Abstract
It is expected that clinically-obtainable fluids that are proximal to organs contain a repertoire of secreted proteins and shed cells reflective of the physiological state of that tissue, and thus represent potential sources for biomarker discovery, investigation of tissue-specific biology, and assay development. The prostate gland secretes many proteins into a prostatic fluid that combines with seminal vesicle fluids to promote sperm activation and function. Proximal fluids of the prostate that can be collected clinically are seminal plasma and expressed prostatic secretion (EPS) fluids. In the current study, MudPIT-based proteomics was applied to EPS obtained from nine men with prostate cancer and resulted in the confident identification of 916 unique proteins. Systematic bioinformatics analyses using publicly-available microarray data of 21 human tissues (Human Gene Atlas), the Human Protein Atlas database and other published proteomics data of shed/secreted proteins were performed to systematically analyze this comprehensive proteome. Therefore, we believe this data will be a valuable resource for the research community to study prostate biology and potentially assist in the identification of novel prostate cancer biomarkers. To further streamline this process, the entire data set was deposited to the Tranche repository for use by other researchers.
Introduction
Proximal fluids are found adjacent to a given tissue or organ and contain a repertoire of secreted proteins and shed cells reflective of the physiological state of that tissue 1. Hence, proximal fluids are rapidly emerging as discovery sources of proteins, metabolites, and genetic biomarkers for cancers. Seminal plasma and expressed prostatic secretion (EPS) are prostate-proximal fluids that can be collected in the clinic. Seminal plasma consists of fluids originating not only from the prostate, but also from several other male accessory glands such as the seminal vesicles, epididymis, and Cowper’s gland. In aging men with prostatic disease, obtaining seminal fluids for diagnostic assays and screening purposes is highly dependent on patient age and disease status, and thus not an attractive assay medium. In contrast, EPS can be readily obtained in the clinic by forced (or expressed) ejection of prostatic fluids into the urethra following prostate massage. For evaluation of some types of prostatitis, a vigorous massage is done to force fluid through the urethra to collect a few drops from the tip of the penis for subsequent use in microorganism cultures. If a more gentle massage is performed in the context of a digital rectal exam (DRE) of the prostate, the EPS is then immediately collected in voided urine post-exam (termed post-DRE urine, or post-DRE EPS urine). Currently, a commercial genetic assay for prostate cancer detection, based on the presence of a non-coding RNA (PCA3), uses post-DRE EPS urines as a source of prostate-derived genetic material 2–4.
In the context of prostate cancer, undiluted EPS can be collected while the patient is under anesthesia, just prior to prostatectomy. In comparison to the clinical DRE massages, a more vigorous digital rectal massage can be done to force prostatic fluids into the urethra with subsequent collection from the penis. For prostate cancer biomarker discovery purposes, direct EPS has several ideal clinical features beyond its nature as a proximal fluid. Since EPS are collected just prior to prostatectomy, clinical information that led to the decision to perform a prostatectomy is readily available 1. Post-prostatectomy pathology staging and grading can also be obtained for comparison with the pre-surgical assessments. When linked with clinical outcome data, these direct EPS fluids thus have the potential to be used for discovery of both diagnostic and prognostic biomarkers for management of prostate cancer. A large number of pre-prostatectomy EPS fluids have been collected by our translational research group 1, which we term ‘direct EPS’ to distinguish them from other EPS designations.
In this study, we applied a MudPIT-based proteomics approach to obtain a detailed inventory of proteins present in pre-prostatectomy direct EPS samples. By using a high mass accuracy Orbitrap MS and rigorous identification criteria, we provide a high quality accounting of prostatic secretions to a degree not previously reported. To further increase the utility of the current proteomic resource, we have systematically integrated our data with a multitude of publicly available databases (i.e. human tissue mRNA microarray, the Human Protein Atlas, alternative proteomics data). Additionally, the entire proteomics data was deposited to the Tranche server (www.proteomecommons.com) for others to utilize. We hypothesize that this comprehensive proteomic analysis of direct EPS will provide an important resource to study prostate biology and/or facilitate the discovery of novel prostate cancer biomarkers.
Materials and Methods
Materials
Ultrapure-grade urea, ammonium acetate, and calcium chloride were from BioShop Canada Inc. (Burlington, ON, Canada). Ultrapure-grade iodoacetamide, DTT and formic acid were obtained from Sigma. HPLC-grade solvents (methanol, acetonitrile and water) were obtained from Fisher Scientific. Mass spectrometrygrade trypsin gold was from Promega (Madison, WI, USA). Solid-phase extraction C18 MacroSpin Columns were from The Nest Group, Inc. (Southboro, MA, USA).
Direct EPS Sample Properties
Direct EPS samples were collected from patients following informed consent and use of Institutional Review Board approved protocols at Urology of Virginia, Sentara Medical Group and Eastern Virginia Medical School between 2007 and 2009 (Table 1). Direct EPS fluids were obtained by vigorous prostate massage from subjects under anaesthesia just prior to prostatectomy as a result of a prostate cancer diagnosis. Each sample (0.2–1 ml) was collected from the tip of the penis in a 10 ml tube, diluted with saline to 5 ml, and stored on ice until transport to the biorepository for processing (within 1 hour of collection). For processing, any particulate material was removed by low-speed centrifugation. Aliquots (1 ml) of the supernatant were stored at −80°C. Tumor grades, staging, prostate volumes and tumor volumes 5 were determined using standard pathological procedures. Patient information was recorded, including demographics, medical history, pathology results, and risk factors, and stored in a Caisis database system. All personal information or identifiers beyond diagnosis and lab results were not available to the laboratory investigators.
Table 1.
Clinical information of the EPS fluids analyzed by MudPIT proteomics
| #1 | #2 | #3 | #4 | #5 | #6 | #7 | #8 | #9 | |
|---|---|---|---|---|---|---|---|---|---|
| Age | 64 | 53 | 67 | 65 | 70 | 50 | 61 | 69 | 59 |
| PSAa | 37.5 | 6.7 | 7.3 | 4.8 | 5.1 | 4.1 | 3.5 | 8.7 | 6.2 |
| Stage | T2b | T1c | T1c | T1c | T1c | T1c | T1c | T1c | T1c |
| Gleasonb | 4+3 | 3+4 | 3+4 | 3+3 | 3+4 | 3+3 | 3+4 | 3+4 | 3+3 |
|
Percent Tumorc |
75 | 25 | 2 | 2 | 5 | 5 | 20 | 40 | 1 |
|
Prostate Volumed |
53 | 41 | 56 | 31 | 96 | 48 | 70 | 42 | 73 |
Serum PSA values, ng/ml
Gleason score determined by pathology post-prostatectomy
Determined by post-prostatectomy pathology
Determine pre-prostatectomy by transurethral ultrasound measurements
Protein digestion and sample preparation
Direct EPS fluid corresponding to 100 µg of total protein (as measured by Bradford assay) were concentrated to ~ 50 µl using an Amicon Ultra spin filter (3kDa cut-off; Millipore) followed by digestion as stated below. Briefly, concentrated EPS corresponding to 100 µg of total protein was diluted with an equal volume of 8M urea, 4mM DTT, 50mM Tris-HCl, pH 8.5 and incubated at 37°C for 1 hour, followed by carbamidomethylation with 10mM iodoacetamide for 1 hour at 37°C in the dark. Samples were diluted with 50mM ammonium bicarbonate, pH 8.5 to ~ 1.5M urea. Calcium chloride was added to a final concentration of 1mM and the protein mixture digested with a 1:30 molar ratio of recombinant, proteomics-grade trypsin at 37°C overnight. The resulting peptide mixtures were solid phase-extracted with C18 MacroSpin Columns from The Nest Group, Inc. (Southboro, MA, USA). Samples were concentrated by speed-vac and stored at −80°C until used for MudPIT analysis.
MudPIT analysis
A fully automated 9-cycle two-dimensional chromatography sequence was set up as previously described 6. Peptides were loaded on a 7cm pre-column (150 µm i.d.) containing a Kasil frit packed with 3.5 cm 5µMagic C18 100 Å reversed phase material (Michrom Bioresources) followed by 3.5 cm LunA® 5µSCX 100Å strong cation-exchange resin (Phenomenex, Torrance, CA). Samples were automatically loaded from a 96-well microplate autosampler using the EASY-nLC system (Proxeon Biosystems, Odense, Denmark) at 3 µl/minute. The pre-column was connected to an 8cm fused silica analytical column (75 µm i.d.; 5µ Magic C18 100 Å reversed phase material (Michrom Bioresources)) via a microsplitter tee (Proxeon Biosystems) to which a distal 2.3 kV spray voltage was applied. The analytical column was pulled to a fine electrospray emitter using a laser puller. For peptide separation on the analytical column, a water/acetonitrile gradient was applied at an effective flow rate of 400 nl/minute, controlled by the EASY-nLC system. Ammonium acetate salt bumps (8µl) were applied at the identical concentrations as above, using the 96-well microplate autosampler at a flow-rate of 3 µl/minute in a vented-column set-up.
Mass spectrometry
All samples were analyzed on a LTQ-Orbitrap XL. The instrument method consisted of one MS full scan (400–1800 m/z) in the Orbitrap mass analyzer, an AGC target of 500,000 with a maximum ion injection of 500 ms, 1 µscan and a resolution of 60,000 and using the preview scan option. Five data-dependent MS/MS scans were performed in the linear ion trap using the five most intense ions at 35% normalized collision energy. The MS and MS/MS scans were obtained in parallel. AGC targets were 10,000 with a maximum ion injection time of 100 ms. A minimum ion intensity of 1,000 was required to trigger a MS/MS spectrum. The dynamic exclusion was applied using a maximum exclusion list of 500 with one repeat count with a repeat duration of 30 seconds and exclusion duration of 45 seconds.
Protein identification and data analysis
Raw data was converted to m/z XML using ReAdW and searched by X!Tandem against a locally installed version of the human UniProt complete human proteome (www.uniprot.org) protein sequence database. The search was performed with a fragment ion mass tolerance of 0.4 Da, a parent ion mass tolerance of ±10 ppm. Complete tryptic digest was assumed. Carbamidomethylation of cysteine was specified as fixed, and oxidation of methionine as variable modification. A target/decoy search was performed to experimentally estimate the false positive rate and only proteins identified with two unique high quality peptide identifications were considered as previously reported 7–9 (1 decoy protein identified; FDR <0.5%). An in-house protein grouping algorithm was applied to satisfy the principles of parsimony 8.
Gene Ontology annotation and data comparisons
Annotation mappings (i.e. GO, predicted transmembrane domains, signal peptides etc.) were performed using the ProteinCenter bioinformatics software (Proxeon Biosystems, Odense, Denmark). Comparison of EPS-detected proteins against other proteomic datasets (prostatic secretions 10, seminal plasma 11, urine 12, condition media 13) was accomplished by ProteinCenter. Proteins were sequence aligned (i.e. BLASTed) against each other and only proteins with at least 95% sequence identity were considered to match (i.e. protein clusters). Comparison to the Human Gene Atlas (mRNA microarray) 14 and the Human Protein Atlas (www.proteinatlas.org) 15 was accomplished via gene mapping using an in-house relational database. For the comparison to data in the Human Protein Atlas, only proteins with available IHC staining in normal or prostate cancer tissues were included. The staining criteria we used for our antibody selection were directly adapted from the Human Protein Atlas website and are based on rigorous multi-step antibody quality control criteria established by this consortium. We only included gene products for our comparisons with a medium to high IHC scoring intensity in either normal prostate or prostate cancer tissue. Detailed information regarding the quality assurance can be found at (http://www.proteinatlas.org/qc.php). Cluster analyses (Euclidean) were performed using the open-source tool Cluster 3.0 (http://bonsai.ims.utokyo. ac.jp/mdehoon/software/cluster/software.htm) and heat maps were displayed using TreeView (http://jtreeview.sourceforge.net).
Results
MudPIT-profiling of EPS from prostate cancer patients
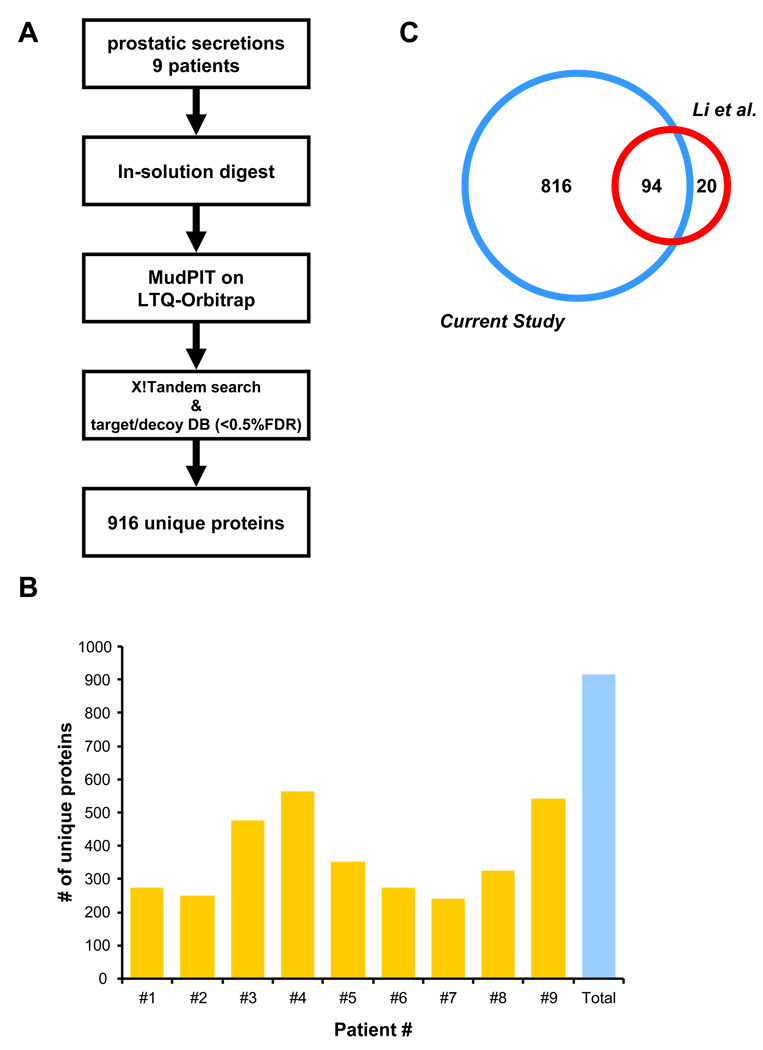
Nine individual direct EPS samples from men with prostate cancer (Gleason 6–7, stages pT2a–pT3b) were selected for MudPIT-based proteomic analysis. To minimize variability introduced by sample handling, EPS samples were directly digested in-solution and analyzed by a 9-step MudPIT on a LTQ-Orbitrap XL mass spectrometer (see Materials and Methods). Rigorous protein identification criteria resulted in the identification of 916 unique proteins with high confidence (Fig. 1A/B), with 230 to 550 unique proteins identified per individual cancer patient sample (Fig. 1B). The list of 916 proteins (SI Tab. 1) obtained from the direct EPS cancer samples were compared to those reported in a recent publication by Lin and colleagues 10 who collected several EPS fluids using a more vigorous prostate massage procedure (Fig. 1C). Only twenty proteins were unique to the study of Li et al. 10 consisting primarily of blood-derived proteins, while the vast majority of proteins were uniquely identified in the current study; 816 unique protein clusters and 94 shared protein clusters (for mapping details see Materials and Methods). Based on the very low levels of hemoglobin and seminogelin proteins detected in our analysis, we conclude that the direct EPS samples have minimal contamination with blood and seminal vesicle derived proteins. Using cumulative spectral counts, albumin and immunoglobulins account for approximately 25% and 14% respectively of protein concentration totals in direct EPS. In relation to known prostate derived proteins, the next most abundant proteins accounting for an additional 25% total abundance were lactoferrin, prostate specific antigen, prostatic acid phosphatise, zinc-α2 glycoprotein and aminopeptidase N. Functional analyses of the EPS proteome using Gene Ontology, predicted transmembrane domains (TMD; TMAP algorithm) and signalling peptides (SP; PrediSi algorithm) revealed a wide variety of functional categories (SI Fig. 1). A significant number of detected proteins were membrane proteins and members of the extracellular region, including 34 CD molecules (SI Tab. 1). This was further supported by the fact that ~ 40% of the detected proteins had at least one predicted TMD and contained a predicted SP, indicative of targeting to the secretory pathway. A smaller number of proteins could be assigned to other intracellular organelles.
Figure 1. Proteomics analyses of direct expressed prostatic secretions.
A) EPS from nine men with confirmed prostate cancer were in-solution digested and individually analyzed by on-line MudPIT on an LTQ-Orbitrap mass spectrometer. Rigorous peptide and protein identification criteria resulted in the identification of 916 unique proteins. B) Number of unique proteins detected in each of the analyzed EPS samples. C) Comparison of our EPS proteome to a previously published EPS dataset.
Integrative data mining and comparison to other public databases
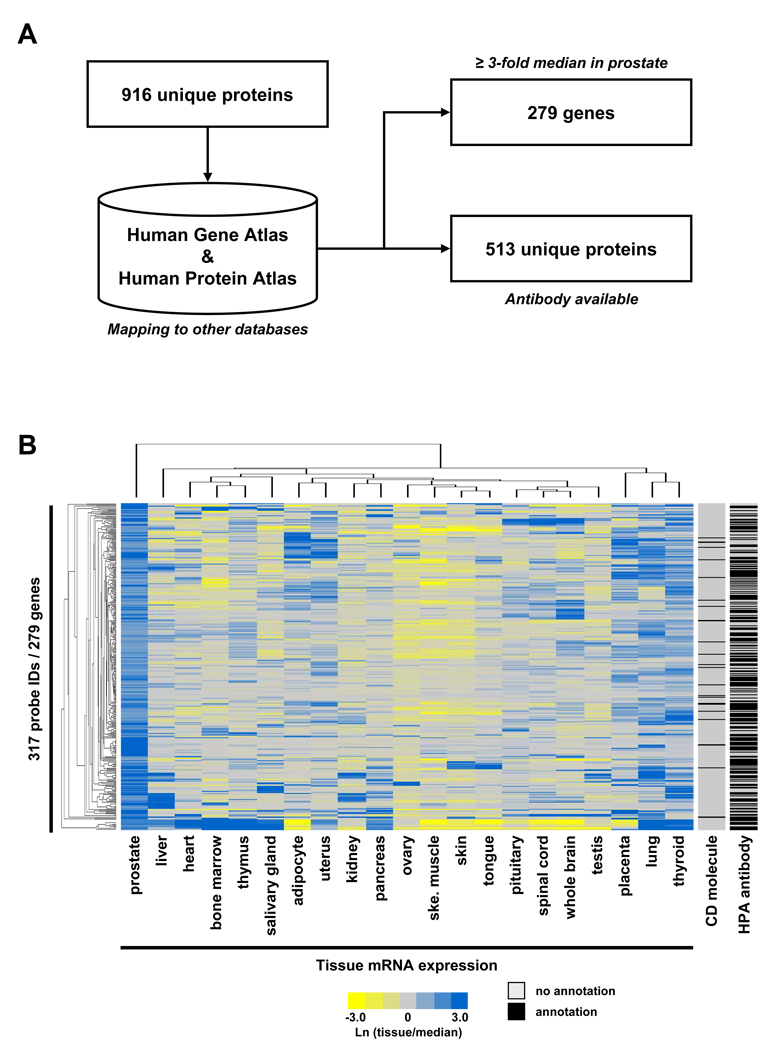
One goal of the current study was to provide the research community with a high quality, well-annotated resource of proteins expressed in EPS. This could improve our understanding of general prostate biology and guide the discovery of novel prostate cancer biomarkers. To accomplish this goal, we mapped our data to two publicly available resources; the Human Gene Atlas 14 which is based on the mRNA microarray expression profile of >50 human tissues, and the Human Protein Atlas (www.proteinatlas.org) 15, 16, a large resource of immunohistochemistry images and available human antibodies (Fig. 2A). The rationale for these comparisons was to identify proteins detected in EPS fluids that are also enriched in prostate tissue relative to other human tissues included in the microarray database. Also, integration of EPS proteins with the Human Protein Atlas database can catalogue the availability of validated antibodies, which could provide researchers with an important tool to further study the biological function of selected proteins.
Figure 2. Integrative data mining and comparison to other databases.
A) The EPS proteome was mapped against the Human Gene Atlas (mRNA microarray) and Human Protein Atlas (antibodies and immunohistochemistry images). B) 279 unique gene products detected in our EPS proteome showed ≥3-fold above median expression in normal prostate tissue on the Human Gene Atlas microarray (detected by 317 probe IDs), as compared to 20 other tissues. Mapping against available Human Protein Atlas antibodies and CD molecules is provided.
Briefly, using only mRNAs with ≥3-fold above median expression in prostate tissue compared to 20 other human tissue types in the Human Gene Atlas, 279 EPS gene products were identified, implying a potential function specifically related to prostate biology. In the Human Protein Atlas, a total of 513 proteins detected in the EPS proteome currently have antibodies with varying staining intensities in human prostate tissue (for details see Materials and Methods). The identities of the EPS proteins mapped to the two public databases are available in SI Tables 2 and 3.
Comparison of the EPS proteome to other published proteomic datasets
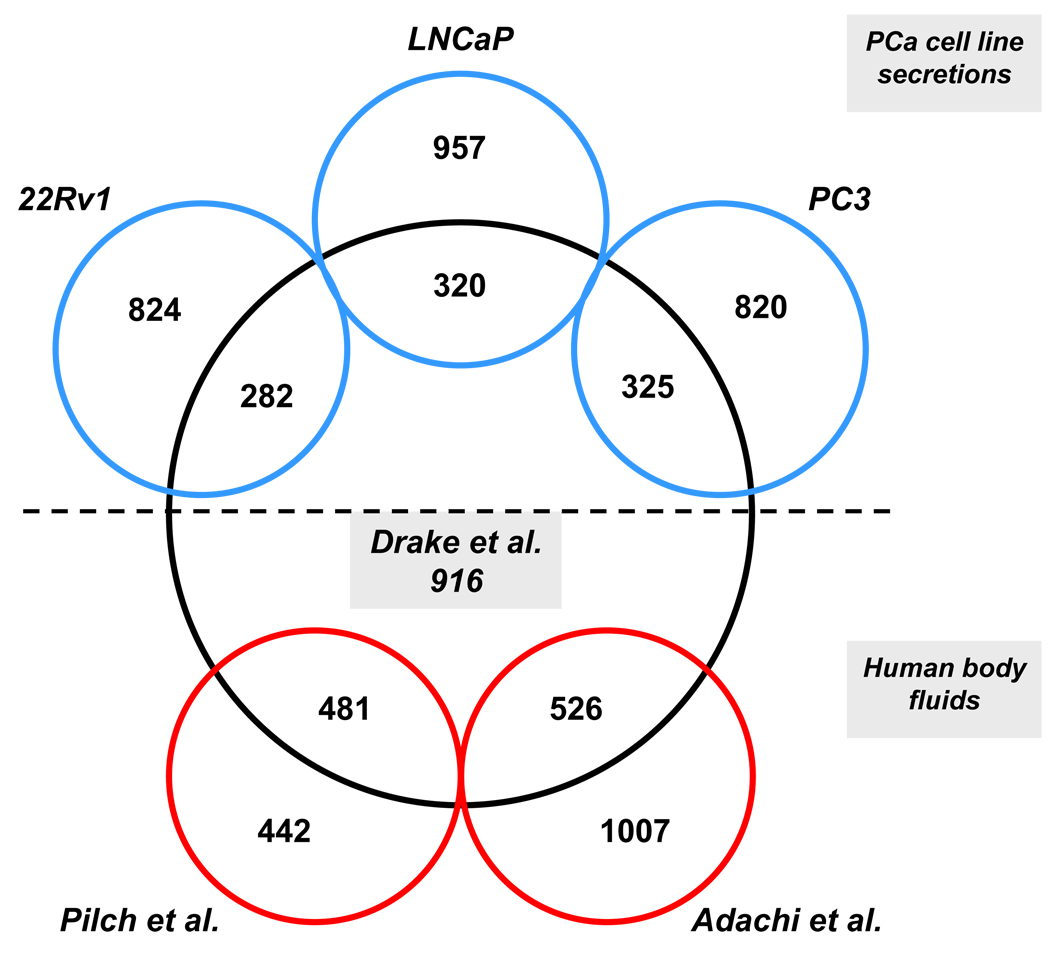
Next, we integrated the EPS proteome with three other published proteomic datasets 11–13. This included a recent publication of proteins secreted into the cell culture media by three established prostate cancer cell lines (PC3, LNCaP, and 22Rv1) 13 and two human body fluid datasets (urine and seminal plasma) directly related to EPS 11, 13. The rationale for the comparison to the cell secretions was to distinguish between proteins readily secreted by prostate cancer cells from proteins being secreted by other cell types of the prostate. This is not an ideal comparison, however, as the cell lines are more reflective of metastatic prostate cancers and the direct EPS samples were from lower grade Gleason 6 and 7 cancers. An additional caveat is that these cells have numerous genomic alterations and have been grown under culture conditions for many passages distant from the primary isolates and might not truly reflect an in vivo setting. Briefly, approximately 300 proteins (Fig. 3) were shared between the EPS proteome and each of the three prostate cancer cell lines (see SI Tables 4–6 for detailed mappings). Likewise, approximately 500 proteins were shared between the EPS proteome and normal human urine and seminal plasma (Fig. 3) (see SI Tables 7 and 8). Several well known prostate markers (i.e. PSA, ACPP) were shared among the EPS proteome and the described prostate cancer cell lines. The data provided in this study could therefore be used to mechanistically investigate proteins identified in vivo (EPS) using well established prostate cancer cell lines. Interestingly, several well known prostate markers (i.e. MSMB, TMPRSS2) were only identified in the EPS proteome and the other two available body fluids (see below), strengthening the rationale of using in vivo obtainable fluids to study prostate/prostate cancer biology. It is likely that proteins exclusively detected in the EPS proteome, as compared to proteins detected in healthy human urine and seminal plasma, are specifically enriched in secretions of the prostate under pathological conditions, and were therefore absent or at a very low concentration, in the other body fluids.
Figure 3. Comparison to other published proteomes.
The EPS proteome was compared to proteins detected in two other published, related body fluid proteomes (Pilch et al. seminal plasma and Adachi et al. urine) and proteins secreted into the condition medium by three established prostate cancer cell lines (Sardana et al.).
Multi-dataset integration
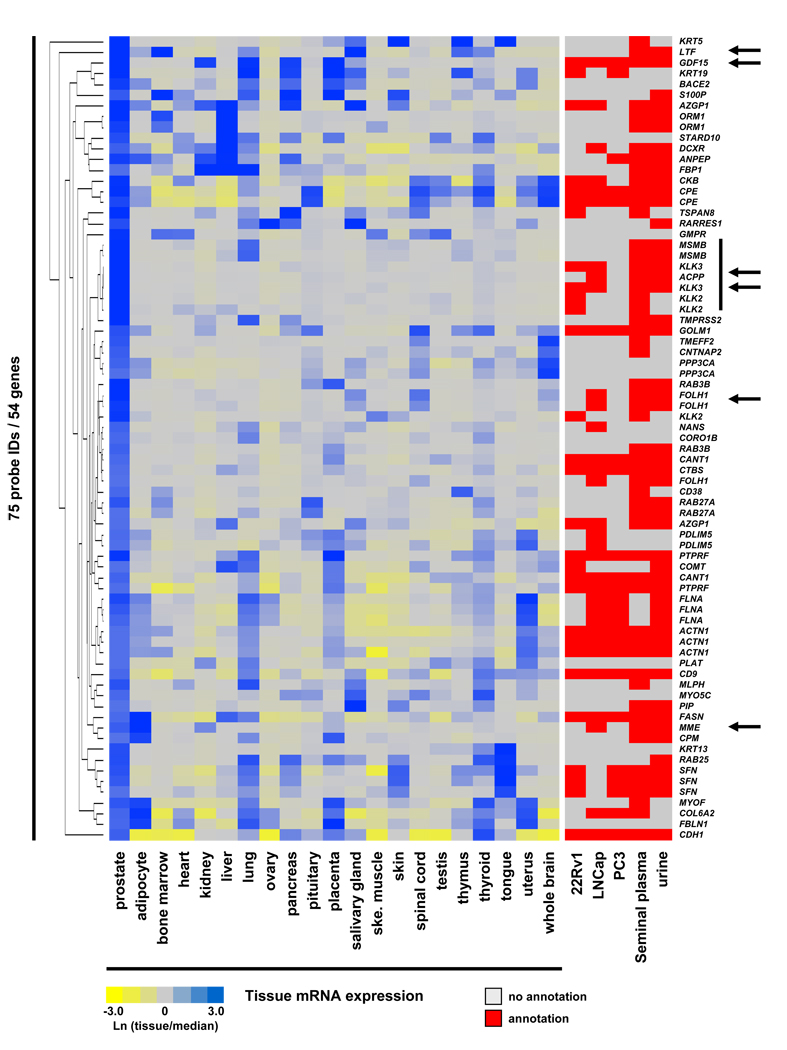
To fully utilize the power of multi-dataset comparison we integrated all publicly available data resources with our current EPS proteome (Fig. 4), as an example of a potential data mining strategy. Briefly, we focused on gene products strongly enriched in normal prostate tissue using the described microarray dataset (10-fold above median expression) that were also detected in our EPS proteome and had antibodies through the Human Protein Atlas database. This integration resulted in 54 unique gene products satisfying all described integration criteria (Fig. 4 and SI Tab. 9). A heat map display of this integration is presented in Figure 4, including available mappings to other public proteome datasets. Although, all gene products are strongly expressed in prostate tissue, most showed additional expression in several other tissues. Known prostate markers (vertical line), such as PSA, ACPP and MSMB were readily identified. Several identified gene products have commercially available ELISAs, indicated by arrows, that we have confirmed for the use in EPS samples (data not shown). Comparison to other published proteomics datasets is also indicated in the presented heat map and this data could be used to further guide potential follow-up analyses. As all 54 proteins displayed in the current heat map have available immunohistochemistry images at the Human Protein Atlas, we systematically screened the available images for both normal prostate tissue and prostate cancer tissue. Several proteins with intriguing staining patters are discussed below, demonstrating an example of how the current data could be used by others.
Figure 4. Integration of multiple data resources with the EPS proteome.
Heat map of 54 unique gene products detected in the EPS proteome with available antibodies in the Human Proteome Atlas database. Selected gene products were enriched in normal prostate tissue as compared to 20 other human tissues using the Human Gene Atlas microarray database (≥10-fold above median expression in prostate). Mappings to other available proteome datasets are indicated. Proteins with commercially available ELISAs that have been tested by us in EPS fluids are indicated by arrows (see SI Methods for details). Well known prostate markers are highlighted by a vertical line.
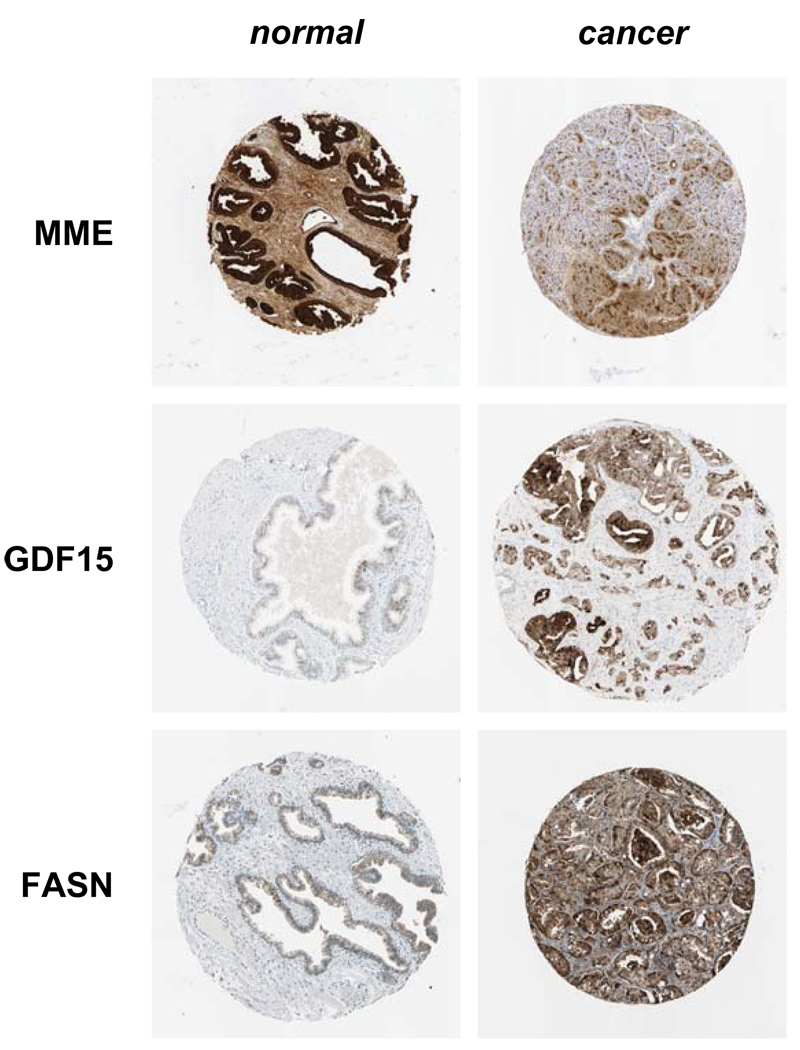
Briefly, several proteins were identified which displayed a trend towards differential expression in normal prostate compared to prostate cancer tissue using available IHC images, although patient variability and image quality differences were also observed (Fig. 5). Neprilysin (MME, CD10) showed a trend towards decreased expression in prostate cancer tissue. Using a commercially available ELISA kit we have recently seen a similar trend using a limited number of EPS samples from aggressive prostate cancer as compared to indolent disease (data not shown). Alternatively, GDF15 and FASN showed a trend towards increased expression in prostate cancer tissues (Fig. 5). As we have now linked our entire data to available mRNA microarray expression, Human Protein Atlas antibody staining and other published, related proteomics datasets, we believe this data will provide an important resource for the prostate biology/cancer community to guide the investigation of other proteins in hypothesis driven experiments.
Figure 5. Immunohistochemistry images as obtained from the Human Protein Atlas.
Selected examples of proteins found in the Human Protein Atlas database that showed trends towards differential expression in normal compared to cancer prostate tissue.
Discussion
The cumulative list of identified proteins in the direct EPS samples currently represents the largest compilation resource of proteins present in prostatic fluid secretions from men with confirmed prostate cancers. In comparison to other proteomics studies aimed at discovering prostate function or prostate cancer biomarkers using cell secretions 13, plasma 17, 18, or tissue 19, 20, the current study used a relevant organ-proximal body fluid for the detection of secreted proteins in vivo. For this comprehensive analysis we used direct EPS fluids, because these fluids most likely contain secreted, prostate-specific proteins in a significantly higher concentration as compared to urine or plasma samples. The relatively high expression of many of these proteins like PSA and ACPP makes the fluid amenable to all current forms of mass spectrometry analysis with reduced front-end processing or purification.
Current early screening for prostate cancer relies on PSA detection in serum 21. Over time, this screening has dramatically reduced the presentation of high-grade disease, yet other studies have indicated that the majority of early-detected prostate cancers are not lethal and may be considered clinically insignificant 22–24. Thus, current methods to establish the risk of progression and prognosis of prostate cancers remain suboptimal, and a large number of patients are over-treated with a significant negative financial impact on health care 22, 23, 25. Thus, new diagnostics are needed that could predict disease course and discriminate indolent tumors from aggressive tumors, allowing for more appropriate treatment options to be offered to newly diagnosed prostate cancer patients. Targeting identification of potential new biomarkers in proximal fluids of the prostate like direct EPS and post-DRE EPS urines could address these clinical needs.
With the availability of modern mass spectrometers and high performance peptide separations like MudPIT 6, 26 we are now able to routinely identify hundreds to thousands of proteins in complex biological samples. This can result in a significant challenge for biomarker validation strategies, as the development of ELISA or MRM-MS assays for all identified proteins is not feasible. Equally challenging is the selection of the most likely candidate biomarkers from the large list of identified proteins. To guide the selection criteria of promising candidate proteins involved in prostate function and prostate cancer biomarker discovery, we have integrated our EPS proteome with a large variety of publicly available datasets. To further streamline MRM-MS assay development, we have deposited the entire MS data to the Tranche database for others to mine, as we have previously reported 7, 8, 27. Cumulatively, this EPS proteome dataset should facilitate the selection and confirmation of individual biomarker candidates in follow up studies by our group and other investigators.
Supplementary Material
Acknowledgements
T.K. is supported through the Canadian Research Chairs Program. This work was supported in parts by grants from Prostate Cancer Canada (2009-454) and the Canadian Institute of Health Research (MOP-93772) to T.K. and J.A.M., and in part by grants from the National Institutes of Health to R.R.D. (R01 CA135087, R21 CA137704) and O.J.S. (U01 CA085067). This research was funded in part by the Ontario Ministry of Health and Long Term Care. The views expressed do not necessarily reflect those of the OMOHLTC. Y.K. is a recipient of a Paul Starita Graduate Student Fellowships.
References
- 1.Drake RR, White KY, Fuller TW, Igwe E, Clements MA, Nyalwidhe JO, Given RW, Lance RS, Semmes OJ. Clinical collection and protein properties of expressed prostatic secretions as a source for biomarkers of prostatic disease. J Proteomics. 2009;72(6):907–917. doi: 10.1016/j.jprot.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deras IL, Aubin SM, Blase A, Day JR, Koo S, Partin AW, Ellis WJ, Marks LS, Fradet Y, Rittenhouse H, Groskopf J. PCA3: a molecular urine assay for predicting prostate biopsy outcome. J Urol. 2008;179(4):1587–1592. doi: 10.1016/j.juro.2007.11.038. [DOI] [PubMed] [Google Scholar]
- 3.Hessels D, Schalken JA. The use of PCA3 in the diagnosis of prostate cancer. Nat Rev Urol. 2009;6(5):255–261. doi: 10.1038/nrurol.2009.40. [DOI] [PubMed] [Google Scholar]
- 4.Wang R, Chinnaiyan AM, Dunn RL, Wojno KJ, Wei JT. Rational approach to implementation of prostate cancer antigen 3 into clinical care. Cancer. 2009;115(17):3879–3886. doi: 10.1002/cncr.24447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McNeal JE. Cancer volume and site of origin of adenocarcinoma in the prostate: relationship to local and distant spread. Hum Pathol. 1992;23(3):258–266. doi: 10.1016/0046-8177(92)90106-d. [DOI] [PubMed] [Google Scholar]
- 6.Taylor P, Nielsen PA, Trelle MB, Horning OB, Andersen MB, Vorm O, Moran MF, Kislinger T. Automated 2D peptide separation on a 1D nano-LC-MS system. J Proteome Res. 2009;8(3):1610–1616. doi: 10.1021/pr800986c. [DOI] [PubMed] [Google Scholar]
- 7.Cox B, Kotlyar M, Evangelou AI, Ignatchenko V, Ignatchenko A, Whiteley K, Jurisica I, Adamson SL, Rossant J, Kislinger T. Comparative systems biology of human and mouse as a tool to guide the modeling of human placental pathology. Mol Syst Biol. 2009;5:279. doi: 10.1038/msb.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gortzak-Uzan L, Ignatchenko A, Evangelou AI, Agochiya M, Brown KA, St Onge P, Kireeva I, Schmitt-Ulms G, Brown TJ, Murphy J, Rosen B, Shaw P, Jurisica I, Kislinger T. A proteome resource of ovarian cancer ascites: integrated proteomic and bioinformatic analyses to identify putative biomarkers. J Proteome Res. 2008;7(1):339–351. doi: 10.1021/pr0703223. [DOI] [PubMed] [Google Scholar]
- 9.Sodek KL, Evangelou AI, Ignatchenko A, Agochiya M, Brown TJ, Ringuette MJ, Jurisica I, Kislinger T. Identification of pathways associated with invasive behavior by ovarian cancer cells using multidimensional protein identification technology (MudPIT) Mol Biosyst. 2008;4(7):762–773. doi: 10.1039/b717542f. [DOI] [PubMed] [Google Scholar]
- 10.Li R, Guo Y, Han BM, Yan X, Utleg AG, Li W, Tu LC, Wang J, Hood L, Xia S, Lin B. Proteomics cataloging analysis of human expressed prostatic secretions reveals rich source of biomarker candidates. Proteomics Clin. Appl. 2008;2:543–555. doi: 10.1002/prca.200780159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pilch B, Mann M. Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 2006;7(5):R40. doi: 10.1186/gb-2006-7-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7(9):R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sardana G, Jung K, Stephan C, Diamandis EP. Proteomic analysis of conditioned media from the PC3, LNCaP, and 22Rv1 prostate cancer cell lines: discovery and validation of candidate prostate cancer biomarkers. J Proteome Res. 2008;7(8):3329–3338. doi: 10.1021/pr8003216. [DOI] [PubMed] [Google Scholar]
- 14.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101(16):6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ponten F, Jirstrom K, Uhlen M. The Human Protein Atlas--a tool for pathology. J Pathol. 2008;216(4):387–393. doi: 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]
- 16.Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, Berglund L, Bergstrom K, Brumer H, Cerjan D, Ekstrom M, Elobeid A, Eriksson C, Fagerberg L, Falk R, Fall J, Forsberg M, Bjorklund MG, Gumbel K, Halimi A, Hallin I, Hamsten C, Hansson M, Hedhammar M, Hercules G, Kampf C, Larsson K, Lindskog M, Lodewyckx W, Lund J, Lundeberg J, Magnusson K, Malm E, Nilsson P, Odling J, Oksvold P, Olsson I, Oster E, Ottosson J, Paavilainen L, Persson A, Rimini R, Rockberg J, Runeson M, Sivertsson A, Skollermo A, Steen J, Stenvall M, Sterky F, Stromberg S, Sundberg M, Tegel H, Tourle S, Wahlund E, Walden A, Wan J, Wernerus H, Westberg J, Wester K, Wrethagen U, Xu LL, Hober S, Ponten F. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4(12):1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Fortin T, Salvador A, Charrier JP, Lenz C, Lacoux X, Morla A, Choquet-Kastylevsky G, Lemoine J. Clinical quantitation of prostate-specific antigen biomarker in the low nanogram/milliliter range by conventional bore liquid chromatography-tandem mass spectrometry (multiple reaction monitoring) coupling and correlation with ELISA tests. Mol Cell Proteomics. 2009;8(5):1006–1015. doi: 10.1074/mcp.M800238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hood BL, Malehorn DE, Conrads TP, Bigbee WL. Serum proteomics using mass spectrometry. Methods Mol Biol. 2009;520:107–128. doi: 10.1007/978-1-60327-811-9_8. [DOI] [PubMed] [Google Scholar]
- 19.Byrne JC, Downes MR, O'Donoghue N, O'Keane C, O'Neill A, Fan Y, Fitzpatrick JM, Dunn M, Watson RW. 2D-DIGE as a strategy to identify serum markers for the progression of prostate cancer. J Proteome Res. 2009;8(2):942–957. doi: 10.1021/pr800570s. [DOI] [PubMed] [Google Scholar]
- 20.Garbis SD, Tyritzis SI, Roumeliotis T, Zerefos P, Giannopoulou EG, Vlahou A, Kossida S, Diaz J, Vourekas S, Tamvakopoulos C, Pavlakis K, Sanoudou D, Constantinides CA. Search for potential markers for prostate cancer diagnosis, prognosis and treatment in clinical tissue specimens using amine-specific isobaric tagging (iTRAQ) with two-dimensional liquid chromatography and tandem mass spectrometry. J Proteome Res. 2008;7(8):3146–3158. doi: 10.1021/pr800060r. [DOI] [PubMed] [Google Scholar]
- 21.Barry MJ. Clinical practice. Prostate-specific-antigen testing for early diagnosis of prostate cancer. N Engl J Med. 2001;344(18):1373–1377. doi: 10.1056/NEJM200105033441806. [DOI] [PubMed] [Google Scholar]
- 22.Draisma G, Boer R, Otto SJ, van der Cruijsen IW, Damhuis RA, Schroder FH, de Koning HJ. Lead times and overdetection due to prostate-specific antigen screening: estimates from the European Randomized Study of Screening for Prostate Cancer. J Natl Cancer Inst. 2003;95(12):868–878. doi: 10.1093/jnci/95.12.868. [DOI] [PubMed] [Google Scholar]
- 23.Esserman L, Shieh Y, Thompson I. Rethinking screening for breast cancer and prostate cancer. Jama. 2009;302(15):1685–1692. doi: 10.1001/jama.2009.1498. [DOI] [PubMed] [Google Scholar]
- 24.Roobol MJ, Zappa M, Maattanen L, Ciatto S. The value of different screening tests in predicting prostate biopsy outcome in screening for prostate cancer data from a multicenter study (ERSPC) Prostate. 2007;67(4):439–446. doi: 10.1002/pros.20545. [DOI] [PubMed] [Google Scholar]
- 25.Lin K, Lipsitz R, Miller T, Janakiraman S. Benefits and harms of prostate-specific antigen screening for prostate cancer: an evidence update for the U.S. Preventive Services Task Force. Ann Intern Med. 2008;149(3):192–199. doi: 10.7326/0003-4819-149-3-200808050-00009. [DOI] [PubMed] [Google Scholar]
- 26.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 27.Elschenbroich S, Ignatchenko V, Sharma P, Schmitt-Ulms G, Gramolini AO, Kislinger T. Peptide separations by on-line MudPIT compared to isoelectric focusing in an off-gel format: application to a membrane-enriched fraction from C2C12 mouse skeletal muscle cells. J Proteome Res. 2009;8(10):4860–4869. doi: 10.1021/pr900318k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.