Abstract
In eukaryotes, 40S and 60S ribosomal subunits are assembled in the nucleus from rRNAs and ribosomal proteins, exported as premature complexes, and processed in final maturation steps in the cytoplasm. Ltv1 is a conserved 40S ribosome biogenesis factor that interacts with pre-40S complexes in vivo and is proposed to function in yeast in nuclear export. Cells lacking LTV1 grow slowly and are significantly impaired in mature 40S subunit production. Here we show that mutation or deletion of a putative nuclear export sequence in LTV1 is strongly dominant negative, but the protein does not accumulate in the nucleus, as expected for a mutation affecting export. In fact, most of the mutant protein is cytoplasmic and associated with pre-40S subunits. Cells expressing mutant Ltv1 have a 40S biogenesis defect, accumulate 20S rRNA in the cytoplasm as detected by FISH, and retain the late-acting biogenesis factor Tsr1 in the cytoplasm. Finally, overexpression of mutant Ltv1 is associated with nuclear retention of 40S subunit marker proteins, RpS2–GFP and RpS3–GFP. We suggest that the proximal consequence of these LTV1 mutations is inhibition of the cytoplasmic maturation of 40S subunits and that nuclear retention of pre-40S subunits is a downstream consequence of the failure to release and recycle critical factors back to the nucleus.
RIBOSOME biogenesis is a major biosynthetic activity of eukaryotic cells and a significant rate-limiting factor in cell growth and proliferation. The pathway appears to be conserved in most respects in eukaryotes from yeast to humans and has been extensively analyzed in the model organism, Saccharomyces cerevisiae. Ribosome biogenesis begins cotranscriptionally in the nucleolus, continues in the nucleoplasm, and is completed in the cytoplasm where final maturation events must occur (for reviews, see Venema and Tollervey 1999; Johnson et al. 2002; Fromont-Racine et al. 2003; Tschochner and Hurt 2003; Granneman and Baserga 2004; Zemp and Kutay 2007; Henras et al. 2008; Johnson 2009). More than 150 trans-acting factors have been identified by genetic and proteomic studies with roles in ribosome biogenesis. Generally, these factors are well conserved through eukaryotic evolution, but the specific function of many of the proteins remains largely unknown.
In yeast, the 18S, 5.8S, and 25S ribosomal RNAs (rRNAs) are transcribed as a single 35S precursor rRNA. About 40 ribosome biogenesis factors, U3 snoRNA, and ribosomal proteins assemble cotranscriptionally on the nascent 35S pre-rRNA to form the 90S pre-ribosome (Dragon et al. 2002; Grandi et al. 2002; Schafer et al. 2003). Cleavage of the 35S pre-rRNA at site A2 releases a pre-40S particle containing 20S pre-rRNA, which then sheds most of the processing factors associated with it (Schafer et al. 2003). Pre-60S biogenesis factors and ribosomal proteins then assemble on the remaining transcript. The pre-60S subunit undergoes extensive remodeling and processing in the nucleolus and nucleoplasm, acquiring new processing factors and incorporating the independently transcribed 5S pre-rRNA before being exported through the nuclear pore to the cytoplasm (reviewed in Fromont-Racine et al. 2003; Tschochner and Hurt 2003). The pre-40S and pre-60S subunits are exported independently of one another, with the pre-40S subunits leaving the nucleus more rapidly than the pre-60S subunits. Both precursor subunits exit the nucleus accompanied by a small contingent of biogenesis factors that are presumed to function either in export or in subsequent cytoplasmic maturation steps. These biogenesis factors, acquired in the nucleus, are released in the cytoplasm and recycled to the nucleus (reviewed in Zemp and Kutay 2007; Johnson 2009).
Nuclear export of both the large and the small ribosomal subunits requires Crm1, the major export karyopherin in yeast, as well as a functional RanGTPase system (Hurt et al. 1999; Moy and Silver 1999; Ho et al. 2000; Gadal et al. 2001; Gleizes et al. 2001; Thomas and Kutay 2003; Trotta et al. 2003). Crm1 exports a broad range of substrates, which contain a leucine-rich nuclear export sequence (NES), a short motif with a loosely conserved pattern of three or four hydrophobic residues (Wen et al. 1995; Fornerod and Ohno 2002; La Cour et al. 2004). Crm1-dependent export of the 60S subunit requires Nmd3, a conserved NES-containing adapter protein (Ho et al. 2000; Gadal et al. 2001; Trotta et al. 2003) that binds Crm1 directly in a Ran–GTP-dependent manner (Thomas and Kutay 2003) and is thought to recruit Crm1 to the pre-60S subunit in the nucleoplasm. For the 40S small subunit, two different yeast nonribosomal proteins, Dim2 and Ltv1, have been proposed to function as adapters for Crm1-mediated 40S export in yeast (Seiser et al. 2006; Vanrobays et al. 2008). Both are late-acting 40S biogenesis factors that shuttle between the nucleus and the cytoplasm. In humans, hRio2 was recently shown to have an NES-like sequence in its C terminus, deletion of which significantly reduced the rate of pre-40S export (Zemp et al. 2009).
Both the pre-40S and the pre-60S subunits undergo final maturation steps in the cytoplasm. For the 60S, this involves a series of ATPase- and GTPase-dependent steps that release late biogenesis and export factors and incorporate cytoplasmic ribosomal proteins necessary for ribosome function (reviewed in Fromont-Racine et al. 2003; Zemp and Kutay 2007; Johnson 2009). For the 40S, final maturation steps include dimethylation of two adenine bases in the 20S pre-rRNA by the 40S biogenesis factor Dim1 (Brand et al. 1977; Lafontaine et al. 1995; Lafontaine et al. 1998) and cleavage of the 20S at site D to produce the mature dimethylated 18S rRNA (Udem and Warner 1973). Cleavage of the 20S rRNA releases a fragment of internal transcribed spacer 1 (ITS1) rRNA, which is then rapidly degraded by the cytoplasmic exonuclease, Xrn1 (Stevens et al. 1991). Two different factors, Nob1 and Fap7, have been proposed to act as the endonuclease responsible for cleaving the 20S pre-rRNA (Fatica et al. 2003; Granneman et al. 2005). Nob1 contains a PiIT N-terminus (PIN) domain, a known nuclease motif, mutation of which blocks 20S processing (Fatica et al. 2003, 2004). Other 40S biogenesis factors required for 20S rRNA cleavage include Rio1, Rio2, Tsr1, and Tsr2 (Gelperin et al. 2001; Vanrobays et al. 2003; Leger-Silvestre et al. 2004). Nob1, Rio2, and Tsr1 were identified, along with Ltv1, Enp1, Hrr25, and Dim2, as components of isolated late pre-40S particles (Schafer et al. 2003). The exact role of these late biogenesis factors in export and/or maturation is for the most part still unclear.
Ltv1 is a conserved, nonessential protein with a role in 40S subunit biogenesis. Yeast cells lacking LTV1 (Δltv1) display a slow-growth phenotype, are hypersensitive to protein synthesis inhibitors, and contain less than half the normal number of 40S subunits (Loar et al. 2004). Ltv1 is a nuclear-cytoplasmic shuttle protein whose export is Crm1 dependent (Seiser et al. 2006) and is a late associating pre-40S factor believed to leave the nucleus in association with the subunit (Schafer et al. 2003). The human homolog of yeast Ltv1, hLtv1, was recently shown to be a component of human late pre-40S particles (Zemp et al. 2009). Consistent with a role in 40S export, two different small subunit ribosomal proteins that assemble into pre-40S subunits in the nucleus are retained in the nucleus in Δltv1 cells relative to wild-type cells. (Seiser et al. 2006). Ltv1 also interacts with Crm1 in a yeast two-hybrid assay (Ito et al. 2001; Neuber et al. 2008; our unpublished results).
We show here that mutation or deletion of a putative NES in Ltv1 is dominant negative (DN), inhibiting the growth of wild-type cells even more than deletion of the gene. Expression of the mutant protein results in a significant 40S biogenesis defect: the number of free 40S subunits is reduced while the number of free 60S subunits is substantially increased. However, contrary to expectations, neither mutation nor deletion of this sequence causes the mutant protein to accumulate in the nucleus. In fact, most of the mutant Ltv1 protein is cytoplasmic and found associated with pre-40S subunits. Cells expressing mutant Ltv1 protein exhibit a striking redistribution of 20S rRNA from the nucleolus to the cytoplasm, indicative of a defect in the cytoplasmic maturation of the pre-40S. In addition, overexpression of mutant Ltv1 is associated with retention of Tsr1 in the cytoplasm. Finally, overexpression of mutant Ltv1 is associated with the nuclear retention of both RpS2–GFP and RpS3–GFP, small subunit ribosomal proteins used as markers for the localization of 40S subunits. We suggest that the nuclear retention of pre-40S subunits is a downstream consequence of the cytoplasmic maturation defects, as failure to release factors required for export from cytoplasmic subunits would lead to their depletion from the nucleus and thus the accumulation of export-incompetent subunits in the nucleus.
MATERIALS AND METHODS
Yeast strains and culture conditions:
Yeast strains used are listed in Table 1. Yeast were grown at 30° in standard YPD (1% yeast extract, 2% bactopeptone, 2% glucose) unless otherwise noted. Strains transformed with expression plasmids using the MET25 promoter were grown in media containing low methionine (134 μm) to derepress the promoter. MNY8 cells used for microscopy were grown in media supplemented with adenine (final concentration of 0.1 mg/ml) to prevent autofluorescence due to the ade2-1 mutation in the strain background. Yeast transformations were performed using the lithium acetate method (Gietz et al. 1992).
TABLE 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| LY134 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Open Biosystems |
| LY136 | MATα his3Δ1 leu2Δ0 ura3Δ0 ltv1Δ∷KanMX | Open Biosystems |
| LY177 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 SIKI-RFP∷G418 | J. Falvo |
| LY247 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 TSR1–GFP∷HIS3MX | A. Johnson |
| LY248 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ENP1–GFP∷HIS3MX | A. Johnson |
| LY251 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 CRM1(T539C-HA) | A. Johnson |
| RKY1977 | MATaade2 ade3 leu2 lys2-801 ura3-52 xrn1Δ | A. Johnson |
| CH1305 | MATaade2 ade3 leu2 lys2-801 ura3-52 | A. Johnson |
| MNY8 | MATaleu2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 crm1Δ∷kanR pDC-CRM1(T539C) (LEU2/CEN) | M. Rosbash |
Construction of plasmids:
Plasmids used are listed in Table 2. Ld57 (PMET–ltv1Δ169-176–GFP) was constructed by QuikChange II site-directed mutagenesis (Stratagene) using Ld47 as template and primer: 5′-CCTGATATGAATCCTGCAGAAGATGAAGCATATGTTG and its complement to generate the 8-amino-acid deletion. Ld58 (PMET–ltv13L-3A–GFP) was generated using Ld57 as template and the primer: 5′-CCTGATATGAATCCTGCAGCAAGAGAGGTGGCTGAAGCCGCAG AAGATGAAGCATATGTTG, and its complement. The presence of the three leucine to alanine mutations as well as the 169–176 deletion were confirmed by DNA sequencing. Ld73, 65, and 74 were constructed by PCR amplification of the LTV1 ORF using Ld47, 57, and 58, respectively, as the template and using gene specific primers for LTV1 that added BamHI and Sal1 sites to the ORF. The PCR products were cleaved with BamHI and SalI and ligated into the multiple cloning site of p415GAL1. The presence of the three leucine to alanine mutations and the 169–176 deletion were confirmed by DNA sequencing.
TABLE 2.
Plasmids
| Plasmid | Relevant markers | References |
|---|---|---|
| Ld104 | pRS316–RIO2–GFP (URA3–CEN) | Schafer et al. (2003) |
| Ld140 | pRS316–RPS2–GFP (URA3–CEN) | Milkereit et al. (2003) |
| Ld70 | PRPS3–RPS3–GFP (URA3–CEN) | This study |
| Ld47 | PMET–LTV1–GFP (HIS3–CEN) | Seiser et al. (2006) |
| Ld57 | PMET–ltv1Δ169-176 GFP (HIS3–CEN) | This study |
| Ld58 | PMET–ltv13L->3AGFP (HIS3–CEN) | This study |
| Ld73 | PGAL–LTV1 (LEU2–CEN) | This study |
| Ld65 | PGAL–ltv1Δ169-176 (LEU2–CEN) | This study |
| Ld74 | PGAL–ltv13L->3A (LEU2–CEN) | This study |
| pUG23 | PMET–GFP | J. Hegemann, Düsseldorf, Germany (personal communication) |
Fluorescence microscopy of living cells:
Cells were harvested by centrifugation, washed three times with sterile water, and resuspended in 50 μl sterile water. Cells were mounted on poly-l-lysine-coated microscope slides and viewed with the ×100 objective of a Zeiss Axioskop epifluorescence microscope using FITC fluorescence filters. Images were collected with a B/W SPOT CCD camera and post-processed using Photoshop (Adobe). Cells treated with leptomycin B (LMB) were incubated at room temperature in 100 ng/μl LMB (dissolved in methanol to prevent ethanol stress) at room temperature for 20–30 min prior to viewing.
In situ hybridization:
Wild-type yeast strain (LY134) transformed with the indicated plasmids was grown overnight at 30° in synthetic complete media minus leucine, then diluted 50-fold into fresh medium containing either 2% glucose or 2% galactose. After 16 hr of additional incubation, cells were fixed in 4% formaldehyde for 30 min and then harvested and washed in buffer containing 0.1 m KPO4 (pH 7.5) and 1.2 m sorbitol. Spheroplast preparation and in situ hybridization of a Cy3-labeled 5′-ITS1 oligonucleotide probe were conducted as previously described (Amberg et al. 1992; Moy and Silver 2002; Seiser et al. 2006). Individual cells were viewed under the ×100 objective of an Olympus BX51 epifluorescence microscope using TRITC fluorescence filters. Images were collected with a Qcolor camera system (Olympus) and post-processed using Photoshop (Adobe).
Polysome analysis:
Wild-type yeast (LY134) containing empty vector (pUG23), PMET–LTV1–GFP (Ld47) or PMET–ltv1Δ169-176–GFP (Ld57) was cultured in synthetic complete media minus histidine. At OD600 0.4–0.5, cycloheximide was added to 100 μg/ml and cells were harvested. Extracts were prepared in 20 mm Tris–HCl (pH 7.5), 20 mm KCl, 5 mm MgCl2, 12 mm β-mercaptoethanol, 100 μg/ml cycloheximide, 1 mm PMSF, 1 μm leupeptin, 1 μm pepstatin by vortexing with glass beads. Nine A260 units were layered onto 7–47% sucrose density gradients in extraction buffer and centrifuged for 3 hr at 36,000 rpm at 4° in a SW41 rotor. Gradients were fractionated with continuous monitoring of absorbance at 254 nm. Fractions were precipitated with 1.7 vol of ethanol at −20° overnight. Samples were resuspended in 1× Laemmli sample buffer and separated on 12% SDS–polyacrylamide gels. The separated proteins were transferred to nitrocellulose membranes and Western blots were carried out using anti-GFP–HRP (Rockland) or anti-RPS3 (M. Seedorf) antibodies.
Northern analysis:
Northern blotting was performed as described previously (Kallstrom et al. 2003). RNAs were separated on a 1.5% agarose–formaldehyge gel, transferred to nylon membrane, and probed with an oligonucleotide specific to 5′-ITS1 (5′−TCTTGCCCAGTAAAAGCTCTCATGC) and SCR1.
RESULTS
Overexpression of mutant Ltv1 is dominant negative and affects 40S export:
We previously suggested that Ltv1 might function as a nonessential adapter protein to facilitate Crm1-mediated export of the small ribosomal subunit in yeast. Both Ltv1 (Seiser et al. 2006) and pre-40S subunits (Moy and Silver 2002; Trotta et al. 2003) accumulate in the nucleus upon inhibition of Crm1, and cells lacking Ltv1 (Δltv1) produce half the number of 40S subunits as wild-type cells (Loar et al. 2004) and retain pre-40S subunits in the nucleus, as measured by RpS3–GFP nuclear retention (Seiser et al. 2006). To test this adapter hypothesis, we mutated a sequence in LTV1, corresponding to residues 169–178, that closely resembles but does not perfectly match the consensus for a leucine-rich NES (Figure 1A). If Ltv1 acts as an adapter and recruits Crm1 via this sequence, mutational inactivation or deletion of the sequence should reduce export of both Ltv1 and pre-40S subunits. Furthermore, by analogy to NES-deficient mutants of the 60S export adapter Nmd3, overexpression of an Ltv1 protein lacking its Crm1-interaction site should be dominant negative; loading of an NES-deficient Ltv1 protein onto pre-40S subunits should prevent recruitment of Crm1 to the subunits, and should thus trap pre-40S subunits in the nucleus as export-incompetent complexes (Belk et al. 1999; Ho et al. 2000).
Figure 1.—
Mutant Ltv1 protein is dominant negative. (A) Sequence of Ltv1 showing NES consensus and mutations. Φ indicates Leu, Ile, Val, Met, or Phe; x indicates any amino acid. Leucine to alanine substitutions are underlined; deleted residues are indicated by an extended line. (B) Serial dilutions (4×) of exponentially growing cultures of strain LY134 (wild type) containing empty vector (pUG23), Ld57 (PMET–ltv1Δ169-176–GFP), or Ld58 (PMET–ltv13L→3A–GFP) were spotted onto Sc–Leu (134 μm methionine) plates and incubated at 30° for 40 hr. (C) Serial dilutions (4×) of cultures of LY134 transformed with Ld73 (PGAL–LTV1), Ld65 (PGAL–ltv1Δ169-176), or Ld74 (PGAL–ltv13L→3A). All cultures were grown into log phase in synthetic complete media lacking leucine (Sc–Leu) with 2% glucose and then spotted onto Sc–Leu with glucose or Sc–Leu with galactose plates and incubated at 30° for 30 hr. (D) Serial dilutions (4×) of exponentially growing cultures of LY136 (Δltv1) transformed with empty vector (pRS415-Gal), Ld65 (PGAL–ltv1Δ169-176), or Ld74 (PGAL–ltv13L→3A). Cells were grown as in C and incubated at 30° for 40 hr.
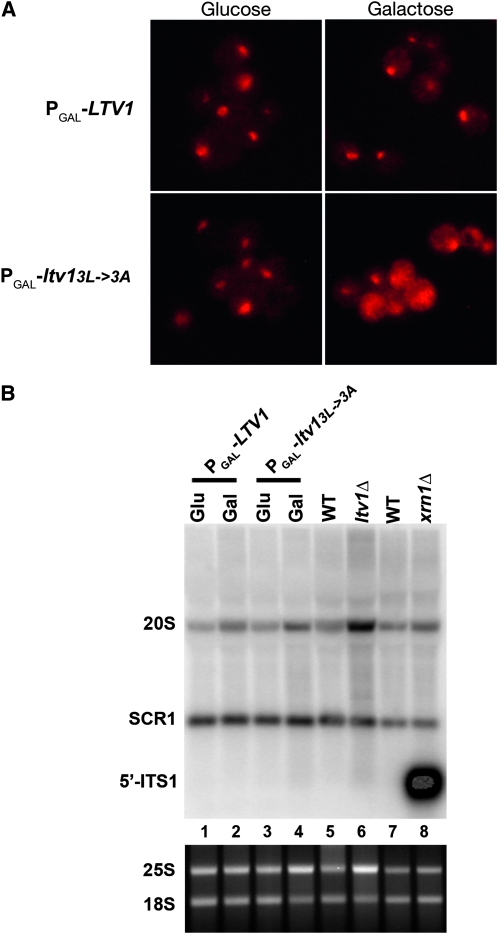
We generated two different LTV1 mutations: deletion of residues 169–176 (ltv1Δ169-176) or mutation of Leu169, 173, and 176 to alanine (ltv13L-3A)(Figure 1A). We then tested whether expression of either mutant protein was dominant negative. As shown in Figure 1B, expression of either ltv13L-3A or ltv1Δ169-176 as GFP fusion proteins in wild-type cells reduced their growth rate relative to cells transformed with an empty vector. To explore this further, we removed the GFP tag and expressed both mutant proteins under the control of the galactose-inducible Gal1 promoter. Overexpression of untagged mutant proteins had a pronounced negative effect on growth (Figure 1C). Thus both mutant proteins are dominant negative, and the toxicity of both proteins is dose dependent. Finally, we found that overexpression of mutant Ltv1 also reduced the growth rate of Δltv1 cells (Figure 1D), which indicates that toxicity of the Ltv1 mutant proteins is not due simply to inactivation of the wild-type protein.
We then asked if mutation or deletion of amino acids 169–176 of Ltv1 blocked its nuclear export. Cells expressing a wild-type GFP-tagged Ltv1 produce a fluorescent signal that is predominantly cytoplasmic, but with fluorescence over both the cytoplasm and the nucleus at steady state (Seiser et al. 2006) (Figure 2). In cells expressing either of the DN mutant Ltv1–GFP proteins, GFP fluorescence remained predominantly cytoplasmic, with no cells exhibiting the nuclear accumulation phenotype characteristic of export-defective proteins (Figure 2). The failure of mutant Ltv1 to accumulate in the nucleus is not due to a defect in nuclear import, as inhibition of Crm1 with leptomycin B (LMB) leads to the nuclear accumulation of the mutant Ltv1–GFP proteins (data not shown). These data suggest that the identified sequence does not function as an NES for Ltv1, as mutation of it does not block Ltv1 export.
Figure 2.—
Mutant Ltv1 protein does not accumulate in the nucleus. Cells expressing nucleolar protein Sik1–RFP (LY177) were transformed with Ld47 (PMET–LTV1–GFP), Ld57 (PMET–ltv1Δ169-176–GFP or Ld58 (PMET–ltv13L→3A–GFP) and grown in synthetic complete media lacking histidine (134 μm methionine) into log phase (OD660 = 0.4). Cells were imaged under both FITC and rhodamine channels. Image level was adjusted and pseudo-colored in Adobe Photoshop CS4.
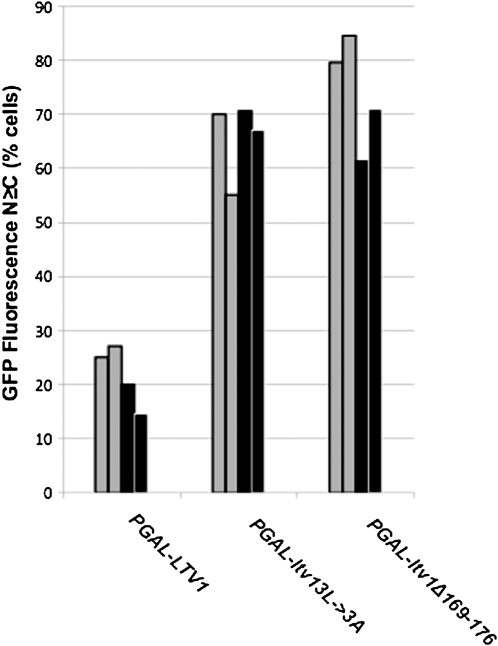
We also tested whether overexpression of either mutant Ltv1 protein impaired pre-40S subunit nuclear export using fluorescence microscopy. GFP-tagged versions of small subunit ribosomal proteins, RpS2 (Milkereit et al. 2003) and RpS3 (Seiser et al. 2006), have both been shown to be functional and incorporated efficiently into pre-40S subunits. In rapidly growing wild-type yeast cells, the fluorescent signal from GFP-tagged Rps2 and Rps3 is almost exclusively cytoplasmic, with most (∼80%) cells exhibiting no signal over the nucleus (Stage-Zimmermann et al. 2000; Milkereit et al. 2003; Schafer et al. 2003; Seiser et al. 2006). This distribution is thought to reflect the abundance and stability of ribosomal subunits in the cytoplasm and the very rapid nuclear export of pre-40S subunits in exponentially growing yeast. If the mutant Ltv1 proteins trapped pre-40S subunits in the nucleus as export-incompetent complexes, then overexpression of the mutants should lead to an increase in the number of cells exhibiting GFP fluorescence in the nucleus. Interestingly, despite the lack of nuclear accumulation of mutant Ltv1 itself, cells overexpressing either of the DN Ltv1 proteins showed a marked change in the distribution of both Rps2–GFP and Rps3–GFP (Figure 3). While only 20% of cells overexpressing wild-type Ltv1 have a nuclear GFP signal that is equal or greater than the cytoplasmic signal (N ≥ C), 60–80% of cells expressing mutant Ltv1 have a nuclear GFP signal, using either Rps2–GFP or Rps3–GFP (Figure 3). Altogether these data suggest that, while residues 169–176 are not required for export of Ltv1 itself, overexpression of Ltv1 proteins with this sequence mutated is dominant negative for growth and appears to reduce pre-40S export (probably through an indirect mechanism; see below).
Figure 3.—
Overexpression of dominant mutant Ltv1 reduces both RpS2 and RpS3 export. LY134 (wild-type) cells were cotransformed with either pRS316–RPS2–eGFP (shaded) or PRPS3–RPS3–GFP (solid) and Ld73 (PGAL–LTV1), Ld65 (PGAL–ltv1Δ169-176) or Ld74 (PGAL–ltv13L→3A). Each transformation was performed twice, and cells from separate transformations were scored independently. In both experiments, all cultures were grown to stationary phase in media containing 2% glucose and then diluted into selective media with 2% galactose and incubated overnight into early log phase (OD660 of 0.1–0.2). Live cells on poly-l-lysine-coated slides were imaged using FITC filters. Cells (n > 30) were scored for the localization of GFP fluorescence primarily to the nucleus (N > C), primarily to the cytoplasm (N < C), or equally distributed in both (N = C), and the percentage of total cells with phenotypes N > C and N = C are represented on the y-axis.
Cells overexpressing mutant Ltv1p have a 40S ribosome biogenesis defect:
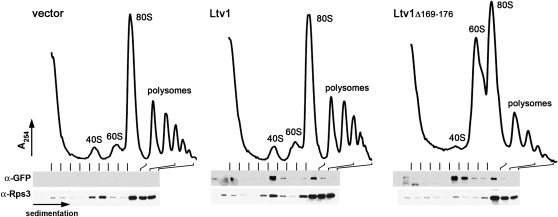
Overexpression of mutant Ltv1 protein could dominantly impair cell growth via any number of mechanisms unrelated to the function of the wild-type protein. To clarify whether the mutant protein is affecting the same pathway as the wild-type protein, we examined polysome profiles from wild-type cells transformed with vectors expressing either wild-type or DN mutant Ltv1 fused to GFP. Cells expressing wild-type Ltv1–GFP have profiles that look identical to cells expressing GFP only (Figure 4, vector). However, expression of either mutant protein results in a significant reduction in the peak of free 40S subunits and a very marked accumulation of free 60S subunits. These changes are similar to those observed in Δltv1 strains (Loar et al. 2004) and other viable 40S biogenesis mutants (Li et al. 2009) and indicate that the DN mutations in Ltv1 affect 40S biogenesis.
Figure 4.—
Dominant-negative mutant Ltv1 is incorporated into subunits and causes a 40S deficit. LY134 (wild-type) cells were cotransformed with empty vector (pUG23), Ld47 (PMET–LTV1–GFP), or Ld57 (PMET–ltv1Δ169-176–GFP). Extracts were prepared and sedimented through sucrose density gradients as described in materials and methods. The presence of wild-type or mutant Ltv1–GFP and Rps3 in individual fractions was determined by Western blotting using anti-GFP and anti-Rps3 antibodies, respectively. Fractions containing 80S and deeper fractions were combined as indicated.
Mutant Ltv1 is efficiently incorporated into 40S subunits:
Ltv1 is a component of late pre-40S complexes (Schafer et al. 2003; Leger-Silvestre et al. 2004) that is apparently released in the cytoplasm and recycled to the nucleus (Seiser et al. 2006). To investigate the mechanism by which the DN mutations in LTV1 interfere with productive 40S subunit biogenesis, we asked whether the mutant protein associates with pre-40S subunits. We performed Western blot analysis of the polysome profiles shown in Figure 4 using antibodies to the small subunit protein RpS3 to identify fractions containing pre-40S subunits (pre-40S subunits co-migrate with mature 40S subunits in these gradients) and anti-GFP antibodies to identify both the wild-type and mutant Ltv1–GFP protein. Cells transformed with the GFP vector do not contain any GFP-reacting protein beyond the uppermost fractions of the gradient. In contrast, we detect a GFP-tagged protein at the position of the pre-40S/40S peak in extracts from cells expressing either the wild-type Ltv1–GFP or the mutant Ltv1Δ169-176–GFP (Figure 4). Expression of mutant Ltv1Δ169-176–GFP leads to a reduced 40S peak and a corresponding reduction in RpS3 signal at that position. Similar results were obtained with the mutant Ltv13L-3A–GFP (data not shown). This result suggests that the mutant protein is readily incorporated into pre-40S subunits and thus likely exerts its effects on 40S ribosome biogenesis in the context of the pre-ribosome.
Overexpression of mutant Ltv1 results in the cytoplasmic accumulation of 20S RNA:
The decrease in 40S subunits in cells overexpressing mutant Ltv1 protein might result from turnover of subunits defective in assembly and export or from defects in cytoplasmic maturation. The predominantly cytoplasmic localization of the DN mutant Ltv1 proteins suggested to us that they likely exert their effect in the cytoplasm. The major cytoplasmic maturation event for the 40S subunit is cleavage of 20S pre-rRNA at site D to generate the mature 18S rRNA. This cleavage releases the ITS1 D-A2 fragment, which is degraded by the cytoplasmic exonuclease Xrn1. Fluorescent in situ hybridization (FISH) with wild-type cells using the ITS1 D-A2 fragment produces a predominantly nucleolar signal, reflecting both the abundance of precursor rRNAs containing this sequence in the nucleolus and the rapid cleavage and degradation of the ITS1 fragment in the cytoplasm (Leger-Silvestre et al. 2004; Seiser et al. 2006). In mutants blocked in the cytoplasmic cleavage of the 20S pre-rRNA or ITS1 degradation, the ITS1 signal becomes predominantly cytoplasmic (Moy and Silver 1999; Leger-Silvestre et al. 2004). We used FISH with the ITS1 D-A2 probe to detect pre-rRNAs containing this sequence in cells overexpressing wild-type or mutant Ltv1. Overexpression of wild-type Ltv1 did not affect the predominantly nucleolar localization of the ITS1 signal observed in wild-type cells (Figure 5A). However, overexpression of mutant Ltv1 protein was associated with a pronounced redistribution of the ITS1 FISH signal from the nucleolus to the cytoplasm (Figure 5A). To clarify the nature of the pre-rRNA hybridizing to this probe, we used Northern blotting of total RNA from cells expressing either the wild-type or mutant Ltv1 protein (Figure 5B). We did not detect any free ITS1 RNA, indicating that mutant Ltv1 is not interfering with Xrn1 function. The fact that the total 20S rRNA detected in cells overexpressing mutant Ltv1 is not increased indicates that the strong cytoplasmic signal we observe by FISH in Figure 5A must result from a redistribution of 20S rRNA from the nucleolus to the cytoplasm. Together, these data suggest that cells overexpressing mutant Ltv1 are able to export pre-40S particles containing 20S rRNA from the nucleus, but once in the cytoplasm, final maturation of the 20S is impeded.
Figure 5.—
Overexpression of dominant mutant Ltv1p results in a redistribution of 20S RNA to the cytoplasm. (A) FISH detection of 20S. Wild-type cells (LY134) were transformed with plasmids Ld73 (PGAL–LTV1) or Ld74 (PGAL–ltv13L→3A). Cultures were grown to stationary phase in selective media containing 2% glucose, diluted into fresh media with 2% glucose or 2% galactose, and then grown overnight before harvesting and fixation. A Cy3-labeled DNA probe complementary to the 5′-ITS1 sequence, an indicator for 20S rRNA, was hybridized with permeabilized cells prior to imaging using TRITC filters. (B) Northern blotting for 20S pre-rRNA. Wild-type (LY134) cells containing Ld73 (PGAL–LTV1) or Ld74 (PGAL–ltv13L→3A) were cultured in the presence of glucose (Glu) or galactose (Gal) to induce expression of the plasmid borne alleles of LTV1. Total RNA was prepared from these cultures as well as from ltv1Δ (LY136) and its isogenic wild-type (LY134) and xrn1Δ (RDKY1977) and its isogenic wild-type CH1305. Top: 10 μg of total RNA was separated on a 1.5% agarose–formaldehyde gel. Northern blot was hybridized with an oligonucleotide probe specific for the 5′-end of ITS1 that detects 20S pre-rRNA and the free ITS1 fragment post cleavage. A probe specific for SCR1 was used as an internal control. Bottom: RNA samples loaded in upper panel (1 μg each) were separated on 1% agarose gel and visualized with ethidium bromide.
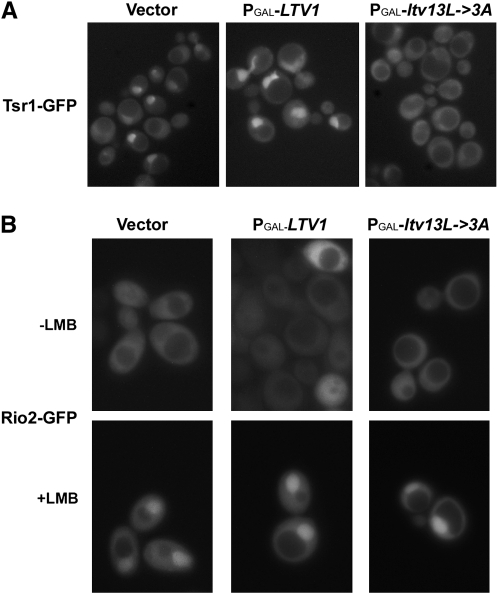
Mutant Ltv1 impedes release of the late 40S biogenesis factor Tsr1:
The mechanisms by which late-acting 40S biogenesis factors are released from the subunit during its maturation in the cytoplasm are not well understood. However, one can predict that a defect in release might manifest as a defect in export, since unreleased factors would accumulate in the cytoplasm and thus fail to recycle to the nucleus. Depletion of a factor from the nucleus that is necessary for subunit export would then lead to the accumulation of export-incompetent pre-40S subunits in the nucleus. To determine whether overexpression of DN mutant Ltv1 affects the release of late 40S biogenesis factors, we used fluorescence microscopy to analyze the cellular locations of GFP-tagged Tsr1, Enp1, and Rio2 following overexpression of mutant Ltv13L-3A protein. Tsr1 and Enp1 are essential ribosome biogenesis factors that associate with the pre-40S, early in the case of Enp1 and later for Tsr1 (Chen et al. 2003; Schafer et al. 2003). Both proteins can be detected in both the nucleus and cytoplasm, but are predominantly nuclear in exponentially growing cells (Schafer et al. 2003; Leger-Silvestre et al. 2004). In cells expressing no excess Ltv1 (vector) or wild-type Ltv1, we observe a Tsr1–GFP signal that is predominantly nuclear with a slight cytoplasmic signal (Figure 6A), consistent with previously published data (Schafer et al. 2003). In contrast, overexpression of mutant Ltv13L-3A was associated with a distinct loss of the nuclear signal and an increase in the cytoplasmic signal of Tsr1–GFP (Figure 6A). In Enp1–GFP-expressing cells, the GFP signal was not significantly altered in cells overexpressing Ltv13L-3A (data not shown). Cytoplasmic retention of Rio2 cannot be assayed in the same way because Rio2, unlike Tsr1 and Enp1, is already predominantly cytoplasmic in exponentially growing wild-type cells. However, if mutant Ltv13L-3A prevents the cytoplasmic release of Rio2 from 40S subunits, then overexpression of this mutant should prevent nuclear accumulation of Rio2–GFP following treatment of cells with the Crm1 inhibitor, LMB, since 40S subunit export is Crm1 dependent. As shown in Figure 6B, Rio2–GFP is predominantly cytoplasmic in cells expressing either wild-type or mutant Ltv13L-3A protein and accumulates strongly in the nucleus following treatment with LMB. These data indicate that overexpression of mutant Ltv13L-3A protein does not affect either Enp1 or Rio2 localization, but does result in the cytoplasmic retention of Tsr1, suggesting that Tsr1 release from cytoplasmic pre-40S subunits is blocked. This result was somewhat unexpected as Ltv1 has been shown to form a stable subcomplex with Enp1 and Rps3 (Schafer et al. 2006), but not Tsr1.
Figure 6.—
Overexpression of mutant ltv1 affects nuclear recycling of late 40S biogenesis factor Tsr1. (A) Cells expressing Tsr1–GFP (LY247) were transformed with vector (p415Gal), Ld73 (PGAL–LTV1), or Ld74 (PGAL–ltv13L→3A). All cultures were grown at 30° to log phase in selective media with 2% glucose and then diluted into selective media with 2% galactose and regrown for 6 hr before harvesting and imaging. (B) A leptomycin B (LMB)-sensitive strain (LY251) was cotransformed with pRS316Rio2–GFP and vector only (p415Gal), Ld73 (PGAL–LTV1), or Ld74 (PGAL–ltv13L→3A). Cells were grown in selective media with glucose and then diluted into selective media with galactose and grown overnight (16 hr) to log phase. Cells treated with LMB (+LMB) were incubated with 100 ng/μl LMB at room temperature for 20 min prior to viewing.
DISCUSSION
We show here that specific mutations in LTV1 impede 40S cytoplasmic maturation. Mutation or deletion of a short hydrophobic sequence in Ltv1 is dominant negative and causes 40S-specific ribosome biogenesis defects. The mutant protein is incorporated into subunits, which are exported from the nucleus, but apparently blocked at cleavage of the 20S rRNA to release ITS1. Expression of the mutant protein results in the abnormal cytoplasmic retention of the biogenesis factor Tsr1, suggesting that the release of Tsr1 from the pre-40S is also blocked. These results indicate that specific mutations in Ltv1 interfere with cytoplasmic maturation of the 40S ribosomal subunit.
The role of Ltv1 in export:
Export of the small subunit requires Crm1, and we suggested previously that Ltv1 might function in export as a Crm1 adapter in the nucleus, binding both RpS3 on the pre-40S subunit and Crm1 (Seiser et al. 2006). Here we show that the sequence previously identified as a putative NES is not in fact necessary for the nuclear export of Ltv1, since neither deletion nor mutation of this sequence leads to nuclear accumulation of Ltv1–GFP. A possible explanation for the lack of nuclear accumulation of mutant Ltv1–GFP is that it is exported as part of the pre-40S via other Crm1 adapter proteins that are functionally redundant to Ltv1. However, we think a more likely interpretation is that the sequence we have identified does not function as an NES. This interpretation implies that there is either another NES sequence in Ltv1 or that Ltv1 does not function directly in export. We favor the former of these explanations, however, since Ltv1 has twice been reported to interact with Crm1 in a two-hybrid assay, once in a genome-wide two-hybrid screen (Ito et al. 2001) and more recently in a directed screen for Crm1-interacting proteins using Crm1 as the bait (Neuber et al. 2008).
The role of Ltv1 in 40S maturation:
Cytoplasmic cleavage of the 20S pre-rRNA at site D is a critical 40S maturation event as this cleavage generates the mature 18S rRNA and functional 40S subunits. A large number of factors are required for this step, most of which must be involved in regulating the cleavage as only two, Nob1 and Fap7, are actually implicated in the cleavage step itself. Depletion of Rio1, Rio2, Tsr1, or Tsr2 all lead to 20S accumulation (Gelperin et al. 2001; Vanrobays et al. 2001, 2003; Geerlings et al. 2003; Peng et al. 2003; Leger-Silvestre et al. 2004). In the case of Rio2 and Tsr1, depletion results in the cytoplasmic accumulation of 20S as measured by FISH (Vanrobays et al. 2003; Leger-Silvestre et al. 2004), indicating that yeast Rio2 and Tsr1 function not in pre-40S export but in the cytoplasmic maturation of the 20SrRNA. Tsr1 is related to the GTP-binding protein, Bms1 (Gelperin et al. 2001), and Rio2 is an ATP-dependent kinase (Vanrobays et al. 2003). We show here that expression of a mutant Ltv1 protein leads to a cytoplasmic redistribution of 20S rRNA signal. In the case of Ltv1, however, it is not depletion of the protein that leads to this phenotype, but expression of a mutated protein, which we show is incorporated into pre-40S subunits and blocks 20S cleavage. Pre-20S rRNA processing could be blocked because the mutated region of Ltv1 fails to recruit essential processing factors. However, if this were true, deletion of the gene should give the same phenotype, which is not the case (Seiser et al. 2006). Alternatively, the mutant protein might fail to release some factor(s) or fail to be released itself, either of which might be necessary to create or expose the target cleavage site. Another possibility is that the mutant Ltv1 protein blocks some conformational transition in the pre-40S that is required for 20S rRNA cleavage. Hrr25-dependent phosphorylation of a trimeric complex consisting of Ltv1, Enp1, and RpS3 has been proposed to create a more flexible beak structure in the pre-40S, perhaps facilitating nuclear export in vivo (Schafer et al. 2006). A subsequent dephosphorylation step is necessary for RpS3 to achieve its final stable association with the 40S, which is crucial for formation of the beak structure (Schafer et al. 2006). It may be that formation of the final beak structure is necessary for 20S pre-rRNA cleavage and that the dominant-negative mutations in Ltv1 prevent this step, either by blocking the conformational change or by inhibiting the release of factors necessary for the conformational change. In this context, two reports published during the preparation of this manuscript showing a synthetic genetic interaction between LTV1 and PRP43 and PAF1 are intriguing (Lebaron et al. 2009; Pertschy et al. 2009). Prp43, an RNA helicase, and Paf1, a G-patch protein that stimulates the helicase activity of Prp43, are both required in a Δltv1 background for cytoplasmic 20S rRNA cleavage. The presence of dominant-negative mutant Ltv1 proteins in pre-40S subunits could interfere either directly or indirectly, as discussed above, with Prp43/Paf1 binding, thus resulting in a 20S cleavage defect.
Maturation requires release of late 40S biogenesis factors:
Maturation of the pre-60S subunit in the cytoplasm requires a number of cytoplasmic GTP- and ATPases that release both export and biogenesis factors to recycle to the nucleus, likely via conformational changes induced by NTP hydrolysis (reviewed in Zemp and Kutay 2007; Johnson 2009; Kressler et al. 2009; Strunk and Karbstein 2009). In contrast to 60S maturation, no ATPases or GTPases have been identified as necessary for cytoplasmic 40S maturation. However, the presence of two different protein kinases, Rio2 and Hrr25, in late pre-40S particles suggests that phosphorylation events may play analogous roles in pre-40S maturation. In human cells, the kinase activity of hRio2 was recently shown to be necessary for the release and recycling of hDim2, hLtv1, and hNob1 from pre-40S particles, and for the cleavage of the human 18S–E rRNA, in an event analogous to yeast 20S rRNA cleavage (Zemp et al. 2009). Release of hEnp1 required hRio2 but not its kinase activity (Zemp et al. 2009), consistent with previous results showing that in yeast, Enp1 phosphorylation requires Hrr25, but not Rio2 (Schafer et al. 2006). We show here that dominant-negative mutant Ltv1p blocks (or reduces) both 20S rRNA cleavage and Tsr1 release, but not Enp1 or Rio2 release. Overexpression of Tsr1 in cells expressing mutant Ltv1 does not suppress the DN slow-growth phenotype of these cells, however (unpublished observations), indicating that Tsr1 is likely not the only biogenesis factor affected by overexpression of mutant Ltv1. Thus in both human cells and yeast, the release of late 40S biogenesis factors appears to be coupled to pre-rRNA cleavage. The fact that expression of mutant Ltv1 allows the release of some but not all of the pre-40S processing factors suggests that the pre-40S is arrested at an intermediate step and may provide insight into the relative order of pre-40S cytoplasmic maturation events.
Reduction in 40S export may be a downstream consequence of a block in maturation:
Overexpression of mutant Ltv1 is also associated with the nuclear retention of two markers of the 40S subunit, RpS2–GFP and RpS3–GFP. This result would seem to be at odds with the strong cytoplasmic ITS1 signal, also a marker of the pre-40S subunit. We note that similar seemingly contradictory results have been reported previously in the literature. For example, depletion of Tsr1 by glucose repression and inactivation of Rio2 by temperature shift of rio2-1ts both lead to cytoplasmic accumulation of pre-40S subunits as measured by ITS1 FISH (Vanrobays et al. 2003; Leger-Silvestre et al. 2004) and to nuclear accumulation of pre-40S subunits as measured by Rps2–GFP fluorescence (Schafer et al. 2003). These apparently contradictory data, for all three proteins, could be explained if the primary consequence of depleting Tsr1 or Rio2, or mutating Ltv1, was a block in the cytoplasmic maturation of the 20S pre-rRNA. If 20S cleavage is required before some of the late biogenesis factors can be released from the pre-40S, these factors would accumulate in the cytoplasm on stalled immature 40S subunits, leading to their nuclear depletion. The nuclear retention of RpS2–GFP and RpS3–GFP that we detect may be a downstream consequence of the failure to execute the 20S rRNA cleavage step, which thus blocks release and recycling of Tsr1 to the nucleus. Similar effects on 60S nuclear export by cytoplasmic maturation factors have been reported for a number of 60S release factors (reviewed in Zemp and Kutay 2007; Johnson 2009). For example, Senger et al. (2001) reported that deletion of the gene for the GTPase Efl1, which is required for the release of the 60S biogenesis factor, Tif6, results in both cytoplasmic retention of Tif6, which is normally localized to the nucleus, and nuclear accumulation of Rpl25–GFP, a large subunit ribosomal protein. Hedges et al. (2005) also showed that expression of a dominant-negative lsg1 mutant led to both cytoplasmic retention of Nmd3 and the nuclear accumulation of Rpl25–GFP. Because export of either the 40S or the 60S subunit requires recycled factors, steps “later” in the cycle (cytoplasmic maturation) can in fact impede steps normally considered “earlier” in the process (nuclear export). We suggest that other ribosome biogenesis mutants with a subunit nuclear accumulation phenotype may in fact be defective in some cytoplasmic maturation event and not in export directly. Further work will be necessary to determine both the order of release of factors from the 40S subunit and which of these are in fact essential for the continued export of additional pre-40S subunits.
Acknowledgments
We are grateful to William Uicker for construction of Ld70 and to undergraduate Maxwell Spector for his help in constructing Ld57 and Ld58. We are also grateful to James Chu for his help with the microscopy of the Sik1 strain. This work was supported by National Institutes of Health (NIH) AREA grant GM-061643-02 to D.E.L. and NIH RO1 grant GM53655 to A.W.J. Undergraduates C.A.F. and B.J.S. were supported by the John S. Rogers Science Research Program and by GM-061643-02.
References
- Amberg, D. C., A. L. Goldstein and C. N. Cole, 1992. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 6 1173–1189. [DOI] [PubMed] [Google Scholar]
- Belk, J. P., F. He and A. Jacobson, 1999. Overexpression of truncated Nmd3p inhibits protein synthesis in yeast. RNA 5 1055–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, R. C., J. Klootwijk, T. J. Van Steenbergen, A. J. De Kok and R. J. Planta, 1977. Secondary methylation of yeast ribosomal precursor RNA. Eur. J. Biochem. 75 311–318. [DOI] [PubMed] [Google Scholar]
- Chen, W., J. Bucaria, D. A. Band, A. Sutton and R. Sternglanz, 2003. Enp1, a yeast protein associated with U3 and U14 snoRNAs, is required for pre-rRNA processing and 40S subunit synthesis. Nucleic Acids Res. 31 690–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher et al., 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica, A., M. Oeffinger, M. Dlakic and D. Tollervey, 2003. Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol. Cell. Biol. 23 1798–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica, A., D. Tollervey and M. Dlakic, 2004. PIN domain of Nob1p is required for D-site cleavage in 20S pre-rRNA. RNA 10 1698–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod, M., and M. Ohno, 2002. Exportin-mediated nuclear export of proteins and ribonucleoproteins. Results Probl. Cell Differ. 35 67–91. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine, M., B. Senger, C. Saveanu and F. Fasiolo, 2003. Ribosome assembly in eukaryotes. Gene 313 17–42. [DOI] [PubMed] [Google Scholar]
- Gadal, O., D. Strauss, J. Kessl, B. Trumpower, D. Tollervey et al., 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerlings, T. H., A. W. Faber, M. D. Bister, J. C. Vos and H. A. Raue, 2003. Rio2p, an evolutionarily conserved, low abundant protein kinase essential for processing of 20 S Pre-rRNA in Saccharomyces cerevisiae. J. Biol. Chem. 278 22537–22545. [DOI] [PubMed] [Google Scholar]
- Gelperin, D., L. Horton, J. Beckman, J. Hensold and S. K. Lemmon, 2001. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA 7 1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, D., A. St Jean, R. A. Woods and R. H. Schiestl, 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleizes, P. E., J. Noaillac-Depeyre, I. Leger-Silvestre, F. Teulieres, J. Y. Dauxois et al., 2001. Ultrastructural localization of rRNA shows defective nuclear export of preribosomes in mutants of the Nup82p complex. J. Cell Biol. 155 923–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss et al., 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10 105–115. [DOI] [PubMed] [Google Scholar]
- Granneman, S., and S. J. Baserga, 2004. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 296 43–50. [DOI] [PubMed] [Google Scholar]
- Granneman, S., M. R. Nandineni and S. J. Baserga, 2005. The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol. Cell. Biol. 25 10352–10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges, J., M. West and A. W. Johnson, 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24 567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras, A. K., J. Soudet, M. Gerus, S. Lebaron, M. Caizergues-Ferrer et al., 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65 2334–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, J. H., G. Kallstrom and A. W. Johnson, 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151 1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt, E., S. Hannus, B. Schmelzl, D. Lau, D. Tollervey et al., 1999. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J. Cell Biol. 144 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori et al., 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A., 2009. Nuclear Export of Ribosomes, pp. 69–84 in Nuclear Transport, edited by R. H. Kehlenbach. Landes Bioscience, Austin, TX.
- Johnson, A. W., E. Lund and J. Dahlberg, 2002. Nuclear export of ribosomal subunits. Trends Biochem. Sci. 27 580–585. [DOI] [PubMed] [Google Scholar]
- Kallstrom, G., J. Hedges and A. Johnson, 2003. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 23 4344–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler, D., E. Hurt and J. Babetaler, 2009. Driving ribosome assembly. Biochim. Biophys. Acta [DOI] [PubMed]
- La Cour, T., L. Kiemer, A. Molgaard, R. Gupta, K. Skriver et al., 2004. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 17 527–536. [DOI] [PubMed] [Google Scholar]
- Lafontaine, D., J. Vandenhaute and D. Tollervey, 1995. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev. 9 2470–2481. [DOI] [PubMed] [Google Scholar]
- Lafontaine, D. L., T. Preiss and D. Tollervey, 1998. Yeast 18S rRNA dimethylase Dim1p: A quality control mechanism in ribosome synthesis? Mol. Cell. Biol. 18 2360–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebaron, S., C. Papin, R. Capeyrou, Y. L. Chen, C. Froment et al., 2009. The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J. 28 3808–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger-Silvestre, I., P. Milkereit, S. Ferreira-Cerca, C. Saveanu, J. C. Rousselle et al., 2004. The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast. EMBO J. 23 2336–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., I. Lee, E. Moradi, N. J. Hung, A. W. Johnson et al., 2009. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 7 e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loar, J. W., R. M. Seiser, A. E. Sundberg, H. J. Sagerson, N. Ilias et al., 2004. Genetic and biochemical interactions among Yar1, Ltv1 and Rps3 define novel links between environmental stress and ribosome biogenesis in Saccharomyces cerevisiae. Genetics 168 1877–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit, P., D. Strauss, J. Bassler, O. Gadal, H. Kuhn et al., 2003. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 278 4072–4081. [DOI] [PubMed] [Google Scholar]
- Moy, T. I., and P. A. Silver, 1999. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 13 2118–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy, T. I., and P. A. Silver, 2002. Requirements for the nuclear export of the small ribosomal subunit. J. Cell Sci. 115 2985–2995. [DOI] [PubMed] [Google Scholar]
- Neuber, A., J. Franke, A. Wittstruck, G. Schlenstedt, T. Sommer et al., 2008. Nuclear export receptor Xpo1/Crm1 is physically and functionally linked to the spindle pole body in budding yeast. Mol. Cell. Biol. 28 5348–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, W. T., M. D. Robinson, S. Mnaimneh, N. J. Krogan, G. Cagney et al., 2003. A panoramic view of yeast noncoding RNA processing. Cell 113 919–933. [DOI] [PubMed] [Google Scholar]
- Pertschy, B., C. Schneider, M. Gnadig, T. Schafer, D. Tollervey et al., 2009. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J. Biol. Chem. 284 35079–35091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer, T., D. Strauss, E. Petfalski, D. Tollervey and E. Hurt, 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22 1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer, T., B. Maco, E. Petfalski, D. Tollervey, B. Bottcher et al., 2006. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature 441 651–655. [DOI] [PubMed] [Google Scholar]
- Seiser, R. M., A. E. Sundberg, B. J. Wollam, P. Zobel-Thropp, K. Baldwin et al., 2006. Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics 174 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger, B., D. L. Lafontaine, J. S. Graindorge, O. Gadal, A. Camasses et al., 2001. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell 8 1363–1373. [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann, T., U. Schmidt and P. A. Silver, 2000. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell 11 3777–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, A., C. L. Hsu, K. R. Isham and F. W. Larimer, 1991. Fragments of the internal transcribed spacer 1 of pre-rRNA accumulate in Saccharomyces cerevisiae lacking 5′−3′ exoribonuclease 1. J. Bacteriol. 173 7024–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk, B. S., and K. Karbstein, 2009. Powering through ribosome assembly. RNA 15 2083–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, F., and U. Kutay, 2003. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 116 2409–2419. [DOI] [PubMed] [Google Scholar]
- Trotta, C. R., E. Lund, L. Kahan, A. W. Johnson and J. E. Dahlberg, 2003. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 22 2841–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner, H., and E. Hurt, 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13 255–263. [DOI] [PubMed] [Google Scholar]
- Udem, S. A., and J. R. Warner, 1973. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 248 1412–1416. [PubMed] [Google Scholar]
- Vanrobays, E., P. E. Gleizes, C. Bousquet-Antonelli, J. Noaillac-Depeyre, M. Caizergues-Ferrer et al., 2001. Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J. 20 4204–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrobays, E., J. P. Gelugne, P. E. Gleizes and M. Caizergues-Ferrer, 2003. Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 23 2083–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrobays, E., A. Leplus, Y. N. Osheim, A. L. Beyer, L. Wacheul et al., 2008. TOR regulates the subcellular distribution of DIM2, a KH domain protein required for cotranscriptional ribosome assembly and pre-40S ribosome export. RNA 14 2061–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema, J., and D. Tollervey, 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33 261–311. [DOI] [PubMed] [Google Scholar]
- Wen, W., J. L. Meinkoth, R. Y. Tsien and S. S. Taylor, 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82 463–473. [DOI] [PubMed] [Google Scholar]
- Zemp, I., and U. Kutay, 2007. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 581 2783–2793. [DOI] [PubMed] [Google Scholar]
- Zemp, I., T. Wild, M. F. O'Donohue, F. Wandrey, B. Widmann et al., 2009. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol. 185 1167–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]