Abstract
Epithelial-mesenchymal transition (EMT), a switch of polarized epithelial cells to a migratory, fibroblastoid phenotype, is considered a key process driving tumor cell invasiveness and metastasis. Using breast cancer cell lines as a model system, we sought to discover gene-expression signatures of EMT with clinical and mechanistic relevance. A supervised comparison of epithelial and mesenchymal breast cancer lines defined a 200-gene EMT signature that was prognostic across multiple breast cancer cohorts. Immunostaining of LYN, a top-ranked EMT signature gene and Src-family tyrosine kinase, was associated with significantly shorter overall survival (P=0.02), and correlated with the basal-like (“triple-negative”) phenotype. In mesenchymal breast cancer lines, RNAi-mediated knockdown of LYN inhibited cell migration and invasion, but not proliferation. Dasatinib, a dual-specificity tyrosine kinase inhibitor, also blocked invasion (but not proliferation) at nanomolar concentrations that inhibit LYN kinase activity, suggesting that LYN is a likely target and invasion a relevant endpoint for dasatinib therapy. Our findings define a prognostically-relevant EMT signature in breast cancer, and identify LYN as a mediator of invasion and possible new therapeutic target (and theranostic marker for dasatinib response), with particular relevance to clinically-aggressive basal-like breast cancer.
Keywords: Breast cancer, epithelial-mesenchymal transition, transcriptional profiling, LYN, dasatinib
INTRODUCTION
In breast cancer, mortality results not from tumor growth per se, but from the tumor invading through normal tissue boundaries and metastasizing to distant sites. To invade and metastasize, breast cancer cells must first dissociate from one another and become motile. These events are reminiscent of epithelial-mesenchymal transition (EMT), a process occurring during tissue patterning in normal embryonic development.
EMT is a coordinated cellular program whereby epithelial cells in layers reversibly or irreversibly convert to mesenchymal cells, fibroblast-like cells loosely embedded in extracellular matrix (1). During EMT, epithelial cells dissociate, acquire a spindly or stellate morphology, and increased motility to carry out orchestrated migrations. EMT is required for normal gastrulation and the formation of the three-layered embryo, and later for the formation of normal tissues and organs, including the heart, musculoskeletal system, and peripheral nervous system (2).
Increasing evidence suggests that in breast cancer, malignant cells co-opt the EMT program (3). EMT provides a pathway by which cancerous layers of epithelial cells (carcinoma in situ) can dissociate and become motile, leading to invasion through the basement membrane into blood vessels or lymphatics, and metastatic spread. As such, targeting EMT represents an important new therapeutic strategy for the prevention or treatment of breast cancer.
A framework of molecular and cellular events underlying EMT has been elucidated (4, 5). In different cell contexts, ligands like HGF (scatter factor), EGF (and related growth factors) and TGFβ can stimulate EMT, acting through signal transduction pathways (including SRC, RAS and PI3K) to alter cell adhesion (through adherens junctions and desmosomes) and cell motility (through cytoskeletal reorganization). Downstream transcriptional regulators, like Snail, Slug and Twist, repress the expression of E-cadherin (a key mediator of epithelial cell-cell adhesion), while activating expression of mesenchymal markers, e.g. vimentin, N-cadherin and smooth muscle actin. However, the pace of recent advances in understanding EMT suggests that much yet remains unknown.
The molecular pathways of EMT have been studied largely in the context of embryogenesis in model organisms, such as Drosophila, Xenopus and mice, and in mammalian cell culture systems (1). The latter include MDCK canine kidney cells, NBII rat bladder carcinoma cells, and NMuMG mouse mammary cancer cells, each which can be stimulated in culture to undergo EMT. However, these canine and rodent cell culture model systems may not faithfully replicate EMT events in human breast cancer.
Breast cancer cell lines display varied morphologies when grown in culture. Some appear epithelial-like, forming cell clusters, while others appear more fibroblast-like (mesenchymal), with dispersed and spindle-shaped cells. The latter cell lines tend to express vimentin, and to be more invasive in vitro, and metastatic in vivo (6, 7), suggesting they have undergone stable EMT conversion. These cell lines provide a useful model for studying the underpinnings of EMT in breast cancer. Here, we set out to explore gene-expression patterns associated with EMT in breast cancer cells in culture, and in particular to discover molecular signatures and biomarkers of EMT with possible prognostic and mechanistic, and therapeutic relevance.
MATERIALS AND METHODS
Specimens
Breast cancer cell lines were obtained directly from the ATCC (Manassas, VA) or DSMZ (Braunschweig, Germany), and grown in RPMI-1640 with 10% FBS and 1% Pen/Strep. Early passage primary breast fibroblasts were prepared from reduction mammoplasties or prophylactic mastectomies. In brief, surgical breast tissue specimens were obtained following informed consent by the Biosample Repository staff and surgery at Fox Chase Cancer Center. De-identified tissue specimens were finely minced and incubated in a collagenase solution (DMEM/media 199, 10% horse serum + collagenase, hyaluronidase, antibiotic/antimycotic, insulin, hydrocortisone) overnight at 37°C in a rotating water bath and then centrifuged at 2,500 rpm for 10 min. The supernatant was decanted to a sterile tube and the residual tissue was rinsed several times, resuspended in culture medium, combined with the supernatant and centrifuged as before. The resulting tissue pellet was resuspended in fibroblast medium (DMEM, 15%FBS, pen/strep, cipro, fungizone, and gentamycin) and was plated in a swine skin gelatin and FBS coated flask. The tissue was permitted to attach to flasks for 24 to 48 hrs. Once this occurred, cells were fed twice a week, increasing medium amounts incrementally. Cells were passaged (at a 1:2 split ratio) until characterized. A tissue microarray (TMA) was constructed from 970 clinically-annotated breast cancer cases (each represented by duplicate 2 mm cores) archived at the Samsung Medical Center (Seoul, Korea). All research was conducted with IRB approval.
Expression profiling
Gene-expression profiling was done using Human Exonic Evidence Based oligonucleotide (HEEBO) arrays obtained from the Stanford Functional Genomics Facility (SFGF) and representing 24,207 human genes. Briefly, 40 µg of sample RNA and 40 µg of “universal” reference RNA were differentially labeled with Cy5 and Cy3, respectively, then co-hybridized onto the microarray in a high volume mixing hybridization at 65°C for 40 hrs. Details of the array processing and sample labeling and hybridization methods have been described (8). The complete microarray data are available at the Stanford Microarray Database1 and Gene Expression Omnibus (accession GSE13915). Some of these microarray data were included in a recent study integrating genomic and transcriptional profiles of breast cancer lines (9).
Microarray data analysis
Background-subtracted fluorescence log2 ratios were globally normalized for each array, and then mean-centered for each gene (i.e., reporting relative to the average log2 ratio across all samples). Subsequent analysis included only the 6,947 well-measured and variably-expressed genes, defined as those with intensities in the Cy5 or Cy3 channel at least 1.5-fold above background in at least 80% of samples, and with at least 3-fold ratio variation from the mean in at least 3 samples. Differentially expressed genes were identified by two-class Significance Analysis of Microarrays (SAM) (10) (False Discover Rate, FDR <5%). An EMT signature was defined by combining the top ranked 100 genes overexpressed in mesenchymal breast cancer lines compared to both epithelial breast cancer lines and normal breast fibroblasts, and in epithelial breast cancer lines compared to both mesenchymal breast cancer lines and normal breast fibroblasts. Gene Ontology (GO) term enrichment was done using FatiGO (11). Clinical relevance of the EMT signature was evaluated using publicly-available microarray data for primary breast tumor cohorts (12, 13, 14). Datasets were mean centered and log transformed, and corresponding EMT signature genes identified by Entrez Gene ID. Breast tumors were then clustered in the space of the EMT signature genes, and the resultant two main sample branches were evaluated by Kaplan-Meier analysis.
Western blot, immunohistochemistry (IHC), and DNA sequencing
Cells were lysed in 1X RIPA Lysis buffer. 40 µg total protein lysate was electrophoresed on a 4–15% polyacrylamide gel, then transferred to PVDF membrane and blocked in TBST-T with 5% dry milk. Anti-LYN, p-LYN (Tyr507), SRC, p-SRC (Tyr527), and p-p130cas (Tyr410) antibodies (Cell Signaling Technology, Danvers, MA) were used at 1:1000 dilution with overnight incubation at 4°C in TBS-T with 5% BSA. Anti-Vimentin, E-cadherin, p130cas, and GAPDH antibodies (Santa Cruz Biotechnology; Sant Cruz, CA) were used at 1:1000 dilution. After incubation with an HRP-conjugated secondary antibody, detection was done using an ECL kit (GE Healthcare, Piscataway, NJ). Band intensities were quantified by densitometry using ImageJ software2. IHC was done using 4 µM sections of the breast cancer TMA. Following heat-induced antigen retrieval, anti-LYN antibody (Santa Cruz Biotechnology) was used at 1:20 dilution, with chromogenic detection by peroxidase-conjugated secondary antibody and DAB reagents (Envision detection kit; DAKO, Carpinteria, CA, USA). Parallel TMA sections were stained with anti-CK5/6 (1:100; D5/16 B4, DAKO), EGFR (1:30; E30, DAKO), HER2 (1:200; CB11, DAKO), ER (1:100; 6F11; Novocastra, Newcastle, U.K.), and PR (1:50; 1A6; Novocastra) antibodies. DNA sequencing of LYN (exons 8–13, inclumbding intron-exon junctions) was done from PCR-amplified genomic DNAs (PCR primers and conditions in Table S1), with Sanger sequencing by Geneway Research (Hayward, CA).
siRNA knockdown and dasatinib treatment
Synthetic 21-nucleotide small inhibitory RNAs (siRNAs) directed against LYN and SRC, and a non-targeting SMART pool, were obtained from Dharmacon (Lafayette, CO) (sequences in Table S2). Briefly, 200,000 cells were seeded in triplicate in six-well plates and transfected with a final concentration of 50 nmol siRNA for 16 hrs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Dasatinib (LC Laboratories, Woburn, MA) was reconstituted in DMSO at 200 mM, and used at concentrations indicated.
Cell proliferation, migration and invasion assays
Effect of gene knockdown or drug treatment on cell proliferation was measured by quantifying the metabolic cleavage of the tetrazolium salt WST-1 (Roche Applied Science, Indianapolis, IN) in viable cells. Motility and invasion were quantified by Boyden chamber assay (BD Biosciences, San Jose, CA). Briefly, 10,000 (migration) or 20,000 (invasion) cells were plated into 24-well inserts using a 0.5% to 5% FBS gradient. Cells were fixed, stained with crystal violet and cells traversing the membrane counted. All assays were performed in triplicate, and mean values and standard deviations reported. IC50 values were determined by fitting sigmoidal (four-parameter logistic) curves with Prism 4.0 software (GraphPad, La Jolla, CA).
RESULTS
Prognostic EMT signature from breast cancer cell lines
Breast cancer cell lines grown in culture display distinct morphologies, with a small subset of lines appearing more “fibroblast-like” (mesenchymal), and being more invasive (6, 7) (and our observations), features suggestive of having undergone EMT. To explore the molecular variation associated with this phenotype, we used whole-genome oligonucleotide microarrays to profile gene expression of 5 mesenchymal breast cancer cell lines (BT549, Hs578T, MDA157, MDA231, MDA436), in comparison to a diverse set of 10 epithelial-like breast cancer cell lines (BT20, BT474, EFM19, MCF7, MDA-361, MDA-453, MDA-468, SKBR3, T47D, UACC893) representing both luminal and basal-like subtypes (9, 15). Molecular pathological features of cell lines and representative morphologies are shown in Fig 1A. We also profiled 5 early-passage primary breast fibroblast cultures, to help identify characteristic profiles of EMT distinct from fibroblasts.
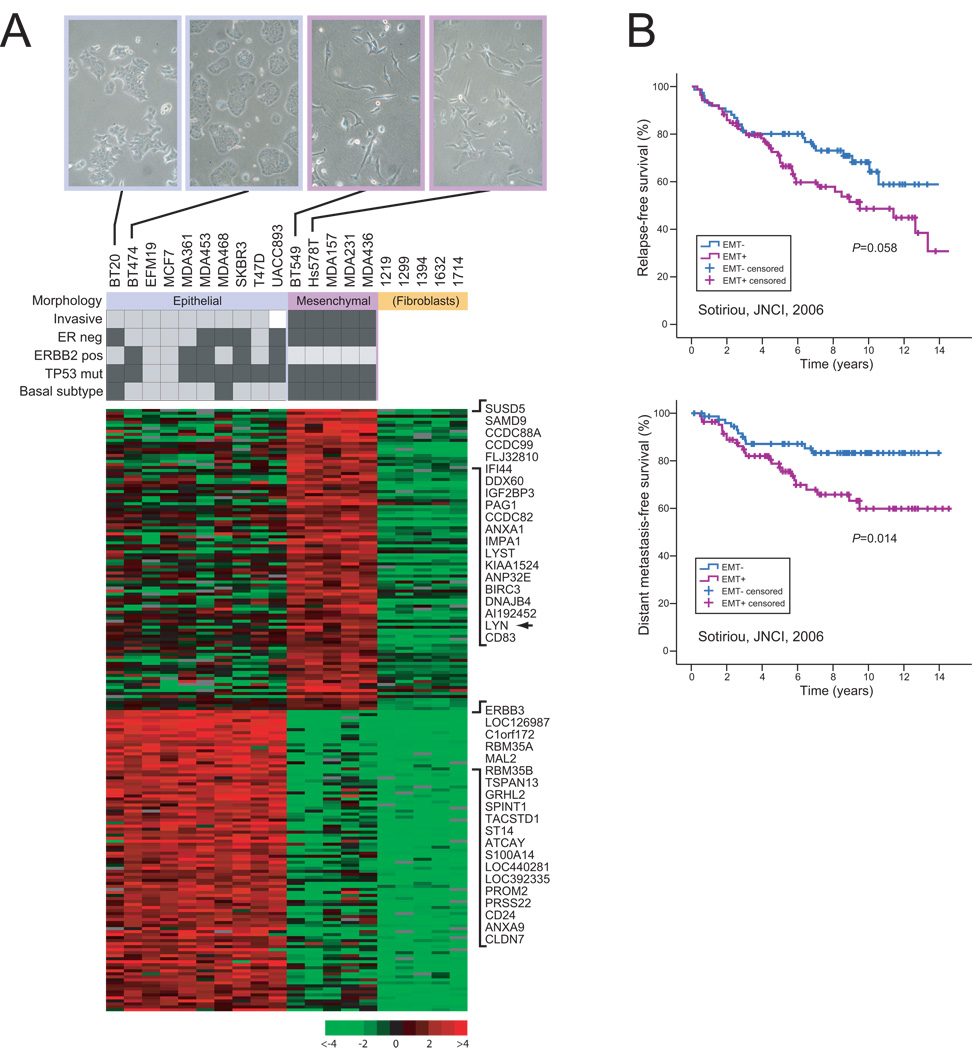
Figure 1. Cell line derived EMT signature shows prognostic relevance.
(A) Expression profiling of morphologically distinct breast cancer cell lines defines signature of EMT. Expression profiles were compared among breast cancer cell lines exhibiting epithelial-like and mesenchymal morphologies, and normal breast fibroblasts. Cell line characteristics (7, 9) are indicated (black box = yes), along with representative photos showing cell culture morphologies (equal cell numbers plated). The EMT signature (heatmap shown) comprises the top ranked 100 genes overexpressed in mesenchymal breast cancer cells compared to epithelial breast cancer cells and normal breast fibroblasts, and in epithelial breast cancer cells compared to mesenchymal breast cancer cells and normal breast fibroblasts. The top 20 genes, including the LYN tyrosine kinase (arrow), are shown. Expression ratio (log2) scale shown. (B) EMT signature is predictive of clinical outcome. Primary breast tumors from a publicly-available microarray dataset (Sotiriou et al.) (12) were clustered in the space of the EMT signature genes, and the two major sample clusters then compared by Kaplan-Meier analysis (P-values shown). The “EMT+” group, associated with EMT genes overexpressed in mesenchymal lines (compared to the converse pattern of genes overexpressed in epithelial lines) showed increased risk of relapse (above) and distant metastasis (below); P-values shown.
Unsupervised cluster analysis of gene expression readily differentiated the three groups (epithelial, mesenchymal, normal fibroblasts) (Fig. S1), indicating robust expression differences. We therefore sought to build a signature that distinguished epithelial from mesenchymal breast cancer cells, distinct from fibroblasts (anticipating stromal “contamination” in subsequently evaluated clinical samples). We defined a 200 gene “EMT signature” by combining the top 100 genes significantly overexpressed (FDR<0.05) in mesenchymal breast cancer lines compared to both epithelial breast cancer lines and fibroblasts, and the top 100 genes significantly overexpressed in epithelial breast cancer lines compared to both mesenchymal breast cancer lines and fibroblasts (Fig. 1A and Fig. S2). Assuringly, the top 100 genes overexpressed in epithelial lines were enriched for the GO term “intercellular junction” (disrupted in EMT) (including genes CLDN3, CLDN4, CLDN7, EVPL, MARVELD2, OCLN, PKP3) (corrected P=0.001), and related GO terms, though no GO term enrichment was identified among the top 100 genes overexpressed in mesenchymal lines.
EMT is thought to underlie aggressive tumor behavior (1). To determine a possible clinical relevance of our EMT signature, we evaluated the signature genes in three different primary breast cancer cohorts using publicly-available microarray data. The first cohort, Sotiriou et al. (12), from the John Radcliff Hospital (United Kingdom) and Uppsala University Hospital (Sweden), comprised 169 cases of invasive ductal carcinoma, profiled with Affymetrix U133A GeneChips. To evaluate the EMT signature, we clustered those samples in the space of the EMT signature genes (Fig. S3), and compared clinical outcomes between the two major sample clusters by Kaplan-Meier analysis. Notably, the sample cluster associated with EMT genes overexpressed in mesenchymal lines (compared to the converse pattern of genes overexpressed in epithelial lines) showed a strong trend towards decreased relapse-free survival (P =0.058), and significantly decreased distant metastasis-free survival (P=0.014) (Fig. 1B). Similar analysis of a second cohort, van de Vijver et al. (13) (295 cases from the Netherlands Cancer Institute (NKI), profiled with Agilent oligonucleotide microarrays) revealed significant association with both metastasis-free (P <0.001) and overall survival (P =0.017) (Fig. S3). Likewise, analysis of a third cohort, Bild et al. (14) (171 cases profiled with Affymetrix U133 2.0 GeneChips), showed significant association with overall survival (P=0.015) (Fig. S3).
Where sufficient clinical annotations were available, we also evaluated the EMT signature in multivariate analysis. In the Sotiriou et al. cohort, the EMT signature was a significant independent predictor of relapse-free and distant metastasis-free survival (Table 1). In the NKI dataset, the signature was significant only when estrogen receptor (ER) status was omitted from the model (data not shown).
Table 1.
Multivariate Analysis
| Variable | Hazard ratio |
95% CI |
P- value |
|
|---|---|---|---|---|
|
Sotiriou, JNCI, 2006 Relapse-free survival |
EMT signature | 2.02 | 1.07–3.81 | 0.029 |
| ER negativity | 1.62 | 0.77–3.39 | 0.20 | |
| Grade (2) | 2.00 | 0.92–4.36 | 0.082 | |
| Grade (3) | 1.13 | 0.53–2.42 | 0.76 | |
| Tumor size (2–5 cm) | 2.66 | 1.38–5.13 | 0.004 | |
| Tumor size (>5 cm) | 5.69 | 1.59–20.4 | 0.007 | |
| Lymph node positivity | 0.73 | 0.30–1.78 | 0.48 | |
|
Sotiriou, JNCI, 2006 Distant metastasis-free survival |
EMT signature | 2.72 | 1.16–6.34 | 0.021 |
| ER negativity | 1.48 | 0.55–3.98 | 0.44 | |
| Histologic grade (2) | 2.20 | 0.79–6.11 | 0.13 | |
| Histologic grade (3) | 1.24 | 0.49–3.14 | 0.65 | |
| Tumor size (2–5 cm) | 5.13 | 1.90–13.8 | 0.001 | |
| Tumor size (>5 cm) | 5.91 | 0.98–35.8 | 0.053 | |
| Lymph node positivity | 1.09 | 0.40–2.94 | 0.87 | |
|
Tissue microarray Overall survival |
LYN positivity | 2.29 | 1.18–4.42 | 0.014 |
| ER negativity | 1.28 | 0.65–2.49 | 0.48 | |
| Tumor size (2–5 cm) | 2.36 | 0.83–6.72 | 0.108 | |
| Tumor size (>5 cm) | 7.45 | 2.47–22.5 | <0.001 | |
| Lymph node positivity | 2.06 | 1.12–3.80 | 0.020 |
Signature gene LYN functions in invasion
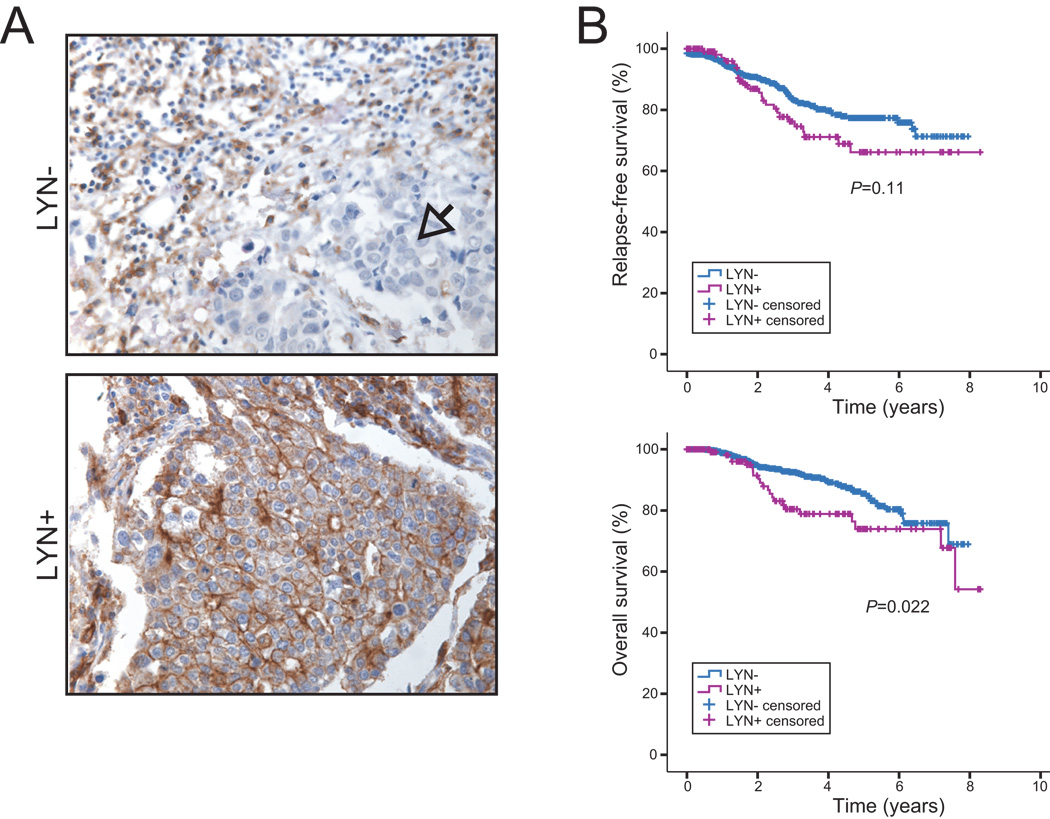
Among highly ranked EMT signature genes overexpressed in mesenchymal lines was LYN (Fig. 1A). LYN is a Src-family kinase, a family of non-receptor tyrosine kinases with roles in signal transduction, often deregulated in cancers and linked to neoplastic transformation (16). LYN was of particular interest because as a kinase it is “druggable”, and might provide a therapeutic opportunity targeting EMT. We first evaluated LYN expression in a large cohort of primary breast cancer cases (archived at the Samsung Medical Center, Seoul, Korea) by IHC on tissue microarrays (TMAs). LYN expression was found in 133 of 939 scorable breast cancer cases (14.2%) (representative images shown in Fig. 2A), and was significantly associated with shorter overall survival (P=0.02) (with a trend for relapse-free survival), most evident between 2 and 6 years after surgery (Fig. 2B). In multivariate analysis, LYN expression was a significant prognostic factor independent of other clinical variables (Table 1). Notably, LYN expression was associated with the triple-negative phenotype (P<0.001; Fisher’s exact test), as defined by IHC (HER2 and ER/PR negative). Forty-six percent of triple negative cases were LYN+, vs. 4% of others, and 79% of LYN+ cases were triple negative, vs. 15% for LYN− cases (Table S3). LYN expression was also associated with basal-marker (CK5/6 and/or EGFR) positivity (P<0.001) (Table S3).
Figure 2. LYN expression is associated with unfavorable outcome.
(A) LYN immunostaining on tissue microarray. Shown are representative breast cancer cases negative (arrow identifies region with tumor cells) or positive for expression of the LYN tyrosine kinase in tumor cells. Note, LYN is expressed in stromal cells in some breast cancer cases (also see Fig. S7). (B) LYN expression in tumor cells predicts unfavorable outcome. Kaplan-Meier analysis comparing LYN positive and negative cases, for relapse-free survival (above) and overall survival (below); P-values shown.
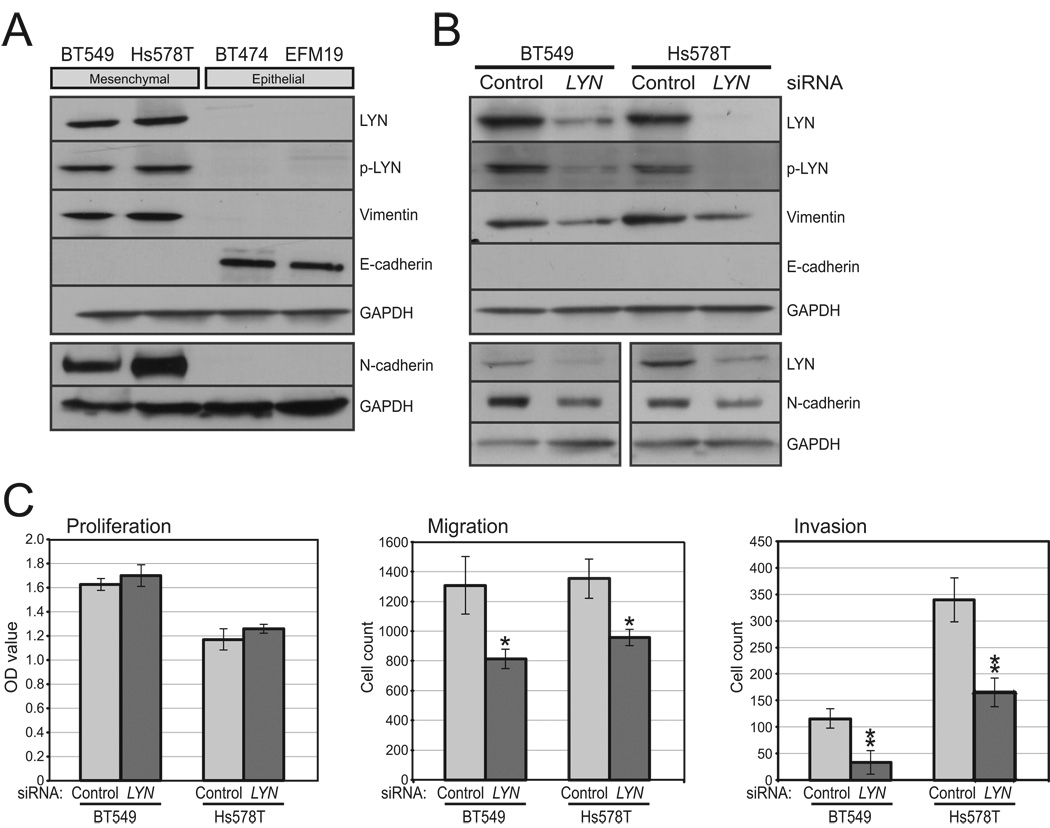
To evaluate a functional connection between LYN and EMT, we first assayed protein levels in breast cancer cell lines by Western blot. Both total LYN and phospho-LYN (Tyr507) (indicative of LYN activation; (17)) were elevated in mesenchymal compared to epithelial breast cancer lines (Fig. 3A). We next used a siRNA pool to knockdown LYN expression in two mesenchymal breast cancer lines (BT549 and Hs578T). Knockdown of LYN (and p-LYN), confirmed by Western blot (75–90% knockdown), led to decreased expression of the mesenchymal markers vimentin (65–75% reduction) and N-cadherin (50–70% reduction), but not to increased E-cadherin (Fig. 3B), nor to observed morphologic changes (Fig. S4). In both cell lines, knockdown of LYN did not alter cell proliferation levels, but led to significantly decreased cell migration and invasion (Fig. 3C). Transfection of individual siRNAs from the pool similarly inhibited invasion (Fig. S5), effectively excluding possible RNAi off-target effects.
Figure 3. LYN overexpression contributes to invasiveness.
(A) LYN exhibits relative overexpression and activation in mesenchymal breast cancer lines. Shown is a Western blot probed with anti-LYN, phospho (activated)-LYN (Tyr507), vimentin and N-cadherin (markers of EMT), E-cadherin (a marker of epithelial morphology) (overnight exposure, positive control not shown), and GAPDH (loading control). (B) Validation of LYN knockdown by siRNA, and effect on vimentin, N-cadherin and E-cadherin levels. Transfected siRNA pools (LYN or control non-targeting siRNA) are indicated. (C) LYN knockdown (compared to non-targeting control) does not alter cell proliferation (left) (measured by WST-1 assay), but leads to significantly decreased cell migration (center) and invasion (right) (measured by Boyden chamber assay). Means and standard deviations shown. *, P<0.05, **, P<0.01; Student’s t-test.
To determine whether LYN overexpression/activation might be driven by DNA amplification, we analyzed our array-based comparative genomic hybridization (aCGH) data for 49 breast cancer cell lines (9) and 172 breast tumors (8). LYN (residing at cytoband 8q12.1) was not found to be focally amplified, though it did reside within broad gains spanning some or all of 8q in 20 of 49 (41%) breast cancer lines (including BT549 and Hs578T), and in 51 of 172 (30%) breast tumors. To determine whether LYN activity might be associated with activating mutations, we sequenced exons 8–13 (corresponding to the P-loop and activation segment of the kinase domain) of LYN from PCR-amplified genomic DNA of the five mesenchymal breast cancer cell lines and 50 breast tumors (LYN+ from the TMA). No mutations were identified.
LYN is likely target of dasatinib
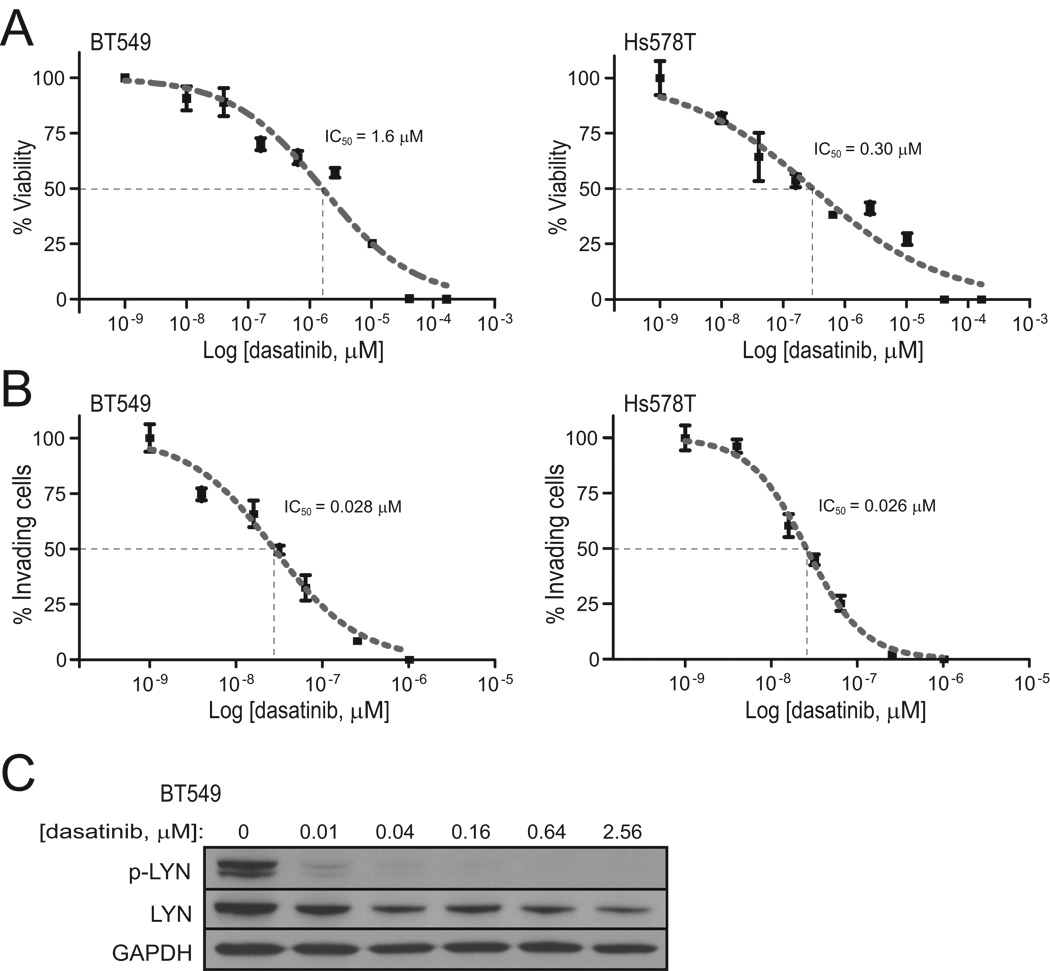
We also sought to determine whether we could inhibit LYN pharmacologically. Dasatinib is a dual-specificity tyrosine kinase inhibitor, active against both ABL and the Src-family tyrosine kinases (of which LYN is a member). Of note, dasatinib was also recently reported to show selective growth inhibition of basal-like breast cancer cell lines (18, 19). In our studies, dasatinib treatment of the mesenchymal (and also basal-like) breast cancer lines BT549 and Hs578T resulted in decreased cell growth/viability (Fig. 4A), with an IC50 (50% inhibitory concentration) of 1.6 µM and 0.30 µM respectively. Notably, these growth inhibitory concentrations were respectively 188- and 35-fold higher than the reported dasatinib IC50 of LYN tyrosine kinase activity in vitro (8.5 nM) (20), suggesting the effect on growth was likely not mediated through LYN. We also measured the effect of dasatinib on cell invasion. In BT549 and Hs578T cells, the IC50 for invasion was 0.028 µM and 0.026 µM (Fig. 4B), or respectively 57-fold and 12-fold lower than the IC50 for cell growth, and more comparable (only 3-fold higher) to the reported IC50 for LYN kinase activity. Western blot confirmed dasatinib inhibition of LYN activity (i.e. p-LYN) at nanomolar concentrations (Fig. 4C).
Figure 4. Dasatinib inhibits cell invasion at concentrations that inhibit LYN.
Dasatinib effect (dose-response curve) on (A) cell viability and (B) cell invasion, for BT549 cells (left) and Hs578T cells (right). IC50 values (indicated) were determined from sigmoidal (four-parameter logistic) curves. (C) Verification in BT549 cells that dasatinib treatment leads to decreased phospho (activated)-LYN, assayed by Western blot.
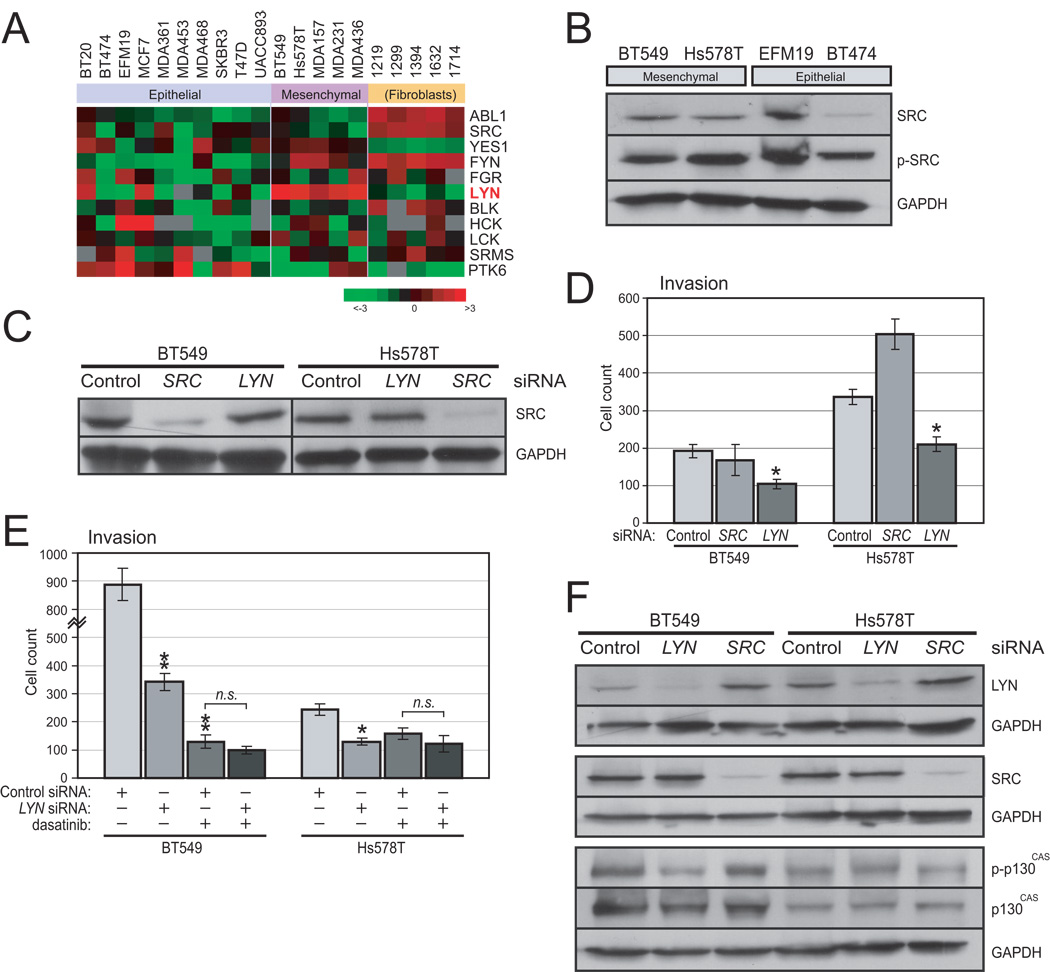
These data are consistent with dasatinib targeting LYN to inhibit cell invasion. However, it remained possible that dasatinib was acting instead through a different Src-family kinase, in particular SRC itself, which has been linked previously to cell invasion (21). Several additional findings argue against this possibility. First, examining transcript levels of all Src-family kinases, we found that LYN but not SRC was relatively overexpressed in invasive, mesenchymal breast cancer lines (P=0.006; Mann-Witney U-test) (Fig. 5A), and more broadly, in basal-like breast lines (P<0.001) (Fig. S6). Consistent with this finding, SRC was also not expressed at higher protein levels (by Western blot) in mesenchymal lines (Fig. 5B). Second, siRNA-mediated knockdown of SRC, confirmed by Western blot (Fig. 5C) did not inhibit BT549 and Hs578 cell invasion (Fig. 5D), suggesting that LYN and not SRC mediates invasiveness. Lastly, siRNA-mediated LYN knockdown and dasatinib treatment did not show additive effects in inhibiting BT549 and Hs578 cell invasion (Fig. 5D), suggesting that LYN siRNA and dasatinib are acting through the same target (i.e. LYN).
Figure 5. Relevant dasatinib target is likely LYN and not SRC.
(A) Heatmap shows microarray expression levels (mean-centered log2 ratios; fold-change indicated) of ABL and Src-family kinase genes. LYN (but not SRC) is significantly overexpressed in mesenchymal (compared to epithelial) breast cancer lines (P=0.006; Mann-Whitney U-test). Note, though, that LYN does appear expressed in the epithelial-like (and not highly invasive) lines BT20 and MCF7, suggesting its role in EMT may be context specific. (B) Western blot shows that SRC is not relatively overexpressed or phosphorylated (Tyr527) in mesenchymal breast lines. GAPDH serves as loading control. (C) Validation of SRC knockdown by siRNA. Transfected siRNA pools (SRC, LYN or control non-targeting siRNA) are indicated; SRC levels assayed by Western blot (GAPDH serves as loading control). (D) Knockdown of SRC (in contrast to LYN) does not inhibit cell invasion in mesenchymal lines BT549 and Hs578T. Note, the apparent augmentation of invasion observed with SRC knockdown in Hs578T cells is not reproducible in replicate experiments. (E) LYN knockdown and dasatinib treatment each significantly inhibits invasion in mesenchymal lines BT549 and Hs578T, but the effect is not additive. Dasatinib was used at 0.03 µM and 0.015 µM for BT549 and Hs578T, respectively. *, P<0.05, **, P<0.01, n.s., not significant; Student’s t-test. (F) Knockdown of LYN (but not SRC) leads to reduced phospho-p130CAS (Tyr410) levels in BT549 cells, assayed by western blot (GAPDH serves as loading control).
The finding that LYN but not SRC promotes invasion in mesenchymal breast cancer lines suggests the possibility that the LYN and SRC tyrosine kinases are phosphorylating distinct target proteins. While there are many known targets of Src-family kinases (22), as a starting point we chose to focus on p130CAS (Crk-associated substrate), previously linked to cell motility/invasion (23, 24). In BT549 (mesenchymal) cells, knockdown of LYN but not SRC led to decreased phospho- p130CAS (Tyr410) levels (65% reduction) (Fig. 5F). Though a similar reduction was not observed in Hs578T cells (Fig. 5F), this finding nevertheless supports a likelihood that the differential effects of LYN and SRC on cell invasion are manifested through the phosphorylation/activation of distinct downstream targets. Future studies should clarify the mechanisms linking LYN to EMT phenotypes.
DISCUSSION
The broad goal of our study was to explore expression patterns of EMT, using breast cancer cell lines as a model system. Comparing mesenchymal and epithelial breast cancer lines (and distinct from normal breast fibroblasts), we defined a 200-gene EMT signature, and that signature was robustly prognostic across three breast cancer microarray datasets representing independent cohorts and different microarray platforms. The EMT signature was a significant predictor independent of clinically-used prognostic factors (tumor size, grade, lymph node and ER status) in the Sotiriou cohort, though not the NKI dataset, where there was some relation with ER status. Nonetheless, the EMT signature does not appear to represent merely an ER+/− or basal/luminal signature (despite having fewer basal-like lines in the epithelial group, due to repository availability at the start of our study). Notably absent among EMT signature genes was ESR1 (ER) itself, as well as any of 89 empirically defined ER target genes (25), or key basal-luminal discriminatory genes (CAV1, CD44, EGFR, MET, ETS1, GATA3, KRT19, MME, MSN) (26).
While the EMT signature was prognostic, eventual clinical utility is less certain. The “prognostic space” for breast cancer is becoming increasingly crowded. Microarray derived prognosticators include a 70-gene outcome signature (“Mammaprint”; (27)), 21-gene “OncotypeDx” signature (28), “Perou-Sorlie” subtypes (29), wound signature (30), hypoxia signature (31), stem cell (CD44+/CD24−/low) signature (32), and stroma signature (33), among several others. Many of these signatures identify the same poor outcome cases, and are likely capturing the same underlying biology (34). Indeed, recent studies support a connection between breast cancer stem cells, the basal-like phenotype, and EMT (35, 36). Irrespective of clinical utility, our finding that the EMT signature identifies aggressive tumors supports a clinical relevance of EMT.
Among the top signature genes, we carried out additional studies of LYN, a Src-family kinase. Members of this family of non-receptor tyrosine kinases function in signal transduction, regulating diverse cellular activities including growth, survival, motility and invasion (21). Src-family kinases, foremost SRC, are also frequently deregulated in cancer, where they have been linked to tumor development and progression (16). LYN itself has been studied mainly in hematopoietic cells, but was recently linked to prostate cancer (where expression was associated with growth, invasion and metastasis) (20, 37, 38), glioblastoma (39) and Ewing’s sarcoma (40). In our studies, we found LYN expressed in 14% of breast cancers, where immunostaining was prognostic, and associated with (though not equivalent to) the triple-negative/basal-like subtype (about half of triple-negatives were LYN+). Of note, in breast cancer cases with positive immunostaining, most (or all) cancer cells expressed LYN, rather than only those cells at the leading edge of invasion. This finding is consistent with the idea that tumor phenotypes like EMT and metastatic-potential might be encoded in the bulk tumor (rather than in a select subpopulation) (41).
Examining cell lines, we confirmed LYN overexpression and increased activity (p-LYN) in mesenchymal breast cancer lines. Knockdown experiments revealed a function of LYN in cell motility and invasion, but not in cell proliferation (i.e. growth, survival). Knockdown of LYN also led to reduced vimentin and N-cadherin (EMT markers), but not increased E-cadherin, suggesting that LYN directs only a portion of the mesenchymal phenotype. Though a single potential LYN activating mutation (D385Y, in activation segment) was reported in a breast cancer case (among 80 samples screened) (42), we did not identify any mutations in 55 samples. Nor did we find focal DNA amplification in breast cancer lines or tumors. Therefore, LYN overexpression/activity is more likely controlled mainly by upstream regulators, which remain to be defined.
Dasatinib is a dual-specificity tyrosine kinase inhibitor, with activity against both ABL and the Src-family tyrosine kinases (43). It is currently used as second line therapy for imatinib (Gleevec)-resistant chronic myeloid leukemia, and efficacy is being explored in solid tumors (44). Indeed, recent studies indicate selective growth inhibition of basal-like breast cancer lines (18, 19), and clinical trials are underway (45). In our study, we found that dasatinib treatment also inhibited cell invasion, and at levels comparable to LYN kinase inhibition and up to ~60-fold lower than required to inhibit cell growth. These findings suggest that LYN is a target of dasatinib, and that invasion is a relevant endpoint for measuring drug response.
While dasatinib is active against other Src-family kinases, in particular SRC, multiple lines of evidence support LYN as the presumptive target. First, LYN (but not SRC) was expressed at higher levels in the invasive, mesenchymal breast cancer lines, and indeed generally at higher levels in basal-like breast cancer lines where dasatinib was previously shown selectively inhibitory (19). Second, knockdown of LYN (but not SRC) in mesenchymal breast cancer lines inhibited invasion (which was also inhibited by dasatinib). Third, the effects of LYN knockdown and dasatinib treatment were not additive, consistent with their sharing the same target. Taken together, our data support LYN as a relevant target of dasatinib in invasive breast cancer cells. Nonetheless, SRC may still function in other aspects of breast cancer pathogenesis. Indeed, SRC was recently shown to support survival of breast cancer cells in the bone marrow (46).
Our studies of LYN have important clinical implications. Foremost, our findings identify LYN as a novel target for therapy in breast cancer, with particular relevance to clinically aggressive basal-like breast cancers. These typically triple-negative tumors are not treatable by standard therapies like ER modulators or HER2 antagonists. Recent studies suggest a promise of poly ADP-ribose polymerase (PARP) inhibitors (47), leveraging probable tumor defects in DNA repair. Dasatinib, targeting Src-family kinases, represents an additional possible treatment (45). Importantly, our findings suggest that LYN immunostaining might be a good “theranostic” biomarker for dasatinib (or similar inhibitor) response, and worthwhile to incorporate into clinical trials. Given its role in invasion (which precedes metastasis), the inhibitory effects on LYN might best be observed in adjuvant studies, which can be lengthy and challenging.
Finally, our studies also underscore the relevance of EMT/invasion (rather than, or in addition to cell growth) as a meaningful biological and clinical endpoint. Currently, most drug screening programs rely on assaying cell growth/cytotoxicity. Indeed, BT549 cells were thus classified as “resistant” to dasatinib (19), though clearly dasatinib inhibits BT549 cell invasion. However, tumor invasion and metastasis, not growth per se, are the real drivers of cancer mortality. Inhibiting tumor cell invasion and metastasis may represent a distinct approach to control (rather than cure) cancer. Our findings highlight the importance of incorporating phenotypes like cell invasion into drug screening, and designing clinical trials that utilize such drugs earlier, in order to block tumor metastasis, thereby effectively managing cancer.
Supplementary Material
ACKNOWLEDGMENTS
We wish to thank the SFGF for microarray manufacture, SMD for database support, and members of the Pollack lab (in particular Keyan Salari) for helpful discussion.
This work was supported by grants from the NIH, CA97139 (J.R.P.), CA113916 (A.K.G), CA09302 (M.B.), and CA130172 (M.B.); NIH contract N01-CN-43309, (A.K.G.); the Department of Defense (BC073467); the California Breast Cancer Research Program, 8KB-0135 (J.R.P.); and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2009-0071010) (Y-L.C.).
Footnotes
The authors have no conflicts of interest to declare.
REFERENCES
- 1.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 2.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincent-Salomon A, Thiery JP. Host microenvironment in breast cancer development: epithelial-mesenchymal transition in breast cancer development. Breast Cancer Res. 2003;5:101–106. doi: 10.1186/bcr578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 5.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 6.Bae SN, Arand G, Azzam H, et al. Molecular and cellular analysis of basement membrane invasion by human breast cancer cells in Matrigel-based in vitro assays. Breast Cancer Res Treat. 1993;24:241–255. doi: 10.1007/BF01833264. [DOI] [PubMed] [Google Scholar]
- 7.Blick T, Widodo E, Hugo H, et al. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin Exp Metastasis. 2008;25:629–642. doi: 10.1007/s10585-008-9170-6. [DOI] [PubMed] [Google Scholar]
- 8.Bergamaschi A, Kim YH, Kwei KA, et al. CAMK1D amplification implicated in epithelial-mesenchymal transition in basal-like breast cancer. Molecular Oncology. 2008;2:327–339. doi: 10.1016/j.molonc.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kao J, Salari K, Bocanegra M, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One. 2009;4:e6146. doi: 10.1371/journal.pone.0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 12.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 13.van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 14.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 15.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Summy JM, Gallick GE. Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev. 2003;22:337–358. doi: 10.1023/a:1023772912750. [DOI] [PubMed] [Google Scholar]
- 17.Donella-Deana A, Cesaro L, Ruzzene M, Brunati AM, Marin O, Pinna LA. Spontaneous autophosphorylation of Lyn tyrosine kinase at both its activation segment and C-terminal tail confers altered substrate specificity. Biochemistry. 1998;37:1438–1446. doi: 10.1021/bi971332s. [DOI] [PubMed] [Google Scholar]
- 18.Finn RS, Dering J, Ginther C, et al. Dasatinib, an orally active small molecule inhibitor of both the src and abl kinases, selectively inhibits growth of basal-type/"triple-negative" breast cancer cell lines growing in vitro. Breast Cancer Res Treat. 2007;105:319–326. doi: 10.1007/s10549-006-9463-x. [DOI] [PubMed] [Google Scholar]
- 19.Huang F, Reeves K, Han X, et al. Identification of candidate molecular markers predicting sensitivity in solid tumors to dasatinib: rationale for patient selection. Cancer Res. 2007;67:2226–2238. doi: 10.1158/0008-5472.CAN-06-3633. [DOI] [PubMed] [Google Scholar]
- 20.Nam S, Kim D, Cheng JQ, et al. Action of the Src family kinase inhibitor, dasatinib (BMS-354825), on human prostate cancer cells. Cancer Res. 2005;65:9185–9189. doi: 10.1158/0008-5472.CAN-05-1731. [DOI] [PubMed] [Google Scholar]
- 21.Parsons SJ, Parsons JT. Src family kinases, key regulators of signal transduction. Oncogene. 2004;23:7906–7909. doi: 10.1038/sj.onc.1208160. [DOI] [PubMed] [Google Scholar]
- 22.Frame MC. Newest findings on the oldest oncogene; how activated src does it. J Cell Sci. 2004;117:989–998. doi: 10.1242/jcs.01111. [DOI] [PubMed] [Google Scholar]
- 23.Brabek J, Constancio SS, Shin NY, Pozzi A, Weaver AM, Hanks SK. CAS promotes invasiveness of Src-transformed cells. Oncogene. 2004;23:7406–7415. doi: 10.1038/sj.onc.1207965. [DOI] [PubMed] [Google Scholar]
- 24.Shin NY, Dise RS, Schneider-Mergener J, Ritchie MD, Kilkenny DM, Hanks SK. Subsets of the major tyrosine phosphorylation sites in Crk-associated substrate (CAS) are sufficient to promote cell migration. J Biol Chem. 2004;279:38331–38337. doi: 10.1074/jbc.M404675200. [DOI] [PubMed] [Google Scholar]
- 25.Lin CY, Strom A, Vega VB, et al. Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol. 2004;5:R66. doi: 10.1186/gb-2004-5-9-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charafe-Jauffret E, Ginestier C, Monville F, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25:2273–2284. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 27.van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 28.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 29.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang HY, Sneddon JB, Alizadeh AA, et al. Gene Expression Signature of Fibroblast Serum Response Predicts Human Cancer Progression: Similarities between Tumors and Wounds. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chi JT, Wang Z, Nuyten DS, et al. Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3:e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu R, Wang X, Chen GY, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 33.Finak G, Bertos N, Pepin F, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 34.Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 35.Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 36.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldenberg-Furmanov M, Stein I, Pikarsky E, et al. Lyn is a target gene for prostate cancer: sequence-based inhibition induces regression of human tumor xenografts. Cancer Res. 2004;64:1058–1066. doi: 10.1158/0008-5472.can-03-2420. [DOI] [PubMed] [Google Scholar]
- 38.Park SI, Zhang J, Phillips KA, et al. Targeting SRC family kinases inhibits growth and lymph node metastases of prostate cancer in an orthotopic nude mouse model. Cancer Res. 2008;68:3323–3333. doi: 10.1158/0008-5472.CAN-07-2997. [DOI] [PubMed] [Google Scholar]
- 39.Stettner MR, Wang W, Nabors LB, et al. Lyn kinase activity is the predominant cellular SRC kinase activity in glioblastoma tumor cells. Cancer Res. 2005;65:5535–5543. doi: 10.1158/0008-5472.CAN-04-3688. [DOI] [PubMed] [Google Scholar]
- 40.Guan H, Zhou Z, Gallick GE, et al. Targeting Lyn inhibits tumor growth and metastasis in Ewing's sarcoma. Mol Cancer Ther. 2008;7:1807–1816. doi: 10.1158/1535-7163.MCT-08-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 42.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lombardo LJ, Lee FY, Chen P, et al. Discovery of N-(2-chloro-6-methyl- phenyl)-2-(6-(4-(2-hydroxyethyl)- piperazin-1-yl)-2-methylpyrimidin-4- ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J Med Chem. 2004;47:6658–6661. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 44.Olivieri A, Manzione L. Dasatinib: a new step in molecular target therapy. Ann Oncol. 2007;18 Suppl 6:vi42–vi46. doi: 10.1093/annonc/mdm223. [DOI] [PubMed] [Google Scholar]
- 45.Kurebayashi J. Possible treatment strategies for triple-negative breast cancer on the basis of molecular characteristics. Breast Cancer. 2009 doi: 10.1007/s12282-009-0111-2. [DOI] [PubMed] [Google Scholar]
- 46.Zhang XH, Wang Q, Gerald W, et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alli E, Sharma VB, Sunderesakumar P, Ford JM. Defective repair of oxidative dna damage in triple-negative breast cancer confers sensitivity to inhibition of poly(ADP-ribose) polymerase. Cancer Res. 2009;69:3589–3596. doi: 10.1158/0008-5472.CAN-08-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.