Lai M, Varma R. Central corneal thickness in Latinos. Invest Ophthalmol Vis Sci 2003; 44: 1508–1512 [DOI] [PubMed] [Google Scholar]
- 7.Lam AK, Douthwaite WA. The corneal-thickness profile in Hong Kong Chinese. Cornea 1998; 17: 384–388 [DOI] [PubMed] [Google Scholar]
- 8.Ehlers N, Hjortdal J. Corneal thickness: measurement and implications. Exp Eye Res 2004; 78: 543–548 [DOI] [PubMed] [Google Scholar]
- 9.Smolin G, Thoft RA. The Cornea: Scientific Foundations and Clinical Practice 3rd ed.Boston: Little, Brown; 1994: xv, 759 [Google Scholar]
- 10.Toh T, Liew SH, MacKinnon JR, et al. Central corneal thickness is highly heritable: the twin eye studies. Invest Ophthalmol Vis Sci 2005; 46: 3718–3722 [DOI] [PubMed] [Google Scholar]
- 11.Alsbirk PH. Corneal thickness. II. Environmental and genetic factors. Acta Ophthalmol (Copenh) 1978; 56: 105–113 [DOI] [PubMed] [Google Scholar]
- 12.Dohadwala AA, Damji KF. Familial occurrence of artefactual ocular hypertension from thick corneas and of primary open angle glaucoma in a French Canadian kindred. Ophthalmic Genet 2000; 21: 1–7 [PubMed] [Google Scholar]
- 13.Asai-Coakwell M, Backhouse C, Casey RJ, Gage PJ, Lehmann OJ. Reduced human and murine corneal thickness in an Axenfeld-Rieger syndrome subtype. Invest Ophthalmol Vis Sci 2006; 47: 4905–4909 [DOI] [PubMed] [Google Scholar]
- 14.Inman DM, Sappington RM, Horner PJ, Calkins DJ. Quantitative correlation of optic nerve pathology with ocular pressure and corneal thickness in the DBA/2 mouse model of glaucoma. Invest Ophthalmol Vis Sci 2006; 47: 986–996 [DOI] [PubMed] [Google Scholar]
- 15.Jester JV, Ghee Lee Y, Li J, et al. Measurement of corneal sublayer thickness and transparency in transgenic mice with altered corneal clarity using in vivo confocal microscopy. Vision Res 2001; 41: 1283–1290 [DOI] [PubMed] [Google Scholar]
- 16.Nissirios N, Goldblum D, Rohrer K, Mittag T, Danias J. Noninvasive determination of intraocular pressure (IOP) in nonsedated mice of 5 different inbred strains. J Glaucoma 2007; 16: 57–61 [DOI] [PubMed] [Google Scholar]
- 17.Puk O, Dalke C, Favor J, de Angelis MH, Graw J. Variations of eye size parameters among different strains of mice. Mamm Genome 2006; 17: 851–857 [DOI] [PubMed] [Google Scholar]
- 18.Schmucker C, Schaeffel F. In vivo biometry in the mouse eye with low coherence interferometry. Vision Res 2004; 44: 2445–2456 [DOI] [PubMed] [Google Scholar]
- 19.Schmucker C, Schaeffel F. A paraxial schematic eye model for the growing C57BL/6 mouse. Vision Res 2004; 44: 1857–1867 [DOI] [PubMed] [Google Scholar]
- 20.Schulz D, Iliev ME, Frueh BE, Goldblum D. In vivo pachymetry in normal eyes of rats, mice and rabbits with the optical low coherence reflectometer. Vision Res 2003; 43: 723–728 [DOI] [PubMed] [Google Scholar]
- 21.Zhou X, Shen M, Xie J, et al. The development of the refractive status and ocular growth in C57BL/6 mice. Invest Ophthalmol Vis Sci 2008; 49: 5208–5214 [DOI] [PubMed] [Google Scholar]
- 22.Zhou X, Xie J, Shen M, et al. Biometric measurement of the mouse eye using optical coherence tomography with focal plane advancement. Vision Res 2008; 48: 1137–1143 [DOI] [PubMed] [Google Scholar]
- 23.Wu Z, Irizarry RA. Preprocessing of oligonucleotide array data. Nat Biotechnol 22: 656–658, 2004; author reply 658 [DOI] [PubMed] [Google Scholar]
- 24.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 2001; 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dennis G, Jr., Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003; 4: P3. [PubMed] [Google Scholar]
- 26.Zhou M, Li XM, Lavker RM. Transcriptional profiling of enriched populations of stem cells versus transient amplifying cells: a comparison of limbal and corneal epithelial basal cells. J Biol Chem 2006; 281: 19600–19609 [DOI] [PubMed] [Google Scholar]
- 27.Chakravarti S, Wu F, Vij N, Roberts L, Joyce S. Microarray studies reveal macrophage-like function of stromal keratocytes in the cornea. Invest Ophthalmol Vis Sci 2004; 45: 3475–3484 [DOI] [PubMed] [Google Scholar]
- 28.Weitkamp JW, Marsden HJ, Berke W, Daijo G. Agreement and repeatability of the Sonogage ultrasound pachometer compared with a Mitutoyo micrometer. Optom Vis Sci 2008; 85: 359–363 [DOI] [PubMed] [Google Scholar]
- 29.Gao F, Toriyama K, Ma H, Nagata T. Light microscopic radioautographic study on DNA synthesis in aging mice corneas. Cell Mol Biol (Noisy-le-grand) 1993; 39: 435–441 [PubMed] [Google Scholar]
- 30.Petkov PM, Ding Y, Cassell MA, et al. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res 2004; 14: 1806–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsang S, Sun Z, Luke B, et al. A comprehensive SNP-based genetic analysis of inbred mouse strains. Mamm Genome 2005; 16: 476–480 [DOI] [PubMed] [Google Scholar]
- 32.Norman B, Davis J, Piatigorsky J. Postnatal gene expression in the normal mouse cornea by SAGE. Invest Ophthalmol Vis Sci 2004; 45: 429–440 [DOI] [PubMed] [Google Scholar]
- 33.Karring H, Thogersen IB, Klintworth GK, Moller-Pedersen T, Enghild JJ. A dataset of human cornea proteins identified by Peptide mass fingerprinting and tandem mass spectrometry. Mol Cell Proteomics 2005; 4: 1406–1408 [DOI] [PubMed] [Google Scholar]
- 34.Lehmann OJ, Tuft S, Brice G, et al. Novel anterior segment phenotypes resulting from forkhead gene alterations: evidence for cross-species conservation of function. Invest Ophthalmol Vis Sci 2003; 44: 2627–2633 [DOI] [PubMed] [Google Scholar]
- 35.Chakravarti S, Paul J, Roberts L, Chervoneva I, Oldberg A, Birk DE. Ocular and scleral alterations in gene-targeted lumican-fibromodulin double-null mice. Invest Ophthalmol Vis Sci 2003; 44: 2422–2432 [DOI] [PubMed] [Google Scholar]
- 36.Swamynathan SK, Katz JP, Kaestner KH, Ashery-Padan R, Crawford MA, Piatigorsky J. Conditional deletion of the mouse Klf4 gene results in corneal epithelial fragility, stromal edema, and loss of conjunctival goblet cells. Mol Cell Biol 2007; 27: 182–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Muijen GN, Ruiter DJ, Franke WW, et al. Cell type heterogeneity of cytokeratin expression in complex epithelia and carcinomas as demonstrated by monoclonal antibodies specific for cytokeratins nos. 4 and 13. Exp Cell Res 1986; 162: 97–113 [DOI] [PubMed] [Google Scholar]
- 38.Bhattacharjee PS, Neumann DM, Foster TP, et al. Effect of human apolipoprotein E genotype on the pathogenesis of experimental ocular HSV-1. Exp Eye Res 2008; 87: 122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verkman AS, Ruiz-Ederra J, Levin MH. Functions of aquaporins in the eye. Prog Retin Eye Res 2008; 27: 420–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levin MH, Verkman AS. Aquaporin-3-dependent cell migration and proliferation during corneal re-epithelialization. Invest Ophthalmol Vis Sci 2006; 47: 4365–4372 [DOI] [PubMed] [Google Scholar]
- 41.Thiagarajah JR, Verkman AS. Aquaporin deletion in mice reduces corneal water permeability and delays restoration of transparency after swelling. J Biol Chem 2002; 277: 19139–19144 [DOI] [PubMed] [Google Scholar]
- 42.Brandt JD, Beiser JA, Kass MA, Gordon MO. Central corneal thickness in the Ocular Hypertension Treatment Study (OHTS). Ophthalmology 2001; 108: 1779–1788 [DOI] [PubMed] [Google Scholar]
- 43.Gordon MO, Beiser JA, Brandt JD, et al. The Ocular Hypertension Treatment Study: baseline factors that predict the onset of primary open-angle glaucoma. Arch Ophthalmol 2002; 120: 714–720, discussion 829–730 [DOI] [PubMed] [Google Scholar]
- 44.Lee BL, Wilson MR. Ocular Hypertension Treatment Study (OHTS) commentary. Curr Opin Ophthalmol 2003; 14: 74–77 [DOI] [PubMed] [Google Scholar]
- 45.Medeiros FA, Sample PA, Weinreb RN. Corneal thickness measurements and frequency doubling technology perimetry abnormalities in ocular hypertensive eyes. Ophthalmology 2003; 110: 1903–1908 [DOI] [PubMed] [Google Scholar]
- 46.Herndon LW, Weizer JS, Stinnett SS. Central corneal thickness as a risk factor for advanced glaucoma damage. Arch Ophthalmol 2004; 122: 17–21 [DOI] [PubMed] [Google Scholar]
- 47.Henderson PA, Medeiros FA, Zangwill LM, Weinreb RN. Relationship between central corneal thickness and retinal nerve fiber layer thickness in ocular hypertensive patients. Ophthalmology 2005; 112: 251–256 [DOI] [PubMed] [Google Scholar]
- 48.Jonas JB, Holbach L. Central corneal thickness and thickness of the lamina cribrosa in human eyes. Invest Ophthalmol Vis Sci 2005; 46: 1275–1279 [DOI] [PubMed] [Google Scholar]