Abstract
Even 30 years after its discovery, the tumor suppressor protein p53 is still somewhat of an enigma. p53's intimate and multifaceted role in the cell cycle is mirrored in its equally complex structural biology that is being unraveled only slowly. Here, we discuss key structural aspects of p53 function and its inactivation by oncogenic mutations. Concerted action of folded and intrinsically disordered domains of the highly dynamic p53 protein provides binding promiscuity and specificity, allowing p53 to process a myriad of cellular signals to maintain the integrity of the human genome. Importantly, progress in elucidating the structural biology of p53 and its partner proteins has opened various avenues for structure-guided rescue of p53 function in tumors. These emerging anticancer strategies include targeting mutant-specific lesions on the surface of destabilized cancer mutants with small molecules and selective inhibition of p53's degradative pathways.
Oncogenic mutations in the p53 tumor suppressor can destabilize it or inhibit DNA binding. Structural studies now allow in silico design of drugs that could rescue p53 activity.
The tumor suppressor p53 is at the hub of a plethora of signaling pathways that control the cell cycle and maintain the integrity of the human genome (Vousden and Prives 2009). It is therefore not surprising that the structure of p53 is of equally intricate complexity. p53 functions primarily as a transcription factor and is biologically active as a homotetramer comprising 4 × 393 amino acid residues. It has a modular domain structure, consisting of folded DNA-binding and tetramerization domains, flanked by intrinsically disordered regions at both the amino- and carboxy-termini, which poses a formidable challenge to the structural biologist (Fig. 1). In the mid-1990s, several groundbreaking studies revealed structural details of individual components of the p53 structure, such as the DNA-binding domain and the tetramerization domain, which laid the framework for understanding the effects of common p53 cancer mutants. But in the decade that followed, comparatively little progress was made in elucidating the structural basis of p53 function or its inactivation in cancer, considering that the scientific literature has been inundated with p53-related publications. Many structural aspects of p53 function have remained elusive. Only in recent years have we begun to grasp how p53 works as a whole by combining classical structural biology, innovative protein engineering techniques, and sophisticated computational methods.
Figure 1.
Domain structure of p53. p53 contains a natively unfolded amino-terminal transactivation domain (TAD), which can be further subdivided into the subdomains TAD1 and TAD2, followed by a proline-rich region (PRR). The structured DNA-binding and tetramerization domains (OD) are connected through a flexible linker region. Similarly to the TAD region, the regulatory domain at the extreme carboxyl terminus (CTD) is also intrinsically disordered. The vertical bars indicate the relative missense-mutation frequency in human cancer for each residue based on the TP53 Mutation Database of the International Agency for Research on Cancer (www-p53.iarc.fr) (Petitjean et al. 2007), showing that most cancer mutations are located in the DNA-binding domain. The structure of the DNA-binding domain (PDB code 1TSR) (Cho et al. 1994) is shown as a ribbon representation and colored with a rainbow gradient from the amino terminus (blue) to the carboxyl terminus (red). Sites of cancer hotspot mutations and essential DNA contacts are shown as stick models. Parts of the figure were adapted from Joerger and Fersht (2008).
STRUCTURE OF THE DNA-BINDING DOMAIN
The structure of the DNA-binding core domain (residues 94-292) consists of a central immunoglobulin-like β-sandwich scaffold and additional structural elements that form the DNA-binding surface (Fig. 1), which include a loop-sheet-helix motif and two large loops (L2 and L3). The architecture of the L2/L3 region is stabilized by a zinc ion, which is tetrahedrally coordinated by Cys176, His179, Cys238, and Cys242 (Cho et al. 1994; Canadillas et al. 2006; Wang et al. 2007). Human p53 core domain is of relatively low intrinsic thermodynamic stability and rapidly unfolds at body temperature with a half-life of 9 minutes (Bullock et al. 1997; Friedler et al. 2003; Ang et al. 2006). Several lines of evidence suggest that the low intrinsic stability of human p53 may be the result of an adaptive evolutionary process (Canadillas et al. 2006; Khoo et al. 2009a; Khoo et al. 2009b), with important implications for protein turnover and binding to partner proteins. Low thermodynamic and kinetic stability may allow for rapid cycling between folded and unfolded states, which could provide an additional layer of regulation of functionally active cellular protein levels, on top of the specific degradation pathways involving ubiquitination and subsequent proteasomal degradation. This low intrinsic stability of the core domain has profound implications with regard to the susceptibility of human p53 to deleterious mutations and cancer development (see later). Low intrinsic stability may also be directly linked with the structural plasticity required to facilitate binding to different partner proteins. The DNA-binding surface, for example, overlaps with the binding sites for the carboxy-terminal domain of ASPP2 (Gorina and Pavletich 1996), the BRCT region of 53BP1 (Derbyshire et al. 2002; Joo et al. 2002), and the large T-antigen of SV40 (Lilyestrom et al. 2006), with significant conformational variability of the L3 loop region (residues 240–250) in the various interfaces (Oldfield et al. 2008). In particular, the L3 loop conformation in the structure of p53 core domain in complex with the large T-antigen of SV40 stands out, reminiscent of the conformation observed in the cancer mutant R249S (Joerger and Fersht 2007).
STRUCTURAL BASIS OF SEQUENCE-SPECIFIC DNA BINDING
The p53 tetramer cooperatively binds to its target duplex DNA in a sequence-specific manner (Weinberg et al. 2004). The target binding sites consist of two decameric motifs (half-sites) of the general form RRRCWWGYYY (R = A, G; W = A, T; Y = C, T), separated by 0–13 base pairs (el-Deiry et al. 1992; Funk et al. 1992). This definition has recently been refined by a genome-wide mapping of p53 binding sites, showing that most p53 response elements have consecutive half-sites (Wei et al. 2006), and by systematic measurements of the effect of every single base-pair substitution within a palindromic half-site on p53 binding (Veprintsev and Fersht 2008). Moreover, an increase in spacer length between the decamer half-sites for a given response element correlates with a decrease in p53 affinity and transactivation, as shown for the TIGAR, Noxa, and p21-5’ response elements (Jordan et al. 2008). Interestingly, a more recent study suggests a set of predictive rules to distinguish between response elements responsible for activation or repression of target genes. p53 response elements for transcriptional repression seem to have characteristic deviations from the canonical response element sequence, for example in the central dinucleotide, that are generally associated with weaker affinity (Wang et al. 2009).
Crystallographic data have provided detailed insights into the structural basis of sequence-specific DNA recognition by p53 tetramers. Two core domains bind to a half-site DNA, forming a symmetrical dimer with a relatively small, self-complementary core domain-core domain interface (e.g., molecules I and II in Fig. 2A), which includes Pro177, His178, Arg181, Met243, and Gly244 (Ho et al. 2006; Kitayner et al. 2006). The L3 loop binds to the DNA minor groove via Arg248, which makes either direct or water-mediated contacts with the DNA backbone. The guanidinium group of Arg249 is essential for stabilizing the hairpin conformation of loop L3 via a network of hydrogen bonds and a salt bridge with Glu171, thus positioning Arg248 for DNA binding. The crucial structural role of Arg249 is highlighted by the deleterious effects of the aflatoxin-induced R249S cancer mutation (Gouas et al. 2009). The structural integrity of the L3 region is severely compromised in this mutant, and the L3 loop becomes highly flexible and favors nonnative conformations, resulting in impaired DNA binding (Joerger et al. 2005; Suad et al. 2009). The side chains of Ser241 and Arg273 make direct contacts with the phosphate backbone. Water molecules are also an integral part of the protein-DNA interface. One structural water molecule in particular forms a total of four hydrogen bonds with the DNA backbone, the main-chain of Cys277, the side chain of Arg280, and the side chain of Asp281, which in turn is stabilized via a salt-bridge network with the guanidinium groups of Arg273 and Arg280 (Fig. 2B). Sequence-specific major-groove contacts are mediated by residues from the carboxy-terminal helix (Ala276, Cys277, and Arg280) and Lys120 from the L1 loop, which shows a small induced-fit movement on DNA binding. Arg280 forms two invariant hydrogen bonds with the highly conserved guanine base, whereas the other residues make varying interactions with base pairs in the major groove, depending on the sequence of the half-site (Kitayner et al. 2006).
Figure 2.
Sequence-specific DNA binding of p53. (A) Cartoon representation of a core domain tetramer bound to DNA, as observed in the crystal structure of human p53 core domain in complex with palindromic half-site DNA (PDB code 2AHI) (Kitayner et al. 2006). Two different views are shown, with individual core domains depicted in different colors: view onto the core domain tetramer, showing core domain-core domain contacts (left) and view along the DNA helix axis (right). (B) Stereo view of the major-groove interaction network of the structure shown in panel A (PDB code 2AHI, chain D). DNA-contact residues are shown as green stick models, and a crucial structural water molecule is shown as a magenta sphere. The polar interaction network involving DNA-contact residues is highlighted with black dashed lines. The orange dashed line indicates hydrophobic interaction between the Cβ atom of Ala276 and a thymine base. Nucleotides are numbered according to their position in the decameric p53 half-site motif GGACA/TGTCC.
In the crystals, the half-site dimers form a tetramer stabilized by base-stacking and protein–protein interactions, mimicking binding of a p53 tetramer to a continuous response element with a spacer of two base pairs between the half-sites (Kitayner et al. 2006). The translational protein interfaces between the half-site dimers are comparatively weak (e.g., contacts between molecules I and III in Fig. 2A) because of the spacer between the half-sites. This interface, and hence the DNA-binding affinity, are likely to change with varying spacer lengths between the half-sites. A computational study modeling p53-DNA complexes predicts significant dimer–dimer contacts for response elements with spacer lengths of 0–2 bp (with the strongest contacts for contiguous half-sites), but significant reduction in binding energy for longer spacers because of weakened dimer–dimer contacts (Pan and Nussinov 2009). A 5-bp spacer, for example, would place the two core-domain dimers on opposite faces of the DNA-double helix, whereas a 10-bp spacer would place the dimers on the same face of the DNA but require significant bending of the DNA for interdimer contacts to occur. Moreover, different spacer lengths may impose different conformational constraints on the linker region between the core and tetramerization domains on binding as a tetramer. In this context, it is also interesting to note that a recently published structure of a mouse p53 core domain tetramer bound to a continuous response element without spacer does not implicate the L1 loop in DNA binding (Malecka et al. 2009). In fact, the L1 loop, which partly adopts a helical conformation, is displaced by about 15 Å compared with the other p53-DNA complexes and points either in the direction of the translational dimer–dimer interface or the solvent. This is particularly interesting in light of recent reports that TIP60 and hMOF-mediated acetylation of Lys120 may play a role in distinguishing between cell-cycle arrest and apoptotic functions of p53 (Sykes et al. 2006; Tang et al. 2006), potentially by modulating protein–protein interactions. Moreover, the L1 loop is only rarely mutated in cancer, suggesting that Lys120 may not be essential for p53's apoptotic functions. However, the structure of the mouse p53-DNA complex has the caveat that the core domain is cross-linked to DNA via Cys274 (equivalent to Cys277 in human p53). The short tether is not detected in the crystal structure because of its flexibility but may perturb the native major-groove interaction network sufficiently to favor alternative binding modes in which the L1 loop is displaced from the DNA-binding interface. In the Caenorhabditis elegans ortholog, CEP-1, the conformation of the L1 loop is significantly different, making it unlikely that it engages in sequence-specific contacts with the major groove as observed in human p53 (Huyen et al. 2004). On the other hand, mutational studies clearly implicate the L1 loop in DNA binding in human p53, showing that the K120A mutation alters the DNA binding properties in vitro and significantly reduces DNA binding in vivo when expressed at normal cellular protein levels (Zupnick and Prives 2006). In addition, many mutations in or next to the L1 loop have been reported that result in altered transactivation patterns (so-called “supertrans” mutants, such as T123A), potentially modulating L1-mediated DNA binding, although the exact mechanisms have not been elucidated yet (Brachmann et al. 1998; Saller et al. 1999; Inga et al. 2001; Inga and Resnick 2001; Zupnick and Prives 2006; Fen et al. 2007). Future studies will undoubtedly shed further light on the enigmatic role of the L1 loop in DNA binding and the finer details of the architecture of p53 tetramers in complex with different DNA targets.
STRUCTURAL BASIS OF TETRAMER FORMATION
The oligomerization state of p53 is regulated via its tetramerization domain (residues 325–355 in human p53). The structure of this domain has been solved by x-ray crystallography and in solution by NMR (Lee et al. 1994; Clore et al. 1995; Jeffrey et al. 1995; Miller et al. 1996; Mittl et al. 1998; Chen and Clore 2000; Mora et al. 2008). The individual subunits consist of a short β-strand followed by an α-helix. These two structural elements are connected by a sharp turn facilitated by a conserved glycine residue (Gly334). Four chains form a tetramer that can be described as a dimer of primary dimers (Fig. 3A). The topology of the tetramer is reflected by its folding pathway, which proceeds via a dimeric intermediate (Mateu et al. 1999), and by studies on p53 biogenesis in vitro, showing cotranslational formation of dimers on the polysome, followed by dimerization of dimers in solution (Nicholls et al. 2002). The primary dimers are stabilized via an antiparallel intermolecular β-sheet and helix-packing interactions (e.g., the red and blue chain in Fig. 3A). Two such dimers assemble in a roughly orthogonal fashion via their hydrophobic helix interfaces to form the tightly packed tetramer, which shows very high thermodynamic stability (Mateu and Fersht 1998; Mora et al. 2008). At the center of this interface, the side chains of Leu344 from all four subunits are in direct contact. Leu344 is part of a leucine-rich nuclear export signal (residues 340–351) (Stommel et al. 1999). This motif is masked in tetrameric p53, as it is buried within the tetramer interface and exposed only after dissociation of the tetramer, suggesting that nuclear p53 levels may be controlled via regulation of its oligomerization state. There is increasing evidence that the oligomerization equilibrium of p53 is modulated via an intricate network of accessory proteins, which can have either positive or negative regulatory effects. Direct binding of apoptosis repressor with caspase recruitment domain (ARC) to the p53 tetramerization domain in the nucleus, for example, inhibits p53 tetramerization and promotes nuclear export. (Foo et al. 2007). Moreover, in vitro binding experiments show that members of the family of calcium-dependent S100 proteins also negatively regulate the oligomerization state of p53 by preferentially binding to tetramerization domain monomers (Fernandez-Fernandez et al. 2008; van Dieck et al. 2009). In contrast, binding of dimeric 14-3-3 proteins to the p53 carboxyl terminus, which is strengthened on phosphorylation of the latter, enhances formation of tetramers in vitro (Rajagopalan et al. 2008).
Figure 3.
p53 family oligomerization domain structures. (A) Crystal structure of the human p53 tetramerization domain (PDB code 1C26) (Jeffrey et al. 1995). (B) Crystal structure of the human p73 tetramerization domain (PDB code 2WQI) (Joerger et al. 2009). (C) Solution structure of the C. elegans p53 ortholog, CEP-1, oligomerization domain dimer (PDB code 2RP5) (Ou et al. 2007). (D) Solution structure of the Drosophila p53 tetramerization domain, Dmp53, (PDB code 2RP4) (Ou et al. 2007). Individual subunits are shown in different colors. The structures of the human p53 and p73 tetramerization domains are shown in two different orientations. The second view is perpendicular to the central dimer–dimer interface, showing differences in the packing angle of the primary dimers between the two structures.
A highly conserved intermolecular salt bridge between Arg337 and Asp352 stabilizes the tetramer at both ends of the primary-dimer helix interface. Molecular dynamics studies suggest that it is part of a fluid salt-bridge cluster together with Arg333 and Glu349 (Lwin et al. 2007). Mutation of Arg337 (R337H) is associated with adrenocortical carcinoma in children from southern Brazil (DiGiammarino et al. 2002) and predisposes to a diverse spectrum of tumors (Achatz et al. 2007). It disrupts the intermonomer salt bridge, resulting in impaired tetramer formation and increased propensity of the mutated tetramerization domain to form amyloid fibrils (DiGiammarino et al. 2002; Galea et al. 2005). Approximately 20% of p53 germline mutations have a mutation at codon 337, whereas the relative frequency of somatic cancer mutations at this site is low (see release R13 of the IARC TP53 Mutation Database at www-p53.iarc.fr) (Petitjean et al. 2007). A recent study shows that the structural integrity of the R337H tetramer can be restored by a designed tetraguanidiniomethylcalix[4]arene ligand, which serves as a template for holding together the four monomers of the mutated tetramerization domain (Gordo et al. 2008).
Most somatic p53 cancer mutations are located in the DNA-binding domain (Fig. 1), and these mutant proteins have, therefore, an intact tetramerization domain. Formation of mixed tetramers of impaired activity between wild-type and mutant p53 is thought to be the molecular basis of the so-called dominant–negative effect of mutant p53 in heterozygous cells (Kern et al. 1992; Chan et al. 2004; Dong et al. 2007; Junk et al. 2008). The extent of the dominant–negative effect (or whether it is observed at all in vivo) depends on the specific mutant (Dearth et al. 2007). The DNA-contact mutant R273H, for example, forms 2:2 heterotetramers with wild-type p53 of weakened DNA-binding affinity in vitro compared with wild-type homotetramers (Natan et al. 2009). The observed subunit exchange between the R273H mutant and the wild type at body temperature is, however, relatively slow compared with the spontaneous denaturation of the proteins.
STRUCTURAL EVOLUTION OF TETRAMERIZATION WITHIN THE P53/P63/P73 FAMILY
A comparison of the tetramerization domain structure of human p53 with that of its homologs provides intriguing insights into how oligomerization has evolved within the p53/p63/p73 family (Fig. 3). Invertebrate species, such as C. elegans and Drosophila, have only one family member. The oligomerization domain of the C. elegans p53 ortholog, CEP-1, is dimeric and has an additional sterile α-motif (SAM) that is important for stabilizing the domain (Fig. 3C). This suggests a potential evolutionary pathway from functional dimers to tetramers (Ou et al. 2007). In contrast, the Drosophila homolog forms a tetramer, but also has additional structural elements, including an additional β-strand at the amino terminus and a second α-helix at the carboxyl terminus. This additional helix packs against the canonical helix from the same subunit but is also involved in dimer–dimer contacts (Fig. 3D). Most interestingly, the center of the dimer–dimer interface is predominantly electrostatic in nature, which is in stark contrast to the hydrophobic contact area in human p53 (Ou et al. 2007).
In vertebrates, three family members, p53, p63, and p73, have evolved from a common, p63/p73-like ancestral protein, as a result of two gene duplication events that occurred after the invertebrate–vertebrate transition (Yang et al. 2002; Belyi and Levine 2009). Interestingly, the oligomerization domains of the p63 and p73 paralogs have a conserved carboxy-terminal extension to the canonical p53 tetramerization motif. Recent structural studies have shown that this region is helical and essential for stabilizing the overall architecture of the tetramer (Coutandin et al. 2009; Joerger et al. 2009). In the p73 tetramer (residues 351-399), the additional helices extend from one dimer subunit to wrap around the adjacent dimer in the tetramer, making extensive intersubunit contacts (Fig. 3B). There are also specific amino-acid substitutions within the canonical tetramerization domain motif; e.g., large-to-small substitutions of key hydrophobic residues in the primary dimer (Phe341 in p53 corresponds to Leu368 in p73) and substitution of the residues that form the highly conserved salt bridge in p53 (Arg337-Asp352). These structural differences rationalize observations that the p53 tetramerization domain does not interact with p63 and p73, whereas the latter two form mixed tetramers in vitro, although the rate of subunit exchange is relatively slow once homotetramers have been formed (Davison et al. 1999; Coutandin et al. 2009; Joerger et al. 2009). Taken together, it appears that in the case of human p53, the tetramerization domain has evolved toward smaller subunits that no longer associate with the corresponding domains of p63 and p73. Consequently, divergent evolution of the tetramerization domain offered a mechanism for uncoupling the p53 pathway from that of its family members (Joerger et al. 2009).
SIGNALING DIVERSITY THROUGH INTRINSIC DISORDER
The amino-terminal transactivation domain (TAD) is essential for p53 transcriptional activity. It connects target gene recognition with target gene expression by direct binding to the transcriptional coactivators p300/CBP and components of the basal transcription machinery (Thut et al. 1995; Lill et al. 1997). The p300 domains Taz2/CH3, Taz1/CH1, Kix, and IBiD bind to the full TAD, with the Taz2/CH3 domain having the highest affinity (Teufel et al. 2007). But the TAD1 subdomain also binds strongly to the negative regulators MDM2 and MDMX that play a crucial role in controlling cellular p53 levels by promoting p53 degradation through the ubiquitin-dependent proteasome pathway (Marine et al. 2006; Toledo and Wahl 2006).
What is the structural basis for promiscuous binding to this diverse set of partner proteins with overlapping binding sites on TAD and, in some cases, antagonistic biological effects? Moreover, what are the mechanisms to provide specificity in response to a particular cellular signal? The classic sequence-structure-function paradigm no longer holds true. Having natively unfolded regions is a recurring motif in proteins at the center of regulatory networks that integrate a multitude of signals (Gsponer and Madan Babu 2009). It is estimated that about 25% of mammalian proteins are intrinsically disordered and that about 75% of proteins involved in signaling contain natively unfolded regions (Dunker et al. 2008). The p53 TAD is intrinsically disordered with two regions of nascent secondary structure (Lee et al. 2000; Rosal et al. 2004; Wells et al. 2008). These regions coincide with conserved hydrophobic residues that are essential for the transactivation function of p53 (Zhu et al. 1998; Venot et al. 1999). TAD1 residues 18-25 become fully helical on binding to MDM2 (Kussie et al. 1996) and MDMX (Popowicz et al. 2008) or the Taz2 domain of the transcriptional coactivator p300 (Feng et al. 2009) (Fig. 4A,B). Regions within TAD2 also fold into amphipathic α-helices on binding to replication protein A (Bochkareva et al. 2005) and the Tfb1 subunit of yeast TFIIH (Di Lello et al. 2006). Intrinsic disorder is a characteristic of transactivation domains of transcription factors (Liu et al. 2006). It is often associated with the presence of so-called molecular recognition features, short sequence motifs that undergo disorder-to-order transition on binding to partner proteins (Mohan et al. 2006; Vacic et al. 2007), thus allowing promiscuous binding of diverse targets at the same site by providing conformational variability and adaptability (Gsponer and Madan Babu 2009).
Figure 4.
Molecular recognition features in p53 that undergo disorder-to-order transition on binding to partner proteins. (A) Overlay of the structure of p53 TAD residues 15–29 in complex with MDM2 (PDB code 1YCR) and MDMX (PDB code 3DAB), showing the three hydrophobic key residues, Phe19, Trp23, and Leu26, that bind to the hydrophobic binding pocket in MDM2 and MDMX. (B) Solution structure of p53 TAD in complex with the Taz2 domain of p300 (PDB code 2K8F). (C) Crystal structure of a peptide derived from the p53 carboxy-terminal regulatory domain in its Lys382-acetylated form bound to the deacetylase sir2 (PDB code 1MA3). (D) Solution structure of a carboxy-terminal p53 peptide bound to the Ca-dependent S100B dimer (PDB code 1DT7).
The interaction properties of p53 TAD are modulated by posttranslational modifications. It has nine phosphorylation sites, seven of which lie within the binding region of regulatory proteins (Ser15, Thr18, Ser20, Ser33, Ser37, Ser46, and Thr55). Phosphorylation of Thr18 significantly reduces the affinity of TAD for MDM2 because of electrostatic repulsion between the phosphate group and a negatively charged patch on the MDM2 surface (Sakaguchi et al. 2000; Schon et al. 2002; Brown et al. 2008). In contrast, phosphorylation at Thr18 and other sites within TAD can significantly enhance the affinity of TAD for p300 and its various subdomains (Ferreon et al. 2009; Jenkins et al. 2009; Lee et al. 2009; Teufel et al. 2009). Multiple phosphorylation of TAD can change its relative affinity for MDM2 and Taz2 in vitro by up to three orders of magnitude in favor of the latter, highlighting how phosphorylation cascades can provide a cellular mechanism to up-regulate p53-dependent transcription and induction of cell-cycle arrest and apoptotic genes (Teufel et al. 2009). As such, changes in the affinity of p53 for competing binding partners in the cell cycle by phosphorylation of TAD allows modulation of p53 activity in response to genotoxic stress.
CHAMELEON BEHAVIOR OF THE EXTREME CARBOXYL TERMINUS
The carboxy-terminal regulatory domain (CTD), which follows the tetramerization domain, is also natively unfolded (Bell et al. 2002). Various posttranslational modifications occur at the six carboxy-terminal lysines (370, 372, 373, 381, 382, and 386), comprising acetylation, methylation, ubiquitination, sumoylation, and neddylation, as well as phosphorylation of serine and threonine residues (e.g., Ser366, Ser378, Thr387, and Ser392). These modifications play diverse and complex roles in the modulation of p53 function and regulation of cellular protein levels (Toledo and Wahl 2006; Kruse and Gu 2009; Vousden and Prives 2009). As in the case of TAD, intrinsic disorder provides binding promiscuity, allowing the carboxy-terminal region to interact with numerous partner proteins and facilitating posttranslational modifications. Similarly to the TAD region, disorder-to-order transition occurs on binding to these partner proteins, but contrary to TAD, the segment undergoing this transition in CTD has the characteristics of a chameleon sequence. Such sequences can adopt a different secondary structure, depending on the structural context (Andreeva et al. 2007). Residues 376–387 form an α-helix when bound to S100B (Rustandi et al. 2000), whereas in complex with sir2, a member of the family of sirtuin deacetylases, parts of the same segment form a β-sheet with flanking strands of the enzyme (Avalos et al. 2002) (Fig. 4C,D). A similar switch of secondary structure between α-helical and β-strand conformation in different structural contexts is observed for an eight-amino-acid chameleon sequence of the homeodomain repressor protein MATα2 (Tan and Richmond 1998). Alternative conformations of the extreme carboxyl terminus of p53 without regular secondary structure or with a β-turn-like conformation are observed in complex with CDK2/cyclin A (Lowe et al. 2002) and the bromodomain of the transcriptional coactivator CBP (Mujtaba et al. 2004), respectively, further exemplifying the conformational adaptability of this region.
THE BIGGER PICTURE: STRUCTURE OF THE FULL-LENGTH PROTEIN
p53 interacts with a very large number of partner proteins. It is a hub protein in a “scale-free” network. As discussed above, most of its interactions are mediated via the disordered amino-terminal transactivation domain, a large number with the disordered C-terminal domain, and a few partners bind to the core domain. The affinity to many of its partners is regulated by phosphorylation of serine and threonine residues or acetylation of lysine residues. Binding partner proteins, such as the negative regulators MDM2, MDMX, and Pirh2, and the transcriptional coactivator p300, are also multidomain proteins that contain structurally disordered regions, and they make multipoint interactions with p53 (Goodman and Smolik 2000; Marine and Jochemsen 2005; Teufel et al. 2007; Sheng et al. 2008). The big challenge is naturally to determine the structures of the complexes of full-length p53 with multidomain partner proteins or individual domains thereof. Not only is this structural biology vital for understanding the role of p53 in the cell cycle and in other activities, but the structures may also help in the design of drugs that affect the cell cycle.
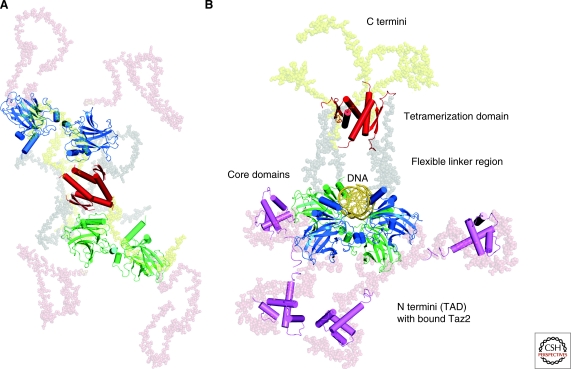
But, there are major obstacles: Full-length p53 is too flexible for crystallographic studies, too big for conventional NMR, but also small enough to be at the limits of cryo-electron microscopy. The solution to this problem is to apply a multitechnique approach, assembling individual jigsaw pieces into a bigger picture, a strategy that may be applicable to an increasing number of multidomain proteins in the cell cycle with intrinsically disordered domains. The arrangement of folded domains within p53 and within its complex with response element DNA was elucidated by using state-of-the-art NMR spectroscopy to detect domain–domain interactions in solution, and small-angle x-ray scattering (SAXS) experiments and computation to reconstruct the structure of p53 and its complexes in solution (Tidow et al. 2007; Wells et al. 2008). Pivotal to these experiments was the design of an engineered stabilized variant of p53 containing four mutations in the core domain (M133L/V203A/N239Y/N268D) that significantly increased the overall stability of the protein (Nikolova et al. 1998; Joerger et al. 2004), making it more amenable to structural studies.
According to these models, full-length p53 in its free state in solution has an open, cross-shaped structure, with the tetramerization domain at its center and a pair of loosely coupled core domain dimers at the ends (Fig. 5A). In this highly dynamic and open conformation, the core domains are accessible for binding to cognate DNA and partner proteins in the cell cycle. On DNA binding, the structure closes around the DNA double helix and becomes more compact (Fig. 5B), with consistent models obtained from SAXS and electron microscopy (Tidow et al. 2007). This arrangement is also in agreement with the crystal structures of the tetrameric core domain-DNA complexes by Kitayner et al. (2006). Domain rearrangements to allow DNA binding are facilitated by the flexible linker region between the core and tetramerization domain. The proline-rich region, linking TAD with the core domain, has the propensity to adopt a stiff polyproline-II conformation (Wells et al. 2008) and may have a predominantly structural role, consistent with mutagenesis studies in mouse models (Toledo et al. 2007). It projects the TAD domains away from the central p53-DNA complex. As a result, the TAD domains are free to interact with coactivators, as shown by the structure of the p53 complex with the Taz2 domain of p300 (Wells et al. 2008) (Fig. 5B). Given the overall structural arrangement of the p53 tetramer and the intrinsic flexibility of the TAD region, such interactions may involve cooperative binding of different subdomains from one partner protein, as proposed for p300 (Teufel et al. 2007), or simultaneous binding of different binding partners. A cryo-electron microscopy study of murine p53 proposes a radically different structural model of the unbound full-length protein (Okorokov et al. 2006). According to this model, the free p53 tetramer adopts a closed structure, reminiscent of a hollow skewed cube. One of the key, and most puzzling, features of this model is the disruption of the tetramerization domain tetramer and formation of α-helix bundles involving parts of the tetramerization domain and the transactivation domain instead, so that the latter is no longer free to interact with partner proteins. It would be interesting to see whether this structure is confirmed by SAXS studies in solution.
Figure 5.
SAXS models of full-length p53. (A) Model of full-length p53 in its unbound state (Tidow et al. 2007). (B) Ternary complex of full-length p53 with cognate DNA and the Taz2 domain (magenta) of p300 (Wells et al. 2008). The p53 DNA-binding domains are shown in blue and green, the tetramerization domain in red.
EXPLOITING STRUCTURAL INFORMATION FOR CANCER THERAPY
Loss of p53 tumor suppressor function is a common feature of human cancers. p53 is inactivated directly by mutation in ∼50% of human cancers and its apoptotic pathways are impaired in the remainder. Pharmacological restoration of p53 function should, therefore, result in tumor regression, as supported by two recent studies on transgenic mice (Ventura et al. 2007; Xue et al. 2007). Although various p53-activating compounds have been reported from screening of chemical libraries, the actual mechanism of action of many of these compounds is, frustratingly, largely unresolved (Issaeva et al. 2004; Krajewski et al. 2005; Romer et al. 2006; Vazquez et al. 2008). A good example is the compound PRIMA-1MET (Bykov et al. 2002; Zache et al. 2008), which has proven antitumor activity and is currently in phase I of clinical trials, yet its mode of action is still unclear. Only recently was it discovered that a hydrolytic degradation product covalently modifies cysteines in the p53 core domain, which may be related to its antitumor function, potentially by modulating the redox state of p53 (Lambert et al. 2009). Structure-guided drug discovery to restore p53 function in tumors has mainly focused on two avenues in recent years: (1) targeting the p53 pathway, in particular negative regulators of p53, and (2) targeting destabilized oncogenic p53 mutants.
TARGETING THE P53 PATHWAY: MDM2, MDMX, SIRTUINS, AND BEYOND
In many cancers with wild-type p53, the negative regulators MDM2 and MDMX are up-regulated, resulting in suppression of p53 function (Marine et al. 2007; Wade and Wahl 2009). Disruption of p53-MDM2/MDMX interaction by small molecules should result in restoration of p53 function in these cancers, making them prime targets for therapeutic intervention. The small-molecule MDM2-antagonists Nutlins activate the p53 pathway and induce apoptosis in cancer cells (Vassilev et al. 2004). These cis-imidazoline analogs target the p53-binding pocket of MDM2 with high affinity by mimicking contacts made by the hydrophobic p53 triad, Phe19, Trp23, and Leu26 (Fig. 6A). Other potent MDM2 antagonists with similar binding modes and desired in vivo activities include spiro-oxindole (Ding et al. 2006; Shangary et al. 2008a; Shangary et al. 2008b) and benzodiazepinedione analogs (Grasberger et al. 2005; Koblish et al. 2006) (Fig. 6B). Because of structural differences between MDM2 and MDMX that alter the shape of the p53-binding pocket, in particular the Leu26 subpocket in the α2'-helix region (Fig. 4A), inhibitors that were specifically designed or screened for MDM2 binding are generally much weaker inhibitors of p53-MDMX interaction (Popowicz et al. 2008). More recent studies have elucidated the binding modes of p53-derived peptides (Czarna et al. 2009; Pazgier et al. 2009) and a chlorinated peptidomimetic (Kallen et al. 2009) that bind to both MDM2 and MDMX with low nanomolar affinities, two to three orders of magnitude tighter than the native p53 peptide. This wealth of novel structural information, which also offers insights into the conformational plasticity within the binding pocket of MDMX, is an excellent starting point for the development of cross-selective MDM2/MDMX inhibitors for use in cancer therapy. Alternative strategies also aim to inhibit the E3-ligase activity of MDM2 directly (Yang et al. 2005). Additionally, the recently solved crystal structure of the MDM2/MDMX RING domain heterodimer suggests that it may be possible to specifically block MDM2/MDMX-mediated p53 degradation by targeting the RING domain interface regions (Linke et al. 2008).
Figure 6.
Structure-guided drug discovery to restore p53 function in tumors. (A) and (B) Binding modes of small-molecule antagonists of MDM2 that inhibit p53-MDM2 interactions. The structure of the imidazoline inhibitor Nutlin-2 (A) (PDB code 1RV1; pink) and a benzodiazepinedione compound (B) (PDB code 1T4E; gray) in complex with MDM2 is superimposed onto that of p53 residues 18–27 bound to MDM2 (PDB code 1YCR; green). The side chains of the hydrophobic p53 triad, FWL, are shown as stick models. The structures of MDM2 are omitted for clarity (see also Fig. 4A). (C) Targeted mutant p53 rescue. Binding mode of the stabilizing small-molecule compound PhiKan083 to a mutation-induced surface crevice in the DNA-binding domain of the p53 cancer mutant Y220C (PDB code 2VUK), which is distant from the functional interfaces of the protein (see Fig. 1).
A new class of compounds, the Tenovins, that specifically inhibits the deacetylases sirtuin-1 (SIRT1) and sirtuin-2 was identified in mammalian cell-based assays for compounds that activate p53 target genes (Lain et al. 2008). Uncontrolled high levels of p53-specific deacetylases have a negative effect on the tumor suppressor function of p53 and may contribute to cancer development. SIRT1 is negatively regulated by the DBC1 protein, which is frequently deleted in breast cancer. DBC1 directly interacts with the deacetylase domain of SIRT1 and inhibits its deacetylase activity (Kim et al. 2008; Zhao et al. 2008). Structural information on SIRT1 and the binding mode of Tenovins should greatly advance the development of potent, class-specific sirtuin inhibitors.
There is an increasing number of reported proteins that regulate p53 activity. Several new E3 ubiquitin ligases that promote degradation of p53 have been discovered, including Pirh2 (Leng et al. 2003; Sheng et al. 2008), Cop1 (Dornan et al. 2004), TOPORS (Rajendra et al. 2004), ARF-BP1 (Chen et al. 2005), CARPs (Yang and El-Deiry 2007), and TRIM24 (Allton et al. 2009; Tai and Benchimol 2009). The interaction pattern of Pirh2 with p53 is fundamentally different from that of MDM2 and MDMX. Pirh2 contains a carboxy-terminal zinc-binding motif that interacts with the tetramerization domain of p53, resulting in preferential ubiquitination of tetrameric p53 (Sheng et al. 2008). In-depth analysis of the structural and functional properties of these novel p53 regulators and their expression levels in cancerous tissues may further expand the arsenal of potential therapeutic targets.
TARGETING P53: THE QUEST FOR THE HOLY GRAIL
Rational drug design targeting p53 itself requires intricate knowledge of the structure and how it responds to mutation. There is a whole spectrum of different p53 cancer mutations, the vast majority of which are located in the DNA-binding domain (Fig. 1); reviewed in Joerger and Fersht (2007). They can be subdivided into “contact” mutations that eliminate an essential DNA contact (e.g., R273H and R248Q) or “structural” mutations that result in structural perturbations, which may include formation of internal (V143A and F270L) or surface cavities (Y220C), and structural distortion in various parts of the DNA-binding surface (G245S, R249S, and R282W). Some 30%–40% of cancer mutations function primarily via destabilizing the protein, which is the mechanistic basis of most structural mutations in the β-sandwich region. These destabilized mutants melt at temperatures close to or below body temperature (Bullock et al. 2000; Mayer et al. 2007). Further, and importantly, they spontaneously denature with half-lives of seconds to a few minutes—the more the destabilization, the shorter the half-life (Friedler et al. 2003; Butler and Loh 2005). But, at low temperature, the natively unfolded mutants are functionally active (Di Como and Prives 1998; Bullock et al. 2000; Dearth et al. 2007) and retain the structural features of the wild-type protein in their folded state (Joerger et al. 2006); they are classical temperature-sensitive mutants. In theory, any small molecule that binds to the native but not the denatured state should shift the folding equilibrium toward the native state according to the law of mass action. p53 is indeed stabilized by binding to cognate DNA (Bullock et al. 1997; Ishimaru et al. 2009) and, as a proof-of-principle, by a designed peptide, CDB3, that targets a natural binding site and could be used in a “chaperone strategy” for rescue of mutant p53 function (Friedler et al. 2002; Issaeva et al. 2003).
Structural studies on the effects of cancer-hotspot mutations have identified a highly “druggable” mutant, Y220C, which offers a noncompetitive strategy for rescue of mutant p53 function. Y220C accounts for approximately 75,000 new cases per year worldwide (Petitjean et al. 2007; Joerger and Fersht 2008). Tyr220 is distal from all the binding interfaces of the core domain (Fig. 1). Its side chain blocks the center of an incipient crevice. The mutation Y220C opens the cavity and lowers the stability of the core domain by 4 kcal/mol as a result, so that it is largely unfolded and inactive at body temperature, but folded at subphysiological temperature (Joerger et al. 2006). In-silico design on the Y220C structure has led to the discovery of a class of molecules, based on a carbazole scaffold, which bind with 100–200 µM affinity to the mutation-induced surface cleft and stabilize the mutant as a result (Boeckler et al. 2008). They raise the melting temperature in vitro, and increase the half-life of the protein. The crystal structure of the Y220C mutant bound to a parent carbazole compound, Phikan083, reveals their binding mode in detail (Fig. 6C). The flat carbazole ring system is sandwiched between hydrophobic residues on both sides of the binding pocket (Val147, Pro151, Pro222, and Pro223), and the ethyl anchor of PhiKan083 occupies the deepest part of the pocket, next to the sulfhydryl group of Cys220. On one side of the pocket, which is hydrophilic, the protonated methanamine moiety of PhiKan083 is hydrogen-bonded to the main-chain carbonyl of Asp228 (Boeckler et al. 2008). Fragment-based screening by NMR and crystallography has identified additional scaffolds and building blocks for targeting different regions of the binding pocket (Joerger and Fersht, unpubl.). Detailed analysis of the dynamic landscape of the cavity and various mutant-ligand interactions will provide a blueprint for the rational and computer-aided design of future generations of compounds.
Studies on Y220C are exceptionally important: Not only is it a worthwhile target in its own right, it is an excellent paradigm, and the best current, for testing and developing p53-stabilizing drugs in general. By targeting a site that is distant from the functional parts of the protein and that can be repaired by small molecules, we can use it to investigate how to rescue p53 in cancer cells, with minimal complicating factors from inhibitory effects of binding. Such studies could act as a paradigm for parallel development of the “Holy Grail” of a generic drug that could treat up to 2–3 million new cases per annum by stabilizing a range of unstable oncogenic mutants. Such drugs may also be adjuncts in the treatment of the large number of cancers in which the level of wild-type p53 is low because of enhancement of its degradative pathways.
WHERE DO WE GO FROM HERE?
The past years have brought significant advances in our understanding of the structure and the dynamic behavior of the p53 protein. Nevertheless, many important aspects of the p53 structure–function relationship remain to be elucidated. Recent technological advances will allow the structure of full-length p53 in complex with its partner proteins to be solved in the future, which should provide further insights into the regulation of p53 function via its intrinsically disordered amino- and carboxyl termini, and the observed cross talk between these regions. An important, and much needed, contribution will come from a systematic characterization of the numerous binding events, accurate determination of binding constants and, crucially, how these are affected by posttranslational modifications. Combining such data from in vitro studies with that of cellular protein levels and their temporal fluctuations will provide a much clearer picture of the “p53 interactome.” Additional insights into the structural basis of p53 function may also come from more in-depth structural studies of p63 and p73, which, being larger, have additional layers of complexity. Detailed structural analysis of common p53 cancer mutants and p53's interaction with its negative regulators has significantly advanced the development of novel anticancer drugs, both conceptually and practically. Design of mutant-specific p53 rescue drugs, for example, has moved from a merely theoretical concept to a realistic proposition in recent years. There is every possibility that drug discovery on p53 and its pathways will result in an effective drug for use in anticancer therapy in the future.
ACKNOWLEDGMENTS
We thank Zippora Shakked, Henning Tidow, Antonina Andreeva, Frank Boeckler, Joel Kaar, Daniel Teufel, Rainer Wilcken, and Caroline Blair for valuable comments on the manuscript.1
Footnotes
All structural figures were prepared using PyMOL (www.pymol.org).
Editors: Arnold J. Levine and David Lane
Additional Perspectives on The p53 Family available at www.cshperspectives.org
REFERENCES
- Achatz MI, Olivier M, Le Calvez F, Martel-Planche G, Lopes A, Rossi BM, Ashton-Prolla P, Giugliani R, Palmero EI, Vargas FR, et al. 2007. The TP53 mutation, R337H, is associated with Li-Fraumeni and Li-Fraumeni-like syndromes in Brazilian families. Cancer Lett 245:96–102 [DOI] [PubMed] [Google Scholar]
- Allton K, Jain AK, Herz HM, Tsai WW, Jung SY, Qin J, Bergmann A, Johnson RL, Barton MC 2009. Trim24 targets endogenous p53 for degradation. Proc Natl Acad Sci 106:11612–11616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A, Prlic A, Hubbard TJ, Murzin AG 2007. SISYPHUS–structural alignments for proteins with non-trivial relationships. Nucleic Acids Res 35:D253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang HC, Joerger AC, Mayer S, Fersht AR 2006. Effects of common cancer mutations on stability and DNA binding of full-length p53 compared with isolated core domains. J Biol Chem 281:21934–21941 [DOI] [PubMed] [Google Scholar]
- Avalos JL, Celic I, Muhammad S, Cosgrove MS, Boeke JD, Wolberger C 2002. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol Cell 10:523–535 [DOI] [PubMed] [Google Scholar]
- Bell S, Klein C, Muller L, Hansen S, Buchner J 2002. p53 contains large unstructured regions in its native state. J Mol Biol 322:917–927 [DOI] [PubMed] [Google Scholar]
- Belyi VA, Levine AJ 2009. One billion years of p53/p63/p73 evolution. Proc Natl Acad Sci 106:17609–17610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH, et al. 2005. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci 102:15412–15417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckler FM, Joerger AC, Jaggi G, Rutherford TJ, Veprintsev DB, Fersht AR 2008. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc Natl Acad Sci 105:10360–10365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann RK, Yu K, Eby Y, Pavletich NP, Boeke JD 1998. Genetic selection of intragenic suppressor mutations that reverse the effect of common p53 cancer mutations. EMBO J 17:1847–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Srinivasan D, Jun LH, Coomber D, Verma CS, Lane DP 2008. The electrostatic surface of MDM2 modulates the specificity of its interaction with phosphorylated and unphosphorylated p53 peptides. Cell Cycle 7:608–610 [DOI] [PubMed] [Google Scholar]
- Bullock AN, Henckel J, DeDecker BS, Johnson CM, Nikolova PV, Proctor MR, Lane DP, Fersht AR 1997. Thermodynamic stability of wild-type and mutant p53 core domain. Proc Natl Acad Sci 94:14338–14342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock AN, Henckel J, Fersht AR 2000. Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: Definition of mutant states for rescue in cancer therapy. Oncogene 19:1245–1256 [DOI] [PubMed] [Google Scholar]
- Butler JS, Loh SN 2005. Kinetic partitioning during folding of the p53 DNA binding domain. J Mol Biol 350:906–918 [DOI] [PubMed] [Google Scholar]
- Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, Bergman J, Wiman KG, Selivanova G 2002. Restoration of the tumor suppressor function to mutant p53 by a low- molecular-weight compound. Nat Med 8:282–288 [DOI] [PubMed] [Google Scholar]
- Canadillas JM, Tidow H, Freund SM, Rutherford TJ, Ang HC, Fersht AR 2006. Solution structure of p53 core domain: Structural basis for its instability. Proc Natl Acad Sci 103:2109–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WM, Siu WY, Lau A, Poon RY 2004. How many mutant p53 molecules are needed to inactivate a tetramer? Mol Cell Biol 24:3536–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Kon N, Li M, Zhang W, Qin J, Gu W 2005. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 121:1071–1083 [DOI] [PubMed] [Google Scholar]
- Chen YW, Clore GM 2000. A systematic case study on using NMR models for molecular replacement: p53 tetramerization domain revisited. Acta Crystallogr D 56:1535–1540 [DOI] [PubMed] [Google Scholar]
- Cho Y, Gorina S, Jeffrey PD, Pavletich NP 1994. Crystal structure of a p53 tumor suppressor-DNA complex: Understanding tumorigenic mutations. Science 265:346–355 [DOI] [PubMed] [Google Scholar]
- Clore GM, Ernst J, Clubb R, Omichinski JG, Kennedy WM, Sakaguchi K, Appella E, Gronenborn AM 1995. Refined solution structure of the oligomerization domain of the tumour suppressor p53. Nat Struct Biol 2:321–333 [DOI] [PubMed] [Google Scholar]
- Coutandin D, Lohr F, Niesen FH, Ikeya T, Weber TA, Schafer B, Zielonka EM, Bullock AN, Yang A, Guntert P, et al. 2009. Conformational stability and activity of p73 require a second helix in the tetramerization domain. Cell Death Differ 16:1582–1589 [DOI] [PubMed] [Google Scholar]
- Czarna A, Popowicz GM, Pecak A, Wolf S, Dubin G, Holak TA 2009. High affinity interaction of the p53 peptide-analogue with human Mdm2 and Mdmx. Cell Cycle 8:1176–1184 [DOI] [PubMed] [Google Scholar]
- Davison TS, Vagner C, Kaghad M, Ayed A, Caput D, Arrowsmith CH 1999. p73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53. J Biol Chem 274:18709–18714 [DOI] [PubMed] [Google Scholar]
- Dearth LR, Qian H, Wang T, Baroni TE, Zeng J, Chen SW, Yi SY, Brachmann RK 2007. Inactive full-length p53 mutants lacking dominant wild-type p53 inhibition highlight loss of heterozygosity as an important aspect of p53 status in human cancers. Carcinogenesis 28:289–298 [DOI] [PubMed] [Google Scholar]
- Derbyshire DJ, Basu BP, Serpell LC, Joo WS, Date T, Iwabuchi K, Doherty AJ 2002. Crystal structure of human 53BP1 BRCT domains bound to p53 tumour suppressor. EMBO J 21:3863–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como CJ, Prives C 1998. Human tumor-derived p53 proteins exhibit binding site selectivity and temperature sensitivity for transactivation in a yeast-based assay. Oncogene 16:2527–2539 [DOI] [PubMed] [Google Scholar]
- Di Lello P, Jenkins LM, Jones TN, Nguyen BD, Hara T, Yamaguchi H, Dikeakos JD, Appella E, Legault P, Omichinski JG 2006. Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol Cell 22:731–740 [DOI] [PubMed] [Google Scholar]
- DiGiammarino EL, Lee AS, Cadwell C, Zhang W, Bothner B, Ribeiro RC, Zambetti G, Kriwacki RW 2002. A novel mechanism of tumorigenesis involving pH-dependent destabilization of a mutant p53 tetramer. Nat Struct Biol 9:12–16 [DOI] [PubMed] [Google Scholar]
- Ding K, Lu Y, Nikolovska-Coleska Z, Wang G, Qiu S, Shangary S, Gao W, Qin D, Stuckey J, Krajewski K, et al. 2006. Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2-p53 interaction. J Med Chem 49:3432–3435 [DOI] [PubMed] [Google Scholar]
- Dong P, Tada M, Hamada J, Nakamura A, Moriuchi T, Sakuragi N 2007. p53 dominant-negative mutant R273H promotes invasion and migration of human endometrial cancer HHUA cells. Clin Exp Metastasis 24:471–483 [DOI] [PubMed] [Google Scholar]
- Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O'Rourke K, Koeppen H, Dixit VM 2004. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429:86–92 [DOI] [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL 2008. Function and structure of inherently disordered proteins. Curr Opin Struct Biol 18:756–764 [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B 1992. Definition of a consensus binding site for p53. Nat Genet 1:45–49 [DOI] [PubMed] [Google Scholar]
- Fen CX, Coomber DW, Lane DP, Ghadessy FJ 2007. Directed evolution of p53 variants with altered DNA-binding specificities by in vitro compartmentalization. J Mol Biol 371:1238–1248 [DOI] [PubMed] [Google Scholar]
- Feng H, Jenkins LM, Durell SR, Hayashi R, Mazur SJ, Cherry S, Tropea JE, Miller M, Wlodawer A, Appella E, et al. 2009. Structural basis for p300 Taz2-p53 TAD1 binding and modulation by phosphorylation. Structure 17:202–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Fernandez MR, Rutherford TJ, Fersht AR 2008. Members of the S100 family bind p53 in two distinct ways. Protein Sci 17:1663–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreon JC, Lee CW, Arai M, Martinez-Yamout MA, Dyson HJ, Wright PE 2009. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc Natl Acad Sci 106:6591–6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo RS, Nam YJ, Ostreicher MJ, Metzl MD, Whelan RS, Peng CF, Ashton AW, Fu W, Mani K, Chin SF, et al. 2007. Regulation of p53 tetramerization and nuclear export by ARC. Proc Natl Acad Sci 104:20826–20831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedler A, Hansson LO, Veprintsev DB, Freund SM, Rippin TM, Nikolova PV, Proctor MR, Rudiger S, Fersht AR 2002. A peptide that binds and stabilizes p53 core domain: Chaperone strategy for rescue of oncogenic mutants. Proc Natl Acad Sci 99:937–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedler A, Veprintsev DB, Hansson LO, Fersht AR 2003. Kinetic instability of p53 core domain mutants: Implications for rescue by small molecules. J Biol Chem 278:24108–24112 [DOI] [PubMed] [Google Scholar]
- Funk WD, Pak DT, Karas RH, Wright WE, Shay JW 1992. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol 12:2866–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galea C, Bowman P, Kriwacki RW 2005. Disruption of an intermonomer salt bridge in the p53 tetramerization domain results in an increased propensity to form amyloid fibrils. Protein Sci 14:2993–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RH, Smolik S 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev 14:1553–1577 [PubMed] [Google Scholar]
- Gordo S, Martos V, Santos E, Menendez M, Bo C, Giralt E, de Mendoza J 2008. Stability and structural recovery of the tetramerization domain of p53-R337H mutant induced by a designed templating ligand. Proc Natl Acad Sci 105:16426–16431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorina S, Pavletich NP 1996. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science 274:1001–1005 [DOI] [PubMed] [Google Scholar]
- Gouas D, Shi H, Hainaut P 2009. The aflatoxin-induced TP53 mutation at codon 249 (R249S): Biomarker of exposure, early detection and target for therapy. Cancer Lett 286:29–37 [DOI] [PubMed] [Google Scholar]
- Grasberger BL, Lu T, Schubert C, Parks DJ, Carver TE, Koblish HK, Cummings MD, LaFrance LV, Milkiewicz KL, Calvo RR, et al. 2005. Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells. J Med Chem 48:909–912 [DOI] [PubMed] [Google Scholar]
- Gsponer J, Madan Babu M 2009. The rules of disorder or why disorder rules. Prog Biophys Mol Biol 99:94–103 [DOI] [PubMed] [Google Scholar]
- Ho WC, Fitzgerald MX, Marmorstein R 2006. Structure of the p53 core domain dimer bound to DNA. J Biol Chem 281:20494–20502 [DOI] [PubMed] [Google Scholar]
- Huyen Y, Jeffrey PD, Derry WB, Rothman JH, Pavletich NP, Stavridi ES, Halazonetis TD 2004. Structural differences in the DNA binding domains of human p53 and its C. elegans ortholog Cep-1. Structure 12:1237–1243 [DOI] [PubMed] [Google Scholar]
- Inga A, Monti P, Fronza G, Darden T, Resnick MA 2001. p53 mutants exhibiting enhanced transcriptional activation and altered promoter selectivity are revealed using a sensitive, yeast-based functional assay. Oncogene 20:501–513 [DOI] [PubMed] [Google Scholar]
- Inga A, Resnick MA 2001. Novel human p53 mutations that are toxic to yeast can enhance transactivation of specific promoters and reactivate tumor p53 mutants. Oncogene 20:3409–3419 [DOI] [PubMed] [Google Scholar]
- Ishimaru D, Ano Bom AP, Lima LM, Quesado PA, Oyama MF, de Moura Gallo CV, Cordeiro Y, Silva JL 2009. Cognate DNA Stabilizes the Tumor Suppressor p53 and Prevents Misfolding and Aggregation. Biochemistry 48:6126–6135 [DOI] [PubMed] [Google Scholar]
- Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, Selivanova G 2004. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med 10:1321–1328 [DOI] [PubMed] [Google Scholar]
- Issaeva N, Friedler A, Bozko P, Wiman KG, Fersht AR, Selivanova G 2003. Rescue of mutants of the tumor suppressor p53 in cancer cells by a designed peptide. Proc Natl Acad Sci 100:13303–13307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey PD, Gorina S, Pavletich NP 1995. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science 267:1498–1502 [DOI] [PubMed] [Google Scholar]
- Jenkins LM, Yamaguchi H, Hayashi R, Cherry S, Tropea JE, Miller M, Wlodawer A, Appella E, Mazur SJ 2009. Two distinct motifs within the p53 transactivation domain bind to the Taz2 domain of p300 and are differentially affected by phosphorylation. Biochemistry 48:1244–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger AC, Allen MD, Fersht AR 2004. Crystal structure of a superstable mutant of human p53 core domain. Insights into the mechanism of rescuing oncogenic mutations. J Biol Chem 279:1291–1296 [DOI] [PubMed] [Google Scholar]
- Joerger AC, Ang HC, Fersht AR 2006. Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc Natl Acad Sci 103:15056–15061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger AC, Ang HC, Veprintsev DB, Blair CM, Fersht AR 2005. Structures of p53 cancer mutants and mechanism of rescue by second-site suppressor mutations. J Biol Chem 280:16030–16037 [DOI] [PubMed] [Google Scholar]
- Joerger AC, Fersht AR 2007. Structure-function-rescue: The diverse nature of common p53 cancer mutants. Oncogene 26:2226–2242 [DOI] [PubMed] [Google Scholar]
- Joerger AC, Fersht AR 2008. Structural biology of the tumor suppressor p53. Annu Rev Biochem 77:557–582 [DOI] [PubMed] [Google Scholar]
- Joerger AC, Rajagopalan S, Natan E, Veprintsev DB, Robinson CV, Fersht AR 2009. Structural evolution of p53, p63, and p73: Implication for heterotetramer formation. Proc Natl Acad Sci 106:17705–17710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo WS, Jeffrey PD, Cantor SB, Finnin MS, Livingston DM, Pavletich NP 2002. Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure. Genes Dev 16:583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan JJ, Menendez D, Inga A, Noureddine M, Bell DA, Resnick MA 2008. Noncanonical DNA motifs as transactivation targets by wild type and mutant p53. PLoS Genet 4:e1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junk DJ, Vrba L, Watts GS, Oshiro MM, Martinez JD, Futscher BW 2008. Different mutant/wild-type p53 combinations cause a spectrum of increased invasive potential in nonmalignant immortalized human mammary epithelial cells. Neoplasia 10:450–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen J, Goepfert A, Blechschmidt A, Izaac A, Geiser M, Tavares G, Ramage P, Furet P, Masuya K, Lisztwan J 2009. Crystal Structures of Human MdmX (HdmX) in Complex with p53 Peptide Analogues Reveal Surprising Conformational Changes. J Biol Chem 284:8812–8821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern SE, Pietenpol JA, Thiagalingam S, Seymour A, Kinzler KW, Vogelstein B 1992. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science 256:827–830 [DOI] [PubMed] [Google Scholar]
- Khoo KH, Andreeva A, Fersht AR 2009a. Adaptive evolution of p53 thermodynamic stability. J Mol Biol 393:161–175 [DOI] [PubMed] [Google Scholar]
- Khoo KH, Joerger AC, Freund SM, Fersht AR 2009b. Stabilising the DNA-binding domain of p53 by rational design of its hydrophobic core. Protein Eng Des Sel 22:421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Chen J, Lou Z 2008. DBC1 is a negative regulator of SIRT1. Nature 451:583–586 [DOI] [PubMed] [Google Scholar]
- Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, Shakked Z 2006. Structural Basis of DNA Recognition by p53 Tetramers. Mol Cell 22:741–753 [DOI] [PubMed] [Google Scholar]
- Koblish HK, Zhao S, Franks CF, Donatelli RR, Tominovich RM, LaFrance LV, Leonard KA, Gushue JM, Parks DJ, Calvo RR, et al. 2006. Benzodiazepinedione inhibitors of the Hdm2:p53 complex suppress human tumor cell proliferation in vitro and sensitize tumors to doxorubicin in vivo. Mol Cancer Ther 5:160–169 [DOI] [PubMed] [Google Scholar]
- Krajewski M, Ozdowy P, D'Silva L, Rothweiler U, Holak TA 2005. NMR indicates that the small molecule RITA does not block p53-MDM2 binding in vitro. Nat Med 11:1135–1136; author reply 1136–1137 [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W 2009. Modes of p53 regulation. Cell 137:609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP 1996. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274:948–953 [DOI] [PubMed] [Google Scholar]
- Lain S, Hollick JJ, Campbell J, Staples OD, Higgins M, Aoubala M, McCarthy A, Appleyard V, Murray KE, Baker L, et al. 2008. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell 13:454–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JM, Gorzov P, Veprintsev DB, Soderqvist M, Segerback D, Bergman J, Fersht AR, Hainaut P, Wiman KG, Bykov VJ 2009. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell 15:376–388 [DOI] [PubMed] [Google Scholar]
- Lee CW, Arai M, Martinez-Yamout MA, Dyson HJ, Wright PE 2009. Mapping the Interactions of the p53 Transactivation Domain with the KIX Domain of CBP (dagger). Biochemistry 48:2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Mok KH, Muhandiram R, Park KH, Suk JE, Kim DH, Chang J, Sung YC, Choi KY, Han KH 2000. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J Biol Chem 275:29426–29432 [DOI] [PubMed] [Google Scholar]
- Lee W, Harvey TS, Yin Y, Yau P, Litchfield D, Arrowsmith CH 1994. Solution structure of the tetrameric minimum transforming domain of p53. Nat Struct Biol 1:877–890 [DOI] [PubMed] [Google Scholar]
- Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S 2003. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112:779–791 [DOI] [PubMed] [Google Scholar]
- Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM 1997. Binding and modulation of p53 by p300/CBP coactivators. Nature 387:823–827 [DOI] [PubMed] [Google Scholar]
- Lilyestrom W, Klein MG, Zhang R, Joachimiak A, Chen XS 2006. Crystal structure of SV40 large T-antigen bound to p53: interplay between a viral oncoprotein and a cellular tumor suppressor. Genes Dev 20:2373–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL 2008. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ 15:841–848 [DOI] [PubMed] [Google Scholar]
- Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK 2006. Intrinsic disorder in transcription factors. Biochemistry 45:6873–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe ED, Tews I, Cheng KY, Brown NR, Gul S, Noble ME, Gamblin SJ, Johnson LN 2002. Specificity determinants of recruitment peptides bound to phospho-CDK2/cyclin A. Biochemistry 41:15625–15634 [DOI] [PubMed] [Google Scholar]
- Lwin TZ, Durant JJ, Bashford D 2007. A fluid salt-bridging cluster and the stabilization of p53. J Mol Biol 373:1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecka KA, Ho WC, Marmorstein R 2009. Crystal structure of a p53 core tetramer bound to DNA. Oncogene 28:325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Dyer MA, Jochemsen AG 2007. MDMX: From bench to bedside. J Cell Sci 120:371–378 [DOI] [PubMed] [Google Scholar]
- Marine JC, Francoz S, Maetens M, Wahl G, Toledo F, Lozano G 2006. Keeping p53 in check: Essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ 13:927–934 [DOI] [PubMed] [Google Scholar]
- Marine JC, Jochemsen AG 2005. Mdmx as an essential regulator of p53 activity. Biochem Biophys Res Commun 331:750–760 [DOI] [PubMed] [Google Scholar]
- Mateu MG, Fersht AR 1998. Nine hydrophobic side chains are key determinants of the thermodynamic stability and oligomerization status of tumour suppressor p53 tetramerization domain. EMBO J 17:2748–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateu MG, Sanchez Del Pino MM, Fersht AR 1999. Mechanism of folding and assembly of a small tetrameric protein domain from tumor suppressor p53. Nat Struct Biol 6:191–198 [DOI] [PubMed] [Google Scholar]
- Mayer S, Rudiger S, Ang HC, Joerger AC, Fersht AR 2007. Correlation of levels of folded recombinant p53 in Escherichia coli with thermodynamic stability in vitro. J Mol Biol 372:268–276 [DOI] [PubMed] [Google Scholar]
- Miller M, Lubkowski J, Rao JK, Danishefsky AT, Omichinski JG, Sakaguchi K, Sakamoto H, Appella E, Gronenborn AM, Clore GM 1996. The oligomerization domain of p53: Crystal structure of the trigonal form. FEBS Lett 399:166–170 [DOI] [PubMed] [Google Scholar]
- Mittl PR, Chene P, Grutter MG 1998. Crystallization and structure solution of p53 (residues 326-356) by molecular replacement using an NMR model as template. Acta Crystallogr D 54:86–89 [DOI] [PubMed] [Google Scholar]
- Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN 2006. Analysis of molecular recognition features (MoRFs). J Mol Biol 362:1043–1059 [DOI] [PubMed] [Google Scholar]
- Mora P, Carbajo RJ, Pineda-Lucena A, Sanchez del Pino MM, Perez-Paya E 2008. Solvent-exposed residues located in the β-sheet modulate the stability of the tetramerization domain of p53–a structural and combinatorial approach. Proteins 71:1670–1685 [DOI] [PubMed] [Google Scholar]
- Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand, Sanchez R, Zeleznik-Le NJ, Ronai Z, Zhou MM 2004. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell 13:251–263 [DOI] [PubMed] [Google Scholar]
- Natan E, Hirschberg D, Morgner N, Robinson CV, Fersht AR 2009. Ultraslow oligomerization equilibria of p53 and its implications. Proc Natl Acad Sci 106:14327–14332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls CD, McLure KG, Shields MA, Lee PW 2002. Biogenesis of p53 involves cotranslational dimerization of monomers and posttranslational dimerization of dimers. Implications on the dominant negative effect. J Biol Chem 277:12937–12945 [DOI] [PubMed] [Google Scholar]
- Nikolova PV, Henckel J, Lane DP, Fersht AR 1998. Semirational design of active tumor suppressor p53 DNA binding domain with enhanced stability. Proc Natl Acad Sci 95:14675–14680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okorokov AL, Sherman MB, Plisson C, Grinkevich V, Sigmundsson K, Selivanova G, Milner J, Orlova EV 2006. The structure of p53 tumour suppressor protein reveals the basis for its functional plasticity. EMBO J 25:5191–5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK 2008. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics 9 Suppl 1:S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou HD, Lohr F, Vogel V, Mantele W, Dotsch V 2007. Structural evolution of C-terminal domains in the p53 family. EMBO J 26:3463–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Nussinov R 2009. Cooperativity dominates the genomic organization of p53-response elements: A mechanistic view. PLoS Comput Biol 5:e1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazgier M, Liu M, Zou G, Yuan W, Li C, Li J, Monbo J, Zella D, Tarasov SG, Lu W 2009. Structural basis for high-affinity peptide inhibition of p53 interactions with MDM2 and MDMX. Proc Natl Acad Sci 106:4665–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M 2007. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat 28:622–629 [DOI] [PubMed] [Google Scholar]
- Popowicz GM, Czarna A, Holak TA 2008. Structure of the human Mdmx protein bound to the p53 tumor suppressor transactivation domain. Cell Cycle 7:2441–2443 [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Jaulent AM, Wells M, Veprintsev DB, Fersht AR 2008. 14-3-3 activation of DNA binding of p53 by enhancing its association into tetramers. Nucleic Acids Res 36:5983–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendra R, Malegaonkar D, Pungaliya P, Marshall H, Rasheed Z, Brownell J, Liu LF, Lutzker S, Saleem A, Rubin EH 2004. Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J Biol Chem 279:36440–36444 [DOI] [PubMed] [Google Scholar]
- Romer L, Klein C, Dehner A, Kessler H, Buchner J 2006. p53–a natural cancer killer: Structural insights and therapeutic concepts. Angew Chem Int Ed Engl 45:6440–6460 [DOI] [PubMed] [Google Scholar]
- Rosal R, Pincus MR, Brandt-Rauf PW, Fine RL, Michl J, Wang H 2004. NMR solution structure of a peptide from the mdm-2 binding domain of the p53 protein that is selectively cytotoxic to cancer cells. Biochemistry 43:1854–1861 [DOI] [PubMed] [Google Scholar]
- Rustandi RR, Baldisseri DM, Weber DJ 2000. Structure of the negative regulatory domain of p53 bound to S100B(ββ). Nat Struct Biol 7:570–574 [DOI] [PubMed] [Google Scholar]
- Sakaguchi K, Saito S, Higashimoto Y, Roy S, Anderson CW, Appella E 2000. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J Biol Chem 275:9278–9283 [DOI] [PubMed] [Google Scholar]
- Saller E, Tom E, Brunori M, Otter M, Estreicher A, Mack DH, Iggo R 1999. Increased apoptosis induction by 121F mutant p53. EMBO J 18:4424–4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon O, Friedler A, Bycroft M, Freund SM, Fersht AR 2002. Molecular mechanism of the interaction between MDM2 and p53. J Mol Biol 323:491–501 [DOI] [PubMed] [Google Scholar]
- Shangary S, Ding K, Qiu S, Nikolovska-Coleska Z, Bauer JA, Liu M, Wang G, Lu Y, McEachern D, Bernard D, et al. 2008a. Reactivation of p53 by a specific MDM2 antagonist (MI-43) leads to p21-mediated cell cycle arrest and selective cell death in colon cancer. Mol Cancer Ther 7:1533–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, Ding K, Wang G, Chen J, et al. 2008b. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci 105:3933–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Laister RC, Lemak A, Wu B, Tai E, Duan S, Lukin J, Sunnerhagen M, Srisailam S, Karra M, et al. 2008. Molecular basis of Pirh2-mediated p53 ubiquitylation. Nat Struct Mol Biol 15:1334–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM 1999. A leucine-rich nuclear export signal in the p53 tetramerization domain: Regulation of subcellular localization and p53 activity by NES masking. EMBO J 18:1660–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suad O, Rozenberg H, Brosh R, Diskin-Posner Y, Kessler N, Shimon LJ, Frolow F, Liran A, Rotter V, Shakked Z 2009. Structural basis of restoring sequence-specific DNA binding and transactivation to mutant p53 by suppressor mutations. J Mol Biol 385:249–265 [DOI] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB 2006. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 24:841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai E, Benchimol S 2009. TRIMming p53 for ubiquitination. Proc Natl Acad Sci 106:11431–11432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Richmond TJ 1998. Crystal structure of the yeast MATα2/MCM1/DNA ternary complex. Nature 391:660–666 [DOI] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W 2006. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 24:827–839 [DOI] [PubMed] [Google Scholar]
- Teufel DP, Bycroft M, Fersht AR 2009. Regulation by phosphorylation of the relative affinities of the N-terminal transactivation domains of p53 for p300 domains and Mdm2. Oncogene 28:2112–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teufel DP, Freund SM, Bycroft M, Fersht AR 2007. Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc Natl Acad Sci 104:7009–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thut CJ, Chen JL, Klemm R, Tjian R 1995. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science 267:100–104 [DOI] [PubMed] [Google Scholar]
- Tidow H, Melero R, Mylonas E, Freund SM, Grossmann JG, Carazo JM, Svergun DI, Valle M, Fersht AR 2007. Quaternary structures of tumor suppressor p53 and a specific p53-DNA complex. Proc Natl Acad Sci 104:12324–12329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Lee CJ, Krummel KA, Rodewald LW, Liu CW, Wahl GM 2007. Mouse mutants reveal that putative protein interaction sites in the p53 proline-rich domain are dispensable for tumor suppression. Mol Cell Biol 27:1425–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Wahl GM 2006. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat Rev Cancer 6:909–923 [DOI] [PubMed] [Google Scholar]
- Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, Dunker AK 2007. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res 6:2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dieck J, Fernandez-Fernandez MR, Veprintsev DB, Fersht AR 2009. Modulation of the oligomerization state of p53 by differential binding of proteins of the S100 family to p53 monomers and tetramers. J Biol Chem 284:13804–13811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844–848 [DOI] [PubMed] [Google Scholar]
- Vazquez A, Bond EE, Levine AJ, Bond GL 2008. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov 7:979–987 [DOI] [PubMed] [Google Scholar]
- Venot C, Maratrat M, Sierra V, Conseiller E, Debussche L 1999. Definition of a p53 transactivation function-deficient mutant and characterization of two independent p53 transactivation subdomains. Oncogene 18:2405–2410 [DOI] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T 2007. Restoration of p53 function leads to tumour regression in vivo. Nature 445:661–665 [DOI] [PubMed] [Google Scholar]
- Veprintsev DB, Fersht AR 2008. Algorithm for prediction of tumour suppressor p53 affinity for binding sites in DNA. Nucleic Acids Res 36:1589–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C 2009. Blinded by the light: The growing complexity of p53. Cell 137:413–431 [DOI] [PubMed] [Google Scholar]
- Wade M, Wahl GM 2009. Targeting Mdm2 and Mdmx in cancer therapy: Better living through medicinal chemistry? Mol Cancer Res 7:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Xiao Z, Ren EC 2009. Redefining the p53 response element. Proc Natl Acad Sci 106:14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Rosengarth A, Luecke H 2007. Structure of the human p53 core domain in the absence of DNA. Acta Crystallogr D 63:276–281 [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. 2006. A global map of p53 transcription-factor binding sites in the human genome. Cell 124:207–219 [DOI] [PubMed] [Google Scholar]
- Weinberg RL, Veprintsev DB, Fersht AR 2004. Cooperative binding of tetrameric p53 to DNA. J Mol Biol 341:1145–1159 [DOI] [PubMed] [Google Scholar]
- Wells M, Tidow H, Rutherford TJ, Markwick P, Jensen MR, Mylonas E, Svergun DI, Blackledge M, Fersht AR 2008. Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc Natl Acad Sci 105:5762–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445:656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Caput D, McKeon F 2002. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet 18:90–95 [DOI] [PubMed] [Google Scholar]
- Yang W, El-Deiry WS 2007. CARPs are E3 ligases that target apical caspases and p53. Cancer Biol Ther 6:1676–1683 [DOI] [PubMed] [Google Scholar]
- Yang Y, Ludwig RL, Jensen JP, Pierre SA, Medaglia MV, Davydov IV, Safiran YJ, Oberoi P, Kenten JH, Phillips AC, et al. 2005. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell 7:547–559 [DOI] [PubMed] [Google Scholar]
- Zache N, Lambert JM, Wiman KG, Bykov VJ 2008. PRIMA-1MET inhibits growth of mouse tumors carrying mutant p53. Cell Oncol 30:411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W 2008. Negative regulation of the deacetylase SIRT1 by DBC1. Nature 451:587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhou W, Jiang J, Chen X 1998. Identification of a novel p53 functional domain that is necessary for mediating apoptosis. J Biol Chem 273:13030–13036 [DOI] [PubMed] [Google Scholar]
- Zupnick A, Prives C 2006. Mutational analysis of the p53 core domain L1 loop. J Biol Chem 281:20464–20473 [DOI] [PubMed] [Google Scholar]