Summary
Colorectal cancer is the second leading cause of cancer related deaths in the United States, with more than 130,000 new cases of colorectal cancer diagnosed each year. Clinical studies have shown that genetic alterations lead to different responses to the same treatment, despite the morphologic similarities of tumors. A molecular test prior to treatment could help in determining an optimal treatment for a patient with regard to both toxicity and efficacy. This article introduces a statistical method appropriate for predicting and comparing multiple endpoints given different treatment options and molecular profiles of an individual. A latent variable-based multivariate regression model with structured variance covariance matrix is considered here. The latent variables account for the correlated nature of multiple endpoints and accommodate the fact that some clinical endpoints are categorical variables and others are censored variables. The mixture normal hierarchical structure admits a natural variable selection rule. Inference was conducted using the posterior distribution sampling Markov chain Monte Carlo method. We analyzed the finite-sample properties of the proposed method using simulation studies. The application to the advanced colorectal cancer study revealed associations between multiple endpoints and particular biomarkers, demonstrating the potential of individualizing treatment based on genetic profiles.
Keywords: Bayesian multivariate regression, Biomarker, Hierarchical model, Interaction, Latent variable, Oncology
1. Introduction
In most cancer clinical trials, some patients respond to chemotherapy very well while others show no sign of response. Likewise, some patients experience more toxicity than others given the same treatment. Recent clinical studies have suggested that patients who possess specific genetic alterations or mutations may respond differently to the same treatment for colorectal cancer (Milano and McLeod, 2000). However, tools for individualizing chemotherapy treatment using genetic profiles are not yet fully developed (McLeod and Murray, 1999).
The objective of a randomized phase III trial (Goldberg et al., 2004), initiated by the Mayo Clinic in 1997, was to compare the effect of combinations of chemotherapy agents in patients with advanced colorectal cancer. At that time, two chemotherapy drugs had been approved by the Food and Drug Administration for treatment of advanced colon cancer: 5-fluorouracil (5-FU) and irinotecan (CPT-11), while oxaliplatin (OXAL), a cisplatinum analogue with activity in colorectal cancer, was an investigational agent in the United States and Canada. Two experimental combinations of regimens, 5-FU+OXAL and OXAL+CPT-11, were compared to the standard regimen, 5-FU+CPT-11, in the trial. We refer to these regimens as arm F, arm G, and the control as arm A, respectively. A total of 1705 patients were included in the study, of which 513 (115 patients in arm A, 292 patients in arm F, and 106 patients in arm G) were genotyped for 23 biomarkers. These biomarkers were selected based on previous reports indicating that they were related to bioactivity of the chemotherapies by direct or indirect mechanisms. Descriptive summaries of the covariates are shown in Table 1.
Table 1. Summary of the variables in the colorectal cancer study.
| Variables | Description | Mean | |||
|---|---|---|---|---|---|
| Arm A (n = 115) |
Arm F (n = 292) |
Arm G (n = 106) |
|||
| Outcomes | Response | 1: Yes, 0: No | 33.9% | 42.8% | 33% |
| Toxicity | 1: Grade > 3 | 12.2% | 23.3% | 20.8% | |
| TTP | Median TTP | 188 | 308 | 249 | |
| status | 0: censor, 1: event | 84.3% | 68.2% | 79.2% | |
| DGV* | AGE | age | 60.835 | 59.685 | 59.179 |
| SEX | 0: Female, 1: Male | 61.7% | 59.6% | 53.8% | |
| Marker | M1 | abcb1/_12 | 75.7% | 83.2% | 79.2% |
| M2 | abcb1/_2677 | 73.9% | 68.5% | 72.6% | |
| M3 | abcb1/_3435 | 80.9% | 77.7% | 84.9% | |
| M4 | abcc1/_14008 | 94.8% | 93.2% | 93.4% | |
| M5 | abcc1/_34215 | 35.7% | 28.4% | 34.9% | |
| M6 | abcc2/_24 | 98.3% | 94.5% | 98.1% | |
| M7 | abcc2/_c1515y | 13.9% | 11.3% | 7.5% | |
| M8 | abcc2/_v417i | 39.1% | 38.7% | 39.6% | |
| M9 | cyp3a4 | 13% | 14% | 9.4% | |
| M10 | cyp3a5 | 16.5% | 18.8% | 15.1% | |
| M11 | dpyd/_5 | 13.9% | 18.8% | 8.5% | |
| M12 | dpyd/_6 | 5.2% | 9.6% | 9.4% | |
| M13 | dpyd/_9a | 42.6% | 41.1% | 41.5% | |
| M14 | ercc2/_k751q | 88.7% | 86.6% | 80.2% | |
| M15 | gstm1/_0 | 47.8% | 49% | 45.3% | |
| M16 | gstp1/_I105v | 54.8% | 57.9% | 52.8% | |
| M17 | gstp/_114 | 5.2% | 16.4% | 15.1% | |
| M18 | mthfr | 20.9% | 18.5% | 26.4% | |
| M19 | tyms/_1494del | 56.5% | 50.3% | 53.8% | |
| M20 | tyms/_tser | 73% | 68.8% | 67% | |
| M21 | ugt1a1/_28 | 57.4% | 50.3% | 49.1% | |
| M22 | xrcc1/_399 | 57.4% | 54.5% | 62.3% | |
| M23 | ABCG2Q141K | 19.1% | 24.7% | 17% | |
DGV = demographic variables.
For binary variables, percentage of 1 was calculated.
In this article, we develop a statistical model, given multiple treatment options and characteristics of an individual, which predicts and compares toxicity and efficacy simultaneously. A direct comparison of treatments will compare the probability that the predicted multiple endpoints for an individual patient or a group of patients fall in a favorable region of treatment outcomes (Δ). This process involves two tasks: (1) building a predictive model with appropriate prognostic and predictive factors and (2) for each treatment, predicting the probability of being in region Δ. To be concrete, let y be a k-dimensional vector of outcomes (y = (y1, …, yk)), x a vector of p predictors, and θ a vector of unknown parameters given model M. The goal is to find Pr(y ∈ Δ ∣ x, θ, M) through the joint distribution p(y1, …, yk ∣ x, θ, M).
Methods that involve Bayesian inference for variable selection, motivated by the seminal work of George and McCulloch (1993), have been applied in many studies (e.g., Brown, Vannucci, and Fearn, 1998; Chen and Dey, 2003; Sha, Tadesse, and Vannucci, 2006). Bayesian methods have the potential to account for small samples and selection of derived covariates, e.g., interaction terms, through a proper specification of the priors. In this article, we propose a statistical method to individualize treatment in the Bayesian framework.
Another challenge to conducting an appropriate analysis is that Δ is a complicated multidimensional space. For example, a patient with a confirmed tumor response may have a longer survival. A multivariate analysis is a reasonable choice when outcomes are not independent, as information is borrowed from the potentially correlated outcomes for variable selection. Furthermore, by estimating the posterior predictive probability of multiple outcomes in region Δ, one can provide a single score to compare treatments from multiple perspectives which may be related, e.g., time to progression, and overall survival. This is of critical importance, because an informed therapeutic decision is often based on consideration of multiple endpoints. When outcomes follow different type of distributions the coefficients of the same regressor across the different outcomes are not directly comparable. In the context of a multivariate regression model, properly scaled coefficients are needed. This can been done using latent variables; in addition, the variance–covariance matrix of such a multivariate regression model is often structured, which dictates additional care when specifying a prior. To address these issues, we propose a method called multivariate Bayesian selection of interactions (MBSI). The MBSI method features joint modeling of categorical and survival responses, a nonconjugate prior for structured variance components, selection of interactions with limited sample size, and a variable selection rule. Each issue has been addressed separately in literature; nevertheless, the particular combination has not been studied.
In Section 2, we describe the hierarchical structure of our MBSI model, and we will show how the decision rules for variable selection are derived. The performance of the MBSI is studied through simulations in Section 3. In Section 4, the method will be illustrated in an analysis of the large phase III colorectal cancer study. Finally, a short discussion is given in Section 5.
2. Method
2.1 Multivariate Regression Model with Latent Variables
We consider the problem of modeling the relationship between K multiple responses (including categorical and survival) and p predictor variables with sample size n in a Bayesian framework. In the motivating example, we are interested in K ≡ 3 outcomes. Let y1 and y2 be the binary outcomes of toxicity and tumor response, y3 be the survival outcome, xi be the p × 1 vector of predictors for the ith sample, and X = (x1, …, xn)T. We assume in this study that the columns of X have been standardized by subtracting their column means and dividing by their column standard deviations. Each response is assumed to follow its own regression model; let β(k) be a p × 1 vector that is a column of the regression coefficients and αk a scalar that is an intercept corresponding to the kth outcome, k = 1, …, 3.
We relate the regressors with the binary responses y1 and y2 through a probit link, which yields
where Φ is the normal cumulative distribution function and k = 1, 2. We introduce a n × 1 vector of latent variable zk (Albert and Chib, 1993). For each individual (i = 1, …, n),
The latent variable has a linear model form: zk = αk + Xβ(k) + εk, where . For reasons of identifiability, we set the scale parameter of the normal distribution to be 1.
With respect to the survival outcome y3, denote the censoring indicator by a n × 1 vector c, where ci = 0 if yi3 is right censored and ci = 1 otherwise (i = 1, …, n). We introduce a n × 1 vector of complete variable z3, defined as
The complete variable has a normal distribution as z3 ∼ N(α3 + Xβ(3), σ2). Note that when c = 0, the variable z3 is unobservable. Our approach is same as the data augmentation Gibbs sampling method used in Bayesian variable selection with log-normal accelerated failure time model in Sha et al. (2006). This results in explicit full conditional distributions of β ≡ [β(1), β(2), β(3)] and Z ≡ [z1, z2, z3], and leads to a more efficient Markov chain Monte Carlo (MCMC) implementation. If a proportional hazards model is preferred, we can specify a conditional hazard function h(y3 ∣ z3) = h0(y3) exp (z3), where z3 ∼ N(α3 + Xβ(3), σ2). This setting changes the relationship between y3 and z3 to a stochastic one. With h0(y3) = λ we obtain the exponential regression model; with , where φ > 0, we obtain the Weibull regression model. Consequently, the full conditional distributions of β and Z are no longer in explicit forms. Thus, the MCMC sampling efficiency decreases greatly for variable selection in a trivariate regression setting.
Having introduced the latent variables (z1, z2) for binary responses and the complete variable (z3) for survival response, we can rewrite the preceding model in a multivariate regression form. Letting α = [α1, α2, α3]T and ε = [ε1, ε2, ε3], we have
| (1) |
Note that ⊗ denotes the Kronecker product, and vec (εT) is the vector obtained by stacking the columns of εT on top of each other. Let ρ = [ρ1, ρ2, ρ3] be the vector of correlation coefficients resulting from the assumption that the three outcomes are mutually correlated.
2.2 Bayesian Variable Selection
We introduce a p × 3 matrix of binary latent variables γ with components γjk = 1 (j = 1, …, p and k = 1, 2, 3) if the jth regressor X(j) is included in the kth model and γjk = 0 otherwise. We assume that the prior distribution of β is a multivariate mixture normal that depends on γ. It takes the form vec(βT) ∣ γ, σ ∼ N3p(0, Σβ), where Σβ a block diagonal matrix with blocks Σβj =
| (2) |
Here Σβ corresponds to an a priori independence assumption for the coefficients. Note that for the survival outcome in equation (2), the variance is adjusted by a quantity σ2. Because the outcomes have different scales, the unadjusted β will be incomparable across the multiple outcomes.
In this prior setting, the variable selection problem is formulated in terms of making inferences regarding γ. A value of c determines the magnitude of the difference between the two mixture normal distributions. George and McCulloch (1993) suggested that choosing c between 10 and 100 tends to work well when implementing MCMC and that computational difficulties can be avoided whenever c ≤ 100. In addition, τ shall be small enough so that βjk is close to zero when γjk = 0.
We assume that all three intercepts are always included in the model. Hence, selection of the intercepts is not performed. We assume a simple diffuse multivariate normal prior for a independent of γ:
Note that Σα is adjusted for the survival outcome in the same manner as in equation (2).
In a Bayesian regression framework, a Wishart distribution is often used as a prior for the variance matrix Σ for convenience. However, in our case the Wishart distribution is not a proper prior due to the constraint on the variance of the binary outcomes y1 and y2. Therefore, we modeled Σ using the separation strategy of Barnard, McCulloch, and Meng (2000).
The prior distributions for σ and ρ = [ρ1, ρ2, ρ3] in equation (1) are specified separately. For σ, an inverse gamma prior is used σ ∼ IG (ν/2, νλ/2). We assume that the correlations ρ1, ρ2, ρ3 are a priori exchangeable. To ensure the positive definiteness of Σ, we need a constraint
| (3) |
Hence, a joint uniform prior for ρ1, ρ2, ρ3 is specified as
with informative marginal distribution
Given the other two elements of ρ, say ρ2 and ρ3, ρ1 has a uniform conditional distribution ρ1 ∣ ρ2, ρ3 ∼ U(L1, U1), where and . The lower and upper bounds L1 and U1 are the roots of the inequality (3). The marginal densities of ρ are symmetric and have more mass close to zero than the tails, which is a plausible assumption because the correlations are rarely very large in real applications.
2.3 Prior for γ
We first assume that the selection of regressors is a priori independent across the outcomes. Second, within each outcome, we assume that the selection of a main effect is dependent on the selection of all its interaction terms. Because interaction terms represent deviation from an additive model, we adopt the convention that a model containing an interaction term should also contain the corresponding main effects (Neter et al., 1996).
Define a p × 3 matrix of indicator variables ξ with components ξjk = 0, (j = 1, …, p, k = 1, 2, 3) if the corresponding regressor is an interaction, and ξjk = 1 if a main effect. Let Ωjk be the set of all the latent variables (γj′k, j′ ≠ j) for the interaction terms that are related to the jth main effect of kth model. Following the above assumptions, the prior for γjk is a Bernoulli distribution and takes the form
To favor parsimonious models or when n < p, the parameters (a, b) in the beta prior can be chosen to force πjk to be small.
2.4 Decision Rules for Variable and Model Selection
It is commonly perceived that the optimal predictive model is the model with highest joint posterior probability, but this is not necessarily the case as discussed in Barbieri and Berger (2004). Our simulation studies also showed that a method based on marginal posterior probabilities was better than that of the highest joint posterior probability method regarding the prediction performance (Table 2 in Section 3.2 and Web Figure 1).
Table 2. True and estimated Pr(ytest ∈ Δ) and 90% CPI coverage.
| ID | True | Highest joint posterior* |
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n = 200 | n = 30 | n = 200 | n = 30 | |||||||
| Est.(SE) | Cvg. | Est.(SE) | Cvg. | Est.(SE) | Cvg. | Est.(SE) | Cvg. | |||
| 1 | 0.056 | 0.074 (0.026) | 0.85 | 0.137 (0.096) | 0.71 | 0.055 (0.023) | 0.88 | 0.079 (0.088) | 0.80 | |
| 2 | 0.311 | 0.284 (0.063) | 0.86 | 0.229 (0.134) | 0.76 | 0.278 (0.074) | 0.82 | 0.227 (0.200) | 0.67 | |
| 3 | 0.524 | 0.499 (0.122) | 0.86 | 0.402 (0.213) | 0.73 | 0.514 (0.139) | 0.82 | 0.481 (0.302) | 0.70 | |
| 4 | 0.757 | 0.744 (0.031) | 0.89 | 0.491 (0.140) | 0.48 | 0.754 (0.028) | 0.94 | 0.606 (0.212) | 0.77 | |
| 5 | 0.933 | 0.905 (0.039) | 0.90 | 0.645 (0.170) | 0.42 | 0.919 (0.033) | 0.95 | 0.819 (0.145) | 0.81 | |
Select model with the highest joint posterior probability.
Select variables satisfying the criteria.
The major differences between choosing a model (using highest joint posterior probability) and choosing important predictors (using marginal posterior probabilities) are the following: (1) The selection domain is p for predictors in contrast to 2p for models, assuming no restriction. For a fixed number of MCMC iterations, it is desirable to search in a small parameter domain space. Determining marginal probabilities is computationally simpler than determining highest joint probabilities (Barbieri and Berger, 2004). (2) The joint prior probabilities are affected by the size of p; whereas the marginal prior probabilities are invariant. (3) The “dilution effect” (George, 1999), caused by multicollinearity, reduces the marginal posterior probabilities allocated to highly correlated variables. This may lead to ruling out potential predictors. To alleviate this problem, if a nearly perfect Pearson's coefficient of correlation (0.9 or greater) between any two variables is observed, the one with smaller sample variance is suggested to be removed when there is no scientific reason to suggest removing the other. (4) When variable selection is performed, a decision of selecting how many predictors is required. We will briefly describe a false discovery rate (FDR)-based decision rule developed by Chen et al. (2008) at the end of this section.
There are two types of errors in the variable selection problem: (1) selecting a variable that in truth is not a predictor (false discovery); and (2) not selecting a variable that in truth is a predictor (false negative). These two errors can be quantified by two complementary Bayesian losses: posterior expected FDR and posterior expected FNR . For the sake of simplicity in this section only, we introduce notation omitting subscript k for the kth outcome, because the definition is the same for all three outcomes. Let dj denote the decision of inclusion (dj = 1) or exclusion (dj = 0) of jth predictor given data, , and p the total number of regressors in consideration. The and are
| (4) |
where νj = P(γj = 1 ∣ Data) is the marginal posterior probability of unobserved truth (γj) of jth predictor. It is estimated by the proportion of times that γj = 1 through the MCMC iterations. In other words, selection of a variable is based on its frequency of occurrences in any model. This is similar to the variable importance measures in the random forest approach, where a variable is considered important if it appears in many trees. The posterior expected losses defined in equation (4) utilize the simple zero-one loss function. To minimize a two-dimensional loss, one can minimize one dimension while controlling the other.
Accounting for the relationship between main effects and interactions, the and will be calculated separately within the set of main effects (s1) and the set of interactions terms (s2). Hence, given αL for main effects and αH for interactions, we have min . The total is controlled at max(αL, αH) (see Lemma 1 in Chen et al. [2008]).
As the decision in s1 is affected by the decision of their higher-order terms in s2, we start with minimizing the followed by minimizing the . After the decisions are made for all the terms in s2, a subset (denoted as ) of the lower-order terms in s1 is forced to be included in the model due to the constraint. Hence, those terms are not involved in the decision to minimize the . Decisions will be made for the remaining terms in the complement of (denoted as ). The following algorithm illustrates the steps to reach these decisions. First, if t is a threshold such that the decision dj = I(νj ≥ t), j ∈ s2 satisfies , the optimal threshold minimizes . Hence, the decisions for the terms in s2 is , j ∈ s2. Second, within s1, we identify a subset so that its related higher-order term is selected in step 1. Set dj =1, . We force these terms to be selected. Third, for the terms in (complement of ), we find an optimal threshold such that the decision minimizes in the same fashion as in step 1.
The choice of αL and αH is different from the conventional significance level for the family-wise error rate. While a high level of (αL, αH) results in more falsely selected regressors, it would not necessarily cause a worse prediction to the outcome but may lead to a less parsimonious model. Therefore, αL and αH can be set according to one's degree of tolerance in the number of the false predictors. In the case of n < p, however, the prior dominates the posterior distribution. Hence, any variable selection method based on the marginal posterior distributions must take the prior assumptions into account. We consider the average posterior to prior probability ratio as the strength of evidence from the data. Similar to Jeffreys' interpretation of Bayes factor (Jeffreys, 1961), we use a ratio larger than 3 (e.g., αH = 1 − 3 × mean of beta hyperprior) as moderate-to-strong evidence. Because the marginal posterior probabilities for main effects are inflated due to the constrained prior structure, a small value of αL should be used to reduce the false discovery among main effects. Yet, αL does not affect the decision of main effects whose interactions are selected.
2.5 Prediction
Let Ml(l = 1, …, L) be a model among L models obtained by varying αL and αH, and θl = {α, β, ρ, σ} be the vector of parameters for model Ml. To predict the probability that ynew, given treatment and patient characteristics data (xnew), belongs to a predefined desirable outcome region Δ, we use the posterior predictive probability . The conditional posterior probability Pr(ynew ∈ Δ ∣ Ml, Data, xnew) in the summation is given by Pr(ynew ∈ Δ ∣ Ml, Data, xnew) = ∫ Pr(ynew ∈ Δ ∣ θl, xnew)p(θl ∣ Ml, Data)dθl.
Analytically evaluating the summation and integral is difficult due to the large model space. An immediate Monte Carlo estimate can be calculated as follows. We obtain draws of the parameters from model Ml. For each set of parameters , t = 1, …, Tl, we calculate the from the multivariate regression model in equation (1). The estimated probability of a new observation ynew belongs to set Δ is . Note that if one model is preferred over Bayesian model averaging, L can be set to 1.
To decide which treatment is best for a given patient with covariates xnew, we computed posterior predictive probabilities of superiority for the three treatments (A, F, and G). For each posterior sample of the model parameters , t = 1, …, T, compute = {Pr(ynew ∣ θA, xnew) > Pr(ynew ∣ θF, xnew) and Pr(ynew ∣ θA, xnew) > Pr(ynew ∣ θG, xnew)}, and analogously compute and . Averaging these indicators over the T posterior samples would yield relative probabilities for superiority (and likewise, pF and pG).
3. Simulation Studies
3.1 Designs
To investigate the performance of the variable selection, the prediction of multiple non-Gaussian outcomes, and the effect of sample size on our proposed method, we performed a series of simulation studies. For each sample size (n = 30 or 200), we set the number of potential predictors p ≡ 55, which consisted of 10 main effects and 45 pairwise interaction terms. We assume that all of the 10 main effects are independent and from a N(0, 1) distribution.
Three responses were simulated: two binary outcomes (with probit link) and one survival outcome (with a lognormal link). Among the 55 potential predictors, the true predictors and their coefficients were set as 0.3 + X1 + X2 + X3 + X4 + X1 X2 + X1X4 for the first binary outcome, −0.3 + X2 + X3 + X4 + X5 + X2X3 + X4X5 for the second binary outcome, and 0.3X3 + 0.3X4 + 0.3X5 − 0.2X3X4 + 0.2X4X5 for the survival outcome. We drew a n × 3 matrix of latent variables Z from the trivariate normal density (1) given these true predictors. The parameters in the variance–covariance matrix of Z in equation (1) were set as ρ1 = 0.1, ρ2 = 0.3, ρ3 = 0.7, and σ = 0.3. For each ith simulated observation (i = 1, …, n), if zik > 0, k = 1, 2, we set yik = 1 and yik = 0 otherwise. This setting generated y1 = 1 approximately 50% and y2 = 1 approximately 40%. For the survival outcome y3, we assume log-normal distribution and noninformative censoring. Random deviates were generated independently from the Unif (0, 4.5) distribution. If y3 = exp(z3) was larger than the corresponding uniform deviate, then that patient was censored at the time of that uniform deviate. This yielded approximately 30% censoring, which matches the percentage of censoring in the colorectal cancer data. We generated 100 replications for each sample size (n = 30 and 200).
For all the simulated data sets, we applied the MBSI method with c = 10 and τ = 0.05. For the hyperparameters in the beta prior, the mode was set to 3/p and the mean was 20% larger than the mode. The length of the MCMC chain was set to be 20,000, from which the first 5000 iterations were discarded. The significance levels (αL, αH) were set to (0.1, 0.2) and (0.3, 0.8), respectively, for the sample sizes 200 and 30. When there was a sufficiently large sample size (n > p), the posterior distribution was not sensitive to the prior assumptions. The setting (αL = 0.1, αH = 0.2) led to at most 20% falsely discovered predictors. However, when the sample size was small (n < p), the marginal posterior distribution was flattened and sensitive to the prior assumptions. We assumed that on average the prior probability to be a true predictor is 0.065 (the mean chosen for the beta prior). The setting αL = 0.3 and αH = 0.8 when n = 30 resulted in an average posterior probability of at least 0.2 among the selected regressors. The average posterior to prior probability ratio for the selected regressors was about 3.
3.2 Prediction Evaluation
We evaluated prediction using the posterior predictive distributions as follows. Let ytest and xtest be the simulated outcomes and covariates independent from the training set. We defined Δ as requiring both binary outcomes be equal to 1 and the survival outcome be larger or equal to 12 months. Let Pr(ytest ∈ Δ) (calculated under model (1)) be the true probability. For each replication, we obtained the posterior predictive distribution p(ytest ∈ Δ ∣ xtest, θl). If the Pr(ytest ∈ Δ) is within 90% of central posterior interval (CPI), we set coverage = 1. The average coverage across all the 100 replications gives the estimate for the 90% coverage for that test case. Five test cases that were scattered in the space of design points were simulated to demonstrate the performance of proposed method.
The true probability Pr(ytest ∈ Δ), the estimated mean and standard error of posterior predictive probability, and the 90% CPI are shown in Table 2. Two model selection approaches are compared in the table. One selects a model with the highest joint posterior probability, while the other selects a model that satisfies the criteria. Graphical illustration of the comparison between the two modeling approaches can be found in Web Figure 1.
When the sample size equals 200, there was no significant difference between the two model selection methods in terms of estimated mean and coverage. When the sample size dropped to n = 30, the prediction uncertainty increased greatly. Test cases 4 and 5 were underestimated using the joint posterior model selection method. The cause of this biased estimation was likely due to the exclusion of the important predictors. The joint posterior probabilities were dominated by the prior distribution, which favors parsimonious models. The predictions for cases 4 and 5 were slightly improved when using the criteria.
4. Colorectal Cancer Study
4.1 Preliminary Analysis
Now we return to the colorectal study described in the Introduction. We dichotomized the 23 biomarkers to mutant (0) or wild type (1). The endpoints observed for each patient in this trial were an indication of toxicities, tumor response, and time to tumor progression (TTP). Several types of toxicities were monitored, e.g., nausea, dehydration, neutropenia, vomiting, diarrhea, febrile neutropenia, and paresthesia. We chose the maximum grade among all types of toxicities and set a binary outcome toxicity (TOX) to 1 if the maximum grade was 4 or 5, which represents life threatening or fatal toxicity on a 5 point scale (National Cancer Institute Common Toxicity Criteria Version 2), and 0 otherwise. If a patient had at least two consecutive complete or partial tumor regressions, the binary outcome tumor response (RSP) was set to 1 and 0 otherwise. The third outcome was the TTP, which was a censored continuous variable. The total number of regressors is 325: 25 main effects (23 biomarkers plus AGE and SEX) plus 300 two-way interactions.
Preliminary investigation of the multiple endpoints (RSP, TOX, and TTP) indicated that they were not independent of each other, as there were positive correlations between RSP and TTP across all the treatment groups. Patients who had a confirmed tumor response consistently had a longer progression time relative to those who had no confirmed tumor response. To check the log-normal assumption for the survival model, we fit a parametric log-normal model to the failure times with no predictors using the survreg function in R. Based on the fit, we plotted the residuals (See Web Figure 2). In all three arms, log-normality appeared to be a tenable assumption.
4.2 Model Building
We applied the proposed MBSI method on each treatment arm and chose c = 10 and τ = 0.05 for the mixture normal prior. This implied that if a coefficient was less than about 2τ = 0.1, we were comfortable excluding that regressor from the model. For the hyperparameters in the beta prior, the mode was set to 0.2 and mean was 0.25, which is 25% larger than the mode.
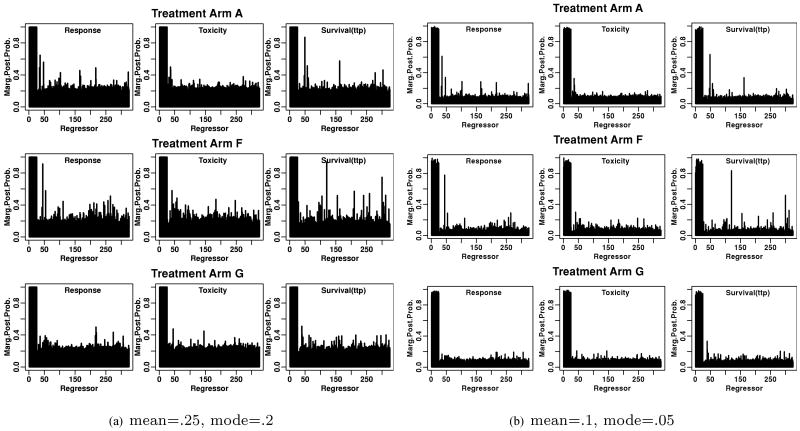
The estimated marginal posterior distributions of the regressors by treatment arms are shown in Figure 1a. The first 25 regressors are main effects and the rest are interaction terms. All the main effects show very high marginal posterior probabilities. The reason is that the marginal probability of a main effect consists of two sources: one is from itself, the other is from its interactions. The more interaction terms, the more the marginal probabilities for the main effects will be inflated. When n > p, there is more separation between signal and noise. See Web Figure 3 for the marginal posterior probabilities in our simulation study. However, when n < p, the beta hyperprior plays an important role. When the anticipated proportion of true predictors (the mean of beta hyperprior) was set to be 0.25, the marginal posterior probabilities for interactions were influenced by this prior accordingly (Figure 1a). To analyze the sensitivity to the choice of beta prior, we used another set of hyperparameters with mode equal to 0.1 and mean 0.05. This setting kept the basic shape of beta distribution unchanged, but the distribution is flatter and skewed more toward smaller values, suggesting that a priori there were fewer true predictors and we would be more uncertain about the number of the true predictors. The estimated marginal posterior probabilities of the interaction terms (Figure 1b) indicated that the posterior probabilities are sensitive to the prior, which is not a surprise as n < p in each treatment group. However, for those interaction terms whose estimated posterior probabilities of being selected are above average, the ordering from the highest to the lowest probabilities is very similar from either settings of beta prior. There is much uncertainty about most regressors, yet the strong signals from several of the interactions are worthy of further investigation.
Figure 1.
Marginal posterior distributions of the regressors by treatment arms in the colorectal cancer study for the sensitivity analysis of beta priors.
To select the important predictors, we set the threshold αL = 0.001 and αH = 0.3 for the main effects and interactions, respectively. The reason to set a low αL in this case is to avoid selecting too many main effects, whose marginal probabilities might be inflated by the large number of interaction terms. The variable selection result was not sensitive to the choice of αH in a range from 0.2 to 0.5. Table 3 shows the estimated coefficients for the selected predictors. The trace plots for the parameters in the model were provided as supplementary material Web Figures 4–6.
Table 3. Est.(SD) of coefficients for the selected predictors.
| Var | Arm A (n = 115) | Arm F (n = 292) | Arm G (n = 106) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| RSP | TOX | TTP | RSP | TOX | TTP | RSP | TOX | TTP | |
| Int. | −0.55 (0.17) | −1.60 (0.37) | 4.95 (0.16) | 0.11 (0.15) | −0.37 (0.11) | 5.85 (0.16) | −0.33 (0.17) | −1.51 (0.53) | 5.41 (0.15) |
| † Age | 0.13 (0.09) | −0.21 (0.10) | 0.24 (0.14) | ||||||
| Sex | −0.12 (0.16) | −0.59 (0.15) | −0.22 (0.23) | −0.49 (0.31) | |||||
| M1 | 0.02 (0.30) | ||||||||
| M2 | 0.51 (0.37) | ||||||||
| M3 | 0.15 (0.14) | 0.44 (0.38) | |||||||
| M5 | 0.60 (0.23) | 0.56 (0.21) | −0.98 (0.26) | ||||||
| M7 | −0.38 (0.30) | 0.05 (0.46) | −0.46 (0.33) | ||||||
| M8 | −0.37 (0.24) | 0.59 (0.23) | |||||||
| M10 | 0.35 (0.27) | −0.40 (0.20) | −0.94 (0.49) | ||||||
| M11 | 0.07 (0.12) | ||||||||
| M12 | 0.10 (0.28) | ||||||||
| M13 | 0.15 (0.23) | −0.05 (0.11) | |||||||
| M14 | 0.73 (0.34) | ||||||||
| M16 | −0.35 (0.13) | ||||||||
| M17 | 0.14 (0.55) | −0.58 (0.40) | |||||||
| M19 | −0.16 (0.13) | −0.5 (0.15) | |||||||
| M21 | 0.21 (0.25) | −0.10 (0.21) | |||||||
| M23 | 0.22 (0.23) | 0.14 (0.14) | −0.04 (0.31) | −0.09 (0.39) | |||||
| Age*M19 | 0.49 (0.13) | ||||||||
| Age*M23 | −0.69 (0.23) | ||||||||
| Sex*M5 | 0.83 (0.32) | ||||||||
| M3*M8 | −0.88 (0.23) | ||||||||
| M5*M13 | −0.90 (0.30) | ||||||||
| M8*M13 | 0.33 (0.20) | ||||||||
| M11*M23 | −0.65 (0.28) | ||||||||
| M16*M19 | 0.52 (0.17) | ||||||||
Variable age was scaled.
The cell is blank if the variable was not selected for an outcome.
Note that the estimated coefficients were not significantly different from 0 for some main effects. For example, in arm F the estimated mean and standard error (SE) of the coefficient were 0.07(0.12) for marker 11 and 0.14(0.14) for marker 23. However, their interaction had estimated mean and SE of −0.65(0.28), which was significantly different from zero. This demonstrates that even though main effects were not significant, their significant joint effect was detected by the MBSI method.
In other examples (i.e., marker 10 and marker 12 in arm F), the coefficients of main effects were not significantly different from zero, and no interactions were selected. This may have one of the three possible explanations. First, fitting all the important variables that are selected due to their high marginal probabilities into a single multiple regression model does not guarantee that all of the coefficients are still significant. The multicollinearity among some important variables may lead to the nonsignificant coefficients. Second, the threshold for procedure could be too low, leading to a high FDR. In our analysis, the threshold for the main effects was set very close to zero, which rules out this possibility. Third, the estimated high marginal probability of a main effect could be due to the falsely selected interactions during the MCMC procedure. A possible remedy for this problem is to apply the MBSI method on the main effects only and review the marginal probabilities of the main effects in question.
The estimated variance components are presented in Web Table 1. A positive correlation between RSP and TTP (ρ2) was found in all the study arms. Across all three arms, arm F resulted in the largest model size (Table 3), which is defined as the total number of distinctive regressors in a multivariate model allowing different regressors for different outcomes. The model size will tend to increase with the size of sample, because the prediction variance will usually be reduced with increased sample size (Miller, 2002).
4.3 Comparing Treatments
To compare the three treatment regimens, we defined a favorable region (Δ) of treatment outcomes as a union of Δ1 (RSP = 0 or 1, TOX = 0, and TTP > 365) and Δ2 (RSP = 1, TOX = 0 or 1, and TTP > 730). The definition of Δ can be very flexible and can change from study to study. A favorable treatment outcome often represents a trade-off between efficacy and toxicity: if a cancer treatment has high efficacy, sometimes patients are willing to accept more toxicity.
In this study, 18 variables, including AGE, were selected. If we select the design points of the continuous variable AGE at age 50 and 70 (20th and 80th percentile of age distribution), together with the other 17 binary variables, the total possible number of different configurations of patient profiles is 218 = 262,144. For each of these 262,144 hypothetical or future patients, we can predict the probability of outcome in region Δ given treatment A, F, or G.
In general, the experimental treatment F had a slightly higher probability of achieving an outcome in the favorable region Δ compared to the standard treatment A, whereas the treatment G had a lower probability (See Web Figure 7). Under treatment A, patients with younger age, mutant markers 5 and 23, and wild-type marker 13 had greater than 50% chance to achieve the outcome region Δ. Patients with mutant marker 7 have worse outcome than those with wild-type marker 7 given treatment G.
4.4 Choice of Tuning Parameter c
Because we are dealing with the n < p situation in this application, further sensitivity analyses of model size were done using different settings of the tuning parameters c and τ (Web Table 2). The range of c where the predictive performance is invariant is narrower in our application (with n < p) than that recommended by George and McCulloch (1993). Possible explanations of this phenomenon are the small transition probabilities and sample size, discussed in Web Appendix A.
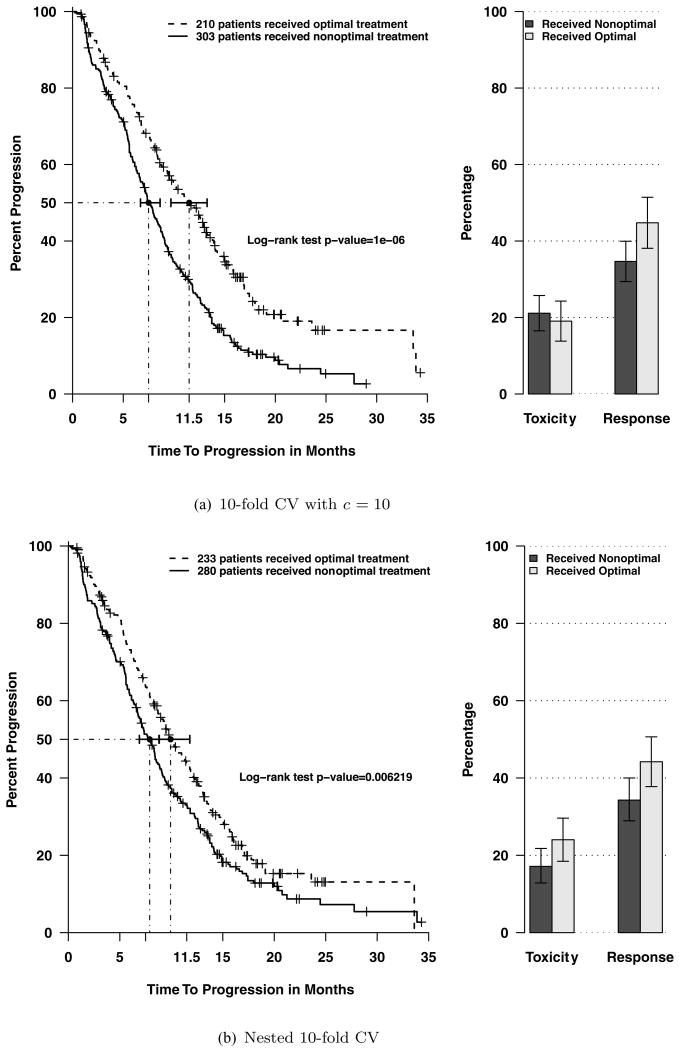
Because of the strong influence of tuning parameter c, we used a 10-fold cross-validation (CV) approach to choose c. We randomly divided data into 10 parts, stratified on treatment arms. In each of 10 subsets, one was set aside as testing data and the remaining nine were used as training set for modeling with different values of c. Optimal treatments for the testing set were predicted (see Section 2.5 for the calculation) using the model trained from the training set. We then separated the 513 patients into two groups: group 1 consisted of patients who received nonoptimal treatment; group 2 consisted of patients who received optimal treatment. When c = 10, the survival curve and percentages of toxicities and responses of each group are shown in Figure 2a. The median time to progression of group 2 was 11.5 months which was 53% greater than that of group 1 (7.5 months). Figures with other c values are shown in Web Figures 8–10. The predictive performance with c = 5 (Web Figure 8) was less than that of c = 10. This is likely due to the smaller model size, which left out a few important predictors. The predictive performance with c = 20 (Web Figure 9) was less than that of c = 10. This is probably due to the large model size, which led to model overfitting. Hence, c = 10 was chosen to have the best prediction performance.
Figure 2.
Comparing treatment outcomes of patients who received optimal treatment versus those who received nonoptimal treatment in the colorectal cancer study.
4.5 Model Validation
To further validate whether or not the proposed individualized therapy model results in any difference in treatment effect, the 10-fold method in Section 4.4 is not appropriate because it used all the data for selecting c. Therefore, to obtain an assessment of prediction, a nested 10-fold CV was performed. We again divided data into 10 parts. For the 9/10 parts that were used for model building, we did inner 9-fold CV to select c – building models for a grid of c values (c = 5, 10, 15, 20) on the 8/9 parts and choosing the one that performed best in predicting the held-out part. Then, whichever c was the best, it was used on predicting the 1/10 part that was not used in selecting c. This was repeated for each of the ten 9/10 splits resulting in a valid assessment of prediction shown in Figure 2b.
The statistical testing and confidence intervals in Figure 2 are for illustration only, and the significance has to be interpreted with caution. Lusa et al. (2007) reported that the CV process results in inflated testing type I error rates. As pointed out by one referee, the only way to truly assess the value of individualized therapy would be to perform a prospective trial in which the patients were treated based on an individualized plan.
5. Concluding Comments
In this work, we proposed a multivariate Bayesian regression model for individualizing cancer treatments. We have addressed three issues: comparing the treatments using a quantitative score derived from predicting the multiple endpoints, modeling jointly non-Gaussian outcomes, and building a regression model with the selection of interactions. The endpoints considered were categorical and censored continuous variables, which are very common in most phase III clinical trial settings. The MBSI procedure which incorporates the selection rule for interaction terms by controlling posterior expected FDR was implemented. This particular selection rule increased the power of detecting interactions, which becomes more and more important in defining useful comprehensive models for complex diseases.
Our simulation study suggests the feasibility of predicting the multiple non-Gaussian outcomes simultaneously. Although the categorical outcomes in the current model were assumed to be binary outcomes, it is straightforward to extend the proposed approach to multilevel outcomes. The survival outcome in the colorectal cancer study was assumed to follow a log-normal accelerated failure time model for the simplicity of computation. We suggest checking this assumption before implementation in the data analysis.
Regarding the priors, although spike and slab mixture priors for variable selection were proposed and applied in the literature, we consider the narrow normal priors for nonsignificant regressors as proposed by George and McCulloch (1993) to be more practical. An important issue is the choice of c. We used CV type of methods to choose c in this application. These are all fairly computationally intensive methods. Further theoretical development is needed in this area.
When applying the MBSI method in the colorectal cancer study, we modeled each arm separately. Because different treatments work through different mechanisms, the possibility of treatment-biomarker interactions exists. Separate models eliminate those interaction terms and result in a less complex model structure and faster computation. As pointed out by one referee, it would be better if the modeling was not completely separate, but rather borrowed strength across treatments in estimating the covariance parameters. A hierarchical model component could be introduced that allowed each treatment to have its own covariance parameters. Nonetheless, the increased model complexity due to the additional level is beyond the scope of this article.
Supplementary Material
Acknowledgments
The authors thank the editor, the associate editor, and the referees for their comments that considerably improved this article. Part of the work was supported by the National Institutes of Health through Karmanos Cancer Institute Support Grant (5 P30 CA022453).
Footnotes
Supplementary Materials: Web Tables and Figures referenced in Sections 3.2 and 4.1–4.4 are available under the Paper Information link at the Biometrics website http://www.biometrics.tibs.org.
References
- Albert JH, Chib S. Bayesian analysis of binary and polychotomous response data. Journal of the American Statistical Association. 1993;88:669–679. [Google Scholar]
- Barbieri MM, Berger JO. Optimal predictive model selection. The Annals of Statistics. 2004;32:870–897. [Google Scholar]
- Barnard J, McCulloch R, Meng XL. Modelling covariance matrices in terms of standard deviations and correlations, with applications to shrinkage. Statistica Sinica. 2000;10:1281–1311. [Google Scholar]
- Brown PJ, Vannucci M, Fearn T. Multivariate Bayesian variable selection and prediction. (B).Journal of the Royal Statistical Society. 1998;60:627–641. [Google Scholar]
- Chen MH, Dey DK. Variable selection for multivariate logistic regression models. Journal of Statistical Planning and Inference. 2003;111:37–55. [Google Scholar]
- Chen W, Ghosh D, Raghunathan TE, Sargent DJ. A false-discovery-rate-based loss framework for selection of interactions. Statistics in Medicine. 2008;27:2004–2021. doi: 10.1002/sim.3118. [DOI] [PubMed] [Google Scholar]
- George EI. Discussion of Bayesian model averaging and model search strategies by M.A. Clyde. In: Bernardo JM, Berger JO, Dawid AP, Smith AFM, editors. Bayesian Statistics. Vol. 6. Oxford: Oxford University Press; 1999. pp. 175–177. [Google Scholar]
- George EI, McCulloch RE. Variable selection via Gibbs sampling. Journal of the American Statistical Association. 1993;88:881–889. [Google Scholar]
- Goldberg RM, Sargent DJ, Morton RF, Fuchs CS, Ramanathan RK, Williamson SK, Findlay BP, Pitot HC, Alberts SR. A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. Journal of Clinical Oncology. 2004;22:23–30. doi: 10.1200/JCO.2004.09.046. [DOI] [PubMed] [Google Scholar]
- Jeffreys H. The Theory of Probability. 3rd. Oxford: Oxford University Press; 1961. [Google Scholar]
- Lusa L, McShane LM, Radmacher MD, Shih JH, Wright GW, Simon R. Appropriateness of some resampling-based inference procedures for assessing performance of prognostic classifiers derived from microarray data. Statististics in Medicine. 2007;26:1102–1113. doi: 10.1002/sim.2598. [DOI] [PubMed] [Google Scholar]
- McLeod HL, Murray GI. Tumor markers of prognosis in colorectal cancer. British Journal of Cancer. 1999;79:191–203. doi: 10.1038/sj.bjc.6690033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milano G, McLeod HL. Can dihydropyrimidine dehydrogenase impact 5FU-based treatment? European Journal of Cancer Prevention. 2000;36:37–42. doi: 10.1016/s0959-8049(99)00211-7. [DOI] [PubMed] [Google Scholar]
- Miller AJ. Subset Selection in Regression. 2nd. London: Chapman & Hall; 2002. [Google Scholar]
- Neter J, Kutner MH, Nachtsheim CJ, Wasserman W. Applied Linear Statistical Models. New York: McGraw-Hill; 1996. [Google Scholar]
- Sha N, Tadesse MG, Vannucci M. Bayesian variable selection for the analysis of microarray data with censored outcomes. Bioinformatics. 2006;22:2262–2268. doi: 10.1093/bioinformatics/btl362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.