Abstract
The Spot 14 (S14; Thrsp) gene has been implicated in supporting regulated lipogenesis in mammals. S14 gene expression in liver is controlled by a wide variety of hormones and dietary factors in parallel with the major lipogenic enzyme genes. In addition, mice deleted for the S14 gene display reduced de novo lipogenesis in the lactating mammary gland. However, no decrease in hepatic lipogenesis was observed in the S14 null mouse. It was postulated that this difference could be due to the expression of a paralogous gene called S14R (S14 related; Mig12) in the liver but not mammary tissue. To test this hypothesis, we used small interfering RNA to simultaneously reduce levels of S14 and S14R in cultured primary hepatocytes. We found that rates of lipogenesis were decreased by approximately 65% in cells treated with insulin and high glucose. This reduction was associated with a decrease in total liver triacylglycerols and an altered morphology of lipid droplets. Expression of either S14 or S14R gene products was sufficient to fully restore normal lipogenesis. No change in the hepatic expression of other major lipogenic enzyme genes occurred during manipulation of S14 and/or S14R levels. These data support the hypothesis that both S14 and S14R are directly involved in supporting hepatic lipogenesis and that the two proteins play overlapping roles in this process.
S14 and S14R are paralogous, low molecular weight polypeptides that play overlapping roles in supporting the process of de novo lipogenesis in hepatocytes.
Spot 14 (S14/Thrsp) is an acidic polypeptide of about 17 kDa identified by two-dimensional gel electrophoresis of rat liver translational products (1). Initial interest in S14 stemmed from its robust and rapid response to thyroid hormone treatment, allowing it to serve as a useful model for exploring the basis of hormone action (2,3,4). Although the physiological role of S14 is unknown, several lines of indirect evidence implicated S14 in some aspect of the lipogenesis process. For example, S14 mRNA is expressed most abundantly in liver, white and brown adipose tissue, and lactating mammary gland, major sites of active, regulated lipogenesis (5). Its expression is induced in liver by feeding of a high-carbohydrate, low-fat diet and in cultured cells by insulin and elevated glucose metabolism (6,7). In this regard, S14 has been identified as a target gene for both the glucose-responsive transcription factor carbohydrate regulatory element binding protein (ChREBP) and the insulin-responsive factor sterol regulatory element-binding protein-1c (8,9,10). In contrast, conditions that inhibit lipogenesis, such as fasting, glucagon, or cAMP, strongly repress its expression (11). Finally, S14 gene expression in the liver is dramatically up-regulated at the time of weaning when pups switch from the high-fat diet of milk to needing to synthesize their own fatty acids (12). Premature weaning of pups causes a corresponding earlier activation of S14 gene expression.
Despite the preponderance of evidence supporting a role in lipogenesis, direct evidence in this regard was not obtained until the generation of a mouse deleted for the S14 gene. Surprisingly, these animals showed no decrease in the rate of lipogenesis in liver or adipose tissue (13). In fact, a modest increase in de novo lipogenesis was observed in liver. However, pups of homozygous null dams were found to be growth retarded during the suckling period (14). This phenotype was traced to a deficiency in triacylglycerol content in the milk produced by the knockout dams. In particular, the milk contained reduced levels of medium-chain length fatty acids, the principal form produced from de novo lipogenesis in the mammary gland. Correspondingly, a 60% decrease in the rate of de novo lipogenesis in mammary tissue from the knockout dams was found. Hence, S14 is important for effective de novo lipogenesis in the mammary gland.
These observations raised the question of why no inhibition in de novo lipogenesis was detected in the livers of S14 knockout animals. One explanation proposed for this paradox was the existence of a paralog to S14, designated S14-related (S14R) or Mig12, in vertebrate genomes (14). The S14R polypeptide is slightly longer than S14 (183 vs. 150 residues) and has 36% amino acid identity with S14 in the overlapping portion. Three regions of 20–33 residues are more highly conserved (40–75%). The S14R gene is expressed in the liver and was recently shown to be a glucose-responsive target of ChREBP in the liver (15). It was postulated that expression of S14R in the liver of knockout mice could compensate for the loss of S14 in these mice to support lipogenesis. In contrast, S14R is expressed at very low levels in the mammary gland and hence might be insufficient to counteract the loss of S14 in mammary lipogenesis. Whereas this presented a plausible explanation, direct evidence supporting this hypothesis has been lacking. We therefore tested the effect of reducing both S14 and S14R expression simultaneously on de novo lipogenesis in cultured primary hepatocytes. These studies revealed a dramatic reduction in de novo lipogenesis and accumulation of triacylglycerols. We also demonstrate the ability of either paralog to rescue the reduced level of de novo lipogenesis. These experiments support the model that S14 and S14R play overlapping and partially redundant roles in the process of de novo lipogenesis.
Materials and Methods
Materials
Effectene reagent and AllStars negative control small interfering RNA (siRNA) were obtained from QIAGEN (Valencia, CA). Triacylglycerol measurements were made using the enzymatic triglyceride kit from Stanbio Laboratories (Boerne, TX). [1-14C]acetic acid was obtained from GE Healthcare BioSciences Corp. (Piscataway, NJ). Cell culture reagents and Trizol were obtained from Invitrogen (Carlsbad, CA) and reagents for RT-PCR from Bio-Rad Laboratories (Hercules, CA). All other reagents were from Sigma-Aldrich (St. Louis, MO).
Culturing and siRNA treatment of hepatocytes
Hepatocytes were isolated from male Sprague Dawley rats (Harlan, Madison, WI) fed ad libitum and weighing 200–300 g by collagenase perfusion (16). All procedures involving animals were approved by the University of Minnesota Institutional Animal Care and Use Committee. Cells were plated in medium 199 containing 11 mm glucose, 23 mm HEPES, 10 nm dexamethasone, 0.1 U/ml insulin, 26 mm Na bicarbonate, 2 mm glutamine, 50 U/ ml penicillin, 50 μg/ml streptomycin, and 10% fetal bovine serum. Cells were immediately transfected with siRNA (1 μg per 106 cells) using the Effectene reagent according to the manufacturer’s protocol. Dicer-substrate siRNA duplexes were obtained from Integrated DNA Technologies (Coralville, IA; S14: NM_012703-10.3; S14R: NM_206950-10.10). The nontargeting siRNA control was AllStars negative control siRNA. After 6 h, cells were switched to media containing 27.5 mm glucose and no fetal bovine serum (high-glucose media) and maintained for 40 h with one change of media. In rescue experiments, cells were transduced with adenovirus expressing either S14 or S14R from the cytomegalovirus enhancer/promoter after the 6-h siRNA treatment. After 2 h of transduction, virus-containing media were removed and cells were incubated in the high-glucose media for 40 h.
Radiolabeling, extraction, and analysis of lipids
Cells were pulsed labeled with 1–10 μCi of 14C-acetate for 30 min. Subsequently cells were collected and lipids were extracted with methanol-chloroform (2:1) as described (17). After extraction, cellular lipids were counted for determination of total incorporation or separated by thin-layer chromatography (TLC) on silica gel G plates for determination of incorporation into different lipid fractions (18). A solvent system comprised of hexane-diethyl ether-acetic acid (80:20:2) was used for separation of fatty acids, triacylglycerols, and phospholipids and a system of benzene-diethyl ether-ethyl acetate-acetic acid (80:10:10:2) was used to distinguish diacylglycerol isomers and cholesterol. Pure standards were used to identify lipid species, which were subsequently scraped and quantified by liquid scintillation counting. Total triacylglycerols were measured using the enzymatic triacylglycerol procedure on lipids extracted as described above. Visualization of lipid droplets in hepatocytes was performed by fixing cells in 4% formaldehyde and staining with 1% Oil Red O for 1 h.
Measurement of mRNA levels
Total cellular RNA was isolated from cells treated with siRNAs for 40 h using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. Specific mRNA levels were determined using a two-step real-time RT-PCR protocol as described previously (9). The results of RT-PCR for each mRNA were normalized to TATA binding protein mRNA levels as an internal standard and then expressed relative to the mRNA levels in cells treated with the negative control siRNA. Primer sequences are available on request.
Statistics
Data are presented as mean ± sd for single experiments and mean ± sem when multiple experiments were pooled. Comparisons between treatments were made by t test for two groups or ANOVA if there were more than two groups. Data were log transformed when necessary to achieve homogeneity of variances between groups. Post hoc comparisons between treatment groups were made using Tukey’s honestly significant difference method in R (R Foundation for Statistical Computing, Vienna, Austria; http://www.r-project.org) and were considered significantly different if the adjusted P value was less than 0.05.
Results
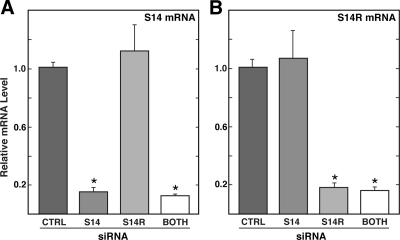
To evaluate the roles of S14 and S14R in the liver, siRNAs were designed to specifically target the 3′-untranslated region of each mRNA. Introduction of these siRNAs into primary hepatocytes resulted in reductions in the mRNA levels of greater than 80% (Fig. 1). No cross-reaction between the two siRNAs that affected the paralogous gene product was observed. Importantly, simultaneous introduction of siRNA targeting both S14 and S14R was as effective as either siRNA alone.
Figure 1.
Effectiveness and specificity of S14 and S14R siRNA. Primary hepatocytes were treated with control (CTRL), S14, S14R, or both S14 and S14R siRNA as indicated for 6 h. Subsequently cells were incubated in high-glucose media for an additional 40 h. Total RNA was isolated and relative levels of S14 mRNA (A) or S14R mRNA (B) were measured by RT-PCR. Values are expressed relative to levels seen in cells treated with control siRNA and represent the means (±sd) of five samples. *, Groups that are statistically different from the control group (P ≤ 0.001).
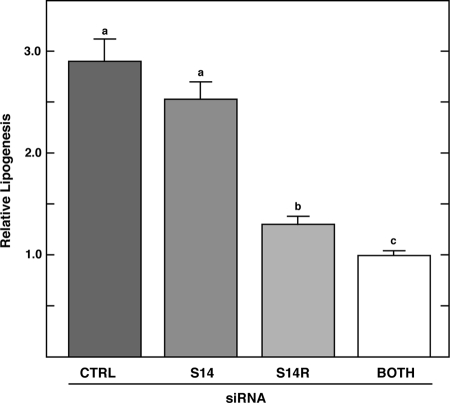
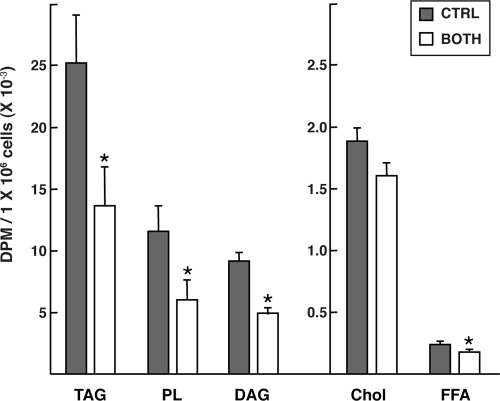
Rates of de novo lipogenesis in cells treated with S14 or S14R siRNAs were measured using 14C-acetate incorporation into lipid (Fig. 2). Cells were treated for 40 h with high glucose plus insulin to stimulate maximal rates of lipogenesis. Knocking down S14 mRNA alone did not result in a significant decrease in the rate of de novo lipogenesis, consistent with earlier observations from the S14 null mouse (13). In contrast, targeting S14R mRNA led to a significant decrease of about 55% in acetate incorporation into lipids. The inhibition of de novo lipogenesis was even stronger in cells treated with both S14 and S14R siRNAs, reaching a level of about 65% inhibition. Analysis of lipids by TLC chromatography demonstrated that the inhibition occurred in all major fractions of fatty acid-derived lipids, including triacylglycerols, phospholipids, and diacylglycerols but not in the cholesterol fraction (Fig. 3). Hence, both S14 and S14R contribute to the process of de novo fatty acid synthesis at a step before fatty acid incorporation into various lipid classes.
Figure 2.
Rates of de novo lipogenesis in S14 and S14R siRNA-treated cells. Primary hepatocytes were cultured and transfected with siRNA as indicated in Materials and Methods. Forty hours after treatment with siRNA, 14C-acetate (1–5 μCi) was added and cells harvested 30 min later for measurement of de novo lipogenesis. The rate of lipogenesis was normalized to that observed in cells transfected with both S14 and S14R siRNA in each experiment. The data were pooled for seven independent experiments and are presented as mean (±sem) with n = 13, 12, 12, or 19 for control (CTRL), S14, S14R, and both S14 + S14R siRNA, respectively. Bars with different letters are significantly different from each other (P < 0.012).
Figure 3.
Knocking down S14 and S14R reduces synthesis of all major fatty acid-derived lipid products. Cells were treated as described in the legend to Fig. 2 except that 10 μCi of 14C-acetate was used for labeling. Lipids were extracted and then separated by TLC as described in Materials and Methods. Results are average (±sd) of four samples for cells treated with the control siRNA (CTRL) or both S14 and S14R siRNAs (BOTH). Values marked with an asterisk are significantly different (P < 0.01) in S14 and S14R siRNA-treated cells compared with the control siRNA-treated cells. TAG, Triacylglycerol; PL, phospholipid; DAG, diacylglycerol; Chol, Cholesterol; FFA, free fatty acid.
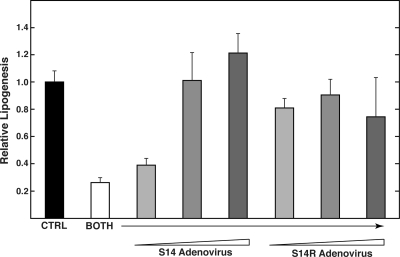
To test the hypothesis that S14 and S14R play overlapping roles in supporting lipogenesis, we tested the ability of each gene product to rescue the double knockdown cells. S14 and S14R coding sequences were inserted into an adenoviral vector behind the cytomegalovirus promoter. These constructs lacked the 3′-untranslated regions from the S14 and S14R mRNAs, which are substituted by a heterologous 3′-untranslated region. Consequently, the expression of the exogenous S14 or S14R is not targeted for inhibition by the siRNAs used in this study. Cells were first treated with siRNA for 6 h and then transduced with either S14- or S14R-expressing adenovirus. We found that either S14 or S14R was capable of fully rescuing the reduced rate of de novo lipogenesis caused by the double knockdown (Fig. 4). Hence, both polypeptides are capable of independently supporting fatty acid synthesis, indicating an overlapping role for these two related gene products.
Figure 4.
Both S14 and S14R are capable of fully rescuing de novo lipogenesis. Primary hepatocytes were treated with either control siRNA (CTRL) or both S14 and S14R siRNA (BOTH) for 6 h. Subsequently cells were treated with increasing amounts of adenovirus expressing either S14 or S14R for 2 h and then incubated for 40 h in high-glucose media. The rates of lipogenesis were measured as described in the legend to Fig. 2 and are expressed relative to that observed in control siRNA-treated cells. Values represent means (±sd) of four samples. In control experiments, transduction of hepatocytes with an adenovirus expressing green fluorescent protein did not affect the rates of lipogenesis.
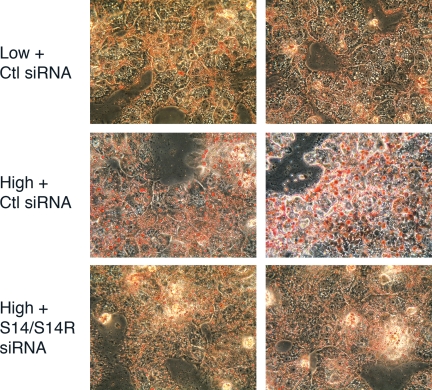
To examine whether the decreased de novo lipogenesis affects the accumulation of triacylglycerols in the cell, measurements were made on triacylglycerol levels in cells treated with both S14 and S14R siRNAs. The level of triacylglycerols in the double-knockdown cells was approximately 80% of that seen in control cells. An even more dramatic effect was observed in staining cells with Oil Red O to detect lipid droplets (Fig. 5). Cells cultured in high glucose plus insulin for 24 h possessed large lipid droplets typical of steatotic liver. However, in the double-knockdown cells, the lipid droplet size was dramatically reduced and resembled the lipid droplets found in cells maintained in low (5.5 mm) glucose. These results demonstrate a clear-cut defect in the process of triacylglycerol formation and storage as a result of simultaneously reducing both S14 and S14R levels in the hepatocytes.
Figure 5.
Accumulation of lipid droplets in hepatocytes treated with S14 and S14R siRNA. Primary hepatocytes were treated with either control (Ctl) siRNA or both S14 and S14R siRNA for 6 h. Subsequently cells were incubated for 40 h in either low- (5.5 mm) or high-glucose media. Cells were stained with Oil Red O to visualize neutral lipids.
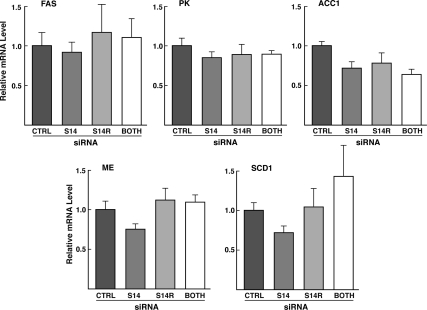
Two general explanations could account for the actions of S14/S14R on lipogenesis. The first is that S14 and S14R polypeptides could play a direct role in the induction of lipogenic enzyme gene products that occurs in conditions of high glucose plus insulin, as proposed by Kinlaw et al. (19). In this case, the reduced rate of lipogenesis would be a consequence of inadequate enzyme levels of key lipogenic enzyme genes. The second possible explanation is that S14/S14R play a direct role in the process of lipogenesis. To distinguish between these possibilities, we measured the mRNA levels of a number of key lipogenic enzyme genes that are known to be induced in hepatocytes under lipogenic conditions. Treatment of cells with S14 and S14R siRNAs did not lead to any significant decreases in the levels of mRNAs for fatty acid synthase, acetyl-CoA carboxylase 1, stearoyl-CoA desaturase 1, L-type pyruvate kinase, or malic enzyme (Fig. 6). Therefore, the actions of S14 and S14R are likely to be directly involved in influencing the process of de novo lipogenesis.
Figure 6.
Measurement of mRNA levels for other lipogenic enzymes in cells treated with S14 and S14R siRNA. Primary hepatocytes were transfected with control siRNA, S14 siRNA, S14R siRNA, or both S14 and S14R siRNA for 6 h. Subsequently cells were incubated in high-glucose media for 40 h. RNA was extracted and specific mRNA levels were measured by RT-PCR. FAS, Fatty acid synthase; PK, L-type pyruvate kinase, ACC-1, acetyl-CoA carboxylase 1; SCD-1, stearoyl-CoA desaturase 1; ME, malic enzyme. Values are means (±sem) of six to 12 samples pooled from two experiments. For each mRNA, ANOVA showed no significant difference between groups. The reduction of S14 and S14R mRNAs in these experiments were greater than 80% (data not shown).
Discussion
The observations of this study strongly implicate both S14 and S14R in supporting the process of elevated fatty acid synthesis and lipogenesis that occurs in conditions of excess carbohydrate consumption. In the presence of high glucose and insulin, the expression of both S14 and S14R is induced in the liver or in cultured hepatocytes. By reducing these products on the order of 80% in these conditions, the levels of each are more comparable to what would be observed in cells maintained in 5.5 mm glucose. These levels are insufficient to support the enhanced rate of de novo lipogenesis that occurs in stimulating conditions. We also examined the effects of knocking down S14 and S14R in hepatocytes cultured in low-glucose conditions. In this case, we found that the rate of lipogenesis was reduced, albeit to a lesser extent (∼30–35%) than seen in cells incubated in high glucose (∼65%). The smaller effect in low glucose could reflect that a greater proportion of acetate is incorporated into cholesterol in these conditions. Nevertheless, it points out that the role of S14 and S14R in supporting fatty acid production may not be limited to conditions of stimulated lipogenesis but instead be more general.
A role for S14 in supporting lipogenesis has been indicated from previous work on the S14 knockout mouse (14). In these animals, reduced S14 levels in the lactating mammary gland are associated with a reduced capacity for de novo lipogenesis and inadequate production of triacylglycerols in the milk. Unlike the liver and adipose tissue, which express both S14 and S14R, the lactating mammary gland expresses almost exclusively S14. This observation may imply a specialized function for S14 in mammary gland lipogenesis. It would be interesting to determine whether S14R could compensate for the reduced rate of de novo lipogenesis observed in mammary glands from S14 knockout mice. S14 has also been implicated in playing a role in breast cancers. The S14 gene is part of an amplicon found in certain forms of breast cancer (20). High levels of S14 expression correlate with poor prognosis (21). In this regard, S14 is similar to fatty acid synthase, which is expressed at high levels in breast cancers with high metastatic potential (22,23). Hence, increased lipogenesis in these tumors in response to S14 and/or fatty acid synthase overexpression appears to provide a significant growth advantage to these transformed cells.
S14 has also been recently implicated in increased lipogenesis induced by activators of the pregnane X receptor in human hepatocytes (24). The S14 gene was shown to be a direct target for the pregnane X receptor, leading to elevated S14 expression in response to xenobiotic and drug activators of this nuclear hormone receptor. Furthermore, overexpression of S14 in human hepatoma HepaRG cells resulted in an increase in de novo lipogenesis.
In contrast to S14, there are few previous data supporting a role of S14R in lipogenesis. S14R was first identified as a protein that associates with Mid1, a microtubule-associated E3 ligase (25). It was proposed to help bundle and stabilize microtubules in the central nervous system during development. We have shown that S14R is induced by feeding a high-carbohydrate diet to mice or elevated glucose levels in cultured primary rat hepatocytes (15). This induction is mediated by the transcription factor ChREBP, which binds to the S14R promoter. Because most targets of ChREBP in the liver are related to the process of lipogenesis (26), these observations are consistent with the possibility that S14R is also involved in supporting fatty acid synthesis. S14R was also recently found to be transiently up-regulated in androgen-stimulated rat prostate, although its role in these cells was not further characterized (27). Consequently, the observations of the current study that expression of S14R could rescue the reduced level of lipogenesis seen in the double-knockdown cells represent the first direct evidence for a role of S14R in the lipogenic process.
In this study, the effect of knocking down S14R mRNA by itself on lipogenesis was significantly greater than that caused by reducing S14 mRNA. There are several possible explanations for the greater effectiveness of the S14R knockdown. First, the pool sizes of S14 and S14R proteins in the liver are unknown, so it is possible that S14R is present at greater levels. Second, it is possible that S14R is more effective in promoting lipogenesis than is S14. However, because the mammary gland expresses predominantly S14, this protein is sufficient to meet the lipogenic needs in that tissue. In the rescue experiment, either S14 or S14R expression led to restoration of lipogenic rates in cells treated with both siRNAs. Hence, both proteins are capable of supporting lipogenesis. However, in this experiment, S14 or S14R mRNA was expressed at significantly higher levels than endogenous mRNA, and hence, the efficiency of each protein in supporting lipogenesis cannot be assessed. Although both proteins supported enhanced lipogenesis individually, the maintenance of two conserved genes in evolution argues that they are optimized for specific functional roles. In this regard, we found that S14R expression is not altered by thyroid hormone treatment of hepatocytes (data not shown), whereas S14 is one of the most highly thyroid-responsive gene products in liver. These differences in regulation of S14 and S14R genes suggest differences in their specific roles. Of the two genes, S14R is conserved in all vertebrate species from teleost fish onward (14). S14 genes are found only with the evolutionary appearance of mammals and the demand for de novo lipogenesis in mammary tissue. But why S14 is expressed as the principal form in this tissue, whereas liver and adipose express both, remains to be determined.
What then is the actual role of these proteins in supporting de novo lipogenesis? The original hypothesis for the function of S14 suggested that it was involved in supporting the increased transcriptional induction of lipogenic genes in response to high-glucose and thyroid hormone stimulation (19). This hypothesis was based on the use of an antisense DNA oligonucleotide introduced into hepatocytes to reduce S14 mRNA levels. This treatment resulted in a reduced rate of lipogenesis and a dramatic inhibition of the induction of lipogenic enzyme genes by glucose and thyroid hormone. However, in the present study, no reduction in lipogenic enzyme gene expression was found as a result of S14 and/or S14R knockdown. Similarly, in the S14 null mice, no decrease in lipogenic enzyme mRNA or activities was found in mammary tissue despite the reduced rates of lipogenesis (14). One explanation for the differences in these studies may be that the antisense S14 study examined its effects on cells treated with both thyroid hormone and glucose. Perhaps the antisense S14 oligonucleotide somehow perturbed the thyroid hormone induction of lipogenic enzyme genes. Alternatively, it is possible that the effects of the antisense DNA on lipogenic gene expression are related to an off-target effect of the particular antisense oligonucleotide used. In this regard, it is notable that induction of type I 5′-deiodinase by thyroid hormone was also blocked by the antisense treatment, despite the fact that this gene product is not directly involved in the lipogenic process (28). In the present study, no effect of S14 and or S14R knockdown on deiodinase mRNA was found (data not shown). In addition, the rescue of the phenotype caused by the siRNA treatment with either S14 or S14R argues a direct, rather than off-target, action. Together we conclude that S14 and S14R are not involved in the transcriptional induction of lipogenic enzyme genes but instead play a direct role in supporting the lipogenic process.
What might this role be? Two observations are relevant in considering their function. First, the synthesis of all major classes of fatty acid-containing lipids was reduced in the double-knockdown animals. This implies a defect upstream from the accumulation of fatty acyl-CoA, the common precursor for synthesis of these various lipids. Second, in lactating mammary glands of S14 knockout mice, it was found that malonyl-CoA levels were approximately 30% higher than in wild-type animals (29). Hence, the defect in lipogenesis likely occurs at some step downstream of malonyl-CoA synthesis by acetyl-CoA carboxylase. The only two steps between these two stages are the fatty acid synthase and acyl-CoA synthetase reactions. We would suggest that S14/S14R might be involved in one of these steps. One possibility is that S14/S14R could help to deliver substrate to or from one of these critical enzymes. Another possibility is that they could help to organize the enzymes involved in the final stages of fatty acid synthesis within the cell. In this regard, it is noteworthy that S14R has previously been found to interact with a microtubule-associated protein, Mid1 (25). Hence, the coupling of the fatty acid synthetic machinery on a microtubule network might facilitate enhanced processing of the products of this pathway in tissues capable of high rates of lipogenesis.
Acknowledgments
We thank Katie Ress for excellent technical assistance.
Footnotes
This work was supported by National Institutes of Health (NIH) Grants DK26919 (to H.C.T.) and P30-DK50456 (to the Minnesota Obesity Center). D.L.A. was supported by NIH Training Grant T32-DK007203 (to C.N.M.).
Disclosure Summary: The authors have nothing to declare.
First Published Online March 16, 2010
Abbreviations: ChREBP, Carbohydrate regulatory element binding protein; S14, Spot 14; siRNA, small interfering RNA; S14R, Spot 14 related; TLC, thin-layer chromatography.
References
- Seelig S, Liaw C, Towle HC, Oppenheimer JH 1981 Thyroid hormone attenuates and augments hepatic gene expression at a pretranslational level. Proc Natl Acad Sci USA 78:4733–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jump DB, Narayan P, Towle H, Oppenheimer JH 1984 Rapid effects of triiodothyronine on hepatic gene expression. Hybridization analysis of tissue-specific triiodothyronine regulation of mRNAS14. J Biol Chem 259:2789–2797 [PubMed] [Google Scholar]
- Liu HC, Towle HC 1994 Functional synergism between multiple thyroid hormone response elements regulates hepatic expression of the rat S14 gene. Mol Endocrinol 8:1021–1037 [DOI] [PubMed] [Google Scholar]
- Mariash CN, Jump DB, Oppenheimer JH 1984 T3 stimulates the synthesis of a specific mRNA in primary hepatocyte culture. Biochem Biophys Res Commun 123:1122–1129 [DOI] [PubMed] [Google Scholar]
- Jump DB, Oppenheimer JH 1985 High basal expression and 3,5,3′-triiodothyronine regulation of messenger ribonucleic acid S14 in lipogenic tissues. Endocrinology 117:2259–2266 [DOI] [PubMed] [Google Scholar]
- Liaw C, Seelig S, Mariash CN, Oppenheimer JH, Towle HC 1983 Interactions of thyroid hormone, growth hormone, and high carbohydrate, fat-free diet in regulating several rat liver messenger ribonucleic acid species. Biochemistry 22:213–221 [DOI] [PubMed] [Google Scholar]
- Topliss DJ, Mariash CN, Seelig S, Carr FE, Oppenheimer JH 1983 Effects of triiodothyronine and glucose on cultured rat hepatocyte gene expression. Endocrinology 112:1868–1870 [DOI] [PubMed] [Google Scholar]
- Jump DB, Thelen AP, Mater MK 2001 Functional interaction between sterol regulatory element-binding protein-1c, nuclear factor Y, and 3,5,3′-triiodothyronine nuclear receptors. J Biol Chem 276:34419–34427 [DOI] [PubMed] [Google Scholar]
- Ma L, Tsatsos NG, Towle HC 2005 Direct role of ChREBP. Mlx in regulating hepatic glucose-responsive genes. J Biol Chem 280:12019– 12027 [DOI] [PubMed] [Google Scholar]
- Stoeckman AK, Ma L, Towle HC 2004 Mlx is the functional heteromeric partner of the carbohydrate response element-binding protein in glucose regulation of lipogenic enzyme genes. J Biol Chem 279:15662–15669 [DOI] [PubMed] [Google Scholar]
- Kinlaw WB, Schwartz HL, Hamblin PS, Mariash CN, Oppenheimer JH 1988 Triiodothyronine rapidly reverses inhibition of S14 gene transcription by glucagon. Endocrinology 123:2255–2260 [DOI] [PubMed] [Google Scholar]
- Perez-Castillo A, Schwartz HL, Oppenheimer JH 1987 Rat hepatic mRNA-S14 and lipogenic enzymes during weaning: role of S14 in lipogenesis. Am J Physiol 253:E536–E542 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Mariash A, Margosian MR, Gopinath S, Fareed MT, Anderson GW, Mariash CN 2001 Spot 14 gene deletion increases hepatic de novo lipogenesis. Endocrinology 142:4363–4370 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Anderson GW, Mucha GT, Parks EJ, Metkowski JK, Mariash CN 2005 The Spot 14 protein is required for de novo lipid synthesis in the lactating mammary gland. Endocrinology 146:3343–3350 [DOI] [PubMed] [Google Scholar]
- Tsatsos NG, Augustin LB, Anderson GW, Towle HC, Mariash CN 2008 Hepatic expression of the SPOT 14 (S14) paralog S14-related (Mid1 interacting protein) is regulated by dietary carbohydrate. Endocrinology 149:5155–5161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry MN, Friend DS 1969 High-yield preparation of isolated rat liver parenchymal cells: a biochemical and fine structural study. J Cell Biol 43:506–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ 1959 A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917 [DOI] [PubMed] [Google Scholar]
- Sapiro JM, Mashek MT, Greenberg AS, Mashek DG 2009 Hepatic triacylglycerol hydrolysis regulates PPAR-α activity. J Lipid Res 50:1621–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinlaw WB, Church JL, Harmon J, Mariash CN 1995 Direct evidence for a role of the “spot 14” protein in the regulation of lipid synthesis. J Biol Chem 270:16615–16618 [DOI] [PubMed] [Google Scholar]
- Moncur JT, Park JP, Memoli VA, Mohandas TK, Kinlaw WB 1998 The “Spot 14” gene resides on the telomeric end of the 11q13 amplicon and is expressed in lipogenic breast cancers: implications for control of tumor metabolism. Proc Natl Acad Sci USA 95:6989–6994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells WA, Schwartz GN, Morganelli PM, Cole BF, Gibson JJ, Kinlaw WB 2006 Expression of “Spot 14” (THRSP) predicts disease free survival in invasive breast cancer: immunohistochemical analysis of a new molecular marker. Breast Cancer Res Treat 98:231–240 [DOI] [PubMed] [Google Scholar]
- Kuhajda FP 2006 Fatty acid synthase and cancer: new application of an old pathway. Cancer Res 66:5977–5980 [DOI] [PubMed] [Google Scholar]
- Swinnen JV, Brusselmans K, Verhoeven G 2006 Increased lipogenesis in cancer cells: new players, novel targets. Curr Opin Clin Nutr Metab Care 9:358–365 [DOI] [PubMed] [Google Scholar]
- Moreau A, Téruel C, Beylot M, Albalea V, Tamasi V, Umbdenstock T, Parmentier Y, Sa-Cunha A, Suc B, Fabre JM, Navarro F, Ramos J, Meyer U, Maurel P, Vilarem MJ, Pascussi JM 2009 A novel pregnane X receptor and S14-mediated lipogenic pathway in human hepatocyte. Hepatology 49:2068–2079 [DOI] [PubMed] [Google Scholar]
- Berti C, Fontanella B, Ferrentino R, Meroni G 2004 Mig12, a novel Opitz syndrome gene product partner, is expressed in the embryonic ventral midline and co-operates with Mid1 to bundle and stabilize microtubules. BMC Cell Biol 5:9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Robinson LN, Towle HC 2006 ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J Biol Chem 281:28721–28730 [DOI] [PubMed] [Google Scholar]
- Nishi N, Shoji H, Miyanaka H, Nakamura T 2008 Transient up-regulation of a novel member of Spot 14 family in androgen-stimulated rat prostate. Biochim Biophys Acta 1780:1004–1009 [DOI] [PubMed] [Google Scholar]
- Brown SB, Maloney M, Kinlaw WB 1997 “Spot 14” protein functions at the pretranslational level in the regulation of hepatic metabolism by thyroid hormone and glucose. J Biol Chem 272:2163–2166 [PubMed] [Google Scholar]
- LaFave LT, Augustin LB, Mariash CN 2006 S14: insights from knockout mice. Endocrinology 147:4044–4047 [DOI] [PubMed] [Google Scholar]