Abstract
Urothelial carcinoma of the bladder (UCC) is a common disease that arises by at least two different molecular pathways. The biology of UCC is incompletely understood, making the management of this disease difficult. Recent evidence implicates a regulatory role for microRNA in cancer. We hypothesized that altered microRNA expression contributes to UCC carcinogenesis. To test this hypothesis we examined the expression of 322 microRNAs and their processing machinery in 78 normal and malignant urothelial samples using realtime rtPCR. Genes targeted by differentially expressed microRNA were investigated using realtime quantification and microRNA knock-down. We also examined the role of aberrant DNA hypermethylation in microRNA down-regulation. We found that altered microRNA expression is common in UCC and occurs early in tumorogenesis. In normal urothelium from patients with UCC 11% of microRNA’s had altered expression when compared to disease-free controls. This was associated with upregulation of Dicer, Drosha and Exportin 5. In UCC, microRNA alterations occur in a tumor phenotype-specific manner and can predict disease progression. High-grade UCC were characterized by microRNA upregulation, including microRNA-21 that suppresses p53 function. In low-grade UCC there was down-regulation of many microRNA molecules. In particular, loss of microRNAs-99a/100 leads to upregulation of FGFR3 prior to its mutation. Promoter hypermethylation is partly responsible for microRNA down-regulation. In conclusion, distinct microRNA alterations characterize UCC and target genes in a pathway-specific manner. These data reveal new insights into the disease biology and have implications regarding tumor diagnosis, prognosis and therapy.
Introduction
Bladder cancer is the fifth commonest malignancy in the United States with an incidence of 67,160 new cases and 13,750 deaths in 2007 (1). The majority of tumors are Urothelial Cell Carcinoma (UCC). Clinical and molecular evidence suggests there are at least two distinct varieties of this tumor. Most UCC belong to a low-grade pathway characterized by FGFR3 mutation, chromosome 9 loss and an indolent clinical phenotype. Around 1/3 of UCC are high-grade in differentiation and arise as lesions initially confined to the bladder mucosa (non-muscle invasive (NMI)). Progression to muscle invasion occurs in around 50% of high-grade lesions and is associated with an ominous prognosis despite radical treatment (2, 3). Whilst numerous genetic and epigenetic alterations are observed in high-grade UCC, loss of p53 function appears most critical (4, 5).
MicroRNAs (miRs) are short non-coding RNA molecules that post-transcriptionally modulate protein expression. They are transcribed as hairpin pri-miRs and processed into pre-miRs by Drosha, an RNAse III endonuclease complexed with DGCR8. Pre-MiRs are exported into the cytoplasm by Exportin 5 before cleavage by Dicer into mature miRs. MiRs are directed to mRNAs with a complementary seed sequence to their first 1-8 nucleotides (6). Alterations in microRNA expression appear important for carcinogenesis (7, 8). Their profile may be used to identify key tumorogenic pathways (9) or clinical outcome (10, 11).
To date, few data detail microRNA in UCC (9, 12, 13). We hypothesized that altered miR expression occurs in UCC and contributes to carcinogenesis. To test this hypothesis we investigated the expression of numerous miRs and their processing molecules in urothelial tissues. Our studies reveal that UCC are characterized by widespread alterations in miR expression. Changes in expression occur early in the tumorogenic pathway, in a phenotype specific manner and are associated with altered expression of their processing machinery. Of particular note, low-grade tumors are characterized by miR mediated FGFR3 upregulation prior to its mutation.
Materials and Methods
Patients, samples and cell lines
We studied 72 urothelial samples and 6 cell lines (Table 1). Tumors were classified using the 2004 WHO/ISUP criteria, and selected from three cancer groups: 1). Low-grade NMI, 2). High-grade NMI and 3). Invasive UCC. Twenty normal urothelial samples were obtained from 10 patients with UCC (NU(UCC) taken distant to any tumor) and 10 disease-free controls (NU(Controls)). Tissues were immediately frozen in liquid nitrogen and histological confirmation obtained prior to use. Patients were treated according to tumor stage and grade following endoscopic resection (14). Adjuvant intravesical chemotherapy was used for low-grade and maintenance intra-vesical BCG for high-grade NMI tumors. Patients underwent surveillance stratified by their disease. Radical cystectomy with pelvic lymphadenectomy was used for invasive or BCG-refractory high-grade NMI tumors (4, 14). We analyzed UCC cell lines representing the three tumor groups (RT4, RT112 and EJ/T24, respectively, purchased from ATCC) grown in Dulbecco’s medium with 10% fetal calf serum, and 3 normal human urothelial (NHU) cell lines maintained in keratinocyte serum-free medium containing bovine pituitary extract, epidermal growth factor (Invitrogen, Paisley, UK) and cholera toxin. Non-immortalized NHU cells were derived from histologically confirmed normal urothelium obtained from patients without a history of UCC, using standard methods (15). DNA Methyltransferase inhibition experiments were performed in quadruplet by adding 2uM 5-azacytidine to the media for 5-7 days before harvesting.
Table 1.
Details of patients and tissue samples analysed in this study
| UCC | Normal Urothelium | |||||
| Low-grade NMI | High-grade NMI | Invasive | Non-UCC | UCC patients | ||
| Total | 22 | 12 | 18 | 10 | 10 | |
| Gender | Male | 14 | 12 | 14 | 10 | 8 |
| Female | 8 | 0 | 4 | 0 | 2 | |
| Age | Mean | 75 | 71.9 | 71.4 | 67 | 66.4 |
| Range | 56-83 | 55-81 | 46-90 | 54-81 | 46-90 | |
| Source of urothelium* | TURP | 4 | ||||
| RP | 6 | |||||
| TURBT | 3 | |||||
| RC | 7 | |||||
| Grade | 1 | 11 | 0 | 0 | 1** | |
| 2 | 11 | 0 | 1 | 4** | ||
| 3 | 0 | 12 | 17 | 5** | ||
| Stage | pTa | 22 | 3 | 0 | 2** | |
| pTis | 0 | 2 | 0 | |||
| pT1 | 0 | 7 | 0 | |||
| pT2-4 | 0 | 0 | 18 | 8** | ||
| Smoker *** | Current | 5 | 1 | 3 | ||
| Ex | 13 | 10 | 12 | |||
| Never | 4 | 1 | 3 | |||
| Recurrence | Yes | 14 | 8 | 5 | ||
| No | 8 | 4 | 13 | |||
| Progression | Yes | 4 | 3 | 11 | ||
| No | 18 | 9 | 7 | |||
| Follow up (months) | Mean | 43.4 | 57.0 | 23.5 | ||
| Range | 4-93 | 15-93 | 2-84 | |||
RC: Radical cystectomy, RP: Radical prostatectomy, TURP/BT: Transurethral resection of prostate/bladder tumor
Details of associated tumour
Ex smoking status defined as stopping > 1 year before tumor
MicroRNA expression profiling
For both normal and malignant samples, ten 10uM sections were microdissected to obtain >90% pure cell populations. Enriched small and total RNA were extracted using the mirVana™ kit (Ambion, TX) according to manufacturer’s protocol. RNA concentrations were measured using a 2100 Bioanalyzer (Agilent, Cheshire, UK). The expression of 354 mature miRs and 3 small nucleolar RNA molecules was determined using real time PCR with pre-printed microfluidic cards (Human microRNA v1.0, Applied Biosystems, Warrington, UK). (16). Initially, reverse transcription using stem loop primers was performed with 50ng RNA, MultiScribe Reverse Transcriptase (Applied Biosystems, Warrington, UK), RNase inhibitor, 100nm dNTPs and nuclease free water. To generate 357 miR and reference rtPCR products, 8 multiplex pools were used per sample (each pool containing ingredients and primers for 48 mature miRs: total volume 10μL (Human Multiplex RT Pool 1-8 v1.0 (Applied Biosystems, Warrington, UK)). The multiplexed RT assays were cooled to 16°C for 30mins, heated to 42°C for 30mins and 86°C for 5mins before diluting 62.5 fold in nuclease free water. From each diluted assay 50μl was mixed equally with 2x Taqman Universal PCR MasterMix and loaded separately into the eight reservoirs of a microfluidic card (Human microRNA v1.0, Applied Biosystems, Warrington, UK). Following careful centrifugation (2mins at 331g) and preparation (well sealing and reservoir removal) the loaded card was analyzed on an ABI 7900HT real time PCR system using the recommended PCR conditions. Each microfluidic card contained MGB-labeled probes specific to 354 mature miRs, 11 duplicate assays, 2 empty wells and 17 replicates of 3 endogenous small nucleolar RNA molecules for relative quantification (RNU6B, RNU44 and RNU48). MiR concentrations were calculated from the cycle number that each reaction crossed an arbitrarily threshold (Ct=0.2). The amplification plots were checked manually (SDS 2.2.1, Applied Biosystems, UK) to confirm the Ct value corresponded with the midpoint of logarithmic amplification. To test the reproducibility of this real time quantification, we analyzed each cell line and 10 tissues samples in duplicate.
Identification of genes targeted by aberrantly expressed microRNA
We identified potential protein targets of miRs using TargetScan 1 (Vsn. 4.2) (17, 18). Target genes were ranked according to context score and the average was used for those with multiple targeting miRs. mRNA transcripts of target and microRNA processing genes were measured in those samples with sufficient material (n=71 (93%)). cDNA synthesis from 100ng whole RNA was performed using random primers, RT buffer, dNTP (100mM), RNase inhibitor and MultiScribe Reverse Transcriptase (cDNA Reverse Transcription kit, Applied Biosystems, Warrington, UK). Realtime quantified PCR using 2uL of synthesized cDNA, gene specific primers with FAM-TAMRA labeled probes (Supplementary table 1), water and 2x Taqman Universal PCR MasterMix (Applied Biosystems, Warrington, UK) in a total volume of 10uL was performed on the ABI 7900HT system according to manufacturers guidelines. Relative mRNA quantification was measured with respect to the mean of GAPDH and β-Actin (and PCNA as a proliferation reference gene).
MicroRNA knock-down in NHU cells
MiRs-99a/100 targeting of FGFR3 was investigated using protein expression following miR knock-down and a Luciferase reporter assay. All experiments were replicated between 3 and 6 times using non-immortalized NHU cell lines at 70% confluence reverse-transfected with anti-miR molecules (Ambion, TX) specific to miR-99a, miR-100 and a negative control (scrambled sequence). To determine target protein expression, NHU cells were transfected with 50nM anti-miR or control sequences using siPORT NeoFX transfection reagent (Ambion). The mixture was dispensed into a 6-well dish, seeded at 2 × 105 cells per well, and incubated for 48h. Transfection was determined by realtime-rtPCR (Applied Biosystems) and FGFR3 expression measured by Western Blotting using anti-FGFR3 primary antibody (1:1000; Cell Signaling Technologies Inc, MA, USA). Cells were lysed in RIPA buffer (20m M Tris·HCl, 135 mM NaCl, 10% glycerol, 1% Igepal, 0.1% SDS, 0.5% deoxycholic acid, 2 mM EDTA) containing protease and phosphatase inhibitors (Complete EDTA-free protease inhibitor cocktail and PhosSTOP phosphatase inhibitor cocktail; Roche, Mannheim, Germany), and protein content was quantified using the DC-protein assay reagent (Bio-Rad, Hercules, CA). Protein lysates (50μg) were loaded onto 8% gels, fractionated, and electroblotted onto nitrocellulose membranes. After blocking with 5% non-fat milk powder and 0.1% Tween, the membranes were incubated overnight with anti-FGFR3 primary antibody (1:1000; Cell Signaling Technologies Inc, MA, USA) at 4°C, washed, and incubated at room temperature for 1h with goat anti-rabbit peroxidase-labelled secondary antibody (1:1000; Cell Signaling Technologies Inc.). β-Actin expression was also measured as a loading control (Sigma, 1:2000). Immune complexes were quantified using ImageJ for band densitometry. To validate direct targeting of FGFR3, NHU cells were co-transfected with either anti-miR-99a, anti-miR-100 or negative control, and the pSSG_3UTR plasmid containing the luc2P Luciferase reporter and either the partial 3′UTR for FGFR3 (from 27 to 1762bp downstream of the gene) or a control (scrambled sequence) (SwitchGear Genomics, CA). Co-transfection was performed in opaque 96 well plates using 40-80% confluent cells and FuGene transfection reagent (Roche, UK) and Opti-MEM serum free media. After incubation for 48 hours, luciferase activity was determined following the addition of Promega Steady-Glo Luciferase Assay Reagent (Promega, UK) in a luminometer. Negative controls using scrambled anti-miR sequences or a scrambled 3′ UTR sequence were included.
Generation of FGFR3 mutant and wild type paired cell lines
Telomerase-immortalized NHU (TERT-NHUC) cells were used to investigate miR expression following FGFR3 mutation (19). Site-directed mutagenesis on FGFR3 IIIb cDNA created the S249C mutation. The presence of this mutation was verified by sequencing. Wildtype and mutant FGFR3 were cloned into a retroviral expression vector (pFB; Stratagene, La Jolla, CA) containing a hygromycin resistance cassette (20). The expression vectors were transfected into Phoenix A cells using siPORT XP-1 (Ambion, Huntingdon, UK). TERT-NHUC were incubated for 8 h with retroviral supernatant containing 8 μg/ml polybrene and selected with hygromycin 48 h after transduction. FGF1 cell stimulation was also performed using 20 ng/ml recombinant human FGF1 and 10 μg/ml heparin (R&D Systems Europe Ltd., Abingdon, UK) after 3 h incubation in depleted medium (KGM2 without added growth factors).
Statistical methods
Relative miR concentrations were calculated with respect to the median of 3 reference RNA molecules (ΔCt = Ct miR − Ct mediancontrol). The median was chosen after analysis revealed variation in concentrations between samples (data not shown). Expression fold changes were computed using 2-ΔΔCt calculations (21). Median data centering was performed prior to analysis with Significance of Analysis of Microarray software (22) and BRB-ArrayTools 2 (Vsn. 3.7) developed by Dr. Richard Simon and BRB-ArrayTools Development Team. Class specific miR signatures were established using Prediction of Microarray software (23). MiR expression and tumor outcome were investigated using the Log rank test and plotted by the Kaplan-Meier method within SPSS (Vsn. 14.0 SPSS Inc.). Tumor progression was defined as the presence of pathological, radiological or clinical evidence of an increase in tumor stage and measured from the time of surgery to the time of proven event. Changes in miR expression and statistical significance were illustrated using Volcano plots. Significant expression difference was defined as both a p<0.05 and a false discovery rate of <0.05. Hierarchical clustering was performed after removal of those miRs with a low detection frequencies in all groups (<20%: defined as ‘noise’) using Cluster 3.0 and visualized in Tree view (Eisen Lab 3). Correlation between variables was assessed using Pearson’s correlation coefficient within SPSS.
Results
MicroRNA expression in normal urothelium
Following assay optimization (Supplementary results) we compared the expression of 322 miRs in normal urothelium from patients with UCC and disease-free controls. Thirty-six miRs were differentially expressed between the two groups (p<0.05 and FDR 0.0, Supplementary table 2) and were always upregulated in the NU(UCC). There was clustering of differential expression according to RNA family, e.g. let-7a/b/c/d/e/g, miRs-382/487b, miRs-17/93, miRs-181b/d, miRs-34a/c, and chromosomal location, e.g. 9q22.32 (let-7a/d), 14q32.31 (miRs-376a/487b/382/412) and 19q13.41 (miRs-520a/b/c/d/e/518a/519e). Unsupervised hierarchical clustering revealed two branches corresponding to the two groups (Supplementary Figure 1). Three miRs were detected only in NU(UCC) (miR-569 (detected in 50% of samples), miR-633 (40%), miR-507(30%)) and 2 by only the NU(controls) (miR-412 (50%), miR551a (30%)) (data not shown).
Differential microRNA expression in malignant urothelium
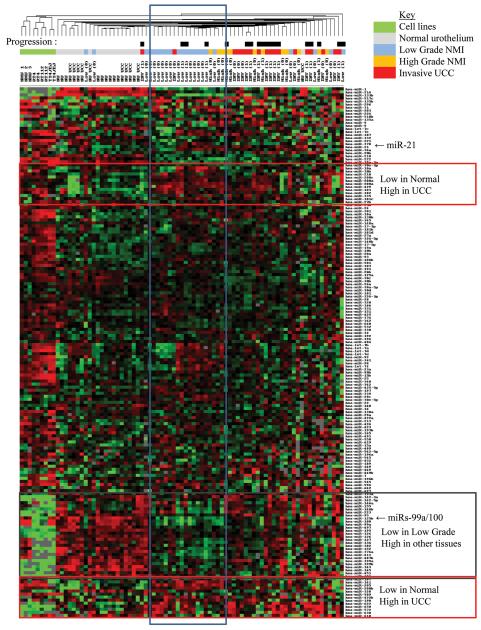
We compared malignant miR expression with that from disease-free normal urothelium (Figure 1). The annotated raw data can be downloaded from our website 4. Twelve miRs were expressed only in the malignant tissues and 5-7 of these could identify cancer with a sensitivity of 90-100% and a specificity of 100-80% (Table 2a), suggesting roles as diagnostic biomarkers. Quantitative analyses revealed that 16 miRs were differentially expressed in the malignant and normal urothelia (p<0.05 and FDR<0.05, Table 2b). Thirteen of these were upregulated in cancer and 7 of these were also aberrantly expressed in the normal urothelium from UCC cases, suggesting early alteration in the disease pathway. Hierarchical clustering revealed miR expression stratified tissues mostly according to phenotype.
Figure 1.
MicroRNA expression in normal and malignant urothelium. Centroid linkage hierarchical clustering was performed after selecting miRs with >90% expression frequency and mean centering data using city block similarity in Cluster 3.0. First dendrogram row indicates subsequent progression (Black box = Progression, White box = No progression (also in brackets after sample ID (0=no, 1=yes)). Boxes outline miRs with similar expression profiles.
Table 2.
Differences in miR expression between UCC and normal urothelium (from control patients (NU(Controls))
Differences in miR expression between normal and malignant urothelium. (a). Absolute expression of a few miRs allows the distinction of malignant and normal urothelium (FN=false negative, FP=false positive, Sens=sensitivity, Spec=specificity). (b) Sixteen MiRs have significant differential expression (p<0.05 and FDR<0.05) in normal and malignant urothelium when compared. The ΔCt mean and standard deviation values are shown, together with fold change expression associated with carcinogenesis (calculated using the 2-ΔΔCt method)
|
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (a) | miR | FN | FP | Sens | Spec | Normal | Low Grade NMI UCC | High Grade | Invasive UCC | |
|
|
||||||||||
| 1 | miR-211 |
|
||||||||
| 2 | miR-549 | |||||||||
| 3 | miR-526b | 10 | 0 | 0.81 | 1.00 | |||||
| 4 | miR-507 | 6 | 0 | 0.88 | 1.00 | |||||
| 5 | miR-147 | 5 | 0 | 0.90 | 1.00 | |||||
| 6 | miR-517a | 1 | 1 | 0.98 | 0.90 | |||||
| 7 | miR-556 | 0 | 2 | 1.00 | 0.80 | |||||
|
|
||||||||||
| (b) | Normal Urothelium ΔCt | UCC ΔCt | Fold | FDR | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MicroRNA | Chr | Family | Present | Mean | St Dev | Present | Mean | St Dev | change | q % | P value | |
| miR-649 | 22 | miR-649 | 90% | 18.59 | 4.11 | 90% | 12.03 | 2.93 | 94.00 | 0.0 | 0.0019 | |
| miR-135b | 1 | miR-135 | 100% | 7.45 | 1.53 | 98% | 3.18 | 2.42 | 19.41 | 0.0 | 0.0037 | |
| miR-133b | 6 | miR-133 | 100% | 0.59 | 1.93 | 98% | 5.41 | 3.05 | 0.04 | 0.0 | 0.0041 | |
| miR-520b | 19 | miR-93* | 20% | 15.00 | 4.94 | 77% | 11.64 | 3.94 | 10.28 | 0.0 | 0.0056 | |
| miR-204 | 9 | miR-204 | 100% | 4.42 | 1.85 | 92% | 9.20 | 3.40 | 0.04 | 0.0 | 0.0063 | |
| miR-601 | 9 | miR-601 | 90% | 14.57 | 3.51 | 94% | 9.92 | 2.79 | 24.98 | 0.0 | 0.0091 | |
| miR-646 | 20 | 70% | 14.58 | 3.39 | 94% | 10.04 | 2.89 | 23.24 | 0.0 | 0.0160 | ||
| miR-639 | 19 | miR-639 | 80% | 13.29 | 2.63 | 90% | 8.82 | 3.16 | 22.18 | 0.0 | 0.0225 | |
| miR-380-5p | 14 | 70% | 4.41 | 4.69 | 75% | 10.50 | 4.34 | 0.01 | 2.6 | 0.0285 | ||
| miR-21 | 17 | miR-21 | 100% | 2.45 | 1.56 | 100% | −0.97 | 2.80 | 10.71 | 0.0 | 0.0296 | |
| miR-644 | 20 | miR-644 | 70% | 17.57 | 3.63 | 92% | 12.66 | 4.05 | 30.18 | 0.0 | 0.0305 | |
| miR-93 | 7 | miR-93* | 100% | 2.38 | 2.05 | 100% | −0.45 | 1.90 | 7.15 | 0.0 | 0.0308 | |
| miR-15a | 13 | miR-15 | 100% | 5.98 | 1.80 | 96% | 2.89 | 2.39 | 8.50 | 0.0 | 0.0354 | |
| miR-520d | 19 | miR-93* | 30% | 21.04 | 1.02 | 75% | 11.47 | 4.58 | 758.94 | 0.0 | 0.0394 | |
| miR-549 | 15 | miR-549 | 0% | 56% | 14.43 | 3.49 | 0.00 | 4.3 | 0.0427 | |||
| miR-449b | 5 | miR-34 | 100% | 12.17 | 2.24 | 90% | 9.11 | 2.69 | 8.36 | 0.0 | 0.0439 | |
miR-93.hd/291-3p/294/295/302/372/373/520
Genes targeted by aberrantly expressed microRNA
With TargetScan we identified potential target genes for the 16 differentially expressed miRs and selected the top 1/3, as this fraction is most associated with altered protein expression (18). Of n=1,095 predicted genes, 276 were targeted by more than one miR; including PLAG1 by 8 miRs, and E2F7 and DMTF1 by 5. Target ranking by context score revealed many carcinogenic genes with high binding affinity (Supplementary figure 2). We selected 9 genes, from those with highest binding affinity and potentially carcinogenic roles, and measured their expression in our urothelial samples. For 6/9 genes there was differential expression between normal and malignant urothelial (Supplementary figure 2c, p<0.02 T test). For 5 of these genes, this differential expression was inverse to that of their targeting miR suggesting they play a regulatory role in expression (17). From UCC gene expression array data (24) we identified 143 putative target genes. For 76 (54%) an inverse relationship between miR and mRNA expression was present and in 23 (11%) this difference reached significance (p<0.05, T Test, Supplementary figure 3).
Expression of microRNA with respect to tumor phenotype
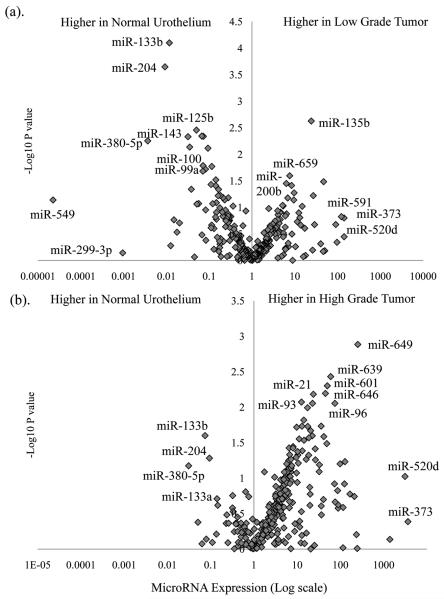
Comparisons of the three tumor groups with disease-free normal urothelium revealed global and specific differential miR expression (Figures 2 and 3). In general, there was a down-regulation of microRNA in the low-grade samples (18/25 differentially expressed miRs) and an upregulation in high-grade and invasive tumors (28/30 and 20/20 miRs respectively, χ2 p<0.0001). Few aberrantly expressed miRs were common to the low-grade and high-grade/invasive cohorts (4/59 (6%)), whilst many were shared between the high-grade and invasive tumors (12/38 (32%), χ2 p=0.003). The magnitude of differential miR expression varied amongst tumor groups (average fold change = 4.32 (low-grade), 28.24 (high-grade NMI) and 50.18 (invasive), ANOVA p=0.006). When the three groups were compared with each other, the low-grade tumors were distinct from the high-grade NMI (n=9 miRs differentially expressed) and invasive cohorts (n=45 miRs differentially expressed), whilst no significant differences in expression were present between the latter two (Figure 3a).
Figure 2.
MicroRNA expression in low and high-grade bladder cancer. In (a) changes in expression (log scale) and statistical significance (SAM P values plotted as -log 10) of low-grade NMI tumors when compared to normal urothelium from non-UCC cases, reveals both up and down-regulation of numerous miRs. In (b) for high-grade NMI and invasive tumors there is a general upregulation in microRNA expression. Whilst changes in some miRs are shared between the two tumor phenotypes (e.g. miR-133b/204) others appear specific (miR-21 or miRs-100/99a). Expression is relative to the median of 3 RNA molecules and linear median centered.
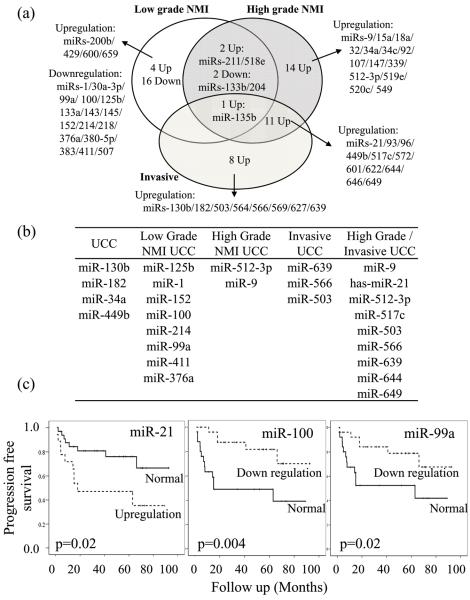
Figure 3.
MicroRNA expression and tumor phenotype. (a). Differentially expressed miRs specific to each phenotype. (b) Signature panels of microRNA for each tissue (c). Progression analysis in all 52 UCC revealed that dichotomous aberrant expression of miRs-21/100/99a was associated with progression to more advanced UCC (Kaplan-Meier method and tested using Logrank analysis).
We defined a specific signature for each tumor group by selecting those miRs that were differentially expressed when compared to both normal urothelium and one or more of the other two tumor groups (Figure 3b). As many miRs were shared between the high-grade NMI and invasive tumor groups, we merged these to create a single defining signature. We also identified 4 miRs that were altered in UCC regardless of tumor phenotype.
Phenotype specific microRNA targets
We were interested by the difference in microRNA expression between the low-grade and high-grade tumors. MicroRNA upregulation was a feature of the high-grade pathway and miRs-21/373 were amongst those with most prominence. Recent data reveal that miR-21 negatively regulates the p53 tumor suppressor pathway leading to a loss of cellular control (25). MiR-373 is known to regulate pro-metastatic pathways (26), possibly through LATS2 suppression (8) (of note, LATS2 was down-regulated in our tumors (Supplementary figure 2)).
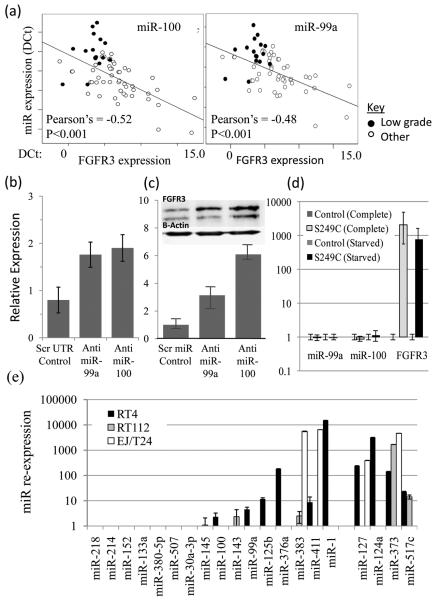
Low-grade tumors were characterized by down-regulation of many miRs. Of the 16 miRs with significant down-regulation, 7 are predicted to target FGFR3 (miRs-99a/100/214/145/30a/125b/507) and 1 is the only highly conserved miR to target H-Ras (miR-218). We focused upon miRs-99a/100 as they target a highly conserved 8mer region within the FGFR3 UTR and their expression was inversely correlated with FGFR3 mRNA (Figure 4a, r=−0.48 and −0.52 respectively, p<0.0004). To investigate whether FGFR3 is a target of miRs-99a/100 we manipulated their expression in NHU cells. These cells represent normal urothelium, have a 200 fold higher miR99a/100 expression than RT4 cells and a low FGFR3 expression (data not shown). Following transfection with anit-miRs, a 70-80% reduction in miR expression (data not shown) resulted in an average 3-fold (miR-99a) and 6-fold (miR-100) upregulation of FGFR3 protein (Figure 4). Direct targeting of the 3′ UTR region of the FGFR3 gene was confirmed by a Luciferase reporter assay (Figure 4). Transfection of NHU cells with anti-miRs to 99a/100 and a plasmid containg Luciferase and a partial FGFR3 3′UTR resulted in a 2-fold (miR-99a) and 3.4-fold (miR-100) increase in Luciferase expression, when compared to cells transfected with a scrambled UTR sequence. To investigate the reverse relationship we produced matching NHU-TERT cell lines with and without the S249C FGFR3 mutation. In each cell line regardless of FGF exposure, confluence or passage number the expression of both miRs was unchanged by the presence of the mutation (Figure 4d), suggesting the epigenetic event is upstream of the gene mutation.
Figure 4.
Analyses of microRNA 99a/100 in UCC. (a) Expression of miRs 100/99a and FGFR3 are inversely correlated (DCT values shown). (b) We found that a 70-80% miR-99a/100 knock-down induced by anti-miRs in NHU cells transcribed with Luciferase and the 3’UTR of FGFR3 was associated with increased Luciferase expression when compared to NHU cells transfected with a scrambled UTR sequence. (c). Upregulation of FGFR3 protein expression was found in NHU cells transfected with anti-miRs to 99a/100 when compared to negative controls (scrambled anti-miR sequence) (Western blot images (FGFR3 is upper band (splice variant seen below)) and photometric relative quantification standardized for B-Actin and measured using ImageJ). (d) The presence of the activating S249C FGFR3 mutation does not alter miR-99a/100 expression (relative miR and mRNA quantification). (e) Reversal of DNA hypermethylation leads to re-expression of 5 down regulated miRs in the low-grade cell line (RT4). Relatively few events were seen in the cell lines representative of high-grade and invasive bladder cancer (RT112 and EJ/T24). Four miRs (mir-127/124a/373/517c) reported to be silenced by DNA hypermethylation are also shown (28, 29).
MicroRNA expression and tumor progression
If miRs-99a/100 and 21/373 characterize low-grade and high-grade UCC, one would expect their expression to be associated with tumor behavior. For each miR there was a significant change in tumor progression rate according to their expression, in a manner similar to their associated genetic traits (Logrank p<0.05, Figure 3c). MiRs-99a/100 down-regulation was associated with a better outcome (analogous to FGFR3 mutation), whilst miRs-21/373 up-regulation was associated with a worse outcome (as with p53 mutation), when compared to contrasting tumors. Multivariate analysis revealed that miR-99a/100 expression was associated with progression when analyzed together with tumor stage, grade and miR-21 expression (Cox MVA p=0.03 and p=0.006, respectively).
Epigenetic regulation of microRNA
The role of aberrant promoter hypermethylation for miR down-regulation in low-grade UCC was analyzed using DNA methyltransferase inhibition. Following 5-azactytidine treatment 8 miRs (Figure 4e) had more than 2-fold increase in expression, including 6 in the low-grade cell line (RT4). The lower rates of miR re-expression in the high-grade tumor lines (RT112 and EJ/T24) suggest that regulation by hypermethylation of these miRs is relatively low-grade specific. Both RT112 and EJ/T24 have higher rates of DNA hypermethylation than RT4 at known tumor suppressor gene promoters (27). For comparison we analyzed 4 hypermethylated miRs reported in UCC (miRs-127/124a/373/517c) (28, 29). For these, re-expression was usually shared between two or more cell lines rather than just isolated to RT4. MiRs-99a/100 are located at 21q21.1 and 11q24.1, respectively, together with two other miRs and one protein coding genes. Neither region contains a canonical CpG island. To investigate long range epigenetic silencing of miRs-99a/100 we looked at the expression of neighbouring genes following 5-azactytidine treatment (Supplementary figure 4). There was an increase in expression of mir-125b (located in duplicate genes, at each locus) but not of either protein coding gene nor the other microRNA in RT4.
Processing molecules and microRNA expression
We analyzed the expression of molecules known to be important for microRNA processing. The expression of each varied significantly between tissue type (Supplementary figure 5, ANOVA p<0.05) but not between tumor phenotype. For Dicer, Drosha (RNASEN) and Exportin 5 there was an upregulation in the normal urothelium from UCC cases which was reversed once malignancy appeared. The largest changes were for Dicer and Drosha (7.4 and 6.1 fold upregulation, p=0.00006 and 0.00001, respectively), whose expression was closely correlated (Pearson’s correlation=0.79, p=7×10-19). For RAN and DGCR8 the opposite was seen, with an initial down-regulation followed by upregulation in the tumors. Concentrations of these two mRNA were almost identical in each tissue (Pearson’s correlation=0.48, p=3×10-6). When expression of these molecules was compared with that of the 322 miRs, several associations were seen. Dicer is known to be targeted by several miRs and expression of miR-130b (r=−0.28, p=0.02) and let-7g (r=−0.25, p=0.05) were inversely correlated with Dicer expression.
Discussion
Here we have demonstrated that altered microRNA expression occurs commonly in UCC, in a phenotype-specific manner and targets key pathways considered ‘hallmarks’ of the disease. Our first finding was that pro-tumorogenic microRNA alterations occur before the histological onset of malignancy, supporting observations of promoter hypermethylation and genetic instability (27, 30). Almost half of our tumor-associated miRs were upregulated in the normal urothelium from patients with the disease, and this was associated with increased Dicer, Drosha and Exportin 5 expression. Dicer overexpression in pre-malignant lesions of the prostate and lung, and reduced Dicer and Drosha expression in ovarian cancer are reported (31-33) supporting our observation. Expression of the microRNA machinery was not associated with specific miRs, suggesting a global microRNA upregulation rather than the selection of tumor-specific species.
When analyzing all UCC cases together, we found the aberrant expression of 16 miRs, including some generic to carcinogenesis (e.g. miR-21 (25), mir-133b (34)). Numerous interesting observations were apparent. For example, reduced miR-204 expression was frequent. MiR-204 is located on Chromosome 9q21.11 within an intron of TRPM3, a gene down-regulated in UCC (35, 36). MiR-204 potentially targets FRS2 an important member of the FGFR3 pathway (37). Observed changes in miR expression occurred in isolation or were clustered. Isolated differences arose from solitary located miRs such as miRs-649/135b or miR-601. The latter is located within intron 1 of DENND1A, an gene upregulated in UCC (38). Isolated differences also arose when a single member of a miR cluster was differentially expressed, including down-regulation of miR-133b (adjacent to mir-206) or upregulation of miR-449b (located with miR-449a) and miR-93 (clustered with miRs-25/106b within the MCM7 gene). Clustered alterations included down-regulation of miRs-133a/1 (located in duplicated clusters on Chr18q11.1 and Chr20q13.33), miRs-99a/125b and miRs-143/145. Familial clustering was also present with 3 members of the miR-93 family (miR-93/520b/520d), miR-133a and 133b, and 3 of the miR-34 family (miR-449b/34c/34a) having aberrant expression. Bioinformatic predictions identified around 1000 genes as putative targets of these miRs. Of these genes, 20% were targeted by more than one miR. We confirmed consistent expression changes for some of these targets including new observations suggesting a carcinogenic role for LATS2, YOD1 and RAB22a in UCC. We identified microRNA as a potential mechanism for SOX4 upregulation in UCC (39).
Whilst the comparison of UCC with normal urothelium revealed interesting data, it did not detail events within the disease well. To obtain a detailed picture it was necessary to analyze microRNA expression between tumor phenotypes. High-grade tumors were characterized by miR upregulation and for most, altered expression occurred before the onset of muscle invasion. Low-grade UCCs were characterized by miR down-regulation and this affected many molecules targeting FGFR3 or H-Ras. Confirmation of FGFR3 microRNA targeting was obtained by the inverse correlation between miR and mRNA expression, and functional miR knockdown experiments. As aberrant expression of miRs-99a/100 was almost ubiquitous in low-grade UCC, occurs more frequently than FGFR3 mutation and the presence of the commonest FGFR3 mutation (S249C) did not alter miR expression, we concluded the microRNA alteration leads to FGFR3 upregulation before the acquisition of gene mutation. This is an intriguing finding and suggests that epigenetic upregulation of FGFR3 facilitates the acquisition of mutation by either increasing cell turnover, reducing regulation to make cells more susceptible to carcinogens or provides a selection advantage for mutant cells. To investigate potential causes of reduced miR expression in low-grade UCC, we examined DNA methylation. For 6 miRs there was significant upregulation in RT4 following DNA methyltransferase inhibition, including miR-1 for which aberrant hypermethylation has recently been reported in hepatocellular carcinoma (40). Our findings with respect to miR-s99a/100 were less clear. There are no canonical CpG islands close to either gene, but two of the four genes clustered around miR-99a or miR-100 loci had increased expression following 5-azacytidine. This finding has been reported by others (40, 41) and suggests long range epigenetic regulation (42). However, the expression of neighboring protein-coding genes did not alter with DNA Methyltransferase inhibition.
In summary we have found new mechanisms related to biology and progression of UCC. These epigenetic events precede histological changes of malignancy or disease-progression and occur in a phenotype-specific manner. These data have implications regarding out understanding of UCC tumor biology, and suggest microRNA as a novel diagnostic or prognostic biomarker, and therapeutic target.
Supplementary Material
Acknowledgements
The authors wish to thank Messrs Anderson, Chapple, Hastie, Hall, Inman, Oakley and Smith for allowing us to study their patients, Sister Louise Goodwin and Leila Ayandi for their help in tissue collection, Richard Bryant and Paul Heath for array advice, and the staff and patients of the Royal Hallamshire hospital.
Funding
This work was supported by a GSK Clinician Scientist fellowship to JWFC and project grants to JWFC from Yorkshire Cancer Research, Sheffield Hospitals Charitable trust and the European Union (Framework 7).
Footnotes
REFERENCES
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.von der Maase H, Sengelov L, Roberts JT, et al. Long-term survival results of a randomized trial comparing gemcitabine plus cisplatin, with methotrexate, vinblastine, doxorubicin, plus cisplatin in patients with bladder cancer. J Clin Oncol. 2005;23:4602–8. doi: 10.1200/JCO.2005.07.757. [DOI] [PubMed] [Google Scholar]
- 3.Herr HW, Dotan Z, Donat SM, Bajorin DF. Defining optimal therapy for muscle invasive bladder cancer. J Urol. 2007;177:437–43. doi: 10.1016/j.juro.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 4.Catto JW, Azzouzi AR, Rehman I, et al. Promoter hypermethylation is associated with tumor location, stage, and subsequent progression in transitional cell carcinoma. J Clin Oncol. 2005;23:2903–10. doi: 10.1200/JCO.2005.03.163. [DOI] [PubMed] [Google Scholar]
- 5.Knowles MA. Molecular subtypes of bladder cancer: Jekyll and Hyde or chalk and cheese? Carcinogenesis. 2006;27:361–73. doi: 10.1093/carcin/bgi310. [DOI] [PubMed] [Google Scholar]
- 6.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–30. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 7.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voorhoeve PM, le Sage C, Schrier M, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–81. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 9.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. Jama. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Gottardo F, Liu CG, Ferracin M, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–92. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 13.Neely LA, Rieger-Christ KM, Neto BS, et al. A microRNA expression ratio defining the invasive phenotype in bladder tumors. Urol Oncol. 2008 doi: 10.1016/j.urolonc.2008.06.006. Epub. [DOI] [PubMed] [Google Scholar]
- 14.Catto JW, Rosario DJ. The road to cystectomy: who, when and why? Eur Urol: EAU Update series. 2005;3:171–9. [Google Scholar]
- 15.Southgate J, Hutton KA, Thomas DF, Trejdosiewicz LK. Normal human urothelial cells in vitro: proliferation and induction of stratification. Lab Invest. 1994;71:583–94. [PubMed] [Google Scholar]
- 16.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 18.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman EJ, Hurst CD, Pitt E, Chambers P, Aveyard JS, Knowles MA. Expression of hTERT immortalises normal human urothelial cells without inactivation of the p16/Rb pathway. Oncogene. 2006;25:5037–45. doi: 10.1038/sj.onc.1209513. [DOI] [PubMed] [Google Scholar]
- 20.Tomlinson DC, L’Hote CG, Kennedy W, Pitt E, Knowles MA. Alternative splicing of fibroblast growth factor receptor 3 produces a secreted isoform that inhibits fibroblast growth factor-induced proliferation and is repressed in urothelial carcinoma cell lines. Cancer Res. 2005;65:10441–9. doi: 10.1158/0008-5472.CAN-05-1718. [DOI] [PubMed] [Google Scholar]
- 21.Schmittgen TD, Lee EJ, Jiang J, et al. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44:31–8. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567–72. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wild PJ, Herr A, Wissmann C, et al. Gene expression profiling of progressive papillary noninvasive carcinomas of the urinary bladder. Clin Cancer Res. 2005;11:4415–29. doi: 10.1158/1078-0432.CCR-05-0259. [DOI] [PubMed] [Google Scholar]
- 25.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–72. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 26.Huang Q, Gumireddy K, Schrier M, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–10. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 27.Dhawan D, Hamdy FC, Rehman I, et al. Evidence for the early onset of aberrant promoter methylation in urothelial carcinoma. J Pathol. 2006;209:336–43. doi: 10.1002/path.1991. [DOI] [PubMed] [Google Scholar]
- 28.Lujambio A, Ropero S, Ballestar E, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–9. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 29.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–43. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 30.Catto JW, Hartmann A, Stoehr R, et al. Multifocal urothelial cancers with the mutator phenotype are of monoclonal origin and require panurothelial treatment for tumor clearance. J Urol. 2006;175:2323–30. doi: 10.1016/S0022-5347(06)00256-4. [DOI] [PubMed] [Google Scholar]
- 31.Chiosea S, Jelezcova E, Chandran U, et al. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–20. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiosea S, Jelezcova E, Chandran U, et al. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–50. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 33.Merritt WM, Lin YG, Han LY, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–50. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandres E, Cubedo E, Agirre X, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blaveri E, Brewer JL, Roydasgupta R, et al. Bladder cancer stage and outcome by array-based comparative genomic hybridization. Clin Cancer Res. 2005;11:7012–22. doi: 10.1158/1078-0432.CCR-05-0177. [DOI] [PubMed] [Google Scholar]
- 36.Dyrskjot L, Kruhoffer M, Thykjaer T, et al. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64:4040–8. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- 37.L’Hote CG, Knowles MA. Cell responses to FGFR3 signalling: growth, differentiation and apoptosis. Exp Cell Res. 2005;304:417–31. doi: 10.1016/j.yexcr.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 38.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–89. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 39.Aaboe M, Birkenkamp-Demtroder K, Wiuf C, et al. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res. 2006;66:3434–42. doi: 10.1158/0008-5472.CAN-05-3456. [DOI] [PubMed] [Google Scholar]
- 40.Datta J, Kutay H, Nasser MW, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–58. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105:13556–61. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–9. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.