Abstract
The escort factor Scap is essential in mammalian cells for regulated activation of sterol regulatory element binding proteins (SREBPs). SREBPs are membrane-bound transcription factors. Cells lacking Scap cannot activate SREBP. They are therefore deficient in the transcription of numerous genes involved in lipid synthesis and uptake; they cannot survive in the absence of exogenous lipid. Here we report that, in contrast to mammalian cells, Drosophila completely lacking dscap are viable. Flies lacking dscap emerge at ∼70% of the expected rate and readily survive as homozygous stocks. These animals continue to cleave dSREBP in some tissues. Transcription of dSREBP target genes in dscap mutant larvae is reduced compared to wild type. It is greater than in mutants lacking dSREBP and remains responsive to dietary lipids in dscap mutants. Flies lacking dscap do not require the caspase Drice to activate dSREBP. This contrasts with ds2p mutants. ds2p encodes a protease that releases the transcription factor domain of dSREBP from the membrane. Larvae doubly mutant for dscap and ds2p exhibit phenotypes similar to those of ds2p single mutants. Thus, dScap and dS2P, essential components of the SREBP activation machinery in mammalian cells, are dispensable in Drosophila owing to different compensatory mechanisms.
THE sterol regulatory element binding proteins (SREBPs) play a central role in the transcriptional coordination of the expression of the genes of lipid synthesis (Horton et al. 2002). All metazoan genomes already sequenced encode SREBP orthologs (Matthews et al. 2009). SREBPs are synthesized as precursor proteins that are anchored to the endoplasmic reticulum (ER) membranes by two membrane-spanning helices that are separated by a small loop projecting into the lumen. Both the amino-terminal transcription factor domain and the carboxy-terminal regulatory domain of SREBP face the cytoplasm. The regulatory domain interacts with the carboxy-terminal domain of the sterol-binding escort factor Scap. The first ∼800 amino acids of Scap contain eight membrane-spanning helices, including a sterol sensing domain (SSD) from helices two through five. The final ∼500 amino acids consist of multiple WD-40 protein–protein interaction motifs (Hua et al. 1996).
When cellular lipid levels are sufficient, the SREBP: Scap complex remains in the ER. When lipid levels decline, a sorting signal on Scap is unmasked; the SREBP:Scap complexes are packaged into COPII vesicles to travel to the Golgi apparatus (Espenshade et al. 2002). Scap then returns to the ER, while SREBP is cleaved in two sequential steps by two different Golgi proteases, site-1 protease (S1P) and site-2 protease (S2P). Cleavage frees the transcription factor to travel to the nucleus and upregulate the transcription of the genes of lipid synthesis. In mammalian cells, Scap is absolutely required for the transport and activation of SREBPs—cells lacking Scap cannot cleave SREBPs and die owing to insufficient production and uptake of cholesterol and unsaturated fatty acids. These cells survive only if their culture medium is supplemented with those products (Goldstein et al. 2002).
Drosophila melanogaster has a single ortholog of SREBP. We designate it dSREBP to distinguish it from its mammalian counterparts with which it shares important similarities but has differences as well. It is also called HLH106 (Theopold et al. 1996). The fly genome encodes other components of the SREBP processing machinery as well, having orthologs of Scap (dScap) and of the site-1 and site-2 proteases (dS1P and dS2P). We have generated null alleles of dscap in D. melanogaster. In contrast to mammalian cells, flies lacking dScap are viable. dSREBP continues to be activated in a subset of tissues in dscap mutants.
We have previously shown that another component of the SREBP processing machinery, dS2P, is also not essential in flies (Matthews et al. 2009). In its absence, the caspase Drice cleaves dSREBP in the juxtamembrane stalk region to release the transcriptionally active amino-terminal domain. Larvae doubly mutant for ds2p and drice phenocopy dSREBP mutants and die at the end of second instar unless their medium is supplemented with fatty acids (Amarneh et al. 2009). Larvae lacking dscap, however, do not require drice, and dscap; drice double mutants survive about as well as either single mutant. Similarly, ds2p is not essential in dscap mutants. Flies lacking both dscap and ds2p exhibit phenotypes similar to those seen with ds2p mutants alone. Thus dscap mutants survive owing to a mechanism distinct from that which permits the survival of ds2p mutants.
MATERIALS AND METHODS
Fly culture media:
Standard culture medium is cornmeal–molasses–agar (1 liter contains 60 g cornmeal, 15 g dry yeast, 80 ml unsulphured molasses, and 12 g agar, 6 ml propionic acid, and 0.1 g tegosept). Semidefined media is as described (Backhaus et al. 1984). One liter contains 80 g baker's yeast, 20 g yeast extract, 20 g peptone, 30 g sucrose, 0.5 g MgSO4·6H2O, 6 ml propionic acid, and 0.1 g tegosept. Supplemented medium (C14:0 + C18:1) is either of these media, as indicated in the figure legends, to which myristate (0.075%, w/v) and oleate (0.15%, w/v) are added as the sodium salts or to which soy lipids (Avanti Polar Lipids) are added (9% w/v) as indicated.
Genetic strains:
All marker mutations and balancer chromosomes are described and referenced by FlyBase Consortium (2003). Other lines are described in (Kunte et al. 2006; Amarneh et al. 2009; Matthews et al. 2009). Crosses were conducted at 25° in vials containing freshly yeasted standard medium. OreR flies serve as wild type. P-element transposon lines KG00745, P{w+, UAS-GFP} (inserted on the third chromosome) and P{ry, hsFlp}; Adv/CyO were obtained from the Bloomington Stock Center. PiggyBac transposon insertion lines PB(WH)f04534 and PB(PB)c00785 were obtained from the Exelixis collection at the Harvard Stock Center (Thibault et al. 2004). Deletion alleles dsrebp189 and ds2p1 and insertion allele ds2p2 were described previously (Kunte et al. 2006; Matthews et al. 2009). The P{w+, GAL4-dSREBPg} transgene is inserted on the third chromosome (Kunte et al. 2006).
FLP–FRT recombination:
Transposons PB(WH)f04534 and PB(PB)c00785 contain the piggyBac vectors (WH) and (PB), respectively (Thibault et al. 2004). Both vectors contain a mini-white marker gene and yeast FLP recombination target (FRT) sequences. In addition, the (WH) vector contains Su(Hw) insulator sequences and a terminal UAS site. Orientation of the transposon insertion was determined by PCR using one primer within the piggyBac transposon and one primer within the genome. The FRT sequences within piggyBac lines f04534 and c00785 are oriented in the same direction with respect to the dscap gene (Figure 1A, open arrows) and therefore, recombination between the two FRT sites deletes the dscap gene. However, the piggyBac (PB) vector in line c00785 contains two FRT sequences flanking a mini-white marker gene. This generates two possible recombination outcomes: (1) A recombined allele containing two mini-white marker genes or (2) a recombined allele containing one mini-white marker gene. To induce FLP–FRT-mediated recombination, we used a P{ry, hsFLP} insertion on the X chromosome (Golic and Lindquist 1989), which was crossed into a PB(WH)f04534 background. Virgin w1118/w1118; PB(PB)c00785/PB(PB)c00785 females were crossed to P{ry, hsFLP}/Y; PB(WH)f04534 /CyO males in bottles. After 2 days, larvae and adults were incubated at 37° for 1 hour to induce recombination as described in Golic et al. (1997). Adults were removed at the end of the day. Larvae were incubated at 37° for 1 hr each day for an additional 4 days. Single virgin P{ry, hsFLP}/+; PB(WH)f04534/PB(PB)c00785 females displaying mosaic eye color were then crossed to balanced w1118/w1118; Sp/CyO males. Single F3 males with solid red eyes were crossed to balanced females and lines were established. Recombinants were verified by Southern blot using restriction enzymes StuI and SphI and PCR using primer sets within the dscap locus.
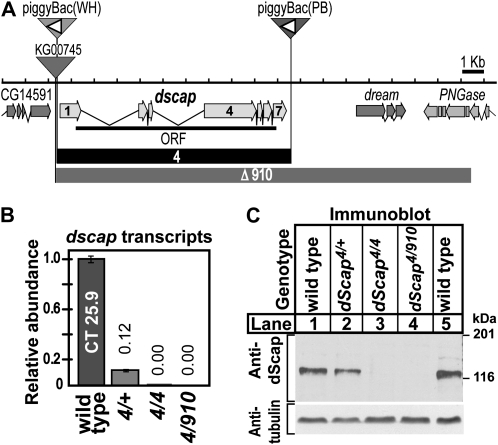
Figure 1.—
(A) Map of dScap locus. The dScap gene comprises seven exons (block arrows with light shading) encoding one protein (ORF, thick solid line). Sites of transposon insertion are indicated by inverted triangles. P-element transposon KG00745 and piggyBac(WH)f04534 are inserted prior to the start of dScap of exon 1 and after CG14591. PiggyBac(PB)c00785 is inserted 88 bp after dscap exon 7. The orientation of the FRT sites within both piggyBac elements are indicated by the open triangles. The extent of dscap4 and dscapΔ910 deletions are indicated by solid boxes and boxes with dark shading, respectively. (B) Quantitative analysis of dScap mRNA from dscap4 homozygous and dscapΔ910/dscap4 transheterozygous larvae compared to wild type (wt = 1). Numbers above bars indicate the relative abundance of transcript. Error bars represent the standard deviation. (C) Immunoblot analysis of whole larval lysates from third instar larvae of the indicated genotype (60 μg total protein/lane). Virgin dscap4/dscap4 females were crossed to either dscap4/CyO, act-GFP or dscapΔ910/CyO, act-GFP males. Embryos were seeded onto dishes containing semidefined media at 10 mg/dish. Larvae were isolated from the food by salt flotation and homozygous larvae were scored for absence of act-GFP fluorescence. The membrane was probed with monoclonal antibody IgG7A8 against dScap TM1-8 (1-min-30-sec exposure), stripped, and reprobed with anti-acetylated tubulin (2-sec exposure).
Analysis of larval growth:
Embryos were collected for 2 hr and plated at 10 mg/plate onto semidefined media as previously described (Kunte et al. 2006). Larval stage was determined by mouth hook and anterior spiracle morphology (Demerec 1950). Representative larvae were selected at each time point and photographed as described (Kunte et al. 2006).
Adult emergence assays:
Embryos were collected overnight and seeded into vials at 1 mg embryo/vial containing standard medium either with no additions or supplemented with 0.075% Na-myristate and 0.15% Na-oleate (Sigma). Larvae were allowed to develop and emerging adults were cleared twice daily from the culture and scored for genotype starting at day 10 after egg laying (AEL).
Lipid mass determination:
For each determination, 60 virgin females of the indicated genotype were collected 3 days posteclosion and homogenized in 3.0 ml extraction reagent [CHCl3:CH3OH:PBS (1:2:0.8)] in a 7-ml Dounce homogenizer. The resulting mixture was separated by centrifugation at 1500 × g for 10 min. The aqueous phase was removed and reextracted with 3.0 ml extraction reagent and reseparated. The pooled organic phase was transferred to a preweighed glass tube and dried under anhydrous nitrogen for 30 min. The mass of lipids was measured by weighing the tube on a Metler-Toledo AX105 DeltaRange analytical balance and subtracting the initial mass of the tube.
Larval imaging:
Larvae were photographed using a Leica MZ16FA fluorescence microscope equipped with an Evolution MP digital camera (Media Cybernetics) and In Focus software (Meyer Instruments, Houston, TX). Contrast was adjusted in Adobe Photoshop CS2 using the autocurves function with 50% fading.
Whole-larval lysis:
Larvae of the indicated age and genotype were homogenized in buffer F (125 mm Tris–HCl, pH 6.8, 8 m urea, and 5% SDS). For first instar larvae, 5 mg of embryos were seeded onto filter paper wet with IPL-41 insect cell culture medium (Life Technologies). Larvae were collected from the paper and homogenized in buffer F. For third instar larvae, larvae were separated from food by salt flotation and homogenized in buffer F. Homogenates were filtered through a 100 μm Nytex mesh at 1000 g for 1 min. Total protein concentration was determined using BCA protein assay (Promega).
Fractionation of adult flies:
On day 0, 24- to 48-hr-old males were fed in vials with fresh yeast. On day one, flies were transferred to vials (50 flies/vial) containing 0.25-inch dacron plugs soaked with “starvation” media (1:3 IPL-41:PBS, made fresh daily). The diluted insect culture medium provides a minimum of nutrition to the adults and greatly improves their survival as compared to starvation on PBS alone. Flies were transferred to vials with fresh starvation media on day three. On day four, dead flies were removed, and the remaining flies were pooled and then divided in two groups. One group was transferred to vials (50 flies/vial) with fresh starvation media, while the second group was transferred to vials (50 flies/vial) with freshly made wet yeast paste. After 24 hr, any dead flies were removed and the remaining flies were harvested for subcellular fractionation as described (Matthews et al. 2009).
Monoclonal antibodies:
IgG-3B2 against Drosophila SREBP is described in Seegmiller et al. (2002). Monoclonal IgG-7A8 was generated as described (Herz et al. 1990) against Drosophila Scap transmembrane domains one through eight. IgG-611B-1 against acetylated tubulin was obtained from Sigma (St. Louis).
Immunoblot analysis:
Protein samples were subjected to standard SDS–PAGE and electroblotted onto nitrocellulose membranes. Membranes were incubated with the indicated primary antibody overnight at 4° and secondary anti-mouse antibody conjugated to horseradish peroxidase for 45 min at room temperature followed by chemiluminescent detection (Super Signal reagent, Thermo Scientific) for the indicated time.
Quantitative analysis of transcripts:
Embryos were collected for 2 hr and plated at 10 mg/plate onto semidefined media. Larvae were collected at the indicated times from one dish per time point and isolated from food by salt flotation (Kunte et al. 2006). Total RNA and cDNA were isolated from 30–100 larvae and real-time quantitative PCR (RT–PCR) analysis was performed using the indicated primers as previously described (Dobrosotskaya et al. 2003; Kunte et al. 2006). The relative abundance of mRNA was normalized to dRP49 and calculated using the comparative CT method. The fold change of transcripts was calculated relative to wild-type RNA levels as indicated in the figure legends. The standard deviation of ΔΔCT was calculated as described in User Bulletin 2 (PE Applied Biosystems).
RESULTS
Generation of dscap null alleles:
To generate deletions in the dscap open reading frame (ORF), we used transposase-mediated P-element excision (Robertson et al. 1988). We targeted transposon KG00745, located 442 bp upstream of the dscap ORF (Figure 1A). We screened 1200 independent excision lines by PCR and Southern blotting analysis and identified 3 lines harboring deletions extending into the dscap ORF. Two of these lines removed the first exon and start of the ORF and produced truncated dscap transcripts; they were not considered further. One deletion, designated dscapΔ910, extends 22 kb, removing the entire dscap gene as well as two downstream genes, dream and PNGase (Figure 1). Therefore, dscapΔ910, which is homozygous lethal, is a deficiency allele.
To delete the entire dscap ORF without disrupting neighboring genes, we induced yeast 2 μ plasmid flipase (FLP). This enzyme mediates recombination between two piggyBac transposon lines containing FRT sequences (Golic and Lindquist 1989; Sadowski 1995; Golic et al. 1997; Thibault et al. 2004). Line f04534 contains a piggyBac(PB) element inserted 814 bp upstream of the dscap ORF and line c00785 contains a piggyBac(WH) element located 88 bp after dscap exon 7 (Figure 1A).
FLP-mediated recombination was performed as described in materials and methods. Candidate lines were analyzed by Southern blot to verify recombination of chromosomes and deletion of dscap. Seventy independent lines successfully recombined between the two FRT containing piggyBac elements to yield a deletion of the entire dscap gene. One line, designated dscap4, was selected for further study. Transcripts for dScap are undetectable in dscap4 homozygotes and dscap4/dscapΔ910 transheterozygotes (Figure 1B). Accordingly, no dScap protein is detected in extracts from these animals by immunoblot analysis (Figure 1C). Therefore, dscap4 is a null allele.
To determine if dScap RNA is maternally loaded into embryos, we measured the relative abundance of dScap RNA in 0- to 2-hr-old embryos and 36-hr larvae from dscap4 heterozygous mothers (supporting information, Figure S1). At 0–2 hr AEL, dScap RNA is present in embryos from dscap4 heterozygous mothers. It is absent in embryos from homozygous mothers. No dScap RNA is detected in homozygous larvae at 36 hr AEL, irrespective of the maternal genotype. To avoid potential consequences of dScap maternally loaded RNA, virgin females homozygous for dscap were used in all experiments.
dscap is not essential in Drosophila:
In Drosophila larvae, transcriptional upregulation of target genes by dSREBP is essential (Kunte et al. 2006). In all systems yet studied, activation of SREBP requires its Scap-mediated transport from the ER to the Golgi (Espenshade and Hughes 2007). We were therefore surprised to find that, rather than exhibiting lethality, flies lacking dscap survive at ∼70% of the expected rate (Figure 2A). Supplementing their food with fatty acids restores full viability. Thus, the reduced survival of dscap homozygotes is a consequence of altered lipid metabolism subsequent to reduced transcription by dSREBP.
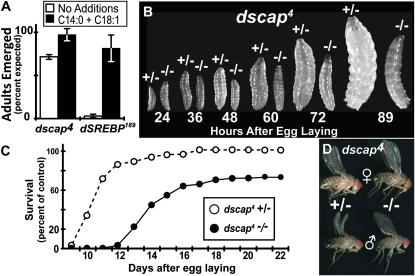
Figure 2.—
(A) Embryos from virgin dscap4/dscap4 females crossed to dscap4/CyO, act-GFP males were collected and set up in vials (n = 10 for each condition) containing standard medium or supplemented medium (C14:0 + C18:1) as described in materials and methods. Starting on day 10 AEL, adults were collected from the vials twice daily and scored for genotype until day 20 AEL, after which no further adults emerged. Shown is the mean of three independent determinations. Error bars represent the SEM. (B) Comparison of larvae homozygous (−/−) or heterozygous (+/−) for dscap4. Virgin dscap4/dscap4 females were crossed to dscap4/CyO, act-GFP males. Embryos were seeded at 10 mg/dish onto dishes containing semidefined media. Larvae were scored for genotype on the basis of GFP fluorescence and photographed at the indicated time points. (C) Emergence of adult flies homozygous (solid circles) or heterozygous (open circles) for dscap4. Embryos from virgin dscap4/dscap4 females crossed to dscap4/CyO, act-GFP males were collected and set up on standard medium and emerging adults scored as described in materials and methods. (D) Comparison of size between dscap4 homozygous (−/−) and heterozygous (+/−) adults reared on standard medium.
Homozygotes lacking dScap develop more slowly than their heterozygous culture mates. Beginning ∼48 hr AEL, dscap4 homozygous larvae are notably smaller than their heterozygous siblings (Figure 2B). This difference in size becomes more pronounced with time. By 89 hr AEL the majority of heterozygotes reach third instar, but most homozygotes remain as second instars. The majority of homozygotes exhibit delayed pupariation and emerge as adults ∼2 days after their heterozygous siblings (Figure 2C). Despite this delay, homozygous adults appear indistinguishable from their heterozygous siblings morphologically (Figure 2D).
dSREBP is processed in dscap mutants:
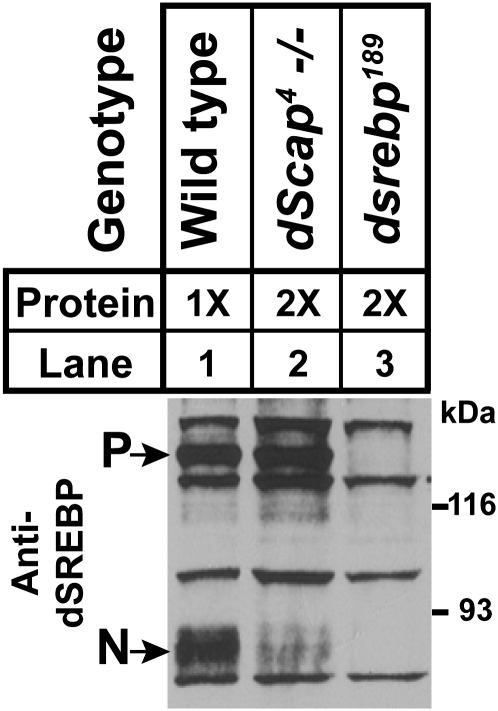
The much greater survival of dscap mutants compared to dSREBP mutants might be owing to continued activation of dSREBP in the absence of dScap. To test this hypothesis, we prepared whole larval lysates from either wild-type or dscap4 homozygous larvae and subjected them to immunoblot analysis using an antibody against Drosophila SREBP (Figure 3). Cross-reacting bands are present in all samples. In addition, lysates from wild-type larvae (Figure 3, lane 1) show bands corresponding to the full-length precursor form (P). They also show a faster migrating band corresponding to the cleaved amino terminus (N). The dscap4 homozygotes harbor a dSREBP fragment that comigrates with cleaved dSREBP from wild type (Figure 3, lane 2). However, the abundance of this product is reduced compared to wild type. Neither precursor nor cleaved dSREBP is detected in dsrebp null mutants (Figure 3, lane 3). Thus, dSREBP is processed in larvae in the absence of dScap, albeit at reduced levels.
Figure 3.—
Immunoblot analysis of whole larval lysates from first instar larvae probed with monoclonal antibody 3B2 against the amino terminus of dSREBP. Wild-type lysates were loaded at 30 μg/ml; dscap4 and dsrebp lysates were loaded at 60 μg/ml. Membranes were exposed to film for 2 min. P, precursor; N, nuclear form.
To determine whether cleaved dSREBP localizes to the nucleus in dscap mutants, we isolated membranes and nuclear extracts from wild-type and dscap4 homozygous adults (Figure S2A). Membranes from dscap4 adults show similar levels of dSREBP precursor (lane 3) compared to wild type (lane 1). The amino-terminal transcription factor domain of dSREBP is detected in the nuclear extracts of both wild-type and dscap4 homozygotes. We do not know why it accumulates somewhat more abundantly in the starved dscap adults than in wild type. Nevertheless, dSREBP is present in the nucleus in flies lacking dScap.
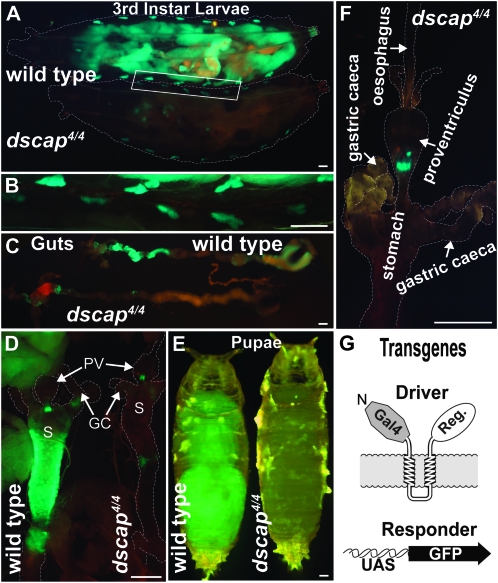
To visualize tissues where dSREBP continues to be cleaved in dscap larvae, we utilized the GAL4-dSREBP; UAS-GFP binary reporter system (Figure 4G). In this system, the transcription factor domain of dSREBP is replaced by a chimeric yeast GAL4-VP16 protein (Kunte et al. 2006). This drives expression from its cognate upstream activating sequence (UAS) that is not otherwise present in the Drosophila genome. Using this system, green fluorescence is only detected in tissues where dSREBP is cleaved. No green fluorescence is observed in animals harboring only the driver or responder transgenes alone (Kunte et al. 2006). In wild-type larvae, GAL4-dSREBP is actively processed in the fat body, midgut, and oenocytes as indicated by GFP fluorescence (Figure 4A, upper larva). In the absence of dScap, GFP fluorescence is greatly diminished in the fat body (Figure 4A, lower larva). Fluorescence is clearly observed in the oenocytes of both homozygous and heterozygous larvae (Figure 4B). Activation of GAL4-dSREBP is strong in the anterior midgut of wild-type larvae but greatly reduced in the absence of dScap (Figure 4, C and D).
Figure 4.—
(A) Ventral views of dSREBP activity in wild-type (upper) and dscap4 (lower) third instar larvae visualized using the GAL4-dSREBP/UAS-GFP reporter system. Fluorescence is readily detected in dscap4 oenocytes, and to a lesser extent in regions of the anterior midgut. Larvae are wild type or homozygous for dscap4 on the second chromosome and homozygous for P{GAL4-dSREBPg}, P{UAS-GFP} on the third chromosome. Dashed lines denote extent of larval bodies. Exposure = 10 sec. (B) Higher magnification view of the region indicated by the white box in A showing wild-type and dscap4 oenocytes. Exposure = 4 sec. (C) Midguts showing reduced fluorescence in dscap4 larvae compared to wild type. Exposure = 8 sec. (D) Anterior midguts from wild-type and dscap4 larvae showing greatly reduced dSREBP activity in dscap4 larvae. GC, gastric caeca; PV, proventriculus; S, stomach; exposure = 4 sec. (E) Wild-type and dscap4pupae (stage P5) (Bainbridge and Bownes 1981). Exposure = 5 sec. (F) Anterior midgut from a dscap4 larva exhibiting fluorescence only at the posterior end of the proventriculus. Exposure = 3 sec. (G) Schematic of the GAL4-dSREBP; UAS-GFP reporter system. Scale bars, 0.2 mm. The variability of dSREBP activity in the midgut of dscap4 larvae is evident among the different individuals shown in C, D, and F (see also Figure 6A).
Examination of mutant larvae revealed some variability in dSREBP activation. Although activation in the midgut in all dscap larvae examined was clearly reduced as compared to wild type (n > 50), some larvae showed patchy domains of fluorescence along the anterior midgut (c.f., Figure 4D, right). In all cases, strong fluorescence was consistently observed in a group of cells at the junction of the proventriculus and stomach (Figure 4, D and F). In mutants, the early pupae show almost no fluorescence as compared to wild type (Figure 4E), except for occasional spots that are likely to be perdurant larval oenocytes. In later pupae, mutants show no activation of dSREBP (not shown). These results demonstrate that dSREBP continues to be cleaved in a subset of larval tissues in the absence of dScap. If this is so, dscap mutants should exhibit transcriptional deficits that are less severe than those of larvae lacking dSREBP itself.
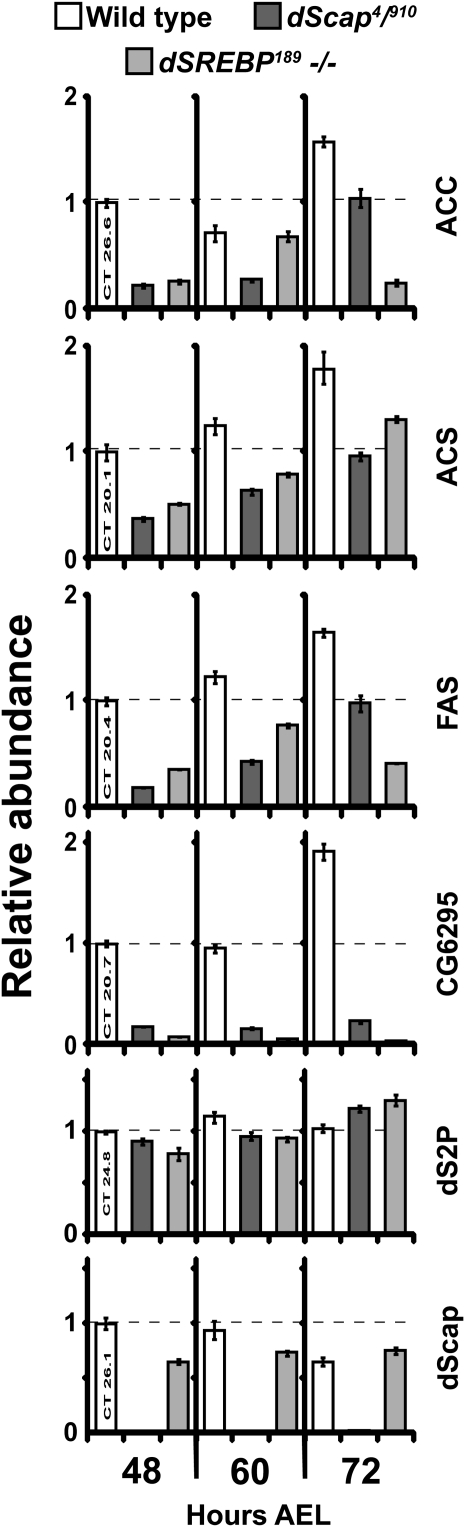
We evaluated the transcript levels of dSREBP target genes at different time points during development in larvae transheterozgyous for dscap4 and dscapΔ910 (Figure 5). As previously described (Seegmiller et al. 2002; Kunte et al. 2006), Drosophila SREBP is one of the transcription factors that activates genes involved in the synthesis of fatty acids. These include acetyl coenzyme A synthase (ACS), acetyl coenzyme A carboxylase (ACC), and fatty acid synthase (FAS). These genes are not wholly dependent on dSREBP for their expression but they are targets of dSREBP. The abundance of these transcripts is relevant to the phenotypes of dSREBP pathway mutants. This is because those phenotypes are ameliorated by supplementing the larval diet with fatty acids (Amarneh et al. 2009; Matthews et al. 2009).
Figure 5.—
Quantitative real-time PCR analysis of mRNAs of dSREBP target genes from dscap4/910 transheterozygous mutant larvae compared to wild-type and dsrebp mutants. These results are representative of three independent determinations. mRNA was isolated from 48, 60, and 72 hr AEL larvae as described in methods and materials. Fold change was calculated relative to 48-hr wild-type RNA levels. Error bars represent the SEM.
The transcript levels of these dSREBP target genes are significantly reduced in dscap transheterozygotes compared to wild-type larvae at 48, 60, and 72 hr AEL. CG6295, which encodes a putative lipase, is a major target of dSREBP and it is almost completely dependent on dSREBP for its transcription (Kunte et al. 2006; Matthews et al. 2009). Its transcripts are very low in dscap homozygotes as compared to wild-type larvae. However, CG6295 transcripts consistently show slightly greater accumulation in dscap as compared to dsrebp mutants (Figure 5 and Figure S1B). Transcript levels for ds2p, which is not a target of dSREBP, are unaffected.
Accumulation of dSREBP target transcripts is responsive to diet in dscap mutants:
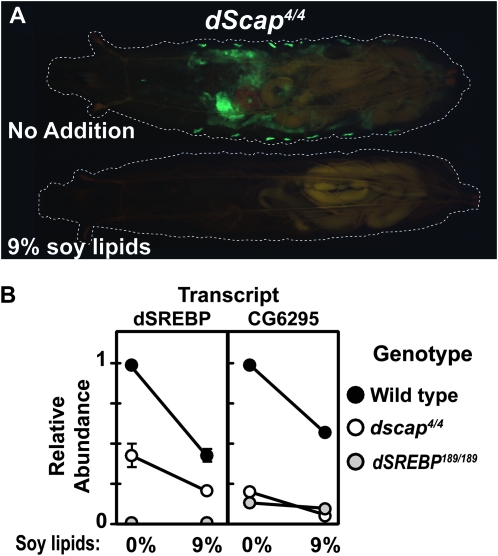
We have previously demonstrated that the membrane-bound GAL4-dSREBP is subject to the same physiological regulation as endogenous dSREBP. This is true both when it is expressed in Drosophila S2 cells or in wild-type larvae (Kunte et al. 2006). To determine the effect of dietary supplementation with lipids on dSREBP activation in the absence of dScap, we raised dscap4 larvae on unsupplemented medium or on medium supplemented with soy lipids (Kunte et al. 2006). In the presence of added soy lipids (mainly phospholipids) no GFP fluorescence is detected in either the midgut or oenocytes (Figure 6A, lower). This contrasts with larvae grown on regular medium (upper).
Figure 6.—
(A) Comparison of GFP fluorescence in dscap4 homozygotes grown on semidefined medium (upper) or the same medium supplemented with 9% soy lipids (lower). Larvae are dscap4/dscap4; P{GAL4-dSREBPg}, P{UAS-GFP}/P{GAL4-dSREBPg}, P{UAS-GFP}. Dashed lines denote extent of larval bodies. Exposure = 2.5 sec. (B) Quantitative RT–PCR analysis of dSREBP and CG6295 transcripts of wild-type (black circles), dscap4 (white circles), and dsrebp189 (gray circles) larvae grown on semidefined medium (0%) or the same medium supplemented with 9% soy lipids (9%). Transcript abundance is relative to wild-type larvae on 0%. Error bars represent the SEM.
In addition to regulation of dSREBP processing, we have previously shown that, in larvae, lipid supplementation results in the decrease of transcript levels for dSREBP itself and dSREBP target genes (Kunte et al. 2006). To determine whether this holds true for larvae lacking dScap, we performed quantitative real-time PCR on 60-hr AEL larvae reared in the presence of 0 or 9% soy lipids (Figure 6B). dSREBP and CG6295 mRNA levels decrease in response to soy lipids both in wild-type larvae and in dscap4 homozygotes. Thus both the accumulation of mRNA for dSREBP and for its target genes are reduced in the presence of additional dietary lipid in dscap mutants.
We next determined whether accumulation of nuclear dSREBP in dscap adults is responsive to the diet as it is in larvae. To induce cleavage of dSREBP, adult males were starved for 3 days prior to refeeding or continued starvation (see materials and methods). After 24 hr, surviving flies were harvested and nuclear and membrane fractions were isolated. The full-length dSREBP precursor form (P) decreases when wild-type and dscap flies are refed (Figure S2A). This decrease in full-length dSREBP protein coincides with the decrease in dSREBP mRNA when flies are fed as compared to starved (Figure S2C). In flies lacking dScap, as in wild-type flies, nuclear dSREBP significantly decreases when flies are refed. At least some of this response likely results from decreased abundance of dSREBP precursor and transcripts.
dscap mutants do not require Drice:
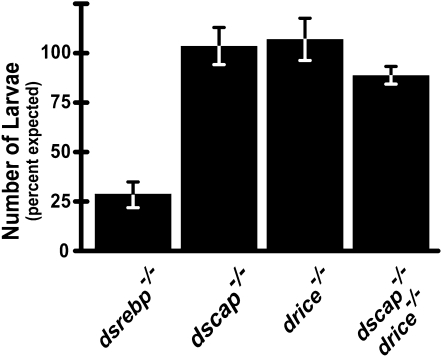
Previously, we generated null alleles of ds2p and determined that larvae lacking ds2p are viable due to an alternative cleavage of dSREBP by the caspase Drice (Matthews et al. 2009). Larvae doubly mutant for ds2p and drice die at the end of second instar owing to a failure to activate dSREBP (Matthews et al. 2009). Drice cleaves dSREBP in only a subset of larval tissues; in ds2p mutants, activation is not detected in all of the tissues where dSREBP is activated in wild-type larvae under the same conditions. Moreover, Drice-mediated cleavage of dSREBP does not confer fully wild-type dSREBP transcriptional activity to ds2p larvae. To test whether survival of dscap larvae results from alternative activation of dSREBP by Drice, we prepared stocks of dscap; drice doubly mutant flies. We evaluated their survival at third instar. This is after lethality owing to loss of dSREBP (at the end of second instar) but before lethality owing to loss of Drice (during pupation). The survival of dscap; drice double mutants did not differ significantly from the survival of either single mutant alone (Figure 7). Unlike the ds2p mutants, flies lacking dScap do not require Drice. Thus their survival is owing to a different mechanism of activating dSREBP that does not require Drice.
Figure 7.—
The caspase Drice is not required for the survival of dscap mutants. For the homozygous lethal dsrebp and drice mutants, crosses were set up using heterozygous males and females balanced with TM3, ser actin-GFP. Homozygous dscap4 females were crossed to dscap4/Cyo, actin-GFP males. For the double mutants, dscap4/dscap4; drice/TM3, ser, actin-GFP virgin females were crossed to dscap4/CyO, twist-GFP; drice/TM3, ser actin-GFP males. Larval survival was assessed as described in materials and methods. Data are presented as the mean of the percentage of expected emergence. The emergence of dscap; drice double mutants does not differ significantly from that of ds2p mutants (P < 0.5) but is significantly different from the emergence of dsrebp homozygotes (P > 0.05) by χ2 tests for independence and goodness of fit. Error bars represent the SEM.
dscap mutants do not require ds2p for survival:
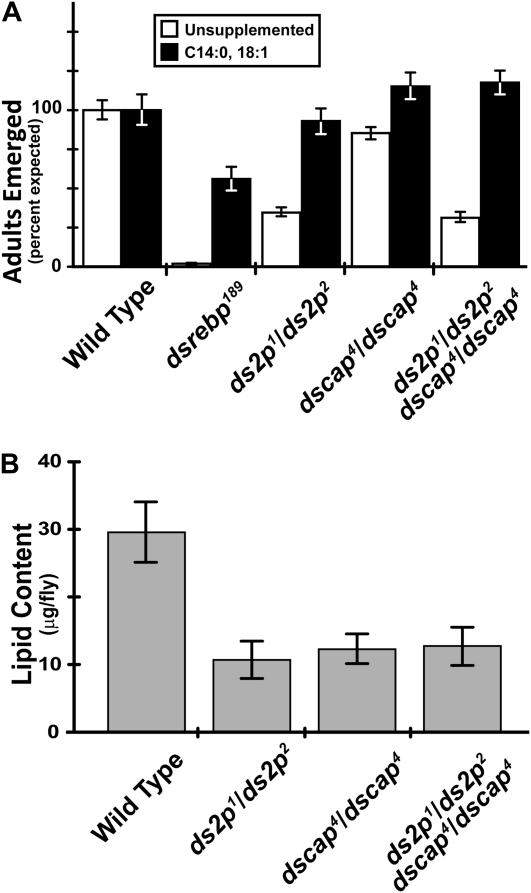
Scap regulates the movement of SREBP from the ER to the Golgi, where SREBP is typically processed by S1P and S2P. Do dscap mutants require dS2P? We prepared doubly mutant animals by recombining the dscap4 allele onto the null-allele ds2p1 and ds2p2 chromosomes. We find that flies lacking both dScap and dS2P are viable and survive about as well as flies lacking only dS2P (Figure 8A, open bars). Like the dscap and ds2p single mutants, survival of the dscap ds2p double mutants is fully restored by supplementation with soy lipids (Figure 8A, solid bars). Examination of the lipid content of adult flies shows that the ds2p, dscap, and dscap ds2p mutants contain markedly less lipid than wild type. The mutants do not differ significantly from one another, however (Figure 8B).
Figure 8.—
Flies doubly mutant for ds2p and dscap exhibit phenotypes similar to ds2p single mutants. Virgin ds2p2/ds2p2 females were crossed to ds2p1/CyO males. Virgin dscap4/dscap4 females were crossed to dscap4/CyO males. For the dscap ds2p double mutants, virgin ds2p2 dscap4/ds2p2 dscap4 females were crossed to ds2p1 dscap4/CyO, actin-GFP males. Heterozgous dsrebp189 males were crossed to females of the same genotype. (A) Rescue of ds2p, dscap double mutants by dietary supplementation. For each experiment, 10 vials were set up at 1 mg embryos/vial on standard medium and an additional 10 vials of each cross were set up on medium supplemented with 0.075% myristate and 0.15% oleate (C14:0 + C18:1). Shown is the mean of three independent experiments. Error bars represent the SEM. A mean of 1415 adults (range, 872–1973) were scored for each genotype and condition. (B) Lipid content of wild-type and mutant flies reared on standard medium. Lipid mass was determined as described in materials and methods. The mean of five independent determinations is shown. Error bars represent the SEM.
dS1P cleaves dSREBP in dscap mutants:
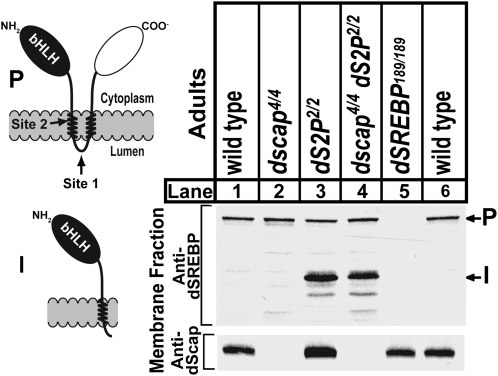
We performed immunoblot analysis on membrane fractions from wild-type adult flies or flies mutant for either dscap, ds2p, or for both (Figure 9). In wild-type flies (lanes 1 and 6), and in dscap4 single mutants (lane 2), only precursor dSREBP is visible. By contrast, in ds2p2 single mutants the intermediate form (I), which is the product of cleavage by S1P, is readily detected (lane 3). In flies doubly mutant for dscap4 and ds2p2 (lane 4), the intermediate form of dSREBP is also detected. Accumulation of the intermediate form in dscap4ds2p2 flies demonstrates that dS1P continues to cleave the precursor in the absence of dScap. In the dscap4 animals, absence of the intermediate form indicates that dSREBP is cleaved and released from their membranes by dS2P.
Figure 9.—
Immunoblot analysis of membrane fractions from adults of the indicated genotypes. The schematic shows SREBP topology. bHLH designates the transcription factor domain. The shaded bar represents the lipid bilayer. Arrows indicate the site 1 protease and site 2 protease cleavage sites. NH2 and COO− designate the amino and carboxy termini. The intermediate form cleavage product (I) is illustrated below. Membrane fractions were prepared as described (materials and methods) and probed with IgG-3B2 against dSREBP. Membranes were then stripped and reprobed with IgG-7A8, against dScap. Sixty micrograms of total protein was loaded per lane. Membranes were exposed for 30 sec. P, precursor; I, intermediate form.
DISCUSSION
Mammalian cells lacking Scap or S2P (or S1P) are auxotrophic for cholesterol and unsaturated fatty acids owing to their failure to activate SREBP (Goldstein et al. 2002). Although mammalian models lacking all forms of SREBP have not been reported, they would presumably also evince lethality (Shimano et al. 1997). The Drosophila genome harbors a single form of SREBP. Null mutants in this gene die at the end of the second instar owing to an insufficiency of fatty acids (Kunte et al. 2006). For mammalian cells that cannot activate SREBP or for dsrebp− mutant flies, survival is restored if the appropriate lipids are added to the culture medium (cholesterol and unsaturated fatty acids in mammalian cells, fatty acids in Drosophila). In contrast to mammalian cells, flies lacking dS2P (Matthews et al. 2009) or dScap (Figure 1) are viable and may be readily maintained as homozygous stocks. Thus, while dsrebp is essential to larvae, components of the classical processing machinery are not.
The present work shows that dScap is dispensable and, in a subset of larval tissues, cleavage of dSREBP continues in its absence (Figures 3 and 4). The tissue-specific pattern of dSREBP activation differs in mutant vs. wild-type animals (Figure 4). This may be explained if different tissues employ different mechanisms to activate dSREBP with only some of the tissues in which dSREBP is normally active (e.g., the fat body) relying on dScap. In the absence of dScap some, but not all, tissues would retain the ability to activate dSREBP. This is what we observe.
The consequences of complete loss of Scap or S2P are not known in whole mammals. Therefore it is also unknown whether any mammalian tissues also utilize alternate means to bypass the classical processing machinery to facilitate normal metabolic responses. It may be that these phenomena are restricted to insects; in mammals, cleavage of SREBPs by caspases has only been observed during apoptosis (e.g., Wang et al. 1996). The fact that cultured mammalian cells require Scap and S2P does not implies that every mammalian cell type has such a requirement. When Drosophila S2 cells are made deficient for dScap via an RNAi strategy, dSREBP is not cleaved. These cells display reduced accumulation of transcripts of dSREBP target genes just as do S2 cells treated with RNAi against dSREBP itself (Seegmiller et al. 2002). Yet the situation in the whole fly is different; some dSREBP continues to be activated in dscap mutants (Figures 3 and 4). These mutants also exhibit deficits in the transcription of dSREBP target genes as compared to wild-type larvae (Figure 5).
dSREBP is one of the transcription factors responsible for the upregulation of transcription of genes involved in fatty acid synthesis (e.g., ACS, ACC, and FAS). Their transcription also depends on other factors in addition to dSREBP. This is indicated by the clear, yet notably reduced accumulation of their transcripts in dsrebp null larvae (Figure 5, bars with light shading). This may be similar to the case in mammalian systems where well-established SREBP targets such as FAS and ACC are also the direct targets of several other transcription factors (Joseph et al. 2002; Iizuka et al. 2004; da Silva Xavier et al. 2006; Matsukuma et al. 2007; Choi et al. 2008). Thus the kinetics of transcript accumulation for ACS, ACC, and FAS are not as simple as seen for CG6295. CG6295 is the gene whose expression is known to be most strongly dependent on dSREBP (Kunte et al. 2006; Matthews et al. 2009). Its expression is consistently very low in dscap larvae but somewhat greater than in dsrebp larvae (Figure 5).
For the ds2p mutants, we have shown that cleavage by the caspase Drice activates dSREBP in larvae and is necessary for their survival (Amarneh et al. 2009). Drice-dependent dSREBP activation in ds2p− larvae is predominately detected in the fat body but little activation is observed in oenocytes (Matthews et al. 2009). This indicates that caspase activation is not significant in these cells. Accordingly, we observe that dSREBP is predominately activated in oenocytes, but significantly reduced in the fat body, of dscap larvae (Figure 4). Caspase activation may, however, explain the variable activation of dSREBP that we observe in the fat body or anterior midgut of dscap larvae. These are tissues where dSREBP continues to be activated by Drice in ds2p mutants. The same process may be active to some extent in dscap larvae as well. However, Drice is dispensable to dscap mutants (Figure 7). Thus Drice cleavage does not explain the survival of the dscap mutants (Figure 7). These data indicate that Drosophila larvae harbor multiple alternative mechanisms that enable activation of dSREBP.
The alternative mechanism at work in the dscap nulls involves cleavage of dSREBP by dS1P and dS2P. In dscap mutants, dS1P continues to cleave dSREBP as indicated by the accumulation of the intermediate form in the membranes of dscap ds2p double mutants (Figure 9, lane 4). The dscap mutants lack the intermediate form (I) in membranes (Figure 9, lane 2). This indicates that in these mutants, the intermediate form is efficiently cleaved by dS2P, just as in wild-type flies (Figure 9, lanes 1 and 6).
In larvae, cleavage of the precursor by S1P and S2P and the accumulation dSREBP and CG6295 transcripts remain responsive to the lipid content of the diet (Figure 6). Owing to differences in feeding behavior, we could not employ precisely parallel nutritional regimens in adults and larvae. Therefore, we devised a starvation/refeeding protocol for adults that exhibits effects similar to lipid supplementation in larvae; in adults, accumulation of nuclear dSREBP and dSREBP transcripts is likewise responsive to the nutritional state (Figure S2, A and C). Some or all of the nutritional responsiveness of dSREBP-mediated transcription in dscap larvae may be due to transcriptional regulation of dsrebp itself rather than cleavage of the precursor by S1P and S2P. In the absence of dScap, flies may no longer be able to regulate the mechanism responsible for bringing dSREBP and dS1P together.
How does membrane-bound dSREBP encounter dS1P and dS2P, which are localized to the Golgi apparatus? One tissue which consistently shows activation of dSREBP in larvae lacking dScap is the oenocytes (Figure 4). Insect oenocytes are thought to be involved in the synthesis and secretion of cuticular hydrocarbons (Demerec 1950). They also exhibit some hepatocyte-like features (Demerec 1950; Gutierrez et al. 2007). They possess a highly elaborated endomembrane system replete with lipid and their ultrastructural morphology is reminiscent of steroidogenic cells in mammals (Locke 1969). Identifying subcellular compartments based on morphology alone is probably insufficient for oenocytes owing to their ultrastructural complexity. Oenocytes might conceivably experience some admixture of ER and Golgi components. This could result in dSREBP being accessible to dS1P without the need for dScap-mediated vesicular transport. Alternatively, in these cells, dSREBP might be packaged into COPII vesicles, perhaps via itself interacting with COPII components. Another possibility is that for dSREBP to move to dS1P (in the Golgi) in the absence of dScap, a different escort factor acting analogously to dScap could be required. The restricted activation of dSREBP in dscap mutants, compared to wild-type larvae, would then be due to tissue-specific expression of this putative escort factor or of specific COPII components interacting with dSREBP (e.g., in oenocytes). We are seeking data that will permit discrimination among these several hypotheses.
The current results, together with those from ds2p mutants (Amarneh et al. 2009; Matthews et al. 2009), show that flies possess at least two alternative means of activating dSREBP. These differ from the classical Scap-S1P-S2P mechanism known from mammals. The first, requiring cleavage of dSREBP by the caspase Drice, explains the survival of flies lacking dS2P. A second novel mode of activation permits dSREBP to be cleaved by dS1P and dS2P in a subset of tissues in the absence of dScap. This alternative activation is sufficient to support the survival of dscap mutants.
Acknowledgments
We are grateful to Indhumathy Subramaniyan, Kaori Tanaka, Denise Parker, Phuong Pham, Praja Lakireddy, Joe Lockridge, and Therese Schindler for excellent technical support; to Jeff Cormier for sequencing and real-time PCR analysis; and to Amit Kunte and Bilal Amarneh for helpful discussions. This work was supported by grants from the National Institutes of Health (R01 GM07145701A1) and the Perot Family Foundation.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.114975/DC1.
References
- Amarneh, B., K. A. Matthews and R. B. Rawson, 2009. Activation of SREBP by the Caspase Drice in Drosophila larvae. J. Biol. Chem. 284 9674–9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhaus, B., E. Sulkowski and F. W. Schlote, 1984. A semi-synthetic, general-purpose medium for Drosophila melanogaster. Drosoph. Inf. Serv. 60 210–212. [Google Scholar]
- Bainbridge, S. P., and M. Bownes, 1981. Staging the metamorphosis of Drosophila melanogaster. J. Embryol. Exp. Morphol. 66 57–80. [PubMed] [Google Scholar]
- Choi, W. I., B. N. Jeon, H. Park, J. Y. Yoo, Y. S. Kim et al., 2008. Proto-oncogene FBI-1 (Pokemon) and SREBP-1 synergistically activate transcription of fatty-acid synthase gene (FASN). J. Biol. Chem. 283 29341–29354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Xavier, G., G. A. Rutter, F. Diraison, C. Andreolas and I. Leclerc, 2006. ChREBP binding to fatty acid synthase and L-type pyruvate kinase genes is stimulated by glucose in pancreatic beta-cells. J. Lipid. Res. 47 2482–2491. [DOI] [PubMed] [Google Scholar]
- Demerec, M. (Editor), 1950. Biology of Drosophila. John Wiley & Sons, New York.
- Dobrosotskaya, I. Y., J. L. Goldstein, M. S. Brown and R. B. Rawson, 2003. Reconstitution of sterol-regulated ER-to-golgi transport of SREBP-2 in insect cells by co-expression of mammalian SCAP and insigs. J. Biol. Chem. 278 35837–35843. [DOI] [PubMed] [Google Scholar]
- Espenshade, P. J., and A. L. Hughes, 2007. Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet. 41 401–427. [DOI] [PubMed] [Google Scholar]
- Espenshade, P. J., W. P. Li and D. Yabe, 2002. Sterols block binding of COPII proteins to SCAP, thereby controlling SCAP sorting in ER. Proc. Natl. Acad. Sci. USA 99 11694–11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FlyBase Consortium, 2003. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 31 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, J. L., R. B. Rawson and M. S. Brown, 2002. Mutant mammalian cells as tools to delineate the sterol regulatory element-binding protein pathway for feedback regulation of lipid synthesis. Arch. Biochem. Biophys. 397 139–148. [DOI] [PubMed] [Google Scholar]
- Golic, K. G., and S. Lindquist, 1989. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59 499–509. [DOI] [PubMed] [Google Scholar]
- Golic, M. M., Y. S. Rong, R. B. Petersen, S. L. Lindquist and K. G. Golic, 1997. FLP-mediated DNA mobilization to specific target sites in Drosophila chromosomes. Nucleic Acids Res. 25 3665–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez, E., D. Wiggins, B. Fielding and A. P. Gould, 2007. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature 445 275–280. [DOI] [PubMed] [Google Scholar]
- Herz, J., R. C. Kowal, Y. K. Ho, M. S. Brown and J. L. Goldstein, 1990. Low density lipoprotein receptor-related protein mediates endocytosis of monoclonal antibodies in cultured cells and rabbit liver. J. Biol. Chem. 265 21355–21362. [PubMed] [Google Scholar]
- Horton, J. D., J. L. Goldstein and M. S. Brown, 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, X., A. Nohturfft, J. L. Goldstein and M. S. Brown, 1996. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell 87 415–426. [DOI] [PubMed] [Google Scholar]
- Iizuka, K., R. K. Bruick, G. Liang, J. D. Horton and K. Uyeda, 2004. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc. Natl. Acad. Sci. USA 101 7281–7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, S. B., B. A. Laffitte, P. H. Patel, M. A. Watson, K. E. Matsukuma et al., 2002. Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J. Biol. Chem. 277 11019–11025. [DOI] [PubMed] [Google Scholar]
- Kunte, A. S., K. A. Matthews and R. B. Rawson, 2006. Fatty acid auxotrophy in Drosophila larvae lacking SREBP. Cell Metab. 3 439–448. [DOI] [PubMed] [Google Scholar]
- Locke, M., 1969. The ultrastructure of the oenocytes in the molti-intermolt cycle of an insect. Tissue Cell 1 103–154. [DOI] [PubMed] [Google Scholar]
- Matsukuma, K. E., L. Wang, M. K. Bennett and T. F. Osborne, 2007. A key role for orphan nuclear receptor liver receptor homologue-1 in activation of fatty acid synthase promoter by liver X receptor. J. Biol. Chem. 282 20164–20171. [DOI] [PubMed] [Google Scholar]
- Matthews, K. A., A. S. Kunte, E. Tambe-Ebot and R. B. Rawson, 2009. Alternative processing of sterol regulatory element binding protein during larval development in Drosophila melanogaster. Genetics 181 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, H. M., C. R. Preston, R. W. Phillis, D. M. Johnson-Schlitz, W. K. Benz et al., 1988. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 118 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski, P. D., 1995. The Flp recombinase of the 2-microns plasmid of Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol. 51 53–91. [PubMed] [Google Scholar]
- Seegmiller, A. C., I. Dobrosotskaya, J. L. Goldstein, Y. K. Ho, M. S. Brown et al., 2002. The SREBP pathway in Drosophila: regulation by palmitate, not sterols. Dev. Cell 2 229–238. [DOI] [PubMed] [Google Scholar]
- Shimano, H., I. Shimomura, R. E. Hammer, J. Herz, J. L. Goldstein et al., 1997. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J. Clin. Invest. 100 2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theopold, U., S. Ekengren and D. Hultmark, 1996. HLH106, a Drosophila transcription factor with similarity to the vertebrate sterol responsive element binding protein. Proc. Natl. Acad. Sci. USA 93 1195–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault, S. T., M. A. Singer, W. Y. Miyazaki, B. Milash, N. A. Dompe et al., 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36 283–287. [DOI] [PubMed] [Google Scholar]
- Wang, X., N. G. Zelenski, J. Yang, J. Sakai, M. S. Brown et al., 1996. Cleavage of sterol regulatory element binding proteins (SREBPs) by CPP32 during apoptosis. EMBO J. 15 1012–1020. [PMC free article] [PubMed] [Google Scholar]