Abstract
The T cell receptor (TCR) is responsible for discriminating between self- and foreign-derived peptides, translating minute differences in amino-acid sequence into large differences in response. Because of the great variability in the TCR and its ligands, activation of T cells by foreign peptides is a quantitative process, dependent on a mix of upstream signals and downstream integration. Accordingly, quantitative data and computational models have shed light on many important aspects of this process: molecular noise in ligand recognition, spatial dynamics in T cell-APC (antigen presenting cell) interactions, graded versus all-or-none decision making by the TCR apparatus, mechanisms of peptide antagonism and synergism, and the tunability and robustness of activation thresholds. Though diverse in their formalism, these studies together paint a picture of how modeling has shaped and will continue to shape understanding of T cell immunobiology.
The response of a T cell receptor to peptide antigens varies significantly, depending on their sequence. Computational models of upstream and downstream signals reveal quantitative details of the activation process.
Deciphering the detailed basis of antigen-specific T cell activation is classically considered to reside in the realm of cell biology and biochemistry, both “wet lab” disciplines. Molecular approaches complemented by increasingly high resolution imaging tools have identified the components and interactions that translate antigen recognition into gene activation. The application of biophysical tools, initially surface plasmon resonance and later, in situ FRET, have added a quantitative element to the pool of data available in this field (Davis et al. 1998). But these advances alone, though critical, have not provided a deep conceptual understanding of how ligand discrimination occurs or the peculiarities of the discrimination and activation processes. In particular, they have not yet yielded a framework that can reliably predict how T cells will respond when molecular components of the transduction machinery are altered in concentration or function, such as in the case of genetic polymorphism or mutation.
Over the past decade, mathematical and computational modeling has emerged as a means of integrating the experimental observations resulting from biochemical, biophysical, and imaging studies into a quantitative view of T cell responses to antigens. In this communication we review the contributions of computer modeling to our understanding of T cell immunobiology. First, we discuss how recent technical developments have yielded more quantitative measurements of immune responses and triggered modeling efforts dealing with the dynamics of T cell activation. We will present the computer modeling tools that have been used and their increasing accessibility to immunobiologists. In a second section, we present different conundrums related to T cell activation and describe how they have been addressed through computer modeling (Fig. 1). Finally, we will discuss why modeling remains only peripherally relevant to experimental immunology, and how this will change through the implementation of better tools and practices.
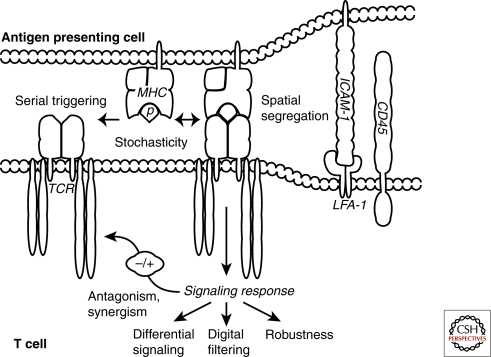
Figure 1.
Aspects of T cell signaling clarified through computational modeling. Stochasticity (molecular noise) has been incorporated into models of the TCR, operating at the level of recognition of just a few antigens. Serial triggering of multiple TCRs by the same antigenic peptide (p) presented in a major histocompatibilty complex protein (MHC) and segregation of bulky phosphatases (CD45) and adhesive molecule pairs (LFA-1 and ICAM-1) from smaller pMHC-TCR complexes have been studied in models quantifying spatial regulation of T cell activation. Antagonism, synergism, differential signaling, digital filtering and robustness, have been examined using models describing the biochemical signaling induced by TCR ligation.
QUANTITATIVE DATA AND APPROACHES TO MODELING
Quantitative characterizations of T lymphocyte biology are providing both the impetus and the raw materials for models correlating biophysical observables with functional outcomes. Indeed, many immunological components have been characterized, from the organ level (e.g., lymph node architecture), to the cellular level (e.g., varied states of differentiation of lymphocytes), down to the molecular level (e.g., expression of signaling proteins, number of secreted molecules). The molecular components of T cell antigen and cytokine signaling have been isolated and purified, and studies of the isolated molecules have precisely determined many of the biophysical parameters governing ligand-receptor interactions.
A prime example of this latter category involves T cell receptor (TCR) interaction with its ligand, a peptide presented by major histocompatibility complex (MHC) surface proteins. Most (but not all) studies have shown a correlation between minute changes in a single kinetic parameter (off-rate for the receptor/ligand complex) and large differences in signaling patterns and functional responses (Davis et al. 1998). As a quantitative explanation for this observation, T. McKeithan first offered a “kinetic proofreading” scheme (Hopfield 1974; Ninio 1975) to account for T cell ligand discrimination (McKeithan 1995). In that scheme, receptors engaging stably binding ligands engender a complete phosphorylation of TCR-associated proteins, whereas receptors engaging transiently binding ligands fail to reach the endpoint required to trigger effective cellular responses (reviewed extensively in Feinerman et al. 2008a). McKeithan's seminal study emphasized that static dose-response equilibrium descriptions of this system insufficiently capture its true behavior. Building on this work, following studies emphasized how dynamic interactions within the TCR apparatus can account for the keen specificity of T cell activation. Most of these have focused on the development of meso-scale biochemical models that typically include 10–30 components such as receptors, adapters, kinases, and phosphatases, representing limited signaling pathways and operating on short timescales (Schoeberl et al. 2002; Faeder et al. 2003; Altan-Bonnet and Germain 2005). They are often well-mixed, deterministic models that rely on classical ordinary differential equations (mass-action laws, Michaelis-Menten or others) to describe protein–protein interaction and enzymological activities.
The principal benefit of such biochemical reaction models is their ability to simulate the behavior of large numbers of molecules by dealing with concentrations rather than individual molecules. The drawback, however, is that the modeler must enumerate all molecular states and transition rates in a series of linked equations for any given signaling pathway. This is reasonably straightforward for a network of reactions in which enzymes and substrates are soluble, do not aggregate, and have a small number of reaction sites. Desktop modeling packages such as CellDesigner (Funahashi et al. 2003), JDesigner (Le Novere et al. 2009), and COPASI (Hoops et al. 2006), which allow schematically specified biochemical networks to be translated into explicit biochemical equations, have expanded the accessibility of such techniques. The behavior of these systems over time can then be simulated with classical deterministic or stochastic methods (Gillespie 2007).
The limitations of this method of model specification become apparent, however, when faced with reactions involving aggregations of components or molecules with large numbers of possible modifications. In these situations, the specification of the network becomes a challenge that has spurred the development of new approaches for defining models. As one example of why this is a central issue in modeling T cell activation, consider the organization of the TCR complex and the earliest biochemical events induced by binding of its ligand, a complex of peptide and MHC (pMHC). The TCR complex (composed of the αβ chains of the TCR as wells as the CD3/ζ chains) has 10 immunoreceptor tyrosine-based activation motifs (ITAMs) that are phosphorylated during TCR activation and provide docking sites for downstream signaling molecules. Each ITAM consists of two tyrosines, for a total of 20 tyrosines in the TCR complex. Hence, there are theoretically 220>106 different phospho-tyrosine states for the TCR complex. Explicitly enumerating all of these phosphorylated states in a model would be very tedious and time-consuming (Blinov et al. 2004). A common simplification is to limit the biochemical models to fewer ITAMs (e.g., the three ITAMs associated with one ζ chain). This approach is problematic, though, as it both underestimates the importance of a full complement of ITAMs (Holst et al. 2008; Malissen 2008) and still allows large models—20 pages in our previous work (Altan-Bonnet and Germain 2005)—that are difficult to proofread and troubleshoot or to expand when multiplexing with other signaling pathways.
Accordingly, the field is applying a new theoretical formalism—known as rule-based modeling (Faeder et al. 2003; Blinov et al. 2004; Hlavacek et al. 2006; Meier-Schellersheim et al. 2006; Lipniacki et al. 2008)—to develop in silico models of signaling cascades in lymphocytes. With rule-based modeling, the modeler articulates single biochemical reactions (e.g., interaction between two proteins or phosphorylation of one tyrosine) as rules; the full network of interactions, including the combinatorial complexity in a given signaling cascade, is then generated automatically using a computational algorithm.
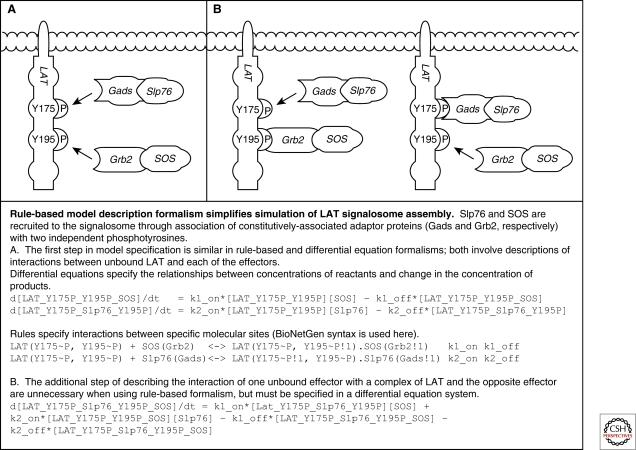
In general, rule-based formalism has the advantage of a simple syntax that translates biochemical cartoons into quantitative models. These are thus more accessible to biologists and easier to troubleshoot. More specifically in the case of T cell signaling, rule-based formalism simplifies the development of models where the combinatorial complexity of the signalosome is daunting (Malissen et al. 2005). Indeed, aggregation of signaling components on phosphorylated ITAMs or on LAT (linker for activation of T cells) downstream of the TCR is automatically accounted for by rules linking individual components, whereas biochemical formalism would require explicit specification of each molecular complex (Fig. 2). Furthermore, cross-talk of the TCR signaling pathway with cytokine- or costimulation-induced signaling can be easily tested: independently constructed and validated rule-based models for signaling pathways are automatically integrated when combined in a comprehensive model.
Figure 2.
Rule-based modeling versus biochemical modeling of LAT signalosome aggregation.
Other avenues of exploration attempted to avoid the issue of complexity altogether. Some investigators have tried to manage the large quantity of qualitative observations and dearth of dynamic parameters in the T cell activation literature by developing a Boolean (logic-based) representation of the T cell signaling network (Saez-Rodriguez et al. 2007). Even without exact kinetic parameters, it is possible to extract further information from current knowledge by building a large-scale logical model, curated from observations in the literature. For example, Saez-Rodriguez et al. used their Boolean model to make a surprising prediction that they subsequently validated experimentally: Cross-linking of CD28 was sufficient to trigger JNK activation without antigen activation. However, because of their inherent on/off nature, Boolean systems are not suited to investigation of phenomena such as ligand discrimination or thymic selection that involve translation of minute differences in biophysical characteristics of pMHC-TCR interaction into discrete functional differences (Altan-Bonnet and Germain 2005; Daniels et al. 2006).
Although the dynamics of molecular pathways downstream of the TCR account for a great deal of antigen recognition, incorporating physical processes that modulate this biochemistry remains a challenge with current methods. In particular, computational time can be forbidding when modeling T cell activation with diffusion of signaling components (Loew and Schaff 2001; Meier-Schellersheim et al. 2006) or with combinatorial complexity in the biochemical network (Hlavacek et al. 2006). More complex phenomena such as membrane deformation and cytoskeletal coupling are even harder computational problems and have been modeled only in frameworks constructed specifically for that purpose (Qi et al. 2001). Thus, there exist many ad hoc computational methods that were specifically designed to address specific biological questions. We expect the field of computational biology to converge on a lingua franca for systems immunology that will make the communication of models more efficient (Le Novere et al. 2009). This will benefit the computational community, as experimental parameters will be more easily shared, and model-building efforts will be cross-validated. The community of immunologists will also benefit as a “reference” model of T cell activation will emerge from computational efforts.
QUESTIONS IN SIGNAL TRANSDUCTION AND T CELL ANTIGEN RECEPTOR SIGNALING THAT HAVE BEEN ADDRESSED BY QUANTITATIVE MODELING
Digital Filtering and the Specificity, Sensitivity, and Speed of TCR Signaling
A striking feature of T cell activation is the specificity of its response to pMHC ligand. Three key observations have set conditions that models of TCR signaling must satisfy (Altan-Bonnet and Germain 2005). First, T cell activation is a very fast process that can be detected within seconds of T:APC contacts (by kinase recruitment [Bunnell et al. 2002; Huse et al. 2007], calcium influx [Delon et al. 1998], TCR phosphorylation [Germain and Stefanova 1999] or others). Second, T cell activation is very sensitive, as one or few ligands are enough to trigger a response (Sykulev et al. 1996; Irvine et al. 2002). Third, T cell activation (beginning at timescales far below a minute) is quite specific: One amino acid variation in an antigenic peptide can ablate its stimulatory function (Davis et al. 1998). The relevance of this specificity has been challenged recently (Zehn et al. 2009), but close examination of these data revealed that minute differences in peptide sequence still yields quantitative difference in T cell proliferation in vivo. Aside from allosteric models (Gil et al. 2002; Schamel et al. 2006; Palmer and Naeher 2009), few of the modeling attempts building primarily on McKeithan's proofreading formalism have managed to simultaneously incorporate all these key features of T cell responses into the output of their model (as reviewed in Feinerman et al. 2008a).
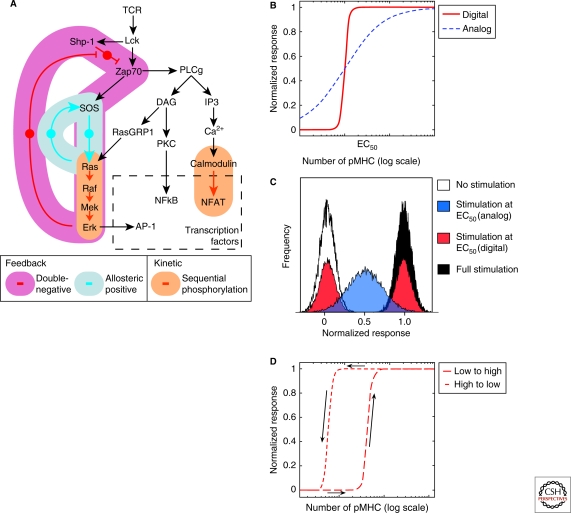
One facet of T cell signaling that has attracted a great deal of theoretical attention is the observation that the signal transduction cascade associated with the TCR translates a spectrum of input strength (quality and quantity of antigens) into all-or-none responses associated with cell differentiation or mitotic progression. Theoretical work has shown that both signaling feedback and allosteric cooperativity have the potential to impart switchlike or “ultrasensitive” behavior (Fig. 3A–C). Analyses of the TCR signaling apparatus have revealed that multiple modules within the signaling cascade can serve as ultrasensitive switches. The ERK mitogen-activated protein kinase (MAPK) pathway contains three interconnected switches, whereas regulation of NFAT provides a fourth, somewhat independent switch (Fig. 3A).
Figure 3.
Digital responses in TCR signaling (A) Many signaling responses in T cells are digital even on short timescales (1 min). Three signaling features contribute to this digital character: Ultrasensitivity through multiple dephosphorylation (cf. NFAT translocation to the nucleus) or through kinase cascade (cf. MAPK activation) or bistability through positive feedback (cf. SOS switch) or double-negative feedback (cf. SHP-1 regulation). (B) Ultrasensitivity yields a sharp dose response that is reversible. (C) The hallmark of digital responses is the bimodal (i.e., all-or-none) distribution of signaling responses: at EC50, 50% of cells have a null response, 50% of the cells have a total response. On the other hand, an analog response yields a gradual increase with a unimodal distribution of response centered at 50% of the maximal response at EC50. (D) Feedback regulation also generates also a sharp dose response similar to that generated by ultrasensitivity but with hysteresis.
The MAPK pathway passes signals from an upstream kinase (such as Ras) to Raf, MEK, then finally ERK. MEK and ERK each have two sites of phosphorylation that are phosphorylated sequentially in two separate enzyme-substrate interactions (Ferrell and Bhatt 1997). Using an analytical model of this cascade, Huang and Ferrell (Huang and Ferrell 1996) showed that the consequence of this pattern of phosphorylation is a sharpening of the input-output dose-response curve at each step of the pathway. Hence, in the absence of feedback, activation of ERK displays ultrasensitivity to input signals from Ras or analogous kinases. This theoretical result was confirmed in vitro by titrating amounts of input signal and demonstrating ultrasensitive phosphorylation of MEK and ERK in Xenopus eggs (Ferrell 2002; Xiong and Ferrell 2003).
Within the TCR apparatus, structural and biochemical findings have pointed to the existence of feedback loops—positive (Das et al. 2009) and double-negative (Stefanova et al. 2003; Altan-Bonnet and Germain 2005)—which are both expected to lead to digital behavior (Ferrell 2002). Positive allosteric feedback is mediated through SOS, a Ras-activating enzyme, which is itself activated by active Ras. In the case of the TCR and B cell receptor (BCR) signaling machinery, Das et al (2009) showed that this positive feedback resulted in bistability. Note that bistability is a hallmark of a positive feedback signaling module, but is absent in ultrasensitive switches. In the same model, the authors showed that a second Ras-activating enzyme (Ras-GRP) that lacks a positive feedback mechanism is able to turn on the MAPK pathway in a graduated, analog manner. Most interestingly, the computer model predicted that bistability in ERK activation as a function of SOS activity should be hysteretic: once the MAPK activity reaches its high state, it is primed to regain that high state on receipt of subsequent, smaller stimuli (Fig. 3D). Hysteresis within the TCR, which the authors confirmed experimentally, may have important implications for T cell antigen recognition in vivo, where T cell-APC contacts are often brief but repeated (Henrickson et al. 2008).
Another signaling module enabling T cell decision-making is a double-negative feedback regulation of TCR signaling. Using computer modeling, we have shown how upstream negative feedback from TCR-induced activation of SHP-1 phosphatase prevents signals from large quantities of subthreshold ligands from initiating full downstream TCR signaling. However, we also found that a downstream feedback mediated by ERK acts to shut down the SHP-1 negative feedback. Such a double-negative feedback drives sustained activation for above-threshold ligands, and stably limits the noise from subthreshold ligands (Feinerman et al. 2008a).
As in the case of ERK, single-cell measurements on T cells showed that nuclear factor of activated T cells (NFAT) is activated in an all-or-none response (Fiering et al. 1990; Macian 2005). Two groups (Salazar and Hofer 2003; Podtschaske et al. 2007) have modeled quantitatively the mechanism by which graded calcium influx drives a graded dephosphorylation of NFAT at many residues, leading to all-or-none nuclear translocation and activation. The proposed mechanism for the ultrasensitive activation of NFAT is therefore a classical scheme of enzymatic activation through multiple modifications (similar to the double phosphorylation of MAPK that triggers its enzymatic activity).
It is worth noting that nuclear localization of NFκB downstream of the TCR is not digital, but rather graded in proportion to stimulation strength (Podtschaske et al. 2007). Consequently, cells may commit to digital responses, such as cytotoxic release of granzymes and perforins, secretion of IL-2 or entry into mitosis, while retaining analog functions such as IFNγ secretion, CD69 expression and others, allowing diverse immune responses to different antigens (Rabinowitz et al. 1996; Itoh and Germain 1997; Hemmer et al. 1998).
The study of analog-to-digital filtering in TCR signaling is a prime example of productive dialogue between experimental and theoretical immunology. Established theoretical groundwork provided explanations for many of the initial descriptions of all-or-none phenotypes seen in T cells and helped to illuminate how sensitivity, specificity and speed are wired into the ligand-sensing signaling network. Further exploration of hallmarks of digital circuits through computational modeling has extended the understanding of T cell activation: Hysteresis, characteristic of positive feedback, indicates that T cells may have short-term memory of stimulation. Double negative feedback, well-understood to suppress noise in near-threshold systems, may also provide insights into the poorly understood phenomena of peptide antagonism and synergism (discussed below). In this way, studies of digital circuits in T cells have provided both understanding of all-or-none decision-making and hints at further phenotypes for experimental investigation.
Antagonism, Synergism
One of the counterintuitive facets of TCR signaling is that pMHC ligands that bind the TCR nearly as well as agonists decrease its sensitivity to agonists when copresented with otherwise stimulatory ligands, whereas peptides that are far from being agonists increase the sensitivity of T cell responses (Krogsgaard et al. 2007). We review here how computer modeling has contributed to our understanding of antagonism and synergism in T cell activation.
Early work to unravel the biochemical mechanism of antagonism showed that it is dependent on signals received through the TCR (Jameson et al. 1993; Racioppi et al. 1996) and is associated with the diffusible spread of the phosphatase SHP-1 (Plas et al. 1996; Dittel et al. 1999; Stefanova et al. 2003). In computational models of this process, briefly binding ligands induce minimal SHP-1 activation, whereas ligands with longer half-lives induce greater tyrosine phosphorylation of SHP-1 and its subsequent association with Lck in the TCR complex. Consequently, as ligand binding time approaches the agonist threshold, induction of negative feedback increases, directly antagonizing signals coming from neighboring stimulatory (agonist) ligands (Altan-Bonnet and Germain 2005). Thus, previous computer models automatically accounted for antagonism by implementing the suppression of substimulatory ligand signaling through SHP-1-mediated negative feedback (Altan-Bonnet and Germain 2005; Wylie et al. 2007; Lipniacki et al. 2008; Owens et al. 2009).
Synergism (coagonist function) was introduced by the Davis group, who showed that endogenous peptides can increase T cell sensitivity to limiting densities of agonist ligands. Evidence for synergism in TCR signaling is extensive (Krogsgaard et al. 2005; Krogsgaard et al. 2007; Yachi et al. 2007) though not unchallenged (Sporri and Reis e Sousa 2002; Ma et al. 2008). One effort to explain and model synergistic effects expanded the TCR-pMHC picture to include intracellular interaction between Lck and the coreceptor (either CD4 or CD8) and extracellular interaction between the coreceptor and the MHC (Li et al. 2004), with the coreceptor acting as a bridging molecule. This model allows even momentary interaction between the TCR and self-pMHC to bring TCR ITAM residues near Lck for phosphorylation by dimerizing self-pMHC and agonist-pMHC molecules. Although this model simulated synergy well, it had a number of weaknesses: it could not account for antagonism, allowed for large numbers of endogenous ligands to trigger signaling, and was not validated beyond its ability to qualitatively show a role for endogenous pMHC-TCR complexes in enhancing signaling from agonist pMHC-TCR complexes. At a conceptual level, however, it showed that cross-linking by the coreceptor could produce lattices of pMHC-TCR complexes and hence may be the key to incorporating synergy into models of TCR triggering.
A model simultaneously accounting for both antagonism and synergism has been attempted (Wylie et al. 2007) but failed to achieve digital activation of the MAPK pathway. However, two existing models of TCR signaling may account for synergism through the “memory” inherent in their signaling cascades. Both our model (Feinerman et al. 2008a), which allows protection of previously unengaged TCR through feedback inhibition of SHP-1 by activated ERK, and the model of Das et al (2009), through hysteretic activation of Ras, may allow weak ligands to contribute in a positive manner to signaling. In these models, crossing of the digital activation threshold by the agonist could allow parallel TCR engagement by weak ligands to drive the analog aspects of the response measured in a synergism assay. Thus existing models may already account for the phenomena of antagonism and synergism, emphasizing the added value of computer modeling efforts to study T cell activation.
Tunability of the T cell Response to Ligand
T lymphocytes must undergo a strict developmental program in the thymus before being released as naïve T cells to the periphery. Hematopoietic progenitors entering the thymus are tested against self pMHC to pass two selecting processes. The absence of responsiveness to self pMHC presented by epithelial cells induces death by neglect, whereas proper responsiveness green-lights further differentiation—so called positive selection. But over-responsiveness toward self pMHC induces apoptosis (negative selection). Ultimately, thymic differentiation leads to the export of mature T cells whose signaling machinery is properly “wired” as indicated by a modest, subactivating level of signaling in response to self-pMHC (Stefanova et al. 2002). T cells endowed mainly with receptors of intermediate affinity for self pMHC ligands pass the filters of positive/negative selection (Alam et al. 1996; Savage et al. 1999; Savage and Davis 2001). Beyond the role of the intrinsic affinity of the TCR for selecting ligands in the maturation process, it is also clear that T cells modify the intracellular components of the signaling apparatus to tune the amplitude of their signaling response induced by self-ligand during thymocyte development, extinguishing responses to weak ligands while not constraining responses to strong ligands (Hogquist et al. 1994; Lucas et al. 1999; Yasutomo et al. 2000; Hogquist 2001; Starr et al. 2003).
This fine-tuning of the activation threshold to diverse ligands is an appealing issue to address with computational models. There is clear evidence for changes at the level of proximal TCR signaling in this regard, based on direct analysis of ZAP-70 and ζ phosphorylation in immature versus mature thymocytes offered ligands of various binding affinities for the TCR (Lucas et al. 1999). Studies involving miR181a (Li et al. 2007) and the ERK cascade (Altan-Bonnet and Germain 2005) as well as the expression of Shp-1 (Plas et al. 1999) suggest that one aspect of the tuning involves a rebalancing of ERK activity relative to SHP-1. Immature cells have fewer ERK phosphatases and higher basal ERK activity, but lower SHP-1 levels than more mature thymocytes; our model predicts that such a state of the feedback control elements would allow weak ligands of the TCR to become stimulatory, as direct biochemical analysis indicates (Li et al. 2007). Loss of the elevated ERK state and increased SHP-1 in more mature cells would extinguish signaling in response to weak (self-) ligands but preserve responses to strong TCR binders as experiments show (Lucas et al. 1999) and our model predicts (Altan-Bonnet and Germain 2005). Examination of how these changes drive positive and negative selection during thymocyte differentiation is a challenge well suited to computer modeling.
Stochasticity
Computer modeling has also been particularly illuminating in dealing with the topic of stochasticity in T cell activation. Early experimental measurements estimated that T cells could respond to one or few pMHC (Sykulev et al. 1996; Irvine et al. 2002; Altan-Bonnet and Germain 2005). Models have pointed out how signaling modules (e.g., MAPK cascade or Ca2+ response) could convert such a minute stimulation into large, all-or-none (digital) effects downstream. The small number of ligands implies that a certain degree of stochasticity (i.e., molecular noise) might be critical when studying the system's dynamics. Unfortunately, the more commonly employed deterministic models predict a fixed and reproducible threshold for activation (e.g., T cells would be activated reproducibly by a certain number of antigenic ligands).
Artyomov and colleagues studied a toy model of competition between positive and negative feedback to argue for the functional relevance of stochasticity in TCR signaling (Artyomov et al. 2007). Indeed, in their differential signaling model, the feedbacks are finely balanced, with similar characteristic times of activation. Stochastic fluctuations then generate the experimentally observed bistability, whereby the system decides between activation and repression depending on which feedback—positive or negative—is activated first. However, given the large fluctuations in signaling protein levels from T cell to T cell and the ensuing variation in signaling dynamics (cf. section 2f), additional mechanisms would be necessary to explain how the system self-regulates to be constantly poised at this tipping point of activation.
More generally, a small amount of random variation in ultrasensitive systems stimulated at near-threshold levels can lead to the system occupying both active and inactive states, either on a cell-by-cell basis (two populations) or temporally switching back and forth between two states (Lipniacki et al. 2008). In such cases, biological stochasticity would generate an immune response that would be qualitatively different from that generated by a deterministic mechanism (Germain 2001). For these, computer models become a critical tool of analysis, without which experimental data may be hard to interpret.
Spatial Regulation of T Cell Activation
Modeling the TCR signaling cascade as a biochemical network in a well-mixed environment has provided great insight into its dynamics and biological function. However, spatial organization of T cell components may be relevant to the regulation of a receptor by its neighbors or by other membrane-signaling proteins within the plasma membrane.
For example, serial engagement of multiple adjacent receptors by a single low-affinity pMHC has been proposed as a mechanism by which few ligands may assemble a robust functional response over time (Valitutti et al. 1995; Itoh and Germain 1997; Hudrisier et al. 1998; Wofsy et al. 2001). Wolfsy et al estimated analytically the competition between ligand binding/debinding, diffusion, and internalization for a given TCR to illustrate how extensive serial engagement of a given ligand could be. One prediction of this model is that there exists an optimal dwell-time for the ligand-receptor interaction below which the T cell truncates its signaling response and above which ligands fail to serially trigger many TCRs because of excessively long engagement times. Some experimental evidence supports the existence of this optimal dwell time (Kalergis et al. 2001). However, these findings are incompatible with the high potency of pMHC engineered for extremely lengthy TCR engagement (Holler et al. 2000).
Settling this issue requires computer models of T cell activation on longer timescales (10 min–1 h) that must also account for well-documented lateral segregation of proteins within the plasma membrane. Initial pMHC-TCR binding results in the steric exclusion of the negative regulatory phosphatase CD45 (Choudhuri et al. 2005) and the concentration of actively signaling TCRs into so-called microclusters (Bunnell et al. 2002; Campi et al. 2005) that move toward the center of the T cell-APC contact (Varma et al. 2006). This dynamic reorganization into an immunological synapse (IS) concentrates TCR-pMHC contacts in the center with a surrounding adhesive ring of LFA-1-ICAM-1 interactions. Theoretical work has shown that self-organization of the membrane components can be sufficient for formation of the IS pattern, even in the absence of active cytoskeletal forces (Qi et al. 2001). This model combined measured values of antigen-receptor interaction and physical properties of the plasma membrane to account for synapse formation. The main prediction of this model—that lower expression of TCR in thymocytes would yield smaller, more transient, multifocal synapse formation—was subsequently confirmed experimentally (Lee et al. 2003).
Further modeling efforts have sought to illuminate how the immune synapse modulates T cell activation by suppressing signaling from strong agonists through increased down-regulation of the TCR (Cemerski et al. 2008), as well as by enhancing signaling from less avid agonist peptides through concentration of signaling components (Chan et al. 2001; Lee et al. 2003a). Other work has probed how exclusion of the bulky CD45 phosphatase could account for TCR triggering—albeit without adequate speed in the models relative to experiment (Burroughs et al. 2006; Burroughs and van der Merwe 2007). This work accounted for experimental observations whereby genetic modification of the volume or heights of receptors modulated the separation between T cell and APC within the contact area, and resulted in varied levels of activation of T cells (Choudhuri et al. 2005). As discussed above, the coupling of local reaction-diffusion with global elastic constraints is a difficult theoretical problem that necessitates specific modeling formalism. Yet, such theoretical effort will certainly illuminate experimental observations on the interplay between TCR signaling and membrane reorganization (currently a field of intense study). Though pharmacological and genetic methods of synapse interruption often have wide-ranging effects that preclude their attribution solely to spatial organization (Lee et al. 2003; Ilani et al. 2009), recent work using micropatterned surfaces of membrane and ligands (Mossman et al. 2005; DeMond and Groves 2007) or using photoactivatable ligands (Huse et al. 2007) enables the direct testing of the effects of geometric constraints on T cell activation. The quantitative and qualitative data from these efforts will in turn allow the revision and validation of models of immune synapse formation and a better understanding of T cell activation on longer timescales (>10 min).
Reconciliation of Robustness and Variability in T Cell Activation
Robustness is a concept that was introduced to describe a salient feature of cell signaling: although cells have fluctuating levels of the many proteins involved in signal transduction, proper physiological function requires predictable responses to extracellular stimuli (Barkai and Leibler 1997). First modeled and experimentally validated in bacteria (Alon et al. 1999; Cluzel et al. 2000; Korobkova et al. 2004), robustness is also critical to T cell discrimination between self and nonself. Quantitative analysis of T cell ligand sensing has shown that the calcium response to a set number of ligands does not vary as much as would be expected from variation in the levels of individual signaling proteins that determine this response (Irvine et al. 2002; Li et al. 2004; Purbhoo et al. 2004; Feinerman et al. 2008a).
To investigate the effects of this variation on the robustness of self/nonself discrimination by T cells, we have systematically investigated the effects of modulating individual species within an experimentally validated computational model of the TCR apparatus (Feinerman et al. 2008b). By varying each component within its physiological range, we characterized the effects of natural variability in signaling components in peripheral T lymphocytes. This analysis predicted that signaling components would fall into three categories: (1) Noncritical signaling proteins whose variation in expression within the observed range does not measurably affect ligand response. (2) Analog regulators whose level of expression fine-tunes the activation threshold in terms of number of ligands required to exceed this threshold. (3) Digital regulators whose level of expression switches a cell between responsive and unresponsive states without affecting activation thresholds in responsive cells (Feinerman et al. 2008b).
These theoretical investigations predicted that ERK-1, despite its crucial role in regulation of ligand discrimination, is a noncritical component because its average expression is in such excess that variation within the physiological range does not affect the T cell response. Other noncritical components are Lck, ZAP70, LAT, Raf, and MEK. In contrast, the CD8 coreceptor was predicted to be an analog-positive regulator, increasing sensitivity with increased expression. SHP-1, on the other hand, was predicted to be a digital negative regulator: For high levels of SHP-1, T cells will not respond even to high doses of antigen. Consequently, our model predicted that even in a clonal population of T cells, the natural range of expression of SHP-1 and CD8 would lead to highly variable sensitivity to antigen.
We then developed an experimental assay to correlate T cell responsiveness with the expression levels of signaling components within individual cells. This assay relies on staining of endogenous proteins (e.g., CD8 and SHP-1) and phospho-proteins (e.g., ppERK), followed by flow cytometry to achieve single-cell resolution. Our experimental measurements validated the predictions from our computer model, demonstrating variability across a population of cells, but also added insight into how the system remains robust: Correlation between expression levels of positive and negative regulators (CD8 and SHP-1, respectively) restricts the actual range of sensitivity to a smaller range than would be predicted from theoretical analysis of uncorrelated protein expression variation. This study shed light on the delicate regulation of ligand discrimination in T cells by showing how it can be robustly defined while remaining tunable and flexible in the face of irreducible cell-to-cell variation in molecular phenotype. In this case, the computational model was not the endpoint of the effort, but a tool to enable more precise selection of experimental approaches.
COMPUTATION AND THE STUDY OF T CELL IMMUNOBIOLOGY: GOING FORWARD
In this review, we have discussed computational models that yielded insights on issues of specificity, antagonism, synergism, stochasticity, spatial regulation, and robustness in T cell activation. The majority of these models served principally as illustrations of complex hypotheses, using quantitative data to explain observed phenomena. However, successful application of modeling requires more than accurate fitting of experimental observations or clarification of purely theoretical points. Models with the ability to generate testable predictions and extend current understanding based on a validated mathematical framework have the greatest potential to contribute to immunology (Qi et al. 2001; Lee et al.; Altan-Bonnet and Germain 2005; Feinerman et al. 2008; Feinerman et al. 2008b). In our experience, the added value of such models is correlated to two simple criteria: model parameters that are tightly constrained by experimental measurements and model predictions that are designed to be testable (and hopefully tested) at the bench.
Proliferation of such added-value models will benefit from current trends in both experimental immunology and modeling software development. From the experimental side, the wider application of quantitative methods, including multiplexed surface plasmon resonance (Lee et al. 2006), absolute quantitative mass spectrometry (Bantscheff et al. 2007), and quantitative flow cytometry (Irish et al. 2004), will continue to restrain the set of unknown parameters, improving the statistical relevance of computer models. From the theoretical side, the rapid dissemination of standardized modeling platforms (particularly Virtual Cell [Loew and Schaff 2001], BioNetGen [Blinov et al. 2004] CellDesigner [Funahashi et al. 2003], and Simmune [Meier-Schellersheim et al. 2006]) will accelerate the exchange of models for further editing, testing, and validation. Thus, one prospect for computer modeling in immunology is more direct participation from immunologists who will both utilize and extend models of T cell signaling in order to develop a more complete model of T cell activation. In the long term, one can envision a time when immunologists will systematically test pre-established models of T cell signaling to assess the significance of new experimental observations and challenge pre-existing mechanisms.
ACKNOWLEDGMENTS
This work was supported in part by funds from the Intramural Research Program, NIAID, National Institutes of Health (NIH). J.C. and G.A-B's research is supported by an NSF grant #0848030 and an NIH grant #R01-AI083408. G.A-B is the incumbent of a Bristol-Myers Squibb / James D. Robinson III junior faculty chair at MSKCC. J.C. and G.A-B wish to thank Matthew Hathorn and Karen Tkach for critical reading of the manuscript.
Footnotes
Editors: Lawrence E. Samelson and Andrey Shaw
Additional Perspectives on Immunoreceptor Signaling available at www.cshperspectives.org
REFERENCES
- Alam SM, Travers PJ, Wung JL, Nasholds W, Redpath S, Jameson SC, Gascoigne NR 1996. T-cell-receptor affinity and thymocyte positive selection. Nature 381:616–620 [DOI] [PubMed] [Google Scholar]
- Alon U, Surette MG, Barkai N, Leibler S 1999. Robustness in bacterial chemotaxis. Nature 397:168–171 [DOI] [PubMed] [Google Scholar]
- Altan-Bonnet G, Germain RN 2005. Modeling T-cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol 3:e356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artyomov MN, Das J, Kardar M, Chakraborty AK 2007. Purely stochastic binary decisions in cell signaling models without underlying deterministic bistabilities. Proc Natl Acad Sci 104:18958–18963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B 2007. Quantitative mass spectrometry in proteomics: A critical review. Anal Bioanal Chem 389:1017–1031 [DOI] [PubMed] [Google Scholar]
- Barkai N, Leibler S 1997. Robustness in simple biochemical networks. Nature 387:913–917 [DOI] [PubMed] [Google Scholar]
- Blinov ML, Faeder JR, Goldstein B, Hlavacek WS 2004. BioNetGen: Software for rule-based modeling of signal transduction based on the interactions of molecular domains. Bioinformatics 20:3289–3291 [DOI] [PubMed] [Google Scholar]
- Bunnell SC, Hong DI, Kardon JR, Yamazaki T, McGlade CJ, Barr VA, Samelson LE 2002. T-cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J Cell Biol 158:1263–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs NJ, van der Merwe PA 2007. Stochasticity and spatial heterogeneity in T-cell activation. Immunol Rev 216:69–80 [DOI] [PubMed] [Google Scholar]
- Burroughs NJ, Lazic Z, van der Merwe PA 2006. Ligand detection and discrimination by spatial relocalization: A kinase-phosphatase segregation model of TCR activation. Biophys J 91:1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campi G, Varma R, Dustin ML 2005. Actin and agonist MHC-peptide complex-dependent T-cell receptor microclusters as scaffolds for signaling. J Exp Med 202:1031–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cemerski S, Das J, Giurisato E, Markiewicz MA, Allen PM, Chakraborty AK, Shaw AS 2008. The balance between T-cell receptor signaling and degradation at the center of the immunological synapse is determined by antigen quality. Immunity 29:414–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, George AJ, Stark J 2001. Cooperative enhancement of specificity in a lattice of T-cell receptors. Proc Natl Acad Sci 98:5758–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri K, Wiseman D, Brown MH, Gould K, van der Merwe PA 2005. T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature 436:578–582 [DOI] [PubMed] [Google Scholar]
- Cluzel P, Surette M, Leibler S 2000. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science 287:1652–1655 [DOI] [PubMed] [Google Scholar]
- Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, Werlen G, Hollander GA, Gascoigne NR, Palmer E 2006. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 444:724–729 [DOI] [PubMed] [Google Scholar]
- Das J, Ho M, Zikherman J, Govern C, Yang M, Weiss A, Chakraborty AK, Roose JP 2009. Digital signaling and hysteresis characterize Ras activation in lymphoid cells. Cell 136:337–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y 1998. Ligand recognition by alpha βT-cell receptors. Ann Rev Immunol 16:523–544 [DOI] [PubMed] [Google Scholar]
- Delon J, Bercovici N, Raposo G, Liblau R, Trautmann A 1998. Antigen-dependent and -independent Ca2+ responses triggered in T-cells by dendritic cells compared with B cells. J Exp Med 188:1473–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMond AL, Groves JT 2007. Interrogating the T-cell synapse with patterned surfaces and photoactivated proteins. Curr Opin Immunol 19:722–727 [DOI] [PubMed] [Google Scholar]
- Dittel BN, Stefanova I, Germain RN, Janeway CA Jr 1999. Cross-antagonism of a T-cell clone expressing two distinct T-cell receptors. Immunity 11:289–298 [DOI] [PubMed] [Google Scholar]
- Faeder JR, Hlavacek WS, Reischl I, Blinov ML, Metzger H, Redondo A, Wofsy C, Goldstein B 2003. Investigation of early events in Fc έRI-mediated signaling using a detailed mathematical model. J Immunol 170:3769–3781 [DOI] [PubMed] [Google Scholar]
- Feinerman O, Germain RN, Altan-Bonnet G 2008a. Quantitative challenges in understanding ligand discrimination by αβ T-cells. Mol Immunol 45:619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan-Bonnet G 2008b. Variability and robustness in T-cell activation from regulated heterogeneity in protein levels. Science 321:1081–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE 2002. Self-perpetuating states in signal transduction: Positive feedback, double-negative feedback and bistability. Current Opinion Cell Biol 14:140–148 [DOI] [PubMed] [Google Scholar]
- Ferrell JE Jr, Bhatt RR 1997. Mechanistic studies of the dual phosphorylation of mitogen-activated protein kinase. J Biol Chem 272:19008–19016 [DOI] [PubMed] [Google Scholar]
- Fiering S, Northrop JP, Nolan GP, Mattila PS, Crabtree GR, Herzenberg LA 1990. Single cell assay of a transcription factor reveals a threshold in transcription activated by signals emanating from the T-cell antigen receptor. Genes Develop 4:1823–1834 [DOI] [PubMed] [Google Scholar]
- Funahashi A, Morohashi M, Kitano H, Tanimura N 2003. CellDesigner: A process diagram editor for gene-regulatory and biochemical networks BioSilico 1:159–162 [Google Scholar]
- Germain RN 2001. The art of the probable: System control in the adaptive immune system. Science 293:240–245 [DOI] [PubMed] [Google Scholar]
- Germain RN, Stefanova I 1999. The dynamics of T-cell receptor signaling: Complex orchestration and the key roles of tempo and cooperation. Ann Rev Immunol 17:467–522 [DOI] [PubMed] [Google Scholar]
- Gil D, Schamel WW, Montoya M, Sanchez-Madrid F, Alarcon B 2002. Recruitment of Nck by CD3ε reveals a ligand-induced conformational change essential for T-cell receptor signaling and synapse formation. Cell 109:901–912 [DOI] [PubMed] [Google Scholar]
- Gillespie DT 2007. Stochastic simulation of chemical kinetics. Annu Rev Phys Chem 58:35–55 [DOI] [PubMed] [Google Scholar]
- Hemmer B, Stefanova I, Vergelli M, Germain RN, Martin R 1998. Relationships among TCR ligand potency, thresholds for effector function elicitation, and the quality of early signaling events in human T-cells. J Immunol 160:5807–5814 [PubMed] [Google Scholar]
- Henrickson SE, Mempel TR, Mazo IB, Liu B, Artyomov MN, Zheng H, Peixoto A, Flynn MP, Senman B, Junt T et al. 2008. T-cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T-cell activation. Nat Immunol 9:282–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavacek WS, Faeder JR, Blinov ML, Posner RG, Hucka M, Fontana W 2006. Rules for modeling signal-transduction systems. Sci STKE 2006:re6. [DOI] [PubMed] [Google Scholar]
- Hogquist KA 2001. Signal strength in thymic selection and lineage commitment. Curr Opin Immunol 13:225–231 [DOI] [PubMed] [Google Scholar]
- Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR 1994. T-cell receptor antagonist peptides induce positive selection. Cell 76:17–27 [DOI] [PubMed] [Google Scholar]
- Holler PD, Holman PO, Shusta EV, O'Herrin S, Wittrup KD, Kranz DM 2000. In vitro evolution of a T-cell receptor with high affinity for peptide/MHC. Proc Natl Acad Sci 97:5387–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst J, Wang H, Eder KD, Workman CJ, Boyd KL, Baquet Z, Singh H, Forbes K, Chruscinski A, Smeyne R et al. 2008. Scalable signaling mediated by T-cell antigen receptor-CD3 ITAMs ensures effective negative selection and prevents autoimmunity. Nat Immunol 9:658–666 [DOI] [PubMed] [Google Scholar]
- Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U 2006. COPASI–a COmplex PAthway SImulator. Bioinformatics 22:3067–3074 [DOI] [PubMed] [Google Scholar]
- Hopfield JJ 1974. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA 71:4135–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Ferrell JE Jr 1996. Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci 93:10078–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudrisier D, Kessler B, Valitutti S, Horvath C, Cerottini JC, Luescher IF 1998. The efficiency of antigen recognition by CD8+ CTL clones is determined by the frequency of serial TCR engagement. J Immunol 161:553–562 [PubMed] [Google Scholar]
- Huse M, Klein LO, Girvin AT, Faraj JM, Li Q-J, Kuhns MS, Davis MM 2007. Spatial and temporal dynamics of T-cell receptor signaling with a photoactivatable agonist. Immunity 27:76–88 [DOI] [PubMed] [Google Scholar]
- Ilani T, Vasiliver-Shamis G, Vardhana S, Bretscher A, Dustin ML 2009. T-cell antigen receptor signaling and immunological synapse stability require myosin IIA. Nat Immunol 10:531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT, Nolan GP 2004. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell 118:217–228 [DOI] [PubMed] [Google Scholar]
- Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM 2002. Direct observation of ligand recognition by T-cells. Nature 419:845–849 [DOI] [PubMed] [Google Scholar]
- Itoh Y, Germain RN 1997. Single cell analysis reveals regulated hierarchical T-cell antigen receptor signaling thresholds and intraclonal heterogeneity for individual cytokine responses of CD4+ T-cells. J Exp Med 186:757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SC, Carbone FR, Bevan MJ 1993. Clone-specific T-cell receptor antagonists of major histocompatibility complex class I-restricted cytotoxic T-cells. J Exp Med 177:1541–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalergis AM, Boucheron N, Doucey MA, Palmieri E, Goyarts EC, Vegh Z, Luescher IF, Nathenson SG 2001. Efficient T-cell activation requires an optimal dwell-time of interaction between the TCR and the pMHC complex. Nat Immunol 2:229–234 [DOI] [PubMed] [Google Scholar]
- Korobkova E, Emonet T, Vilar JM, Shimizu TS, Cluzel P 2004. From molecular noise to behavioural variability in a single bacterium. Nature 428:574–578 [DOI] [PubMed] [Google Scholar]
- Krogsgaard M, Juang J, Davis MM 2007. A role for “self” in T-cell activation. Sem Immunol 19:236–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogsgaard M, Li Q-j, Sumen C, Huppa JB, Huse M, Davis MM 2005. Agonist/endogenous peptide-MHC heterodimers drive T-cell activation and sensitivity. Nature 434:238–243 [DOI] [PubMed] [Google Scholar]
- Le Novere N 2006. Model storage, exchange and integration. BMC Neurosci 7:S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novere N, Hucka M, Mi H, Moodie S, Schreiber F, Sorokin A, Demir E, Wegner K, Aladjem MI, Wimalaratne SM et al. 2009. The systems biology graphical notation. Nat Biotechnol 27:735–741 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Hori Y, Chakraborty AK 2003b. Low T-cell receptor expression and thermal fluctuations contribute to formation of dynamic multifocal synapses in thymocytes. Proc Natl Acad Sci 100:4383–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Nedelkov D, Corn RM 2006. Surface plasmon resonance imaging measurements of antibody arrays for the multiplexed detection of low molecular weight protein biomarkers. Anal Chem 78:6504–6510 [DOI] [PubMed] [Google Scholar]
- Lee K-H, Dinner AR, Tu C, Campi G, Raychaudhuri S, Varma R, Sims TN, Burack WR, Wu H, Wang J et al. 2003a. The immunological synapse balances T-cell receptor signaling and degradation. Science 302:1218–1222 [DOI] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P et al. 2007. miR-181a is an intrinsic modulator of T-cell sensitivity and selection. Cell 129:147–161 [DOI] [PubMed] [Google Scholar]
- Li Q-J, Dinner AR, Qi S, Irvine DJ, Huppa JB, Davis MM, Chakraborty AK 2004. CD4 enhances T-cell sensitivity to antigen by coordinating Lck accumulation at the immunological synapse. Nat Immunol 5:791–799 [DOI] [PubMed] [Google Scholar]
- Lipniacki T, Hat B, Faeder JR, Hlavacek WS 2008. Stochastic effects and bistability in T-cell receptor signaling. Journal of Theoretical Biology 254:110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loew LM, Schaff JC 2001. The Virtual Cell: A software environment for computational cell biology. Trends Biotechnol 19:401–406 [DOI] [PubMed] [Google Scholar]
- Lucas B, Stefanova I, Yasutomo K, Dautigny N, Germain RN 1999. Divergent changes in the sensitivity of maturing T-cells to structurally related ligands underlies formation of a useful T-cell repertoire. Immunity 10:367–376 [DOI] [PubMed] [Google Scholar]
- Ma Z, Sharp KA, Janmey PA, Finkel TH 2008. Surface-anchored monomeric agonist pMHCs alone trigger TCR with high sensitivity. PLoS Biol 6:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macian F 2005. NFAT proteins: Key regulators of T-cell development and function. Nat Rev Immunol 5:472–484 [DOI] [PubMed] [Google Scholar]
- Malissen B 2008. CD3 ITAMs count! Nat Immunol 9:583–584 [DOI] [PubMed] [Google Scholar]
- Malissen B, Aguado E, Malissen M, Frederick W. Alt KFATKFMJWU, Emil RU 2005. Role of the LAT Adaptor in T[hyphen (true graphic)]Cell Development and Th2 Differentiation. Advances in Immunology, pp. 1–25 Academic Press; [DOI] [PubMed] [Google Scholar]
- McKeithan TW 1995. Kinetic proofreading in T-cell receptor signal transduction. Proc Natl Acad Sci 92:5042–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Schellersheim M, Xu X, Angermann B, Kunkel EJ, Jin T, Germain RN 2006. Key role of local regulation in chemosensing revealed by a new molecular interaction-based modeling method. PLoS Comput Biol 2:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman KD, Campi G, Groves JT, Dustin ML 2005. Altered TCR signaling from geometrically repatterned immunological synapses. Science 310:1191–1193 [DOI] [PubMed] [Google Scholar]
- Ninio J 1975. Kinetic amplification of enzyme discrimination. Biochimie 57:587–595 [DOI] [PubMed] [Google Scholar]
- Owens ND, Timmis J, Greensted A, Tyrrell A 2010. Elucidation of T-cell signalling models. J Theor Biol 262:452–470 [DOI] [PubMed] [Google Scholar]
- Palmer E, Naeher D 2009. Affinity threshold for thymic selection through a T-cell receptor-co-receptor zipper. Nat Rev Immunol 9:207–213 [DOI] [PubMed] [Google Scholar]
- Plas DR, Johnson R, Pingel JT, Matthews RJ, Dalton M, Roy G, Chan AC, Thomas ML 1996. Direct regulation of ZAP-70 by SHP-1 in T-cell antigen receptor signaling. Science 272:1173–1176 [DOI] [PubMed] [Google Scholar]
- Plas DR, Williams CB, Kersh GJ, White LS, White JM, Paust S, Ulyanova T, Allen PM, Thomas ML 1999. Cutting edge: the tyrosine phosphatase SHP-1 regulates thymocyte positive selection. J Immunol 162:5680–5684 [PubMed] [Google Scholar]
- Podtschaske M, Benary U, Zwinger S, Hofer T, Radbruch A, Baumgrass R 2007. Digital NFATc2 activation per cell transforms graded T-cell receptor activation into an all-or-none IL-2 expression. PLoS ONE 2:e935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purbhoo MA, Irvine DJ, Huppa JB, Davis MM 2004. T-cell killing does not require the formation of a stable mature immunological synapse.[erratum appears in Nat Immunol. 2004 Jun;5(6):658]. Nat Immunol 5:524–530 [DOI] [PubMed] [Google Scholar]
- Qi SY, Groves JT, Chakraborty AK 2001. Synaptic pattern formation during cellular recognition. Proc Natl Acad Sci 98:6548–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, Beeson C, Wulfing C, Tate K, Allen PM, Davis MM, McConnell HM 1996. Altered T-cell receptor ligands trigger a subset of early T-cell signals. Immunity 5:125–135 [DOI] [PubMed] [Google Scholar]
- Racioppi L, Matarese G, D'Oro U, De Pascale M, Masci AM, Fontana S, Zappacosta S 1996. The role of CD4-Lck in T-cell receptor antagonism: Evidence for negative signaling. Proc Natl Acad Sci 93:10360–10365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Rodriguez J, Simeoni L, Lindquist JA, Hemenway R, Bommhardt U, Arndt B, Haus U-U, Weismantel R, Gilles ED, Klamt S et al. 2007. A logical model provides insights into T-cell receptor signaling. PLoS Comput Biol 3:e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar C, Hofer T 2003. Allosteric regulation of the transcription factor NFAT1 by multiple phosphorylation sites: A mathematical analysis. J Mol Biol 327:31–45 [DOI] [PubMed] [Google Scholar]
- Savage PA, Davis MM 2001. A kinetic window constricts the T-cell receptor repertoire in the thymus. Immunity 14:243–252 [DOI] [PubMed] [Google Scholar]
- Savage PA, Boniface JJ, Davis MM 1999. A kinetic basis for T-cell receptor repertoire selection during an immune response. Immunity 10:485–492 [DOI] [PubMed] [Google Scholar]
- Schamel WW, Risueno RM, Minguet S, Ortiz AR, Alarcon B 2006. A conformation- and avidity-based proofreading mechanism for the TCR-CD3 complex. Trends Immunol 27:176–182 [DOI] [PubMed] [Google Scholar]
- Schoeberl B, Eichler-Jonsson C, Gilles ED, Muller G 2002. Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat Biotechnol 20:370–375 [DOI] [PubMed] [Google Scholar]
- Sporri R, Reis e Sousa C 2002. Self peptide/MHC class I complexes have a negligible effect on the response of some CD8(+) T-cells to foreign antigen. Eur J Immunol 32:3161–3170 [DOI] [PubMed] [Google Scholar]
- Starr TK, Jameson SC, Hogquist KA 2003. Positive and negative selection of T-cells. Annu Rev Immunol 21:139–176 [DOI] [PubMed] [Google Scholar]
- Stefanova I, Dorfman JR, Germain RN 2002. Self-recognition promotes the foreign antigen sensitivity of naive T lymphocytes. Nature 420:429–434 [DOI] [PubMed] [Google Scholar]
- Stefanova I, Hemmer B, Vergelli M, Martin R, Biddison WE, Germain RN 2003. TCR ligand discrimination is enforced by competing ERK positive and SHP-1 negative feedback pathways. Nat Immunol 4:248–254 [DOI] [PubMed] [Google Scholar]
- Sykulev Y, Joo M, Vturina I, Tsomides TJ, Eisen HN 1996. Evidence that a single peptide-MHC complex on a targeT-cell can elicit a cytolytic T-cell response. Immunity 4:565–571 [DOI] [PubMed] [Google Scholar]
- Valitutti S, Muller S, Cella M, Padovan E, Lanzavecchia A 1995. Serial triggering of many T-cell receptors by a few peptide-MHC complexes.[see comment]. Nature 375:148–151 [DOI] [PubMed] [Google Scholar]
- Varma R, Campi G, Yokosuka T, Saito T, Dustin ML 2006. T-cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity 25:117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wofsy C, Coombs D, Goldstein B 2001. Calculations show substantial serial engagement of T-cell receptors. Biophys J 80:606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie DC, Das J, Chakraborty AK 2007. Sensitivity of T-cells to antigen and antagonism emerges from differential regulation of the same molecular signaling module. Proc Natl Acad Sci 104:5533–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Ferrell JE Jr 2003. A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision.[see comment]. Nature 426:460–465 [DOI] [PubMed] [Google Scholar]
- Yachi PP, Lotz C, Ampudia J, Gascoigne NR 2007. T-cell activation enhancement by endogenous pMHC acts for both weak and strong agonists but varies with differentiation state. J Exp Med 204:2747–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasutomo K, Lucas B, Germain RN 2000. TCR signaling for initiation and completion of thymocyte positive selection has distinct requirements for ligand quality and presenting cell type. J Immunol 165:3015–3022 [DOI] [PubMed] [Google Scholar]
- Zehn D, Lee SY, Bevan MJ 2009. Complete but curtailed T-cell response to very low-affinity antigen. Nature 458:211–214 [DOI] [PMC free article] [PubMed] [Google Scholar]