Abstract
Hypoxia is an important ecological, evolutionary, and biomedical stressor. While physiological acclimatization of mammals to hypoxia is well studied, the variation in gene expression that underlies acclimatization is not well studied. We acclimatized inbred mice for 32 days to hypoxic conditions that simulated altitudes of 1400, 3000, and 4500 m. We used oligonucleotide microarrays to measure changes in steady-state abundance of mRNA in the livers of these mice. Mice exposed to more severe hypoxia (simulated altitude of 4500 m) were smaller in mass and had higher hematocrit than mice exposed to less severe hypoxia. ANOVA and false discovery rate tests indicated that 580 genes were significantly differentially expressed in response to chronic hypoxia. Few of these 580 genes have previously been reported to respond to hypoxia. In contrast, many of these 580 genes belonged to same functional groups typically respond to acute hypoxia. That is, both chronic and acute hypoxia elicit changes in transcript abundance for genes involved in angiogenesis, glycolysis, lipid metabolism, carbohydrate metabolism, and protein amino acid phosphorylation, but the particular genes affected by the two types of hypoxia were mostly different. Numerous genes affecting the immune system were differentially expressed in response to chronic hypoxia, which supports recently proposed hypotheses that link immune function and hypoxia. Furthermore, our results discovered novel elevated mRNA abundance of genes involved in hematopoiesis and oxygen transport not reported previously, but consistent with extreme hematocrits found in hypoxic mice.
Keywords: microarray, hematopoiesis, leptin receptor, Mus musculus, immune system
the survival of an organism often depends on its ability to acclimatize to environmental stressors. Acclimatization results from remodeling of the organism's physiological and anatomical phenotype in an attempt to accommodate an environmental stressor. A major mechanism underlying physiological remodeling is the regulation of genes and their products (84). Hence, physiologists are increasingly utilizing genomic tools to investigate physiological change. In particular, high-throughput studies of steady-state mRNA expression with DNA microarrays have proven useful in generating hypotheses concerning the mechanisms of acclimatization (34, 35).
Acclimatization and adaptation of mammals to high altitude hypoxia [reduced partial pressure of oxygen (Po2)] have received much attention due to its biomedical and ecological relevance (46, 51, 70, 72–74, 79, 92, 93, 103). Mammals experiencing chronic hypoxia undertake diverse responses aimed at maintaining oxygen delivery despite a reduced driving force (partial pressure gradient) for delivering oxygen. These responses include: increased hemoglobin production with a concomitant increase in hematocrit and blood oxygen carrying capacity; increased production of 2,3 bisphosphoglycerate, an allosteric effector of hemoglobin-oxygen binding affinity; pulmonary vasoconstriction; increased lung and liver mass; increased left ventricular mass, which leads to increased stroke volume; increased tidal volume and ventilation rate; increased capillary density; and anorexia and subsequent weight loss (1, 6, 12, 33, 64, 87). In addition, while the above physiological changes are taking place, many mammals alter their metabolism to decrease their demand for oxygen. These metabolic alterations may include increased anaerobic glycolysis and glucose utilization, decreased whole animal metabolic rate, and decreased body temperature (32, 45, 86, 90).
While much is known about the physiological acclimatization to high-altitude hypoxia, few studies have focused on the underlying molecular genetic changes. Most studies of the effects of hypoxia on gene and protein expression have focused on responses of cells to acute hypoxia (seconds to hours of exposure). Acute responses to cellular hypoxia include changes in a number of transcription factors, such as AP-1, NF-κB, p53, Erg-1, and the Myc family of proteins. These transcription factors respond to a variety of stressors, and they are essential for regulating cell development, apoptosis, cell proliferation and differentiation, and inflammation (20, 54). Hypoxia-inducible factor (HIF) is the best understood of the transcription factors that responds to hypoxia. HIF is a heterodimer of HIF-1β and one of three tissue-specific, hypoxia-regulated dimers, HIF-1α, HIF-2α, or HIF-3α. The HIF-α subunits rapidly accumulate during hypoxic conditions, dimerize with HIF-1β, and induce the transcription of >70 hypoxia-responsive genes (40, 42, 52). Many of these genes are associated with physiological changes observed in response to hypoxia (81). For example, in response to hypoxia, HIF causes upregulation of 1) erythropoietin (EPO), which is involved in erythropoiesis (increasing red blood cell production), 2) transferrin, a key to iron transport and metabolism, 3) genes associated with angiogenesis, such as vascular endothelial growth factor (VEGF), and 4) genes associated with many metabolic substrates, such as GAPDH and pyruvate kinase, which increase glucose uptake and anaerobic glycolysis (11, 85). Indeed, a deficiency of HIF-1α, one of the HIF subunits, dramatically inhibits long-term physiological responses to hypoxia (107). Thus, the HIF pathway plays an important role in both acute responses and physiological acclimation to hypoxia. However, the relationship between acute and chronic hypoxic responses remains unexplored.
A limitation of many studies investigating genomic responses to hypoxia is that they are often conducted using in vitro cell lines. Focusing on the cellular response to hypoxia removes the cells from their normal environment of the tissue within the organism. In recent years, increased attention has been placed on the effect of hypoxia on tissues in vivo (21, 25, 47). From the few studies that have employed multiple in vivo tissues, it appears that changes in gene expression are likely to vary with how the tissues experience hypoxia and with their metabolic role within the organism (8, 36, 80, 94, 105). Whereas previous studies have focused primarily on acute or intermittent hypoxia, none have compared genome-wide gene expression of mammals acclimated to constant hypoxia for extended periods of time (i.e., weeks) with unacclimated controls.
Herein we document changes in steady-state mRNA expression in the livers of house mice acclimated to normobaric hypoxia for 32 days. Much of what we know of the cellular response to hypoxia was established with hepatocyte cell lines (4, 27, 33, 86, 102), and the liver is known to alter is gene expression profile in response to acute hypoxia in vivo (21, 52). The response of the liver to hypoxia is not surprising given that the liver plays a central role in regulating energy balance, substrate metabolism, detoxification, and a small role in erythropoiesis (23). All of these processes are affected by hypoxia, and they may be critical for acclimatizing to hypoxia. For example, acute hypoxia stimulates cells to switch from oxidative phosphorylation to anaerobic glycolysis, while perhaps decreasing fatty acid utilization. Increased gluconeogenesis performed by the liver can supply these hypoxic tissues with the glucose that they require (16) while increasing lipid storage of circulating fatty acids (13, 76). In contrast, during chronic hypoxia liver substrate preference appears to switch from glucose and anaerobic glycolysis toward fatty acid metabolism (68). Under normal circumstances, the liver produces small amounts of EPO compared with the kidney, but under extreme hypoxia, production of EPO by the liver increases substantially (75). Patients suffering from chronic obstructive pulmonary disease have decreased drug clearance ability, and this response has been linked to the decreased expression of various cytochrome P450 isoforms by the hypoxic liver (28). Finally, the liver is one of the few tissues that expresses all three HIF isoforms (HIF-1α, HIF-2α, and HIF-3α), which suggests that it is important in responding to hypoxic conditions (76). Because the liver is a key mediator of metabolic process and because it plays an important role in acclimatization to hypoxic, we focused our study on the liver (52).
MATERIALS AND METHODS
Treatments and animal handling.
We obtained 36 male mice of the inbred strain C57BL/6 from Charles River Laboratories (Hollister, CA). After acclimating to our vivarium in Reno, Nevada, for 1 wk, the 10 wk old mice were randomly assigned to one of three simulated altitude treatments. The treatments approximated the Po2 found at 1,400 m (18.0 kPa), 3,000 m (14.5 kPa), and 4,500 m (11.5 kPa). The Po2 were achieved by adding appropriate amounts of nitrogen to air using thermal mass flow controllers, and supplying this mixed air to each of three 150 l environmental chambers. Each treatment consisted of one chamber with 12 mice. Reno, NV, is located at 1,400 m above sea level, and the ambient barometric pressures averages 86.1 kPa. The 1,400 m treatment received ambient air. The 3,000 m group received air with 15.3% oxygen, and the 4,500 m group received air with 11.8% oxygen. Because animals inside the environmental chamber consume oxygen, the concentration of oxygen leaving the environmental chambers (i.e., the excurrent concentration) will be lower than the concentration entering the chamber (incurrent concentration). The faster the flow rate through the chamber is, the lower the oxygen concentration difference will be between the inlet and outlet air. Hence, faster flow rates are good for minimizing the oxygen concentration difference, but they also require more nitrogen and more frequent changing of the gas cylinders providing the nitrogen. As a compromise between these two constraints, we calculated the flow rate necessary to keep the incurrent and excurrent concentration within 1% O2 using the formula V̇o2 = FRi (O2Ci − Oe)/(1 − O2Ce), where V̇o2 = oxygen consumed, FR = flow rate, O2C = fractional oxygen concentration (e.g., O2C for ambient air ≈0.2095), the subscripts i and e stand for incurrent and excurrent, respectively (106). To be conservative, we estimated total V̇o2 in the chamber as 24 times the average daily oxygen consumption reported by Jackson Laboratories (Bar Harbor, Maine, http://www.jax.org) for adult male C57BL/6 mice. That is, we calculated the flow rate needed to keep the incurrent and excurrent oxygen concentrations within 1% O2 even if the V̇o2 of the 12 mice per chamber was double the normal V̇o2. Based on this calculation, we estimated that a flow rate of ∼2.5 l/min was needed, although the flow delivered to each chamber ranged from 2.5 to 2.8 l/min. Periodic checks over the course of the experiment indicated that the average oxygen concentrations of excurrent air were 20.2, 14.5, and 11.2% for the 1,400, 3,000, and 4,500 m treatments, respectively. These values compare with average incurrent concentrations of 20.9, 15.3, and 11.8%, respectively. We acknowledge that imperfect mixing may have resulted in some variation in Po2 within each chamber and that the slight pressure differential needed to ensure airflow through each chamber may have resulted in achieved Po2s that were slightly different than those that would be calculated from our data on oxygen concentration and ambient barometric pressure.
Within each 150 l chamber, the 12 mice were individually housed in 30 × 12 × 8 cm standard rodent cages and provided with food and water ad libitum for 32 days. The chambers were maintained in a room with 12 h light-12 h dark cycle and at ∼21°C (i.e., room temperature that reflected ambient building condition, not a precisely regulated thermal environment). Data from other related experiments indicate that temperatures within the chamber did not vary significantly from the ambient temperature of the room. The chambers were opened briefly on days 9, 17, 24, and 30 to change cages, add food and water, and weigh the mice.
After 32 days, 2 days after the final body mass measurements, we removed the mice, took two blood samples from the infraorbital sinus with microhematocrit tubes, and killed the mice via cervical dislocation. We immediately harvested the liver and preserved a small tissue sample in RNALater (Ambion). The time span from the chamber opening to tissue preservation was <30 min. Within 3 h of collecting the blood samples we centrifuged the microhematocrit tubes. Hematocrit was measured as the percentage of red blood cells in the column of blood. The two samples for each mouse were averaged before subsequent analyses were performed. Variations in body mass and hematocrit were analyzed with a one-way ANOVA followed by the Ryan-Einot-Gabriel-Welsch multiple-range test for pair-wise tests among the means of the three groups (104). All procedures outlined above were approved by the University of Nevada, Reno Institutional Animal Care and Use Committee.
Liver samples were stored at −80°C until the time of RNA purification. After homogenizing 30 mg of liver tissue using a liquid nitrogen cooled hammer and QiaShredder vials (Qiagen,), we used a Qiagen RNA Extraction Minikit (Qiagen) to purify RNA from the tissue samples following the procedures outlined in the Qiagen RNA Extraction Minikit instruction manual. The quality and quantity of the RNA were evaluated using an ABI Bioanalyzer (ABI Systems), and all samples were found to be of acceptable quality for microarray hybridization.
Microarray experiment.
For the purpose of minimizing individual variation, RNA samples obtained from mice were pooled. Each pooled sample contained RNA from four mice. With 12 mice in each treatment, this design resulted in three microarrays per treatment, and a total of nine microarrays in the experiment. Because of the documented reliability of Affymetrix products, we used the Affymetrix GeneChip Mouse Genome 430 2.0 Array (22, 57). The RNA sample preparations and microarray hybridization procedures were carried out by the University of Nevada Genomics Center according to the procedures set forth by Affymetrix. Aside from the microarray platform used, these procedures are identical to that described in Grimplet et al. (38).
GeneChip data processing and analysis.
The Affymetrix GeneChip arrays were first inspected using a series of quality control steps. Images of all arrays were examined, and no obvious scratches or areas of spatial variation were observed. A visual inspection of the distributions of raw perfect match (PM) probe values for the nine arrays showed one outlying array (the third replicate of the 3,000 m experiment). Digestion curves describing trends in RNA degradation between the 5′ end and the 3′ end of each probe set were examined, showing a notable outlying trend in the same outlying array, with irregular degradation between the sixth and eleventh probe pairs. The three Affymetrix quality control metrics for noise and background of this array were greater than two standard deviations away from the mean rates across all nine arrays, which led us to discard this array from further study.
For the 22,226 probe sets that were detected in at least one of the eight remaining arrays, raw intensity values were processed and normalized by RMA (robust multiarray average) (48). Specifically, expression values were computed from raw CEL files by first applying the RMA model of probe-specific correction of PM probes. These corrected probe values were then normalized via quantile normalization, and a median polish was applied to compute one expression measure from all probe values. Resulting RMA expression values were log2-transformed. The RMA expression values and raw microarray data have been submitted to the Gene Expression Omnibus database (series no. GSE15891).
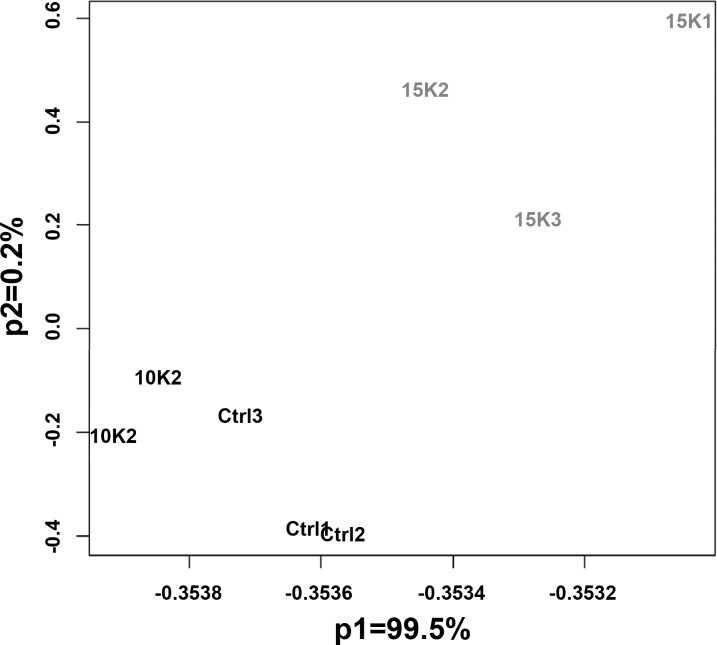
Distributions of expression values processed via RMA of the eight arrays were similar with no apparent outlying arrays. Pearson correlation coefficients and Spearman rank coefficients were computed on the RMA expression values for each set of biological replicates, with all coefficients ranging between 0.992 and 0.997. A principal components analysis showed a clear separation between the three experimental conditions, with >99% of the variability of the experiment attributed to differences between expression measures of the highest elevation and the two lower elevations (Fig. 1).
Fig. 1.
Principal component analysis of the 8 Affymetrix arrays. Note that 99.5% of the experimental variation is associated with the first principal component, and it indicates the separation between the highest altitude group and the other 2 groups.
To determine whether genes were differentially expressed between the three simulated altitudes, an ANOVA was performed on the RMA expression values. Data were fit to a simple linear model for a one-way ANOVA, ANOVA was performed on each probe set using the linear model, and contrasts were based on differences between the three experimental conditions (57). A multiple testing correction method [false discovery rate (FDR)] was applied to the P values of the F-statistic (6). Genes identified with an adjusted P value P < 0.05 were extracted for further inspection and analysis. The z-scored values (expression values normalized across all eight arrays) of these probe sets were clustered using a simple hierarchical clustering procedure with the Pearson correlation coefficient as distance metric and the average agglomerative method.
The genes associated to the resulting list of 580 differentially expressed probe sets were compared with previous studies on hypoxia gene expression, and a list of genes common to previous studies was compiled. Gene annotation was gathered from the Affymetrix NetAffx Analysis Center (http://www.affymetrix.com/analysis/index.affx). Genes were categorized by their biological functions as classified by Gene Ontology (GO) (3), and these categories were analyzed using a series of Fisher's exact tests to determine whether functional categories of the selected genes were over- or underrepresented. GO functional groups were also compared with other investigations in hypoxia induced gene expression. Gene Set Enrichment Analysis (GSEA) (63) was performed as recommended by the GSEA User's Guide (http://www.broad.mit.edu/gsea/doc/GSEAUserGuideFrame.html) on the pathways including at least 15 probe sets from the significant gene list (Supplementary Table S11). Pathways were defined by the Broad Institute's Molecular Signatures Database C2 collection of curated gene sets from online pathway databases, PubMed, and domain experts. One thousand trials were performed, in which gene set labels were randomly permuted to estimate P values for each pathway. The P values were adjusted using the FDR method, where the FDR is the estimated probability that a gene set represents a false positive finding. In the case of small sample sizes, the recommended FDR threshold is 5%. Thus, we considered only those pathways with an FDR of at most 0.05.
Confirmatory real-time qRT-PCR.
We selected six genes showing significant differential expression between simulated altitudes to verify the microarray experiment. β-Actin (NM_007393) was chosen as our endogenous control gene. Previously published primer pairs for each of these seven genes were selected from Primerbank (Table 1). The efficacy of each primer pair was verified prior to performing the qRT-PCR experiment. The source RNA used for amplification was derived from the same samples used for microarray hybridization. Aliquots of the same 36 RNA samples were pooled in an identical manner to that used for microarray hybridization such that we had nine samples representing the three hypoxia treatments. These nine pooled samples were then reverse transcribed into cDNA using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA).
Table 1.
Selected genes and primers used for qRT-PCR
| Ref Sequence ID | Common Gene Name | Forward Primer Sequence | Reverse Primer Sequence |
|---|---|---|---|
| NM_007468 | apolipoprotein A-IV | CAACAGGCTGAAGGCTACGAT | CGATTTTTGCGGAGACCTTGG |
| NM_010704 | leptin receptor | GTCTTCGGGGTTGTGAATGTC | ACCTAAGGGTGGATCGGGTTT |
| NM_009063 | regulator of G protein signaling 5 | ATGGATTTGCCAGCTTCAAAAGT | GAAGTGGTCAATGTTCACCTCTT |
| NM_008218 | hemoglobin-α | CACCACCAAGACCTACTTTCC | CAGTGGCTCAGGAGCTTGA |
| NM_024406 | fatty acid binding protein 4 | AAGGTGAAGAGCATCATAACCCT | TCACGCCTTTCATAACACATTCC |
| NM_007822 | cytochrome P450 4,a,14 | GTCTCTCGGGGAGCAATATACG | ACCAATCCAGGGAGCAAAGAA |
| NM_007393 | β-actin | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
For each gene, qRT-PCR was performed in duplicate on each cDNA sample. Genes were amplified using 1 ng of total cDNA, 150 nM of each primer (forward and reverse), and the iTaq SYBR Green Supermix with ROX reagents per manufacturer's direction (Bio-Rad). PCR plates were designed such that two genes and the endogenous control gene (β-actin) along with the corresponding standard curves were run on each plate. Amplifications were performed using an ABI 7000 PRISM Sequence Detection System (Applied Biosystems, Foster City, CA) through 40 cycles 95°C for 3 s and 58°C for 30 s. We analyzed the data using comparative cycle threshold method (ΔΔCT method). The ΔCT was calculated as CT target − CT β-actin. The ΔΔCT was ΔCT 4,500 − ΔCT 1,400 for each gene. The fold difference was expressed as 2−ΔΔCT (60).
RESULTS
Physiological responses.
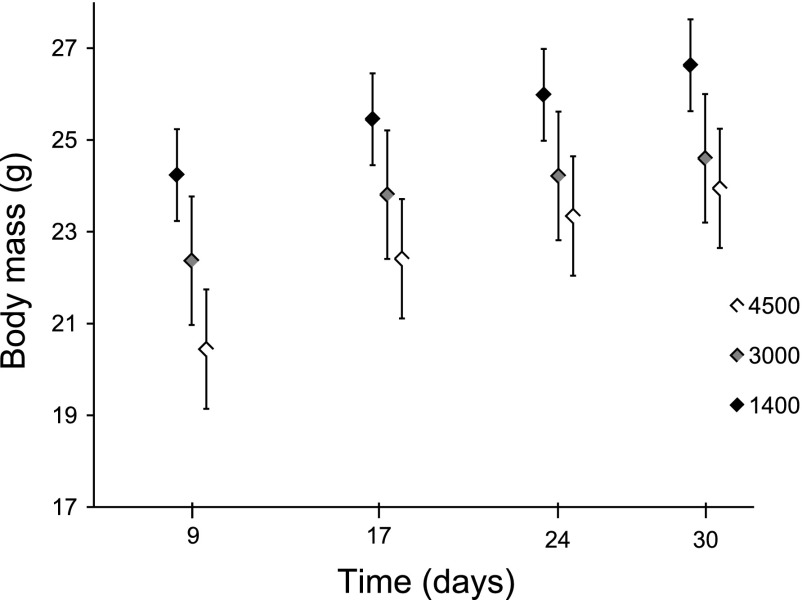
After 4 wk of exposure to hypoxia, the body masses of mice differed significantly (P < 0.001) among the three treatments. Mice from the two most hypoxic treatments (3,000 and 4,500 m) were significantly (P < 0.05) smaller in mass than mice from the 1,400 m treatment. This difference appeared to have occurred during the first week of exposure to hypoxia. Data for the initial starting masses are unavailable. However, because these mice are from an inbred strain and because they were randomly assigned to treatments, it is unlikely that the starting masses were significantly different. As illustrated in Fig. 2, variation in body mass was established within the first week by either a loss of body mass or a slower growth rate of hypoxic mice. The growth rates thereafter appear to be comparable among the treatments.
Fig. 2.
Mean body mass of mice over 30 days of residence in hypoxia chambers simulating 3 altitudes. By day 9, 4,500 m mice were significantly smaller (20.38 ± 1.20 g) than 3,000 m mice (22.37 ± 1.35 g), and both 4,500 m and 3,000 m mice were significantly smaller than 1,400 m mice (24.24 ± 1.35 g) (P < 0.001). This pattern remained until the end of the experiment, but the differences between the 3,000 and 4,500 m experiments were no longer significant. After 30 days, mice in the 4,500 m (23.9 ± 0.91 g) and 3,000 m (24.6 ± 1.40 g) chambers were significantly smaller than mice in the 1,400 m chamber (26.6 ± 1.34 g) (P < 0.001).
Whether the body mass of the 3,000 and 4,500 m mice differed depended on the inclusion of two potential outlier mice. During the course of the experiment, two of the mice that were in the 4,500 m chamber were much smaller (>3.5 g) than the other mice in that group. If those mice are excluded from the body mass analyses, then the mice in the 3,000 and 4,500 m chambers are not significantly different from each other (mean masses of 24.6 and 23.9 g, respectively), but both those groups of mice are significantly different in mass (P < 0.05) from the 1,400 m group (mean mass of 26.6 g, Fig. 2). If the two unusually small mice (16.9 and 19.1 g) from the 4,500 m are included in the analyses, the residuals from the analysis of variance deviate significantly from a normal distribution, which might cast doubt on that analysis. However, the nominal P values from the analyses including all 36 mice suggest that the body mass of all three groups differed significantly (P < 0.05) from one another. Figure 2 depicts the mean body mass over the 4 wk with the two outlier mice excluded from the data set.
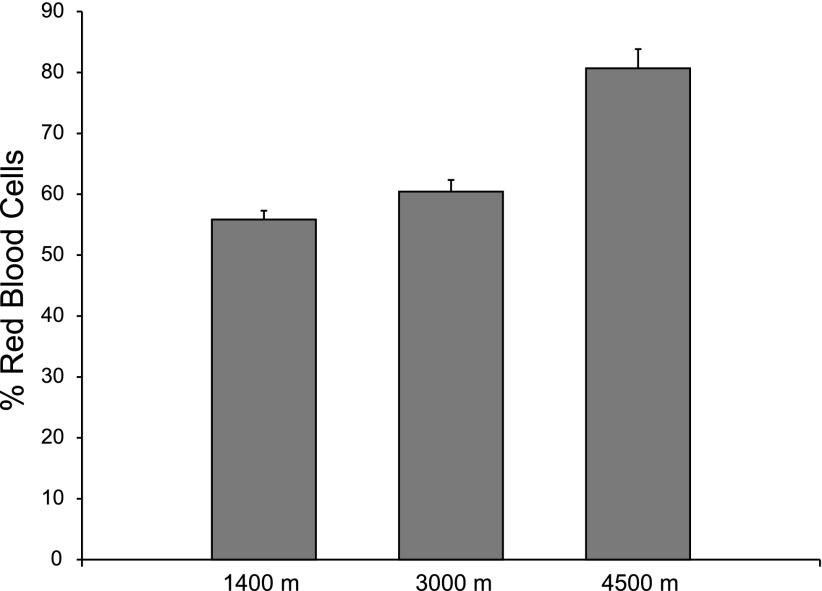
Hematocrit differed significantly (P < 0.001) among the three treatments. Mice housed in the 1,400 m chamber had a mean hematocrit of 56.2% red blood cells (RBC), the 3,000 m treatment had a mean hematocrit of 60.6% RBC, and mice in the 4,500 m treatment had an incredibly high mean hematocrit of 80.7% RBC (Fig. 3).
Fig. 3.
Mean hematocrits (Hct) of mice after 32 days in hypoxia chambers. All Hct are significantly different (P < 0.001) with the 4,500 m mice having a mean hematocrit of 80.9% RBC (± 3.4%), the 3,000 m group having a mean Hct of 60.6% RBC (± 1.9%), and the 1,400 m group having a Hct of 56.1% (± 1.4%). RBC, red blood cell.
Microarray results.
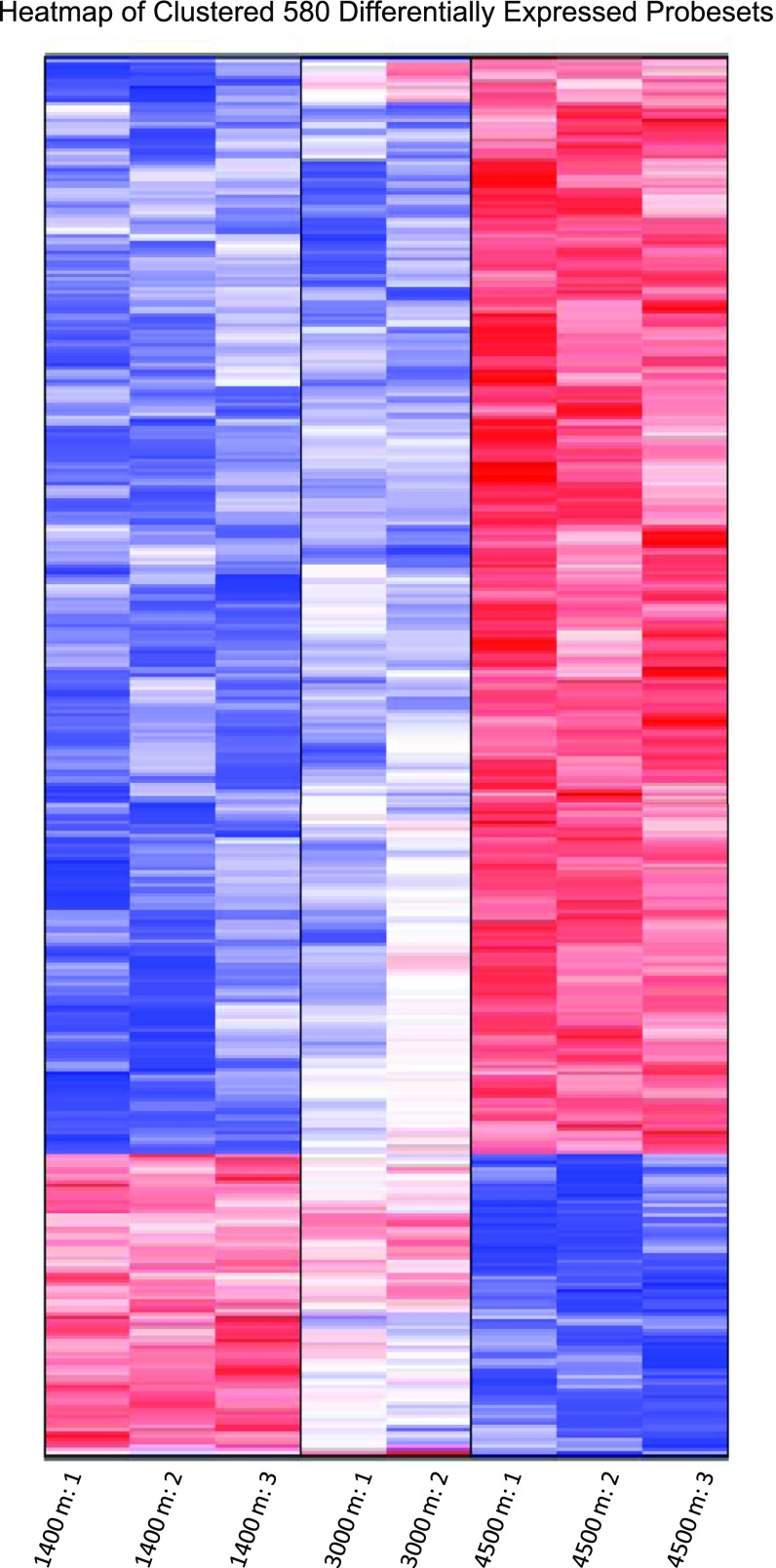
Of the 45,101 probe sets on the array, 22,226 probe sets were detected in at least one of the eight useable arrays. Upon the application of ANOVA and the control of the FDR, 580 probe sets showing significant (P < 0.05 after multiple testing correction) differential expression across the three experimental conditions. Of these probe sets, 455 showed an increase in expression in the 4,500 m treatment compared with the 1,400 m treatment, with the remaining 125 having a decrease in expression in 4,500 m treatment vs. the 1,400 m treatment. A heat map representation of gene expression patterns revealed a trend of distinct differences between the high and low elevation, with ∼85% of genes showing intermediate transcript abundances in the 3,000 m group (Fig. 4). Because of this trend, we focused further investigation on the comparison between 1,400 and 4,500 m groups.
Fig. 4.
Heat map of 580 differentially expressed probe sets. Values are normalized across the 8 arrays and clustered via a simple hierarchical clustering procedure.
To determine whether a specific functional group was overrepresented in the set of 580 probe sets that showed significant differences in expression, a series of Fisher's exact tests was performed on 21 GO functional categories, each of which contained at least five probe sets. After a multiple testing correction, only one category other than the “Undefined Function” category showed a significant difference (decrease) in the percentage of occurrence of functionally assigned probe sets among all probe sets in the analysis (n = 14,107), and those in our list of interest (n = 389). This category, defined as “transcription,” indicated that genes involved in transcription are underrepresented in hypoxic mice (4,500 vs. 1,400 m). Other than this group, the lack of under- or overrepresentation of a particular functional group may be due to the small number of probe sets with significant differences in expression.
Independent of the GO functional analysis above, we compiled a list of biological functional groups with more than five representative probe sets that show significant differential expression between the 1,400 and 4,500 m treatments. These functional groups included those associated with the immune response, angiogenesis and blood vessel development, oxygen transport, oxidation reduction (electron transport), glycolysis, protein amino acid phosphorylation, carbohydrate metabolic process, fatty acid and lipid metabolic processes, and other metabolic processes (Table 2). While the representations of these functional groups were not statistically significant in our selected subset of interest, many of these functional groups have previously been reported to be important to responses to hypoxia.
Table 2.
Genes differentially expressed by chronic hypoxia arranged in functional classes
| Functional Class | Common Gene Name | Probe Set ID | Log2-fold Change | P Value |
|---|---|---|---|---|
| Immune System* | ||||
| apolipoprotein A-IV | 1417761_at | 1.801 | 0.012 | |
| apolipoprotein A-IV | 1436504_x_at | 1.415 | 0.005 | |
| ATP-binding cassette, subfamily C (CFTR/MRP), member 9 | 1420408_a_at | 1.252 | 0.040 | |
| vascular cell adhesion molecule 1 | 1448162_at | 1.247 | 0.010 | |
| CD24a antigen | 1448182_a_at | 0.994 | 0.032 | |
| CD38 antigen | 1450136_at | 0.980 | 0.020 | |
| serine (or cysteine) peptidase inhibitor, clade A, member 3G | 1424923_at | 0.861 | 0.012 | |
| C-type lectin domain family 4, member n | 1425951_a_at | 0.833 | 0.011 | |
| CD68 antigen | 1449164_at | 0.833 | 0.019 | |
| eosinophil-associated, ribonuclease A family, member 1 | 1422411_s_at | 0.794 | 0.002 | |
| cytotoxic T lymphocyte-associated protein 2α | 1448471_a_at | 0.779 | 0.011 | |
| defensin-β1 | 1419491_at | 0.751 | 0.019 | |
| CD55 antigen | 1460242_at | 0.711 | 0.010 | |
| CD163 antigen | 1419144_at | 0.706 | 0.035 | |
| interleukin 10 receptor, β | 1419455_at | 0.704 | 0.004 | |
| CD24a antigen | 1416034_at | 0.698 | 0.018 | |
| BCL2/adenovirus E1B interacting protein 3-like | 1448525_a_at | 0.695 | 0.032 | |
| Fc receptor, IgG, low affinity III | 1448620_at | 0.692 | 0.011 | |
| C-type lectin domain family 4, member n | 1419627_s_at | 0.684 | 0.013 | |
| interleukin 13 receptor, α1 | 1427164_at | 0.662 | 0.015 | |
| collectin subfamily member 10 | 1457871_at | 0.661 | 0.009 | |
| CD48 antigen | 1427301_at | 0.558 | 0.023 | |
| leukocyte immunoglobulin-like receptor, subfamily B, member 4 | 1460258_at | 0.558 | 0.011 | |
| interleukin 6 signal transducer | 1460295_s_at | 0.557 | 0.019 | |
| eosinophil-associated, ribonuclease A family, member 2 | 1449846_at | 0.551 | 0.035 | |
| attractin | 1421166_at | 0.517 | 0.034 | |
| serine (or cysteine) peptidase inhibitor, clade A, member 3K | 1424923_at | 0.501 | 0.035 | |
| interleukin 6 signal transducer | 1452843_at | 0.480 | 0.028 | |
| ATP-binding cassette, subfamily C (CFTR/MRP), member 9 | 1435751_at | 0.409 | 0.009 | |
| Angiogenesis and Blood Vessel Development† | ||||
| heme oxygenase (decycling) 1 | 1448239_at | 1.959 | 0.022 | |
| connective tissue growth factor | 1416953_at | 1.385 | 0.013 | |
| ELK3, member of ETS oncogene family | 1448797_at | 0.774 | 0.026 | |
| forkhead box O1 | 1416981_at | 0.615 | 0.046 | |
| vascular endothelial growth factor B | 1451803_a_at | 0.606 | 0.024 | |
| endomucin | 1425582_a_at | 0.600 | 0.020 | |
| EGF-like domain 7 | 1451428_x_at | 0.573 | 0.018 | |
| platelet-derived growth factor, α | 1418711_at | 0.571 | 0.031 | |
| matrix metallopeptidase 14 (membrane-inserted) | 1448383_at | 0.515 | 0.021 | |
| endoglin | 1432176_a_at | 0.493 | 0.050 | |
| reticulon 4 | 1421116_a_at | 0.467 | 0.027 | |
| cadherin 5 | 1433956_at | 0.416 | 0.035 | |
| Electron Transport | ||||
| cytochrome P450, family 4, subfamily a, polypeptide 14 | 1423257_at | 1.970 | 0.009 | |
| cytochrome P450, family 17, subfamily a, polypeptide 1 | 1417017_at | 1.800 | 0.005 | |
| cytochrome P450, family 39, subfamily a, polypeptide 1 | 1418780_at | 1.576 | 0.002 | |
| flavin containing monooxygenase 2 | 1453435_a_at | 1.485 | 0.006 | |
| flavin containing monooxygenase 2 | 1435459_at | 1.449 | 0.007 | |
| P450 (cytochrome) oxidoreductase | 1416933_at | 0.808 | 0.012 | |
| STEAP family member 4 | 1425829_a_at | 0.672 | 0.039 | |
| cytochrome P450, family 4, subfamily a, polypeptide 10 | 1424853_s_at | 0.648 | 0.019 | |
| ERO1-like (S. cerevisiae) | 1419030_at | 0.647 | 0.040 | |
| cytochrome b-245, β-polypeptide | 1422978_at | 0.641 | 0.043 | |
| glutaredoxin | 1416593_at | 0.617 | 0.009 | |
| cytochrome P450, family 2, subfamily c, polypeptide 38 | 1452501_at | 0.569 | 0.012 | |
| cathepsin B | 1417491_at | 0.562 | 0.022 | |
| glutaredoxin | 1416592_at | 0.499 | 0.022 | |
| Oxygen Transport | ||||
| hemoglobin-α, adult chain 1 | 1417714_x_at | 1.865 | 0.000 | |
| hemoglobin-α, adult chain 1 | 1428361_x_at | 0.641 | 0.019 | |
| hemoglobin, β-adult major chain | 1417184_s_at | 0.593 | 0.015 | |
| hemoglobin-α, adult chain 1 | 1452757_s_at | 0.536 | 0.020 | |
| Protein Amino Acid Phosphorylation | ||||
| wee 1 homolog (S. pombe) | 1416774_at | 1.426 | 0.025 | |
| Rap guanine nucleotide exchange factor (GEF) 4 | 1421622_a_at | 1.159 | 0.015 | |
| wee 1 homolog (S. pombe) | 1416773_at | 0.818 | 0.015 | |
| endothelial-specific receptor tyrosine kinase | 1418788_at | 0.745 | 0.013 | |
| serum/glucocorticoid-regulated kinase 3 | 1420918_at | 0.739 | 0.031 | |
| protein kinase C, η | 1422079_at | 0.655 | 0.033 | |
| transient receptor potential cation channel, subfamily M, member 7 | 1416800_at | 0.639 | 0.038 | |
| serine/threonine kinase 3 (Ste20, yeast homolog) | 1418513_at | 0.523 | 0.034 | |
| cyclin-dependent kinase 4 | 1422440_at | 0.506 | 0.035 | |
| tyrosine kinase receptor 1 | 1416238_at | 0.502 | 0.026 | |
| protein kinase N2 | 1437296_at | 0.484 | 0.033 | |
| receptor (TNFRSF)-interacting serine-threonine kinase 1 | 1419508_at | 0.392 | 0.040 | |
| Metabolic Processes Glycolysis | ||||
| pyruvate kinase, muscle | 1417308_at | 0.750 | 0.016 | |
| 2,3-bisphosphoglycerate mutase | 1448119_at | 0.731 | 0.016 | |
| 2,3-bisphosphoglycerate mutase | 1415864_at | 0.629 | 0.016 | |
| aldolase 1, A isoform /// | 1439375_x_at | 0.494 | 0.041 | |
| aldolase 1, A isoform | 1434799_x_at | 0.482 | 0.024 | |
| pyruvate dehydrogenase E1 α1 | 1449137_at | 0.463 | 0.040 | |
| Metabolism‡ | ||||
| leptin receptor | 1456156_at | 3.051 | 0.002 | |
| leptin receptor | 1425644_at | 2.934 | 0.001 | |
| CD38 antigen | 1450136_at | 0.980 | 0.002 | |
| ATPase, class VI, type 11C | 1442367_at | 0.935 | 0.006 | |
| ATPase, class VI, type 11C | 1442367_at | 0.935 | 0.006 | |
| ectonucleotide pyrophosphatase/phosphodiesterase 1 | 1419276_at | 0.838 | 0.030 | |
| leptin receptor | 1425875_a_at | 0.807 | 0.013 | |
| RIKEN cDNA A230097K15 gene | 1454799_at | 0.676 | 0.032 | |
| carbonyl reductase 1 | 1460196_at | 0.656 | 0.009 | |
| glutathione S-transferase, mu 3 | 1427473_at | 0.633 | 0.013 | |
| retinol dehydrogenase 9 | 1427963_s_at | 0.624 | 0.022 | |
| phosphoribosyl pyrophosphate amidotransferase | 1452831_s_at | 0.589 | 0.018 | |
| arsenic (+3 oxidation state) methyltransferase | 1431980_a_at | 0.548 | 0.013 | |
| dehydrogenase/reductase (SDR family) member 7 | 1426440_at | 0.495 | 0.017 | |
| NAD kinase | 1416249_at | 0.455 | 0.029 | |
| RIKEN cDNA A530057A03 gene | 1456208_at | 0.453 | 0.043 | |
| Carbohydrate and Glucose Metabolism§ | ||||
| 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | 1416432_at | 1.673 | 0.021 | |
| glucokinase | 1419146_a_at | 0.979 | 0.009 | |
| glucokinase | 1419146_a_at | 0.979 | 0.009 | |
| glucokinase | 1425303_at | 0.849 | 0.010 | |
| glucokinase | 1425303_at | 0.849 | 0.010 | |
| preimplantation protein 4 | 1429639_at | 0.784 | 0.046 | |
| protein phosphatase 1, regulatory (inhibitor) subunit 2 | 1417341_a_at | 0.624 | 0.024 | |
| preimplantation protein 4 | 1429144_at | 0.566 | 0.028 | |
| pyruvate dehydrogenase kinase, isoenzyme 1 | 1423748_at | 0.509 | 0.037 | |
| membrane interacting protein of RGS16 | 1418444_a_at | 0.489 | 0.035 | |
| N-acetylneuraminate pyruvate lyase | 1424265_at | 0.340 | 0.039 | |
| Fatty Acid and Lipid Metabolic Processa | ||||
| cytochrome P450, family 39, subfamily a, polypeptide 1 | 1418780_at | 1.576 | 0.002 | |
| lipin 1 | 1426516_a_at | 1.280 | 0.009 | |
| lipin 1 | 1418288_at | 1.262 | 0.007 | |
| very low density lipoprotein receptor | 1434465_x_at | 1.134 | 0.007 | |
| N-acylsphingosine amidohydrolase 3-like | 1451355_at | 0.994 | 0.003 | |
| ELOVL family member 6, elongation of long chain fatty acids (yeast) | 1417404_at | 0.967 | 0.016 | |
| ELOVL family member 6, elongation of long chain fatty acids (yeast) | 1417403_at | 0.844 | 0.024 | |
| acyl-CoA synthetase long-chain family member 4 | 1427595_at | 0.807 | 0.031 | |
| cytochrome P450, family 4, subfamily a, polypeptide 10 | 1424853_s_at | 0.648 | 0.019 | |
| platelet-activating factor acetylhydrolase, isoform 1b, α2 subunit | 1422793_at | 0.632 | 0.026 | |
| acetyl-coenzyme A carboxylase-α | 1427595_at | 0.450 | 0.045 |
Containing Gene Ontology (GO) Biological Functional designations immune response, inflammatory response, acute phase response, defense response, chemotaxis.
Containing GO Biological Functional designations angiogenesis, blood vessel development, blood vessel maturation.
Containing GO Biological Functional designations metabolic process, regulation of metabolic process, one-carbon metabolic process).
Containing GO Biological Functional designations carbohydrate metabolism, glucose metabolic process.
Containing GO Biological Functional terms for fatty acid metabolism or lipid metabolism.
In addition to using GO term-based functional analyses, we used GSEA to investigate the over- and underrepresentation of functional groups within the genes of interest (95). GSEA is a method that tests whether gene sets, rather than individual genes or probe sets, are associated with experimental conditions or classes in a given study. Definitions of gene sets are based on published and curated information on biochemical pathways. The GSEA method yielded several pathways having a statically significant (FDR value of <0.05) association with the upregulation in the 4,500 m group vs. the 1,400 m group. Of particular interest was the Hematopoietic Stem Cell and Adult Progenitors gene set, which is a set of genes found to be expressed during hematopoietic stem cell and hematopoietic progenitor cell development (49). The list of genes belonging to this gene set show increased expression in the 4,500 m treatment and is presented in Table 3.
Table 3.
List of genes belonging to the Hematopoietic Stem Cell and Progenitors gene set that show increased expression in 4,500 m treatment
| Common Gene Name | Probe Set ID | Log2-fold Change | Adjusted P Value |
|---|---|---|---|
| Ubiquitin-specific peptidase 2 | 1417168_a_at | 1.643 | 0.013 |
| Regulator of G protein signaling 3 | 1425701_a_at | 1.509 | 0.019 |
| C-type lectin domain family 14, member a | 1419467_at | 1.273 | 0.001 |
| Coiled-coil domain containing 85B | 1435589_at | 1.112 | 0.002 |
| Rap guanine nucleotide exchange factor (GEF) 5 | 1455137_at | 1.110 | 0.009 |
| EGF, latrophilin seven transmembrane domain containing 1 | 1418059_at | 1.095 | 0.002 |
| Peptidylprolyl isomerase C | 1416498_at | 0.983 | 0.005 |
| ELOVL family member 6, elongation of long chain fatty acids (yeast) | 1417403_at | 0.967 | 0.016 |
| Ankyrin repeat domain 37 | 1436538_at | 0.948 | 0.007 |
| Rho-related BTB domain containing 1 | 1429206_at | 0.921 | 0.019 |
| Tissue factor pathway inhibitor | 1452432_at | 0.842 | 0.020 |
| Ectonucleotide pyrophosphatase/phosphodiesterase 1 | 1419276_at | 0.838 | 0.030 |
| FYVE, RhoGEF and PH domain containing 5 | 1460578_at | 0.760 | 0.040 |
| Endothelial-specific receptor tyrosine kinase | 1418788_at | 0.745 | 0.013 |
| Cyclin D2 | 1434745_at | 0.693 | 0.010 |
| Programmed cell death 6 interacting protein | 1426184_a_at | 0.692 | 0.021 |
| Coiled-coil domain containing 80 | 1424186_at | 0.674 | 0.009 |
| Protein kinase C, η | 1422079_at | 0.655 | 0.033 |
| Gap junction membrane channel protein α1 | 1438945_x_at | 0.616 | 0.048 |
| Vascular endothelial growth factor B | 1451803_a_at | 0.606 | 0.024 |
| MAM domain containing 2 | 1453152_at | 0.540 | 0.015 |
| Thymosin, β10 | 1436902_x_at | 0.521 | 0.022 |
| Dynamin 1-like | 1452638_s_at | 0.415 | 0.037 |
| N-acetylneuraminate pyruvate lyase | 1424265_at | 0.340 | 0.039 |
| Aldehyde dehydrogenase 1 family, member L1 | 1424400_a_at | −0.411 | 0.036 |
| ATP-binding cassette, subfamily B (MDR/TAP), member 11 | 1449817_at | −0.438 | 0.020 |
| Endothelial cell growth factor 1 (platelet-derived) | 1432181_s_at | −0.499 | 0.023 |
Based on previous studies of how gene expression responds to acute or cellular hypoxia, we compiled a list of genes that we suspected would be affected by chronic systemic hypoxia. Many of these genes are regulated by the HIF pathway. However, few of these genes appeared to be significantly differentially expressed in this experiment. Indeed, in our list of 580 probe sets, only seven are known to be regulated by HIF (Table 4). When we compared our data for genes not regulated by HIF with data from a study looking at effects of acute hypoxia on hepatocytes (89), we found three genes in common with two genes showing concordant responses to hypoxia and one gene being expressed in the opposite direction in response to hypoxia. Furthermore, only a handful of the 580 genes that were differentially expressed in our experiment have previously been reported to respond to hypoxia in any study (Table 5).
Table 4.
Transcripts known to be regulated by HIF-1α that show significant differences in abundance between 1,400 and 4,500 m treatments after 32 days
| Common Name | Probe Set ID | Log2-fold Change | P Value |
|---|---|---|---|
| 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | 1416432_at | 1.67 | 0.021 |
| Heme oxygenase (decycling) 1 | 1448239_at | 0.97 | 0.022 |
| Lectin, galactose binding, soluble 3 | 1426808_at | 0.83 | 0.015 |
| Pyruvate kinase, muscle | 1417308_at | 0.75 | 0.016 |
| Insulin-like growth factor binding protein 3 | 1458268_s_at | 0.54 | 0.029 |
| Aldolase 1, A isoform | 1434799_x_at | 0.48 | 0.024 |
| Presenilin 2 | 1425869_a_at | −0.68 | 0.009 |
HIF, hypoxia-inducible factor.
Table 5.
Transcripts showing differential expression in response to chronic hypoxia that have previously been reported to respond to hypoxic stimulus
| Common Gene Name | Probe Set ID | Log2-fold Change | P Value | Reference No. |
|---|---|---|---|---|
| Cytochrome P450, family 17, subfamily a, polypeptide 1 | 1417017_at | 1.800 | 0.005 | (56) |
| CD24a antigen | 1416034_at | 0.994 | 0.011 | (83) |
| Heat shock protein 110 | 1423566_a_at | 0.772 | 0.009 | (89) |
| CD24a antigen | 1448182_a_at | 0.70 | 0.019 | (82) |
| Superoxide dismutase 3, extracellular | 1417633_at | 0.657 | 0.012 | (96) |
| ATPase, Na+/K+ transporting, β1 polypeptide | 1439036_a_at | 0.585 | 0.033 | (53) |
| Caveolin, caveolae protein 1 | 1449145_a_at | 0.541 | 0.036 | (82) |
| Pyruvate dehydrogenase kinase, isoenzyme 1 | 1423748_at | 0.509 | 0.037 | (89) |
| Caspase 1 | 1449265_at | 0.450 | 0.027 | (110) |
| Protein phosphatase 1, regulatory (inhibitor) subunit 3C | 1425631_at | −0.937 | 0.044 | (89) |
| Protein phosphatase 1, regulatory (inhibitor) subunit 3C | 1433691_at | −1.096 | 0.013 | (89) |
Several genes unique to hypoxia studies were expressed >1.5-log2-fold when comparing 4,500 and 1,400 m simulated altitudes. A probe set representing leptin receptor showed the most dramatic difference in expression with a log2-fold increase of 3.05 in 4,500 m mice compared with 1,400 m mice. Two other probe sets representing the leptin receptor also showed significant differences in expression with log2-fold changes of 2.93 and 0.807, respectively. Consistent with the hematocrit results, a transcript representing α-globin showed a 1.97-log2-fold increase in expression in the 4,500 m vs. the 1,400 m mice. Two other probe sets representing α-globin and one probe set representing β-globin also showed significant differences in gene expression, though none over 1.5-log2-fold. Several genes encoding three families of cytochrome p450 and several genes involved in the G protein-coupled receptor protein signaling pathway were among the genes with >1.5-log2-fold changes (Table 6).
Table 6.
List of genes showing >1.5-log2-fold increased transcript abundance in response to chronic hypoxia
| Common Name | Probe Set ID | Log2-fold Change | Adjusted P Value |
|---|---|---|---|
| Leptin receptor | 1456156_at | 3.05 | 0.002 |
| Leptin receptor | 1425644_at | 2.93 | 0.001 |
| Regulator of G protein signaling 16 | 1426037_a_at | 2.26 | 0.024 |
| Regulator of G protein signaling 5 | 1420941_at | 2.24 | 0.000 |
| Fatty acid binding protein 4, adipocyte | 1417023_a_at | 2.21 | 0.000 |
| G protein-coupled receptor 98 | 1425314_at | 2.19 | 0.000 |
| Regulator of G protein signaling 16 | 1451452_a_at | 2.07 | 0.027 |
| — | 1417466_at | 2.01 | 0.000 |
| Cytochrome P450, family 4, subfamily a, polypeptide 14 | 1423257_at | 1.97 | 0.009 |
| Hemoglobin-α, adult chain 1 | 1417714_x_at | 1.87 | 0.000 |
| Fatty acid binding protein 4, adipocyte | 1451263_a_at | 1.86 | 0.004 |
| Synuclein-α | 1436853_a_at | 1.86 | 0.000 |
| Predicted gene, ENSMUSG00000073738 | 1439816_at | 1.84 | 0.000 |
| Regulator of G protein signaling 5 | 1420940_x_at | 1.84 | 0.000 |
| Apolipoprotein A-IV | 1417761_at | 1.80 | 0.005 |
| Cytochrome P450, family 17, subfamily a, polypeptide 1 | 1417017_at | 1.80 | 0.005 |
| ATP-binding cassette, subfamily D (ALD), member 2 | 1438431_at | 1.76 | 0.005 |
| Major facilitator superfamily domain containing 2 | 1428223_at | 1.73 | 0.019 |
| ATP-binding cassette, subfamily D (ALD), member 2 | 1419748_at | 1.71 | 0.008 |
| 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | 1416432_at | 1.67 | 0.021 |
| Ubiquitin-specific peptidase 2 | 1417168_a_at | 1.64 | 0.013 |
| Von Willebrand factor homolog | 1435386_at | 1.63 | 0.004 |
| Synuclein-α | 1418493_a_at | 1.58 | 0.010 |
| Cytochrome P450, family 39, subfamily a, polypeptide 1 | 1418780_at | 1.58 | 0.002 |
| — | 1417466_at | 1.57 | 0.000 |
| Regulator of G protein signaling 3 | 1425701_a_at | 1.51 | 0.019 |
As mentioned earlier, relatively few genes showed decreased transcript abundance in the hypoxic conditions compared with genes that showed increased transcript abundance. Table 7 lists 25 genes that had a decreased log2-fold change of >1.0 in the 4,500 m treatment vs. the 1,400 m treatment. These genes represent a diverse set of functions, without any readily discernable pattern.
Table 7.
List of genes showing >1.0-log2-fold decreased in transcript abundance in response to chronic hypoxia
| Common Gene Name | Probe Set ID | Log2-fold Change | P Value |
|---|---|---|---|
| Immunoglobulin heavy chain 6 (heavy chain of IgM) | 1427351_s_at | −1.60 | 0.028 |
| Immunoglobulin heavy chain 6 (heavy chain of IgM) | 1427329_a_at | −1.58 | 0.030 |
| Tubulin, β2a | 1427347_s_at | −1.57 | 0.009 |
| Macrophage activation 2 like | 1438676_at | −1.46 | 0.005 |
| Macrophage activation 2 like | 1447927_at | −1.44 | 0.003 |
| Oligodendrocyte transcription factor 1 | 1416149_at | −1.28 | 0.007 |
| Elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 | 1420722_at | −1.27 | 0.002 |
| UDP glucuronosyltransferase 2 family, polypeptide B38 | 1423397_at | −1.25 | 0.011 |
| Early growth response 1 | 1417065_at | −1.22 | 0.011 |
| RIKEN cDNA 2310043N10 gene | 1428083_at | −1.11 | 0.031 |
| RIKEN cDNA 1810046K07 gene | 1453547_at | −1.10 | 0.003 |
| Protein phosphatase 1, regulatory (inhibitor) subunit 3C | 1433691_at | −1.10 | 0.013 |
| Aquaporin 8 | 1417828_at | −1.10 | 0.001 |
| cDNA sequence BC015286 | 1457619_at | −1.04 | 0.011 |
| Carboxylesterase 2 | 1424245_at | −1.02 | 0.001 |
Confirmatory qRT-PCR.
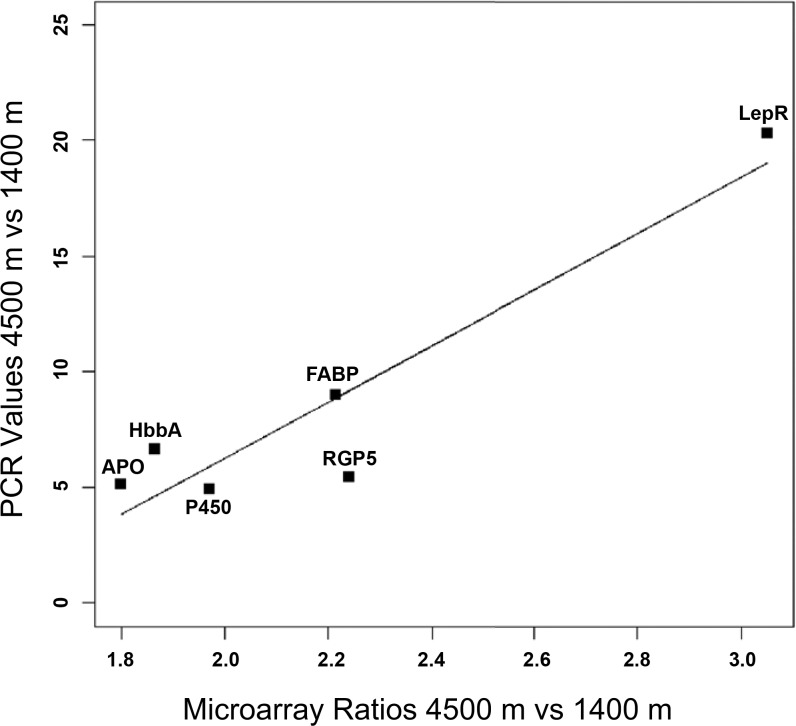
To validate the expression profiles obtained from the Affymetrix GeneChip Mouse 430 expression set array, quantitative real-time PCR was performed on six target genes and one endogenous control gene. Each of these genes was covered by at least one probe set that appeared in our list of the 20 most differentially expressed transcripts and had known biological function. We compared transcript abundance between the 4,500 and 1,400 m for all six genes. We performed a linear regression analysis to measure the relationship between the PCR data and the microarray data, which resulted in a very strong correlation of 0.935, and a P value of P = 0.006. This result indicates a high correlation and confirmation of our microarray results (Fig. 5).
Fig. 5.
Confirmatory real-time qRT-PCR of 6 transcripts. Comparison between the expression values from the Affymetrix GeneChip Mouse Expression Set 430 and expression values from quantitative real-time PCR. LepR, leptin receptor; FABP, fatty acid binding protein; RGP5, regulator of G protein signaling 5; P450, cytochrome P450 family 4, subfamily a, protein 14; HbbA, hemoglobin-α; APO, apolipoprotein.
DISCUSSION
Hypoxia is a significant physiological stressor associated with high altitude environments. Although most mammals have limited evolutionary history with hypoxic environments, mammals display various characteristic physiological responses when acclimatizing to high altitudes. This study utilized DNA microarrays 1) to help understand the gene expression changes in the liver that may underlie physiological responses following chronic exposure and 2) to identify other potential physiological effects of hypoxia that may have been overlooked previously by acute studies.
The experiment was conducted at 1,400 m, not sea level. While there are some effects of this altitude on physiology (for instance, a hematocrit of 56.2 as opposed to 52% as is average for sea level C57BL/6 mice), this elevation is not considered to be extreme. The Po2 found in Reno is not expected to elicit many of the characteristic responses to high altitude that we typically see >2,000 m (103). That being said, it is important to appreciate that the variation in gene expression we report is not a comparison of hypoxia vs. normoxia, but of varying degrees of hypoxia, from mild to extreme.
Based on the observed differences in body mass and hematocrit among our treatments, the hypoxic conditions were effective in eliciting significant physiological responses. However, the mRNA expression in the liver after 32 days of hypoxia was different from expression patterns reported in studies of acute hypoxia studies (37, 89, 99). While several previous studies reported similar numbers of genes with differential expression, most of the genes in this experiment exhibited increased transcript abundance, but most studies of acute or cellular hypoxia report a general trend of decreased transcript abundance (89). Thus, it appears that after more prolonged periods of hypoxia, relative steady-state transcript abundance changes from a trend of being reduced to a trend of being increased. This result is consistent with data collected from murine hearts (25).
Few genes known to be regulated by HIF are included among the 580 genes that were significantly differential expressed between our high- and low-altitude treatments. This result indicates that, after 32 days, the previously described stress pathways, and in particular the HIF pathway, are no longer significant factors in regulating and maintaining physiological acclimation to hypoxia. Indeed, other studies have shown that HIF-1α abundance peaks after 4–5 h in the liver then decreases back to normal over the course of several hours (94). These findings indicate that, while HIF is an important transcription factor in regulating the initial responses to acute hypoxia, it is not as important in the maintenance of acclimatization to chronic hypoxia stress, at least in the liver. There are two nonmutually exclusive explanations for this result. One is that as animals physiologically acclimate to hypoxic environments, their tissues return to a state of oxygen homeostasis and various stress responses and the HIF pathways are no longer activated. The second is that as hypoxic exposure moves from an acute to chronic state, other genetic pathways and transcription factors are activated that regulate the specific processes associated with maintaining the acclimatized state.
Few genes that are typically reported to be responsive to hypoxia showed differential expression in our study. A possible explanation for this discrepancy is that we studied an organ in vivo, whereas most other studies have been in vitro. However, the patterns of gene expression reported here are different than those reported in in vivo studies of murine livers exposed to either acute systemic or intermittent hypoxia (21, 56). Both of those studies of murine livers reported significant changes in lipid biosynthesis and sterol metabolism. While some genes related to these processes are differentially expressed in this study, there are not as many, nor are they the same genes. This finding indicates that chronic hypoxia induces different gene expression patterns in liver in vivo than either acute or intermittent hypoxia. Indeed, recent findings confirm that intermittent and chronic hypoxia stimulate different gene expression patterns in cardiac tissue (25, 47).
The most dramatic result of this study was the large increase in hematocrit in the 4,500 m mice. While modest increases in hematocrit are generally seen as being beneficial acclimations to hypoxia (10), an increase to 80.7% RBC is extreme and much greater than expected. A hematocrit this high leads to viscous blood and can put a significant strain on the heart. Consistent with this response was the increased expression of both α-globin and β-globin, the two protein subunits of hemoglobin. However, what is unclear is why these genes are expressed in the liver. There are two possible explanations for this phenomenon. One is the presence of immature erythrocytes within the liver tissue, which would express both the α-globin and β-globin mRNAs. Indeed, polycythemia at the level reported here is often accompanied by increased circulation of immature erythrocytes. However, it is unclear whether there would be sufficient numbers of immature erythrocytes in the liver to elicit this response. An alternative explanation is that portions of the liver were converted to hematopoietic tissue to support the massive production of RBCs. This hypothesis is supported by the overrepresentation of differentially expressed genes belonging to the hematopoietic stem cell gene set, as indicated by the GSEA. The phenomenon of extramedullary hematopoiesis is well documented in cases of pathological anemia, such as severe thalassemia or myelofibrosis (62, 63), and it can occur when anemia is induced artificially (77). However, there is little, if any, information on this phenomenon in relation to systemic hypoxia or exposure to high altitude environments. Therefore, the incidence of extramedullary hematopoiesis and its consequences for high-altitude acclimation have yet to be seriously considered. This result highlights the dynamic nature of the liver and the importance of in vivo studies to chronic hypoxia. The result deserves additional study. While we were unable to conduct additional confirmatory histological studies to confirm that extramedullary hematopoiesis took place in the liver, it is one possible explanation for our results.
The transcript showing the most dramatic increase in expression in the 4,500 m treatment vs. the 1,400 m treatment encoded a leptin receptor. This result is intriguing given the known responsiveness of the cytokine leptin to hypoxic stimulus. Circulating levels of leptin, a pleiotropic cytokine released by fat cells, are increased by acute hypoxia, and transcription appears to be regulated by HIF-1 (39, 61, 87, 88). However, in contrast, humans exposed to chronic high-altitude hypoxia had decreased levels of circulating leptin (108). It is possible that regulation of leptin receptor is another important mechanism of regulating the leptin pathway. Indeed, leptin receptor mRNA abundance in the liver increases in response to short-term fasting and increased circulating leptin (17).
While our results indicate that the leptin pathway may be an important response to hypoxia, the pleiotropic nature of this pathway makes it difficult to interpret the physiological significance of increased leptin receptor mRNA abundance in the liver. The leptin pathway is best known for its inhibitory effect on appetite, and an obvious hypothesis would be to link the leptin pathway with hypoxia-induced anorexia. However, despite increased expression during hypoxia, leptin does not appear to regulate hypoxia-induced anorexia (88, 98). Leptin signaling also has a significant positive effect on organismal metabolic rate and body temperature (2, 31, 59, 69, 91). In contrast, acute hypoxia typically leads to decreased metabolic rate and body temperature, at least in small mammals (12, 29, 32, 64, 65, 90). While we did not measure metabolic rate or body temperature in our mice, it seems likely that they became hypometabolic and anapyrexic when initially exposed to hypoxia (41). However, the effects of prolonged hypoxic exposure on metabolism are less obvious as there are comparatively fewer data on the effects of chronic hypoxia. Nonetheless, we hypothesize that at least in some strains and species, hypometabolism and anapyrexia may be transient responses to hypoxia (1, 67, 78). As an animal acclimates to hypoxia and attempts to regain oxygen homeostasis via increased oxygen delivery to its tissues, it seems plausible that metabolic rate would return toward normal. Perhaps leptin and its receptors help regulate this process? Furthermore, responses to hypoxia can be species specific (40, 42). For example, compared with effects on metabolic rates of rats, hamsters may be less affected by hypoxia and mice may be more affected (30, 41). Accordingly hypoxia-induced changes in gene expression may also be species specific, and as for any study of a single species of mammal, our results may not be generally applicable to other mammals. We therefore conclude that the effects of chronic hypoxia on the leptin pathway and its relationship to body temperature and metabolism is an area in need of further research.
Other physiological processes that are affected by leptin include glucose and fatty acid metabolism, hematopoiesis, and immune function (2, 31, 100). These physiological processes also appear to be important in acclimation to hypoxia. Previous studies have demonstrated that fatty acid metabolism is enhanced in rats exposed to chronic hypobaric hypoxia (68), and indeed several genes related to fatty acid metabolism were differentially expressed in our experiment. While it is unknown whether fatty acid metabolism was increased in our mice, it is possible that altering fatty acid metabolism is important for acclimating to hypoxia and that the leptin pathway plays a role in this response. Another important function of leptin is its role in hematopoiesis. Leptin receptor can be expressed by hematopoietic stem cells, and indeed, the leptin pathway is an important progenitor of hematopoiesis (7, 26). The dramatic increased expression of leptin receptor in our 4,500 m liver samples may be another indication that extramedullary hematopoiesis is taking place.
We sorted transcripts significantly affected by hypoxia into their functional classes (Table 2). These biological functional groups include transcripts involved in angiogenesis, oxidation reduction, carbohydrate metabolism, fatty acid and lipid metabolism, glycolysis, and protein amino acid phosphorylation. Many of these functional groups typically respond to acute or intermittent hypoxia, and thus these results are not surprising (37, 56, 89). However, the particular genes representing these functional groups are not typical of previous studies of hypoxia. This result indicates that these biological functions are important in acclimation to hypoxia but that the particular transcripts that are involved in adjusting to hypoxia vary temporally with duration of hypoxic exposure.
The metabolic activities of the liver, which often generate carbon dioxide, also play an important role in acid-base homeostasis. This process is important to hypoxia acclimatization in that the hypoxic ventilatory response (HVR) typically leads to respiratory alkalosis. While increased ventilation remains during long periods of acclimatization (71), the pH of the blood typically returns close to normal via metabolic and blood regulation of bicarbonate (1, 67, 103). However, because we did not measure either ventilatory rate or blood pH, it is impossible to say whether alterations in the above transcript abundances could be contributing to maintenance of acid-base homeostasis or be affected by the slightly higher blood pH typically found in mammals acclimatized to hypoxia. To add to the complexity, two transcripts that have been implicated in maintaining acid-base balance had decreased expression in the 4,500 m mice: aquaporin 8 (log2-fold change of −1.09) and carbonic anhydrase 14 (log2-fold change of −0.95) (44, 50, 58). So while we are unable to draw direct conclusions regarding the relationship between liver transcript abundance and acid-base balance in response to HVR, we encourage the reader to keep this process in mind.
In this study we found an abundance of differentially expressed genes related to the immune system. Unlike the functional classes described above, the immune system is not typically viewed as important in responding to chronic, systemic hypoxia. However, there is increasing evidence that hypoxia may alter the immune system. For example, several studies of humans in high-altitude environments indicate perturbations of the immune system, particularly with respect to changes in circulating leukocytes (5, 14, 15, 24). Numerous explanations have been proposed to explain why sojourners to high altitude experience immune changes, including changes in diet, stress, etc. (5). More recent evidence indicates that hypoxia in and of itself has an influence on the circulating immune system. In particular, hypoxia has a proinflammatory effect on macrophages, neutrophils and other leukocytes and an anti-inflammatory effect on some lymphocytes (9, 18, 55, 66, 97, 101). Furthermore, there is growing evidence that HIF is a key regulator of many immunological processes (19, 43, 109). Yet again, the particular genes regulated by HIF pathway are not differentially expressed in our study. This result is similar to that for the other biological functional groups reported above. Namely, the immune system appears to respond to both acute and chronic hypoxia, but the particular transcripts involved in the immune systems responses vary with length of exposure to hypoxia.
The liver is a dynamic organ that not only is composed of hepatocytes but also houses many immune cells, blood cells, and, perhaps in this case, hematopoietic tissue. Unfortunately, it is not possible to distinguish which responses in gene expression are attributable to which cell types or processes. This may be a major source of variation between this study and other reports of gene expression in response to hypoxia. However, this study indicates that many of the biological functional groups regulated in response to acute hypoxia are also important in maintaining acclimation to chronic systemic hypoxia. Therefore, we hypothesize that regulatory pathways, and thus the representative genes expressed from a functional group, change with time and exposure to systemic hypoxia. So, while one set of genes achieves initial acclimatization to hypoxia, different but related groups of genes are likely involved in maintaining acclimatization. Understanding these differences will be important to better understanding the physiological effects of hypoxia and acclimatization to high altitudes.
GRANTS
This work was supported by a graduate student fellowship to M. Baze from the National Science Foundation EPSCoR Integrated Approaches to Abiotic Stress program (#0132556) and a core incentive use grant from Nevada IDeA Network of Biomedical Research Excellence (Nevada INBRE). K. Schlauch and the Nevada Genomics Center are also supported by National Institutes of Health Grant Number P20 RR-016464 from the INBRE Program of the National Center for Research Resources.
DISCLOSURES
No conflicts of interest are declared by the authors.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jennifer Gworek and Debra Russell for assistance with handling the mice, and Richard Tillett for technical assistance with real-time qRT-PCR. We also thank Lee Weber, Kenneth Hunter, John Cushman, Robert Donovan, Cynthia Downs, Marta Labocha, Mike Sears, and Bernard Wone for support in all phases of this study.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Aaron EA, Powell FL. Effect of chronic hypoxia on hypoxic ventilatory response in awake rats. J Appl Physiol 74: 1635–1640, 1993. [DOI] [PubMed] [Google Scholar]
- 2. Ahima RS, Flier JS. Leptin. Annu Rev Physiol 62: 413–437, 2000. [DOI] [PubMed] [Google Scholar]
- 3. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baker M, Candinas D, Stroka DM. Activation of HIF-1 in primary human hepatocytes in response to hypoxia and cytokine stimulation. Hepatology 38: 590A–591A, 2003. [Google Scholar]
- 5. Basnyat B, Cumbo TA, Edelman R. Infections at high altitude. Clin Infect Dis 33: 1887–1891, 2001. [DOI] [PubMed] [Google Scholar]
- 6. Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc Ser B Method 57: 289–300, 1995. [Google Scholar]
- 7. Bennett BD, Solar GP, Yuan JQ, Mathias J, Thomas GR, Matthews W. A role for leptin and its cognate receptor in hematopoiesis. Curr Biol 6: 1170–1180, 1996. [DOI] [PubMed] [Google Scholar]
- 8. Bianciardi P, Fantacci M, Caretti A, Ronchi R, Milano G, Morel S, von Segesser L, Corno A, Samaja M. Chronic in vivo hypoxia in various organs: Hypoxia-inducible factor-1 alpha and apoptosis. Biochem Biophys Res Commun 342: 875–880, 2006. [DOI] [PubMed] [Google Scholar]
- 9. Bosco MC, Puppo M, Santangelo C, Anfosso L, Pfeffer U, Fardin P, Battaglia F, Varesio L. Hypoxia modifies the transcriptome of primary human monocytes: Modulation of novel immune-related genes and identification of CC-chemokine ligand 20 as a new hypoxia-inducible gene. J Immunol 177: 1941–1955, 2006. [DOI] [PubMed] [Google Scholar]
- 10. Bozzini CE, Barcelo AC, Conti MI, Martinez MP, Alippi RM. Enhanced erythropoietin production during hypobaric hypoxia in mice under treatments to keep the erythrocyte mass from rising: Implications for the adaptive role of polycythemia. High Alt Med Biol 6: 238–246, 2005. [DOI] [PubMed] [Google Scholar]
- 11. Bracken CP, Whitelaw ML, Peet DJ. The hypoxia-inducible factors: key transcriptional regulators of hypoxic responses. Cell Mol Life Sci 60: 1376–1393, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Branco LGS, Gargaglioni LH, Barros RCH. Anapyrexia during hypoxia. J Therm Biol 31: 82–89, 2006. [Google Scholar]
- 13. Bruder ED, Lee PC, Raff H. Metabolic consequences of hypoxia from birth and dexamethasone treatment in the neonatal rat: comprehensive hepatic lipid and fatty acid profiling. Endocrinology 145: 5364–5372, 2004. [DOI] [PubMed] [Google Scholar]
- 14. Chohan IS, Singh I. Cell-mediated immunity at high-altitude. Intl J Biometeorol 23: 21–30, 1979. [DOI] [PubMed] [Google Scholar]
- 15. Chohan IS, Singh I, Balakrishnan K, Talwar GP. Immune-response in human subjects at high-altitude. Intl J Biometeorol 19: 137–143, 1975. [DOI] [PubMed] [Google Scholar]
- 16. Choi JH, Park MJ, Kim KW, Choi YH, Park SH, An WG, Yang US, Cheong J. Molecular mechanism of hypoxia-mediated hepatic gluconeogenesis by transcriptional regulation. FEBS Lett 579: 2795–2801, 2005. [DOI] [PubMed] [Google Scholar]
- 17. Cohen P, Yang GQ, Yu XX, Soukas AA, Wolfish CS, Friedman JM, Li C. Induction of leptin receptor expression in the liver by leptin and food deprivation. J Biol Chem 280: 10034–10039, 2005. [DOI] [PubMed] [Google Scholar]
- 18. Conforti L, Petrovic M, Mohammad D, Lee S, Ma Q, Barone S, Filipovich AH. Hypoxia regulates expression and activity of Kv1.3 channels in T lymphocytes: a possible role in T cell proliferation. J Immunol 170: 695–702, 2003. [DOI] [PubMed] [Google Scholar]
- 19. Cramer T, Yamanishi Y, Clausen BE, Forster I, Pawlinski R, Mackman N, Haase VH, Jaenisch R, Corr M, Nizet V, Firestein GS, Gerber HP, Ferrara N, Johnson RS. HIF-1 alpha is essential for myeloid cell-mediated inflammation. Cell 112: 645–657, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cummins EP, Taylor CT. Hypoxia-responsive transcription factors. Pflügers Arch 450: 363–371, 2005. [DOI] [PubMed] [Google Scholar]
- 21. Dolt KS, Karar J, Mishra MK, Salim J, Kumar R, Grover SK, Pasha MAQ. Transcriptional downregulation of sterol metabolism genes in murine liver exposed to acute hypobaric hypoxia. Biochem Biophys Res Commun 354: 148–153, 2007. [DOI] [PubMed] [Google Scholar]
- 22. Dumur CI, Nasim S, Best IM, Archer KJ, Ladd AC, Mas VR, Wilkinson DS, Garrett CT, Ferreira-Gonzales A. Evaluation of quality-control criteria for microarray gene expression analysis. Clin Chem 50: 1994–2002, 2004. [DOI] [PubMed] [Google Scholar]
- 23. Eckardt KU, Ratcliffe PJ, Tan CC, Bauer C, Kurtz A. Age-dependent expression of the erythropoietin gene in rat-liver and kidneys. J Clin Invest 89: 753–760, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Facco M, Zilli C, Siviero M, Ermolao A, Travain G, Baesso I, Bonamico S, Cabrelle A, Zaccaria M, Agostini C. Modulation of immune response by the acute and chronic exposure to high altitude. Med Sci Sports Exerc 37: 768–774, 2005. [DOI] [PubMed] [Google Scholar]
- 25. Fan CH, Iacobas DA, Zhou D, Chen QF, Lai JK, Gavrialov O, Haddad GG. Gene expression and phenotypic characterization of mouse heart after chronic constant or intermittent hypoxia. Physiol Genomics 22: 292–307, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fantuzzi G, Faggioni R. Leptin in the regulation of immunity, inflammation, and hematopoiesis. J Leukoc Biol 68: 437–446, 2000. [PubMed] [Google Scholar]
- 27. Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol 16: 4604–4613, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fradette C, du Souich P. Effect of hypoxia on cytochrome P450 activity and expression. Curr Drug Metab 5: 257–271, 2004. [DOI] [PubMed] [Google Scholar]
- 29. Frappell P, Lanthier C, Baudinette RV, Mortola JP. Metabolism and ventilation in acute-hypoxia - a comparative-analysis in small mammalian species. Am J Physiol Regul Integr Comp Physiol 262: R1040–R1046, 1992. [DOI] [PubMed] [Google Scholar]
- 30. Frappell PB, Mortola JP. Hamsters vs. rats - metabolic and ventilatory response to development in chronic hypoxia. J Appl Physiol 77: 2748–2752, 1994. [DOI] [PubMed] [Google Scholar]
- 31. Friedman JM, Halaas JL. Leptin and the regulation of body weight in mammals. Nature 395: 763–770, 1998. [DOI] [PubMed] [Google Scholar]
- 32. Gautier H. Interactions among metabolic rate, hypoxia, and control of breathing. J Appl Physiol 81: 521–527, 1996. [DOI] [PubMed] [Google Scholar]
- 33. Gopfert T, Gess B, Eckardt KU, Kurtz A. Hypoxia signaling in the control of erythropoietin gene expression in rat hepatocytes. J Cell Physiol 168: 354–361, 1996. [DOI] [PubMed] [Google Scholar]
- 34. Gracey AY. Interpreting physiological responses to environmental change through gene expression profiling. J Exp Biol 210: 1584–1592, 2007. [DOI] [PubMed] [Google Scholar]
- 35. Gracey AY, Cossins AR. Application of microarray technology in environmental and comparative physiology. Annu Rev Physiol 65: 231–259, 2003. [DOI] [PubMed] [Google Scholar]
- 36. Gracey AY, Troll JV, Somero GN. Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. Proc Natl Acad Sci USA 98: 1993–1998, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Greijer AE, van der Groep P, Kemming D, Shvarts A, Semenza GL, Meijer GA, van de Wiel MA, Belien JAM, van Diest PJ, van der Wall E. Up-regulation of gene expression by hypoxia is mediated predominantly by hypoxia-inducible factor 1 (HIF-1). J Pathol 206: 291–304, 2005. [DOI] [PubMed] [Google Scholar]
- 38. Grimplet J, Deluc LG, Tillett RL, Wheatley MD, Schlauch KA, Cramer GR, Cushman JC. Tissue-specific mRNA expression profiling in grape berry tissues. BMC Genomics 8: 23, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grosfeld A, Andre J, Mouzon SHd, Berra E. Hypoxia-inducible Factor 1 transactivates the human leptin gene promoter. J Biol Chem 277: 42953–42957, 2002. [DOI] [PubMed] [Google Scholar]
- 40. Hammond KA, Chappell MA, Kristan DM. Developmental plasticity in aerobic performance in deer mice (Peromyscus maniculatus). Comp Biochem Physiol A Mol Integr Physiol 133: 213–224, 2002. [DOI] [PubMed] [Google Scholar]
- 41. Haouzi P, Bell HJ, Notet V, Bihain B. Comparison of the metabolic and ventilatory response to hypoxia and H2S in unsedated mice and rats. Respir Physiol Neuro 167: 316–322, 2009. [DOI] [PubMed] [Google Scholar]
- 42. Haouzi P, Notet V, Chenuel B, Chalon B, Sponne I, Ogier V, Bihain B. H2S induced hypometabolism in mice is missing in sedated sheep. Respir Physiol Neuro 160: 109–115, 2008. [DOI] [PubMed] [Google Scholar]
- 43. Hellwig-Burgel T, Stiehl DP, Wagner AE, Metzen E, Jelkmann W. Hypoxia-inducible factor-1 (HIF-1): a novel transcription factor in immune reactions. J Interf Cytokine Res 25: 297–310, 2005. [DOI] [PubMed] [Google Scholar]
- 44. Hilvo M, Supuran CT, Parkkila S. Characterization and inhibition of the recently discovered carbonic anhydrase isoforms CA XIII, XIV and XV. Curr Topics Med Chem 7: 893–899, 2007. [DOI] [PubMed] [Google Scholar]
- 45. Hochachka PW, Buck LT, Doll CJ, Land SC. Unifying theory of hypoxia tolerance: Molecular metabolic defense and rescue mechanisms for surviving oxygen lack. Proc Natl Acad Sci USA 93: 9493–9498, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hochachka PW, Somero GN. Biochemical Adaptation. New York: Oxford University Press, 2002, p. 466. [Google Scholar]
- 47. Iacobas DA, Fan C, Iacobas S, Haddad GG. Integrated transcriptomic response to cardiac chronic hypoxia: translation regulators and response to stress in cell survival. Funct Integr Genomics 8: 265–275, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264, 2003. [DOI] [PubMed] [Google Scholar]
- 49. Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science 298: 601–604, 2002. [DOI] [PubMed] [Google Scholar]
- 50. Juel C, Lundby C, Sander M, Calbet JAL, van Hall G. Human skeletal muscle and erythrocyte proteins involved in acid-base homeostasis: adaptations to chronic hypoxia. J Physiol 548: 639–648, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Julian CG, Wilson MJ, Moore LG. Evolutionary adaptation to high altitude: a view from in utero. Am J Hum Biol 21: 614–622, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jungermann K, Kietzmann T. Oxygen: modulator of metabolic zonation and disease of the liver. Hepatology 31: 255–260, 2000. [DOI] [PubMed] [Google Scholar]
- 53. Kasai K, Yamashita T, Yamaguchi A, Yoshiya K, Kawakita A, Tanaka H, Sugimoto H, Tohyama M. Induction of mRNAs and proteins for Na/K ATPase alpha l and beta 1 subunits following hypoxia/reoxygenation in astrocytes. Mol Brain Res 110: 38–44, 2003. [DOI] [PubMed] [Google Scholar]
- 54. Kenneth NS, Rocha S. Regulation of gene expression by hypoxia. Biochem J 414: 19–29, 2008. [DOI] [PubMed] [Google Scholar]
- 55. Kojima H, Gu H, Nomura S, Caldwell CC, Kobata T, Carmeliet P, Semenza GL, Sitkovsky MV. Abnormal B lymphocyte development and autoimmunity in hypoxia-inducible factor 1 alpha-deficient chimeric mice. Proc Natl Acad Sci USA 99: 2170–2174, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li JG, Grigoryev DN, Ye SQ, Thorne L, Schwartz AR, Smith PL, O'Donnell CP, Polotsky VY. Chronic intermittent hypoxia upregulates genes of lipid biosynthesis in obese mice. J Appl Physiol 99: 1643–1648, 2005. [DOI] [PubMed] [Google Scholar]
- 57. Lipshutz RJ, Fodor SPA, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet 21: 20–24, 1999. [DOI] [PubMed] [Google Scholar]
- 58. Liu K, Nagase H, Huang CG, Calamita G, Agre P. Purification and functional characterization of aquaporin-8. Biol Cell 98: 153–161, 2006. [DOI] [PubMed] [Google Scholar]
- 59. Luheshi GN, Gardner JD, Rushforth DA, Loudon AS, Rothwell NJ. Leptin actions on food intake and body temperature are mediated by IL-1. Proc Natl Acad Sci USA 96: 7047–7052, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Luu-The V, Paquet N, Calvo E, Cumps J. Improved real-time RT-PCR method for high-throughput measurements using second derivative calculation and double correction. Biotechniques 38: 287–293, 2005. [DOI] [PubMed] [Google Scholar]
- 61. Meissner U, Hanisch C, Ostreicher I, Knerr I, Hofbauer KH, Blum WF, Allabauer I, Rascher W, Dotsch J. Differential regulation of leptin synthesis in rats during short-term hypoxia and short-term carbon monoxide inhalation. Endocrinology 146: 215–220, 2005. [DOI] [PubMed] [Google Scholar]
- 62. Mesa RA, Barosi G, Cervantes F, Reilly J T, Tefferi A. Myelofibrosis with myeloid metaplasia: disease overview and non-transplant treatment options. Best Practice Res Clin Haematol 19: 495–517, 2006. [DOI] [PubMed] [Google Scholar]
- 63. Mohamed N, Jackson N. Severe thalassaemia intermedia: clinical problems in the absence of hypertransfusion. Blood Rev 12: 163–170, 1998. [DOI] [PubMed] [Google Scholar]
- 64. Mortola JP. How newborn mammals cope with hypoxia. Respir Physiol 116: 95–103, 1999. [DOI] [PubMed] [Google Scholar]
- 65. Mortola JP. Implications of hypoxic hypometabolism during mammalian ontogenesis. Respir Physiol Neuro 141: 345–356, 2004. [DOI] [PubMed] [Google Scholar]
- 66. Murdoch C, Muthana M, Lewis CE. Hypoxia regulates macrophage functions in inflammation. J Immunol 175: 6257–6263, 2005. [DOI] [PubMed] [Google Scholar]
- 67. Olson EB, Jr, Dempsey JA. Rat as a model for humanlike ventilatory adaptation to chronic hypoxia. J Appl Physiol 44: 763–769, 1978. [DOI] [PubMed] [Google Scholar]
- 68. Ou LC, Leiter JC. Effects of exposure to a simulated altitude of 5500 m on energy metabolic pathways in rats. Respir Physiol Neuro 141: 59–71, 2004. [DOI] [PubMed] [Google Scholar]
- 69. Pelleymounter MA, Cullen MJ, Baker MB, Hecht R, Winters D, Boone T, Collins F. Effects of the obese gene-product on body-weight regulation in ob/ob mice. Science 269: 540–543, 1995. [DOI] [PubMed] [Google Scholar]
- 70. Powell FL. Functional genomics and the comparative physiology of hypoxia. Annu Rev Physiol 65: 203–230, 2003. [DOI] [PubMed] [Google Scholar]
- 71. Powell FL, Milsom WK, Mitchell GS. Time domains of the hypoxic ventilatory response. Respir Physiol 112: 123–134, 1998. [DOI] [PubMed] [Google Scholar]
- 72. Raguso CA, Guinot SL, Janssens JP, Kayser B, Pichard C. Chronic hypoxia: common traits between chronic obstructive pulmonary disease and altitude. Curr Opin Clin Nutr Metab Care 4: 411–417, 2004. [DOI] [PubMed] [Google Scholar]
- 73. Ramirez G, Bittle PA, Rosen R, Rabb H, Pineda D. High altitude living: Genetic and environmental adaptation. Aviat Space Environ Med 70: 73–81, 1999. [PubMed] [Google Scholar]
- 74. Ramirez JM, Folkow LP, Blix AS. Hypoxia tolerance in mammals and birds: from the wilderness to the clinic. Annu Rev Physiol 69: 113–143, 2007. [DOI] [PubMed] [Google Scholar]
- 75. Rankin EB, Biju MP, Liu QD, Unger TL, Rha J, Johnson RS, Simon MC, Keith B, Haase VH. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J Clin Invest 117: 1068–1077, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rankin EB, Rha J, Selak MA, Unger TL, Keith B, Liu QD, Haase VH. Hypoxia-inducible factor 2 regulates hepatic lipid metabolism. Mol Cell Biol 29: 4527–4538, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Redondo PA, Alvarez AI, Diez C, Fernandezrojo F, Prieto JG. Physiological-response to experimentally-induced anemia in rats - a comparative-study. Lab Anim Sci 45: 578–583, 1995. [PubMed] [Google Scholar]
- 78. Reeves SR, Gozal E, Guo SZ, Sachleben LR, Brittian KR, Lipton AJ, Gozal D. Effect of long-term intermittent and sustained hypoxia on hypoxic ventilatory and metabolic responses in the adult rat. J Appl Physiol 95: 1767–1774, 2003. [DOI] [PubMed] [Google Scholar]
- 79. Rhodes J. Comparative physiology of hypoxic pulmonary hypertension: historical clues from brisket disease. J Appl Physiol 98: 1092–1100, 2005. [DOI] [PubMed] [Google Scholar]
- 80. Rossignol F, Solares M, Balanza E, Coudert J, Clottes E. Expression of lactate dehydrogenase A and B genes in different tissues of rats adapted to chronic hypobaric hypoxia. J Cell Biochem 89: 67–79, 2003. [DOI] [PubMed] [Google Scholar]
- 81. Sarkar S, Banerjee PK, Selvamurthy W. High altitude hypoxia: an intricate interplay of oxygen responsive macroevents and micromolecules. Mol Cell Biochem 253: 287–305, 2003. [DOI] [PubMed] [Google Scholar]
- 82. Sbaa E, DeWever J, Martinive P, Bouzin C, Frerart F, Balligand JL, Dessy C, Feron O. Caveolin plays a central role in endothelial progenitor cell mobilization and homing in SDF-1-driven postischemic vasculogenesis. Circ Res 98: 1219–1227, 2006. [DOI] [PubMed] [Google Scholar]
- 83. Scheurer SB, Rybak JN, Rosli C, Neri D, Elia G. Modulation of gene expression by hypoxia in human umbilical cord vein endothelial cells: a transcriptomic and proteomic study. Proteomics 4: 1737–1760, 2004. [DOI] [PubMed] [Google Scholar]
- 84. Schlichting CD, Smith H. Phenotypic plasticity: linking molecular mechanisms with evolutionary outcomes. Evol Ecol 16: 189–211, 2002. [Google Scholar]
- 85. Semenza GL. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J Appl Physiol 88: 1474–1480, 2000. [DOI] [PubMed] [Google Scholar]
- 86. Semenza GL, Roth PH, Fang HM, Wang GL. Transcriptional regulation of genes encoding glycolytic-enzymes by hypoxia-inducible factor-1. J Biol Chem 269: 23757–23763, 1994. [PubMed] [Google Scholar]
- 87. Shukla V, Singh SN, Vats P, Singh VK, Singh SB, Banerjee PK. Ghrelin and leptin levels of sojourners and acclimatized lowlanders at high altitude. Nutr Neurosci 8: 161–165, 2005. [DOI] [PubMed] [Google Scholar]
- 88. Simler N, Grosfeld A, Peinnequin A, Guerre-Millo M, Bigard AX. Leptin receptor deficient obese Zucker rats reduce their food intake in response to hypobaric hypoxia. Am J Physiol Endocrinol Metab 290: E591–E597, 2006. [DOI] [PubMed] [Google Scholar]
- 89. Sonna LA, Cullivan ML, Sheldon HK, Pratt RE, Lilly CM. Effect of hypoxia on gene expression by human hepatocytes (HepG2). Physiol Genomics 12: 195–207, 2003. [DOI] [PubMed] [Google Scholar]
- 90. Steiner AA, Branco LGS. Hypoxia-induced anapyrexia: implications and putative mediators. Annu Rev Physiol 64: 263–288, 2002. [DOI] [PubMed] [Google Scholar]
- 91. Steiner AA, Krall CM, Liu E. A reappraisal on the ability of leptin to induce fever. Physiol Behav 97: 430–436, 2009. [DOI] [PubMed] [Google Scholar]
- 92. Storz JF, Moriyama H. Mechanisms of hemoglobin adaptation to high altitude hypoxia. High Alt Med Biol 9: 148–157, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Storz JF, Runck AM, Sabatino SJ, Kelly JK, Ferrand N, Moriyama H, Weber RE, Fago A. Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc Natl Acad Sci USA 106: 14450–14455, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Stroka DM, Burkhardt T, Desbaillets I, Wenger RH, Neil DAH, Bauer C, Gassmann M, Candinas D. HIF-1 is expressed in normoxic tissue and displays an organ-specific regulation under systemic hypoxia. FASEB J 15: 2445–2453, 2001. [DOI] [PubMed] [Google Scholar]
- 95. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Suliman HB, Ali M, Piantadosi CA. Superoxide dismutase-3 promotes full expression of the EPO response to hypoxia. Blood 104: 43–50, 2004. [DOI] [PubMed] [Google Scholar]
- 97. Thake CD, Mian T, Garnham AW, Mian R. Leukocyte counts and neutrophil activity during 4 h of hypocapnic hypoxia equivalent to 4000 m. Aviat Space Environ Med 75: 811–817, 2004. [PubMed] [Google Scholar]
- 98. Vats P, Singh VK, Singh SN, Singh SB. High altitude induced anorexia: Effect of changes in leptin and oxidative stress levels. Nutr Neurosci 10: 243–249, 2007. [DOI] [PubMed] [Google Scholar]
- 99. Vengellur A, Phillips JM, Hogenesch JB, LaPres JJ. Gene expression profiling of hypoxia signaling in human hepatocellular carcinoma cells. Physiol Genomics 22: 308–318, 2005. [DOI] [PubMed] [Google Scholar]
- 100. Vishesh A, Arora S. Leptin and its metabolic interactions - an update. Diabetes Obes Metab 10: 973–993, 2008. [DOI] [PubMed] [Google Scholar]
- 101. Walmsley SR, Print C, Farahi N, Peyssonnanx C, Johnson RS, Cramer T, Sobolewski A, Condliffe AM, Cowburn AS, Johnson N, Chilvers ER. Hypoxia-induced neutrophil survival is mediated by HIF-1 alpha-dependent NF-kappa B activity. J Exp Med 201: 105–115, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wang GL, Semenza GL. General involvement of hypoxia-inducible factor-i in transcriptional response to hypoxia. Proc Natl Acad Sci USA 90: 4304–4308, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ward MP, Milledge JS, West JB. High Altitude Medicine and Physiology. London: Arnold, 2000. [Google Scholar]
- 104. Welsch RE. Stepwise multiple comparison procedures. J Am Stat Assoc 72: 566–575, 1977. [Google Scholar]
- 105. Wiener CM, Booth G, Semenza GL. In vivo expression of mRNAs encoding hypoxia-inducible factor 1. Biochem Biophys Res Commun 225: 485–488, 1996. [DOI] [PubMed] [Google Scholar]
- 106. Withers PC. Design, calibration and calculation for flow-through respirometry systems. Austr J Zool 49: 445–461, 2001. [Google Scholar]
- 107. Yu AY, Shimoda LA, Iyer NV, Huso DL, Sun X, McWilliams R, Beaty T, Sham JSK, Wiener CM, Sylvester JT, Semenza GL. Impaired physiological responses to chronic hypoxia in mice partially deficient for hypoxia-inducible factor 1 alpha. J Clin Invest 103: 691–696, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zaccaria M, Ermolao A, Bonvicini P, Travain G, Varnier M. Decreased serum leptin levels during prolonged high altitude exposure. Eur J Appl Physiol 92: 249–253, 2004. [DOI] [PubMed] [Google Scholar]
- 109. Zarember KA, Malech HL. HIF-1 alpha: a master regulator of innate host defenses? J Clin Invest 115: 1702–1704, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zhang WH, Wang X, Narayanan M, Zhang Y, Huo CF, Reed JC, Friedlander RM. Fundamental role of the Rip2/caspase-1 pathway in hypoxia and ischemia-induced neuronal cell death. Proc Natl Acad Sci USA 100: 16012–16017, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.