Abstract
The reliable identification of protein interaction partners and how such interactions change in response to physiological or pathological perturbations is a key goal in most areas of cell biology. Stable isotope labeling with amino acids in cell culture (SILAC)-based mass spectrometry has been shown to provide a powerful strategy for characterizing protein complexes and identifying specific interactions. Here, we show how SILAC can be combined with computational methods drawn from the business intelligence field for multidimensional data analysis to improve the discrimination between specific and nonspecific protein associations and to analyze dynamic protein complexes. A strategy is shown for developing a protein frequency library (PFL) that improves on previous use of static “bead proteomes.” The PFL annotates the frequency of detection in co-immunoprecipitation and pulldown experiments for all proteins in the human proteome. It can provide a flexible and objective filter for discriminating between contaminants and specifically bound proteins and can be used to normalize data values and facilitate comparisons between data obtained in separate experiments. The PFL is a dynamic tool that can be filtered for specific experimental parameters to generate a customized library. It will be continuously updated as data from each new experiment are added to the library, thereby progressively enhancing its utility. The application of the PFL to pulldown experiments is especially helpful in identifying either lower abundance or less tightly bound specific components of protein complexes that are otherwise lost among the large, nonspecific background.
Many biological processes are mediated by the action and regulation of multiprotein complexes and large molecular machines rather than by individual protein molecules. Protein functions are often controlled by, and dependent upon, specific associations with one or more interaction partners, which can control subcellular localization, catalytic activity, and/or substrate specificity. Multiprotein complexes also interconnect to form functional networks that are highly dynamic and reflect the temporal and spatial complexity of cellular activity (for a review, see Ref. 1). Exploring the dynamics of protein complexes during biological responses, rather than describing static snapshots of protein interactions under unique physiological conditions, will be essential to move from a descriptive catalogue to a more functional pathway analysis. Hence, a key goal in cell biology involves identifying specific protein interaction partners and characterizing the dynamics of protein complexes and how they interconnect.
Protein complexes can include both stable, long term interactions between core components and transient and dynamic interactions that are often regulated in response to specific stimuli or signaling events. Components within multiprotein complexes can thus interact with a range of different affinities, resulting in differential loss of specific subunits during isolation or purification. In addition, not all protein subunits are present in equal stoichiometry, increasing the difficulty of reliably identifying specific but lower affinity and/or lower abundance interaction partners when characterizing protein complexes.
Various biochemical techniques have been used to identify protein-protein interactions. The most common include yeast two-hybrid screens and affinity purification procedures, either using antibodies to endogenous proteins or more frequently using exogenous expression of tagged recombinant protein baits. Recently, because of its high sensitivity, MS has become established as the method of choice for identifying purified proteins. This has been facilitated both by the improvements in MS technology and by on-line access to total genome sequences for many model organisms, including human (2). The resulting successful combination of different affinity purification techniques with MS has thus become widely used as a sensitive method for characterizing and comparing protein complexes (for reviews, see Refs. 3–5). This can be applied in high throughput and used to characterize large interaction networks or “interactomes”. For example, recent studies exploited a combined affinity purification-MS approach for the global analysis of protein complexes in yeast, reporting identification of a core set of more than 2,700 proteins organized into 491 and 547 distinct complexes, respectively (6, 7).
The high sensitivity of MS technology increases the total number of proteins identified in each pulldown experiment. However, the majority of these proteins usually represent contaminants, including proteins that bind nonspecifically to the affinity matrix. Thus, despite many technical improvements made in recent years, the unambiguous discrimination between genuine protein interaction partners, either stable or transient, and co-purifying contaminants remains one of the major challenges in the field.
Most researchers have sought to identify specific protein interactors by reducing or eliminating the background of nonspecific proteins through either biochemical or data analysis strategies. For example, at the experimental level, the buffer stringency can be increased to reduce binding of low affinity contaminants, and a two-step tandem affinity purification method can be used rather than a one-step procedure (8, 9). However, this can decrease the yield of protein recovered and risks losing low abundance and/or lower affinity specific protein interaction partners. Alternatively, on the data analysis level, several approaches have been used to identify and thereby discard the putative contaminants that are recovered after purification. For example, bioinformatics can be used to measure “confidence scores” by comparing the results of interaction studies with either predicted protein-protein interaction data or previous results described in the literature (10) or by integrating different properties of the interaction network generated by the analysis, e.g. interaction bidirectionality, etc. (11, 12).
The combination of quantitative MS and differential labeling of proteins with heavy isotopes, especially stable isotope labeling with amino acids in cell culture (SILAC)1 (13, 14), can also help to distinguish between specific and nonspecific binding proteins in a co-immunoprecipitation (co-IP) experiment. This is achieved through the inclusion of an internal negative control, which allows for direct comparison between the relative levels of each protein present in the control and experimental samples (see Fig. 1). SILAC thus objectively identifies proteins that can bind nonspecifically, e.g. to the affinity matrix and/or the fusion tag, and highlights by comparison proteins that bind specifically to the bait protein (for reviews, see Refs. 15 and 16). We and others have used this isotope-based, quantitative MS approach to characterize both tagged and endogenous protein complexes in mammalian cells (17–20). Related differential isotope-based labeling strategies, combined with MS, have also been used to analyze specific binding proteins (21, 22).
Fig. 1.
Overview of triple SILAC-based analysis of protein interaction partners. A, metabolic labeling of cells in culture using the triple SILAC approach can be used to detect specific protein interaction partners and dynamic changes in protein interactions under different biological conditions. Examples include comparing control conditions with (i) treatment with chemical inhibitors/stress etc., (ii) effect of mutations in the bait protein, or (iii) isoform-specific interactions. Light medium refers to normal environmental isotopes of carbon, nitrogen, and hydrogen, i.e.“unlabeled” 12C, 14N, and 1H, whereas medium and heavy media refer to cells grown in medium containing heavy isotope-labeled arginine (R) and lysine (K) as follows: medium, [13C6]arginine (R6) and 4,4,5,5-D4-lysine (K4); heavy, [13C6,15N4]arginine (R10) and [13C6,15N2]lysine (K8). B, overview showing the work flow in a representative triple SILAC analysis of protein interactions and their response to inhibitor treatment for either GFP-tagged or endogenous cell proteins. C, diagram illustrating the SILAC principle of differential labeling and how specific interacting proteins have higher ratios of heavy isotope-labeled peptides as compared with nonspecific contaminants. D, example of MS spectra for representative peptides illustrating a specific protein interaction partner (top), an internal contaminant binding nonspecifically to the beads (middle), and an external environmental contaminant, e.g. keratins (bottom). CTL, control; prot., protein; Ab, antibody; 1D, one-dimensional.
However, relying upon isotope labeling ratios alone does not entirely solve the contaminant problem. Indeed it is often impossible to establish a threshold ratio level in these experiments that eliminates all of the contaminating proteins without discarding, en passant, genuine interaction partners of lower abundance and/or lower binding affinity. We previously addressed this issue by systematically identifying proteins that frequently occur in pulldown experiments. These proteins were documented in a “bead proteome,” which provided a filter to help discriminate between specific interaction partners and the inevitable nonspecific background (20). However, the bead proteome approach is not a general solution to the problem because it is a static list of putative contaminants that is not updatable and that is directly relevant only to a certain set of experimental conditions.
In this study, we present a methodology for the reliable identification of specific protein interaction partners and the characterization of protein complex dynamics that overcomes limitations with our previous bead proteome approach. We drew on data analysis strategies from the field of business intelligence (BI) and applied them to integrate complex data sets arising from MS pulldown experiments. We used this to generate a protein frequency library (PFL) that can be customized to the conditions of specific experiments and continually updated. We demonstrate its use both as a specificity filter to discriminate specific protein interactions and as a tool to normalize data sets and hence facilitate comparison of separate experiments.
EXPERIMENTAL PROCEDURES
A step-by-step procedure for triple labeling SILAC-based affinity purification experiments and data analysis work flow, applied to the co-IP of either GFP-RNA polymerase II subunit C (Pol2C) or endogenous RNA polymerase II subunit A (Pol2A), is described below. Both tag-based and endogenous pulldown experiments have advantages and disadvantages. Tagged baits have been used successfully in a large number of MS-based affinity purification studies and provide a scalable and general method to identify specific protein interaction partners. In particular, the GFP tag can be used in a dual strategy combining both fluorescence microscopy and MS-based proteomics, which allow comparison of both the dynamics of localization and composition of protein complexes, respectively (19, 23). GFP has proven to be an effective tag for MS-based affinity purification procedures because of its low background of nonspecific interactions and now also because of the efficient recovery possible using recently developed “GFP-TRAP” affinity matrices (20, 24, 25). All tags, however, can potentially affect protein structure, resulting in alteration of both protein function and association with binding partners. In contrast, co-IP of endogenous proteins avoids several problems associated with the use of tags because cellular physiology is not perturbed by overexpression of a fusion protein or through structural changes induced by the tag in the bait protein. However, this strategy relies on the availability of a specific and high affinity antibody, which isolates the endogenous bait protein efficiently, that is often not available.
Tissue Culture
Parental U2OS and stable GFP-Pol2C U2OS cells were grown in custom-made Dulbecco's modified Eagle's medium (minus arginine and lysine; Invitrogen) supplemented with 10% dialyzed fetal calf serum (Invitrogen) and penicillin/streptomycin (Invitrogen). l-Arginine (R0) (84 mg/ml; Sigma) and l-lysine (K0) (146 mg/ml; Sigma) were added to the “light” (L), l-[13C6]arginine (R6) and l-4,4,5,5-D4-lysine (K4) (Cambridge Isotope Laboratories) were added to the “medium” (M), and l-[13C6,15N4]arginine (R10) and l-[13C6,15N2]lysine (K8) (Cambridge Isotope Laboratories) were added to the “heavy” (H) media. The amino acid concentrations are based on the formula for normal Dulbecco's modified Eagle's medium (Invitrogen). Once prepared, the SILAC medium was mixed well, filtered through a 0.22-μm filter (Millipore) using a suction pump, and stored at 4 °C. U2OS cells were grown in 140-mm diameter culture dishes, and five dishes were used per SILAC condition. Cells were passaged in SILAC media for at least five to six cell doublings prior to harvesting to ensure complete incorporation of isotopic amino acids (for reviews, see Refs. 26 and 27). PBS-based non-enzymatic cell dissociation buffer (Invitrogen) was used to passage cells as trypsin-EDTA-free solutions may contain amino acids. Prior to harvesting, U2OS cells stably expressing GFP-Pol2C that were grown in the heavy medium were treated with α-amanitin (5 μg/ml) for 16 h. In the case of endogenous Pol2A co-IP, parental U2OS cells that were grown in the heavy medium were treated with α-amanitin (5 μg/ml) and leptomycin B (7 nm) for 16 h.
Preparation of Cellular Extracts
Cells were trypsinized, pelleted, and resuspended in 5 ml of ice-cold buffer (20 mm Tris, pH 7.5, 10 mm KCl, 3 mm MgCl2, 0.1% Nonidet P-40, 10% glycerol, and Complete protease inhibitor mixture (Roche Applied Science)) for 10 min. After centrifugation at 750 × g for 10 min at 4 °C, cells were subjected to a second extraction step for 10 min at 4 °C in 1× RIPA buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, and Complete protease inhibitor mixture (Roche Applied Science)). The baits, were mostly detected in the cellular extracts resulting from the second extraction which were thus used for the subsequent steps. Extracts were briefly sonicated on ice (3 × 10 s at full power) and then cleared by centrifuging at 2,800 × g (3,500 rpm; GH3.8 rotor, Beckman Coulter GS-6) for 10 min at 4 °C, and total protein concentrations were measured using a Bradford assay.
Immunoaffinity Purification of GFP-tagged and Endogenous Proteins
The type of beads used for each pulldown is an issue that is worth considering as the efficiency and cleanliness of different types of beads may vary according to the cell type and the type of extract used. In our experience, Dynabeads (Invitrogen) work well for nuclear extracts, whereas Sepharose and agarose beads (GE Healthcare) can give lower backgrounds when used with cytoplasmic extracts and whole cell extracts (20).
Prior to endogenous Pol2A IP, monoclonal anti-Pol2A (Euromedex) and control anti-HA 3F10 (Roche Applied Science) antibodies were covalently coupled to protein G-Sepharose beads (GE Healthcare) at 1 mg/ml. The beads were incubated with antibody for 1 h at 4 °C and then washed twice with 10 volumes of 0.1 m sodium borate, pH 9. Next the beads were incubated with 10 volumes of borate buffer containing 20 mm dimethyl pimelimidate (Sigma) for 30 min at room temperature. The beads were pelleted and resuspended with 10 volumes of freshly prepared 20 mm dimethyl pimelimidate in borate buffer for an additional 30-min incubation. The beads were washed twice with 10 volumes of ice-cold 50 mm glycine, pH 2.5 to remove unbound antibody and then washed several times with PBS or RIPA buffer for use and/or storage at 4 °C.
For the GFP-Pol2C pulldown experiment, extracts from each cell line were precleared by incubation with protein G-Sepharose beads alone for 30 min at 4 °C and then mixed in a 1:1:1 ratio based on total protein concentration. GFP-Pol2C was affinity-purified by incubation with GFP-TRAP_A affinity matrix (Chromotek) (20, 25) for 1 h at 4 °C (equivalent of 50 μl of beads/extract). As described above, to recover transient and dynamic interaction partners that may exchange between the L, M, and H extracts during the incubation, it is advisable to keep incubation times as short as possible (less than 1 h).
In the case of GFP pulldowns, it is also possible to perform SILAC IPs by mixing extracts from control and experimental cell cultures after the affinity purification step (see Fig. 1B). This method can be used to preserve transient interactions as it has been shown that an exchange can occur between transient and dynamic protein interaction partners from different extracts during the incubation with the affinity matrix. This has been used to compare dynamic and stable interaction partners using either purification after mixing-SILAC or mixing after purification-SILAC (28, 29). These studies emphasized that the identification of specific but transient and dynamic interaction partners can be challenging.
For endogenous Pol2A IP, cellular extracts were precleared as described above, and separate IPs were performed in parallel by incubating the L extracts with the control anti-HA-Sepharose beads and the M and H extracts with the specific anti-Pol2A-Sepharose beads (50 μl of beads/extract). Again incubation times were limited to 1 h. Beads were mixed after the IP and then washed.
After the affinity purification step, the affinity matrix was washed five times with 1× RIPA buffer. To ensure efficient elution of bound proteins, a bead-equivalent volume of 1% SDS was added, the matrix was boiled for 10 min, and then a 4× volume of dH2O added. The matrix was vortexed, and the solution removed and reduced to the original bead-equivalent volume (and 1% SDS concentration) using a SpeedVac. Proteins were reduced and alkylated in this solution, first by the addition of 10 mm DTT (boiled for 2 min) and then by the addition of 50 mm iodoacetamide (incubated at room temperature in the dark for 30 min). A small aliquot of 4× lithium dodecyl sulfate sample buffer (Invitrogen) was added, and proteins were separated by one-dimensional SDS-PAGE by running halfway down NuPAGE 12% Bis-Tris gels (Invitrogen). Gels were stained with SimplyBlueTM SafeStain solution (Invitrogen), which is compatible with MS, for 1 h at room temperature and washed in dH2O overnight prior to excision of equal slices (five bands per gel lane, cut in 3 × 1-mm pieces). Gel bands were destained in dH2O and 20 mm NH4HCO3 followed by in gel-digestion using trypsin in 20 mm NH4HCO3 (Trypsin Gold, Promega) essentially as described (30). Peptides were extracted from gel pieces using CH3CN and 1% formic acid, then vacuum-dried, and resuspended in 1% formic acid solution for analysis by MS. There are alternative methods for protein digestion including (i) in-solution digestion and (ii) filter-aided sample preparation, which combines advantages from the two other techniques (31).
Liquid Chromatography-Tandem Mass Spectrometry
Trypsin-digested peptides were separated using an Ultimate U3000 (Dionex Corp.) nanoflow LC system. 10 μl of sample (a total of 2 μg of protein) was loaded with a constant flow of 20 μl/min onto a PepMap C18 trap column (0.3-mm inner diameter × 5 mm; Dionex Corp.). After trap enrichment, peptides were eluted onto a PepMap C18 nanocolumn (75 μm × 15 cm; Dionex Corp.) with a linear gradient of 5–35% solvent B (90% acetonitrile with 0.1% formic acid) over 65 min with a constant flow of 300 nl/min. The HPLC system was coupled to a linear ion trap-orbitrap hybrid mass spectrometer (LTQ-Orbitrap XL, Thermo Fisher Scientific Inc.) via a nanoelectrospray ion source (Proxeon Biosystems). The spray voltage was set to 1.2 kV, and the temperature of the heated capillary was set to 200 °C. Full-scan MS survey spectra (m/z 335–1800) in profile mode were acquired in the Orbitrap with a resolution of 60,000 after accumulation of 500,000 ions. The five most intense peptide ions from the preview scan in the Orbitrap were fragmented by collision-induced dissociation (normalized collision energy, 35%; activation Q, 0.250; and activation time, 30 ms) in the LTQ after the accumulation of 10,000 ions. Maximal filling times were 1,000 ms for the full scans and 150 ms for the MS/MS scans. Precursor ion charge state screening was enabled, and all unassigned charge states as well as singly charged species were rejected. The lock mass option was enabled for survey scans to improve mass accuracy. Data were acquired using the Xcalibur software.
Quantitative Data Analysis
The raw mass spectrometric data files obtained for each experiment were collated into a single quantitated data set using MaxQuant (version 1.0.12.31) (32, 33) and the Mascot search engine (Matrix Science, version 2.2.2) software. Enzyme specificity was set to that of trypsin, allowing for cleavage N-terminal to proline residues and between aspartic acid and proline residues. Other parameters used were: (i) variable modifications, methionine oxidation and protein N-acetylation; (ii) fixed modifications, cysteine carbamidomethylation; (iii) database: target-decoy human MaxQuant (ipi.HUMAN.v3.52.decoy) (containing 148,380 database entries); (iv) heavy labels: R6K4 and R10K8; (v) MS/MS tolerance, 0.5 Da; (vi) maximum peptide length, 6; (vii) top MS/MS peaks per 100 Da, 6; (viii) maximum missed cleavages, 2; (ix) maximum of labeled amino acids, 3; and (x) false discovery rate, 5%. In addition to the false discovery rate, proteins were considered to be identified if they had at least one unique peptide, and they were considered quantified if they had at least one quantified SILAC pair, although data quality (e.g. number of unique peptides and number of quantification events) was an essential parameter considered for the confidence given to results. It is important to keep in mind that SILAC analysis quantifies peptides, whereas the analysis of interaction partners specifically compares data for proteins. Because there can be considerable variation in the number of peptides identified and the accuracy of the quantitation for each peptide, not all protein values being compared may be equally robust or reliable. This difference in data quality is also reflected in the percentage of protein sequence coverage by the peptides identified for different proteins. In addition, it is possible that peptides identified can be assigned to specific proteins incorrectly, which can occur either for proteins expressed as multiple isoforms or where a peptide arises from a motif or domain shared by more than one protein. The use of MaxQuant software, as described above, has significantly improved the reliability and accuracy of peptide quantitation and assignment to proteins (32, 33). Nonetheless the successful interpretation of data from SILAC analyses must include awareness of the quality and confidence scores for data concerning every peptide assigned to each protein identified.
A total of 709 and 696 protein groups were identified in GFP-Pol2C and Pol2A affinity purification experiments, respectively. Proteins labeled as _REV (non-real proteins from the reverse database) were automatically discarded as well as proteins that did not show any SILAC M/L, H/L, and H/M ratio. This yielded 604 protein groups for the GFP-Pol2C pulldown and 618 protein groups for the Pol2A endogenous IP. Average SILAC ratios for each remaining protein group were plotted in several ways to assess ratio distribution (see Fig. 2B) and changes in interactions between different conditions tested (see Fig. 2C).
Fig. 2.
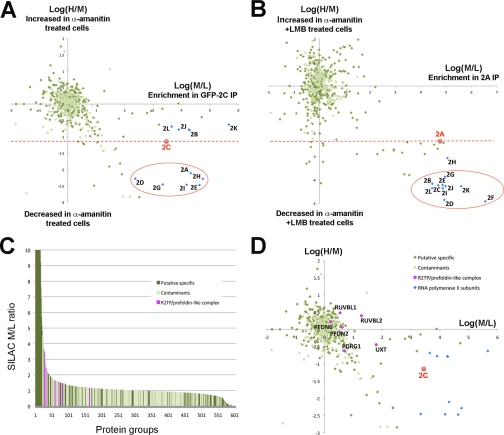
Visualization of contaminant profiles and threshold levels. A representative example of a triple SILAC co-IP experiment using GFP-Pol2C as bait in cells either with or without α-amanitin treatment2 was used to generate the graphs shown. A, graphs showing median SILAC ratios for every protein group identified and quantified by MaxQuant (604 distinct protein groups) with each protein group plotted on the x axis and the median SILAC value for that protein group plotted on the y axis. Two arbitrarily chosen thresholds are illustrated (black horizontal lines in left and right panels). B, representative ratio distribution plots. Data are plotted as a histogram with log2 SILAC ratios on the x axis and number of proteins for a given ratio on the y axis. Nonspecific contaminants reproducibly cluster in a Gaussian (normal) distribution centered at ∼0 (left panel), although the exact mean can deviate from 0 due to experimental variability as seen for the GFP-Pol2C data set (right panel). C, data from the GFP-Pol2C data set plotted with log2(M/L) SILAC ratio on the x axis and log2(H/M) SILAC ratio on the y axis with each point corresponding to the ratio value for a specific protein group. The bait protein is shown in red. Putative experimental contaminants (Experim. contamin.) cluster around the origin.
Sun Model and Multidimensional On-line Analytical Processing (OLAP) Analysis
The steps involved in creating the sun model and OLAP cube are detailed below. Initially, a data environment (PepTracker) was created to manage the experimental data sets generated by MaxQuant. PepTracker shall be described, in detail, separately. The data sets were collated in a relational database implemented using MySQL and used to generate a PFL. In addition, extensive metadata were also recorded that describe the conditions under which experiments were carried out and parameters associated with the experiments, such as cell type, organism, extract, type of beads, machine, date, user, antibody, treatment, etc. The relational database modeled reality by breaking the data into one or more sets, each of which represented a class of real world entity, for example protein, experiment, user, etc. This data store was optimal for storage and transaction-based operations.
Because of the complexity of the analysis and the large volumes of data involved, a new approach was required. The alternative approach we adopted made use of BI principles, which include methods of leveraging data to provide an informed platform for decision making. The BI method of analyzing data includes OLAP, which can use a multidimensional data model that alleviates problems inherent in a relational database by making it easier to select, navigate, and explore data. It is also able to provide increased query performance in comparison with a relational database because the structure supports preaggregation of the data. Almost all query result times benefit from this type of precomputation.
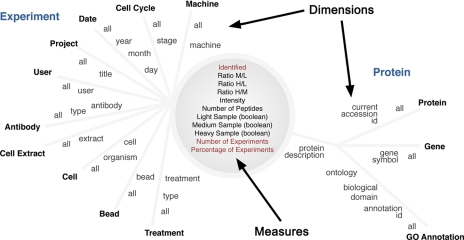
When designing the multidimensional structure, the user model, which is defined by the users' understanding and perception of the data, was translated into a logical model. This logical model contained measures and dimensions. The measures are numerical values from the experimental data that are of interest to researchers, e.g. ratio, intensity, etc. The dimensions define the various groupings (often hierarchical) by which users can aggregate the measures, e.g. treatment, date, and cell cycle. The logical model was then represented as a sun model (see Fig. 3), which shows the measures in the center of the diagram and dimensions radiating from the center. The hierarchies in a dimension are symbolized by the levels marked along a dimension line. For example, the date dimension is hierarchical and has year, month, and day levels. In this study, we made use of the dimensions “bead type” and “cell extract” as filters to obtain customized PFLs.
Fig. 3.
Sun diagram and logical model of SILAC data. A logical model is presented in the form of a sun diagram illustrating the relationship between Measures and Dimensions captured in a SILAC experiment. The measures are typically numerical values from the experimental data, e.g.“number of peptides.” The dimensions define the various groupings (often hierarchical) by which users can aggregate the measures, e.g. cell type, date, cell extract, etc. id, identification.
The data for the analysis came from three sources: the relational database within PepTracker described earlier and local versions of the IPI human and gene ontology databases. To ensure high data quality and consistency of format, the data were extracted from these systems, transformed appropriately, and loaded into a central repository (data warehouse). During this process, appropriate tables were created to store the data. The measures were incorporated into a fact table, and the dimensions each became a dimension table. The fact table maintained a link to all of the related dimension tables, creating a star schema whereby the dimension tables relate to a central fact table producing a star shape. This extract, transform, and load process is characteristic of most BI systems as is the creation of a data warehouse. The data in the data warehouse are not updated, but rather new data sets are appended to the data when they become available. This method created a data warehouse containing historical experimental data that are subject-orientated, non-volatile, and well integrated, existing separately from the operational environment of the original data.
In terms of the IP data sets, there were a number of decisions that had to be considered carefully to decide how best to transform the data into an accurate representation of the user model. This involved making the determination that proteins should only be included if they were both identified and quantified in a SILAC experiment. In addition, it was decided that proteins should be identified via IPI accession number as this is the identifier type used by the MaxQuant suite to make protein identifications. Furthermore, because the IPI accession identifiers are continuously updated, it was decided that proteins should be mapped to the most current identifier. Thus, multiple occurrences of the same protein accession number in a single experiment should only be allocated the weighting of a single identification and quantification in the frequency value of any generated protein library annotation.
The next step involved converting the logical model, designed for the PFL, to a physical model using the SQL Server Analysis Services (SSAS) component of Microsoft SQL Server 2005. This physical model, a multidimensional database, is often known as an OLAP cube. The OLAP cube was generated from the consistent, clean data stored in the data warehouse. When generating the OLAP cube, the data source, fact tables, dimension tables, relationships between fact and dimension tables, and hierarchies had to be specified. Using this information, the cube was then processed with all of the data aggregated at defined levels within the multidimensional structure. With the use of the OLAP cube and its modeling of a range of measures and dimensions, it was possible to perform a variety of query and analysis tasks. To extract data from the OLAP cube, a powerful analytical query language (multidimensional expressions (MDX)) is available that allows very complex analytical queries to be expressed with ease. In this study, we used Excel to connect to the OLAP cube and extract the required data for the PFL. Leveraging the dimensions within the cube, we were also able to extract subsets of data for customized PFLs (see Fig. 5).
Fig. 5.
Filtering of PFL using experimental parameters (dimensions). A, using a web-based interface, any individual dimensions within the data model (corresponding to experimental parameters recorded in the database) can be used in conjunction with the OLAP cube to create a customized PFL. This is illustrated here for the dimensions cell extract (cytoplasmic and nuclear) (B) and affinity matrix (Sepharose beads) (C) used for pulldown experiments.
RESULTS
Multiplex SILAC Identification of Specific Protein Interactions
A standard work flow for triple labeling SILAC-based pulldown experiments in mammalian cells is summarized in Fig. 1. This triple labeling strategy enables both the identification of specific protein interactions and the analysis of changes occurring in these protein interactions between two different conditions. This is done by the use of three separate growth conditions to label cells with different isotopes that can be resolved and quantitated by MS. The same strategy can be applied to the pulldown of both tagged and endogenous protein baits (19, 20). A triple SILAC GFP pulldown is shown in Fig. 1A as an example. An internal control is provided by cells grown in light (L), i.e. unlabeled (12C, 14N, 1H) medium, whereas cells stably expressing the GFP-bait fusion protein are grown either with medium (M) or with heavy (H) isotope-labeled arginine or lysine amino acids (Fig. 1A). The cells grown in medium and heavy media are used to compare changes in specific protein interaction partners between, for example, (i) control conditions and treatment with chemical inhibitors/stress etc., (ii) wild-type and mutant forms of the bait protein, or (iii) two different isoforms of the same protein (19) (Fig. 1A). Several SILAC experiments can be performed in parallel to analyze dynamics of protein interactions under more than two conditions.
A typical experimental procedure for both GFP and endogenous co-IP triple SILAC experiments is described in Fig. 1B (for details, see “Experimental Procedures”). In brief, cell lysates are prepared from each of the L, M, and H cell cultures, and the bait protein and associated interaction partners are immunoaffinity-purified. Eluted proteins are then in-gel digested with trypsin (or other proteases), and peptides are analyzed by LC-MS/MS and quantified using MaxQuant (32, 33).
All affinity purification methods are inevitably linked with the co-purification of “contaminants” that bind nonspecifically to the beads and/or to the fusion tag. The SILAC principle of using differential isotope labeling is shown in Fig. 1C. In a triple SILAC pulldown experiment, each identified peptide thus shows a typical MS spectrum with three main peaks that correspond to the light, medium, and heavy isotopic forms, respectively (Fig. 1D). The relative abundance of each distinct peak area can be efficiently quantified by MaxQuant (32, 33), which indicates three median ratios (M/L, H/L, and H/M) for each peptide as well as the median value for all peptides quantitated for a specific protein. In this strategy, both the bait protein itself and its specific interaction partners are expected to have a higher M/L and/or H/L ratio than nonspecific contaminants. In contrast, experimental contaminants, e.g. proteins that bind nonspecifically to beads, are expected to have M/L and H/L ratios close to 1. Proteins that show M/L and H/L ratios significantly lower than 1 are mostly external contaminants, such as keratins, with no incorporation of heavy isotopes (Fig. 1D). In summary, the analysis of triple isotope labeling SILAC co-IP data theoretically allows for (i) the discrimination between contaminants and genuine interaction partners and (ii) the characterization of changes in protein complexes under specific biological conditions.
Discriminating Specific from Nonspecific Interaction Partners: Contaminant Profiles and Establishing Thresholds
Fig. 2 shows an example of data analysis from a representative SILAC pulldown experiment in which GFP-tagged Pol2C was affinity-purified from U2OS cells. In this case, after in-gel digestion, the MS analysis identified and quantitated over 4,000 peptides that were assigned by MaxQuant to 604 human protein groups. For each protein group (x axis), a median M/L SILAC ratio was calculated from all of the individual peptide values determined and is shown plotted on the y axis (Fig 2A). This shows that a minor group of proteins has a high SILAC M/L ratio (>2), whereas ∼80% of the proteins (i.e. over 480 of a total of 604 protein groups) have a SILAC ratio <1.4. As described above, the former are strong candidates to be specific interaction partners (Fig. 2A, green columns), whereas the latter are more likely to be nonspecific interaction partners. However, experience has shown that some bona fide specific interaction partners can have SILAC ratios lower than abundant contaminants (e.g. in the range of ∼0.6–1.4). Thus, when setting the threshold to an arbitrary value, it is important to understand that if the selected threshold is high, although most (or all) contaminants will be eliminated, low abundance and/or low affinity genuine interaction partners will be lost. Conversely, if the chosen threshold value is low with the aim of identifying all low abundance and/or low affinity partners, a larger number of contaminants will remain (Fig. 2A, compare left and right panels). It is therefore not possible to use a specific ratio value as a threshold that consistently and unambiguously separates the specific from the nonspecific interaction partners.
Another way of visualizing the same SILAC pulldown data is to plot the ratio distribution as a histogram. Thus, for either M/L, H/L, or H/M SILAC ratios, the number of proteins with each ratio value is plotted on the y axis against log2 SILAC ratio values on the x axis (Fig. 2B). Here, nonspecific, experimental contaminants reproducibly cluster in a Gaussian (normal) distribution centered at the log2 ratio of ∼0 (which corresponds to a SILAC ratio of ∼1) (Fig. 2B, left panel). Theoretically, the normal distribution should be centered on a log2 value of exactly 0, but in practice, this varies between individual experiments, and the actual mean can be either higher or lower even for the separate M/L, H/L, and H/M ratios measured within a single triple SILAC experiment (Fig. 2B, right panel). In contrast, putative interaction partners are expected to show log2 ratio values greater than the mean of the Gaussian curve, whereas environmental (external) contaminants always have values lower than the central mean value (Fig. 2B, left panel). The Gaussian curve can be useful to help refine the analysis of predicted specific interacting proteins using a mathematical description of the protein distribution. However, there is still no single ratio value to reliably distinguish specific from nonspecific proteins.
Fig. 2C shows a third way of visualizing the data, i.e. by plotting log2(H/M) (y axis) versus log2(M/L) (x axis) SILAC ratio values for all proteins identified in the triple SILAC co-IP experiment using GFP-Pol2C as bait. This visualization provides an indication of both the specificity of the interaction (M/L ratio) and the changes occurring between the two conditions tested (H/M ratio). From this graph, it is evident that most proteins have SILAC ratio values that cluster around the origin (Fig. 2C, circled proteins). As these proteins have log2(M/L) and log2(H/M) ratios of approximately 0, they have a high probability of being contaminants. Because of small variations in each experiment, e.g. volume differences when mixing extracts, the contaminants typically cluster around values that can, however, deviate from 0 (Fig. 2, B, right panel, and C). In contrast, putative specific interaction partners are present in the right side of the graph. But as described above and regardless of how the SILAC pulldown data are visualized, the problem remains that a significant overlap invariably exists between the SILAC ratio values of specific interaction partners and contaminating background proteins. Thus, although the SILAC approach is a powerful approach to identify stable interaction partners, we have observed that relying upon SILAC ratios alone is often not enough to reliably identify bona fide interaction partners of lower abundance and/or lower binding affinity. To address this problem, we sought to add an additional objective criterion to the analysis. Thus we developed a strategy based upon systematically annotating each protein in the proteome with its frequency of detection in a database of independent co-IP experiments, creating what we term a PFL. Hence, the PFL provides a probability estimate for each protein to be a contaminant that is independent of the information given by SILAC ratios and, therefore, can be applied to analyze both SILAC and label-free data.
Sun Model and Protein Frequency Library
To generate the PFL, a data environment was created (PepTracker) that will be described in more detail in a future publication. This data environment manages MS-based proteomics data, including the experimental data sets generated by MaxQuant, along with consistent and reliable metadata descriptors. This records parameters including cell type, organism, extract type, type of affinity matrix, mass spectrometer, date, user, etc. Furthermore, analytical functionality was built into the system, enabling it to generate a library of protein annotations. Because of the high complexity of the analysis and large volumes of data involved, our approach takes advantage of BI principles designed for rapid interactive responses (34) combined with OLAP, which makes use of a multidimensional data model in preference to a relational database structure.
An OLAP cube manages data in a cubelike structure in which the edges of the cube represent dimensions, and the measures are contained within the cube. Data are then extracted from the cube by traversing the edges. Using the hierarchies within the dimensions, users can both drill down and drill up to the required level of detail and make use of “slice and dice” operations to change the set of dimensions being viewed. Although an OLAP cube suggests modeling of only three dimensions, in reality data of n dimensions can be modeled by OLAP in a hypercube structure.
To quantify the analytical requirements that typify quantitative SILAC pulldown experiments, a logical model was constructed. This takes form as a “sun model” (Fig. 3), which shows the “measures” (e.g. SILAC ratio values, number of peptides identified, etc.) in the center of the diagram and the “dimensions” (e.g. type of affinity matrix, type of extract, date, user, etc.) radiating from the center. The hierarchies that can exist within each dimension (e.g. date can include year, month, day, etc.) are symbolized by the levels marked along a dimension line. This logical model was then concerted to an OLAP cube implementation (see “Experimental Procedures” for details of implementation). The PFL was extracted using the logical model combined with the OLAP cube, focusing on the measure called “identified.”
The measure called identified signifies whether a given protein was identified and quantified in a particular co-IP experiment that is currently in the data repository. The data repository used here contains 38 SILAC co-IP experiments, but it is important to note that the PFL can also be generated using data from non-SILAC, label-free experiments. Proteins were identified via IPI accession number, which provides a comprehensive description that is consistent with the output from MaxQuant. We note that, because of the continuous updating of IPI identifiers, proteins were mapped to the most current identifier, and thus multiple occurrences of the same protein accession number in a single experiment were only allocated the weighting of a single identification and quantification in the frequency value of any generated protein library annotation. Using the measure called identified, the number of times each protein appeared in all 38 experiments in the database was calculated, giving rise to a deduced “frequency of detection” for each of the 10,623 IPI numbers described by the data sets. This value was used in the generation of the PFL.
The PFL graph presented in Fig. 4A shows a visualization of the frequency of detection plotted against all proteins that were identified and quantified in any of the 38 experiments. In this graph, each protein is shown sorted from the highest to the lowest percentage. Hence, the proteins appearing nearest the origin of the graph have the highest probability of being contaminants.
Fig. 4.
Protein frequency library construction and validation. A, the sun diagram was used in conjunction with an OLAP cube to analyze the frequency of protein detection in a database containing data from 38 separate SILAC co-IP experiments. The graph illustrates the frequency of detection (y axis) for 10,623 separate IPI numbers (x axis). This defines a PFL. B, comparison of data from the current PFL and a previously determined list of bead proteome contaminants (20). C, correlation between the bead proteome coverage (20) and the PFL with PFL proteins ranked from highest to lowest detection frequency (left to right). X-ref is cross reference. D, comparison, for each 10% PFL segment, measuring the number of bead proteome proteins (20) found in that segment versus the total number of proteins found in that segment.
We compared the PFL with the previously characterized bead proteome, which contains 3,400 separate human IPI numbers that were frequently found in 27 independent SILAC pulldown experiments (20). The bead proteome includes many abundant factors, such as histones and cytoskeleton and heat shock proteins, and was thus extrapolated to include most members of these large protein families. Although they all potentially can behave as common contaminants, not all are either expressed or detected in every cell type or pulldown experiment. An overlap of 64% was observed between the static bead proteome and the PFL as shown in the Venn diagram (Fig. 4B). The 36% of bead proteome proteins that were not present in the PFL were mostly additional members of large protein families that did not appear in this set of 38 pulldown experiments.
Further comparison shows that most of the common proteins listed in both the bead proteome and PFL appear in the top 2,000 of 10,623 IPI numbers of the PFL when proteins are ranked from highest to lowest detection frequency. In contrast, only a small fraction of the bead proteome proteins are found in the bottom (low frequency) end of the PFL (Fig. 4C). This shows that most contaminants identified in the bead proteome are associated with a high frequency in the PFL. In addition, we compared, for each sequential PFL “10%” segment, i.e. all proteins associated with a PFL frequency range between 90 and 100%, 80 and 90%, etc., the number of bead proteome proteins versus the total number of proteins found in that segment. This shows that almost all proteins with a high frequency of detection in the PFL (>60%) are also listed in the bead proteome, whereas most proteins with a low frequency of detection in the PFL (<20%) are not (Fig. 4D). These data underline the positive correlation between the PFL and the bead proteome and validate the utility of the PFL approach for predicting contaminant proteins. A major advantage of the PFL, as compared with the previous “static” bead proteome, is that it provides an annotation of proteins that is both customizable to reflect the details of individual experiments and updatable. Hence, it will increase in accuracy as new data are added to the data repository.
Filtering of Protein Frequency Library Using Experimental Parameters
The use of the OLAP cube and its range of measures and dimensions provide a dynamic list of contaminants that can be customized for individual experiments. Fig. 5A shows an example of an interface in PepTracker that can be used to flexibly specify the parameters (in principle, drawing on all of the dimensions that were incorporated into the cube) on which the library could be filtered so that an analysis can be customized to the detailed conditions used for a specific pulldown experiment. Here, we filtered the PFL using the dimensions cell extract (Fig. 5B) and bead type (i.e. type of affinity matrix) (Fig. 5C). Thus, among all 38 SILAC pulldown experiments in the data repository, only the ones that were performed with either a specific type of extract (e.g. cytoplasmic or nuclear extract) (Fig. 5B) or a specific type of bead (e.g. Sepharose beads) (Fig. 5C) were used to generate a customized PFL. This customization feature of the PFL avoids the need to have a large set of control experiments that exhaustively cover every possible experimental parameter analyzed by combining the different parameters associated with each experiment in the data repository and thus increasing the value of each individual data set. The PFL is thus applicable also for the analysis of low throughput co-IP experiments when high throughput bioinformatics analysis techniques aiming to discard contaminants cannot be applied.
Application of PFL Filter to Analysis of Multiprotein Complexes
The PFL was applied to analyze the SILAC data from the GFP-Pol2C pulldown experiment (Fig. 6A). As this was performed with Sepharose beads, we filtered the PFL to generate a Sepharose PFL as in Fig. 5C. A subset of the Sepharose PFL library is shown where only proteins that were identified in the GFP-Pol2C data set are displayed, i.e. a cross-reference between the Pol2C data set and the Sepharose PFL, which gives a total of 2,973 IPI numbers. A continuous color coding (from red to green) was applied to the graph, representing proteins with highest detection frequency (red) to the proteins with lowest detection frequency (green) (Fig. 6A, left panel). The same high (red) to low (green) color coding was then applied to the log2(H/M) against log2(M/L) ratio plot (Fig. 6A, right panel). Proteins with high frequency of detection (red) cluster around the origin, whereas the proteins with lower frequency of detection (green) spread further across the graph. This illustrates the strong positive correlation between proteins that show a high frequency of detection and proteins that cluster around the origin in this IP experiment, which is the expected behavior of contaminant proteins.
Fig. 6.
Application of PFL data in identification of specific protein interactors. A, cross-reference (x-ref) between the customized “Sepharose” PFL data (as in Fig. 5C) and the GFP-Pol2C data set. Continuous color coding from red (highest) to green (lowest) is used to depict frequency of protein detection (left panel). In the right panel, the same color coding is applied to the log2(H/M) against log2(M/L) SILAC ratio plot of GFP-Pol2C data set (plot as shown in Fig. 2C). B, comparison of arbitrary threshold values (80, 60, 40, and 20% detection frequency in PFL) to visualize all proteins in the data set that show a frequency of detection above that threshold (highlighted in red) on the log2 plot of SILAC ratios for the same pulldown experiment shown in A. Lower threshold values result in highlighting of larger number of proteins. C, the graph shows the PFL frequency (y axis) plotted against the SILAC M/L ratio (x axis) for each protein group in the Pol2C data set. A red line is drawn indicating the minimum suitable PFL threshold that includes all protein groups with a high M/L ratio in the likely set of putative interaction partners.
Next we used the Sepharose PFL to isolate within the GFP-Pol2C data set a group of proteins predicted to include predominantly contaminants. This was done by (i) establishing a threshold value for protein detection frequency and (ii) highlighting all proteins in the data set that show a frequency of detection above that threshold. A threshold value of 100% corresponds to only those proteins detected in every data set in the library. A threshold value of 0 instead would include every protein identified in any data set. We therefore investigated four intermediate frequency thresholds, corresponding to 80, 60, 40, and 20% frequency of detection. Each was applied to log2(H/M) versus log2(M/L) ratio plots of the GFP-Pol2C data set and compared (Fig. 6B). As the threshold value for the frequency of detection decreases, the number of proteins included in the subset of putative contaminants (highlighted in red on the graphs) increases. The majority of these proteins cluster either at the origin or on the left quadrants of the graph, exactly as expected if they are indeed contaminants. External contaminants, such as keratins, are always present in the left-hand quadrants. At lower threshold values, the probability that some specific interacting proteins are also highlighted is increased. By plotting the PFL frequency value against the M/L SILAC ratio for every protein in the data set (Fig. 6C), a threshold value of 40% was chosen because it retains the main stable interaction partners of the bait and selects a suitable subset of clustered contaminants for further normalization of the GFP-Pol2C data set as described below. The choice of an optimal frequency threshold may vary for different experiments. However, the threshold value used is expected to become lower as the number of experiments used to generate the PFL increases. Although it is currently not possible to calculate accurately the minimal number of independent experiments required to provide a reliable PFL, based upon our current experience, we estimate that at least 10–15 independent pulldown experiments using different baits constitute a basic requirement.
Use of PFL to Normalize Data Sets
Ideally, samples for SILAC analysis are prepared identically with no variability in experimental conditions and with precisely equal amounts of labeled samples mixed before MS, which should lead to a normal distribution of SILAC ratios centered on exactly 0. However, in practice, slight variations in experimental conditions, e.g. pipetting accuracy, etc., are unavoidable, resulting in minor variations in SILAC ratios and hence in a ratio distribution whose mean deviates from 0 (Fig. 2, B and C). Although this generally does not compromise the interpretation of data within a given experiment, it can complicate the accurate comparison of separate data sets, i.e. either biological replicates or independent experiments. Accurate comparison of separate experiments thus requires that data sets are normalized objectively to compensate for intrinsic variations in SILAC ratios.
The MaxQuant software provides a method of data normalization that is based on the whole data set in a specific experiment being analyzed, and this assumes that most proteins should not change between conditions. However, in a SILAC pulldown experiment, it is expected that specific interacting proteins should change between the three conditions (L, M, and H). Thus, we make use of the PFL to normalize data sets by isolating a group of proteins that can confidently be predicted as mostly contaminants and, hence, whose log SILAC ratios should be exactly 0.
The normalization process is illustrated for the GFP-Pol2C data set (Fig. 7A). Using the Sepharose PFL with a threshold value set to 40% frequency, the resulting proteins with frequency above 40% were isolated within the data set (Fig. 7A, middle graph), and their median value of SILAC ratios was calculated for all three conditions (i.e. M/L, H/L, and H/M). MaxQuant non-normalized SILAC ratios were used, and the SILAC ratios of external contaminants, e.g. keratins, were excluded from the normalization process. SILAC ratio values for all proteins in the data set, including putative contaminants and specific interactors, were then divided by the corresponding median value. This normalizes the median log ratio value for the predicted contaminant group to exactly 0 (Fig. 7A, right panels). The cluster of contaminants is thereby centered on the origin of the graph (Fig. 7A, bottom graph). This normalization process does not alter the positions of proteins relative to each other within this experiment but rather globally affects the ratio values of all the proteins in the data set. The same normalization process can be applied to any data set, including co-IP analysis of an endogenous protein, as shown for the data set from SILAC affinity purification of endogenous Pol2A (Fig. 7B).
Fig. 7.
Normalization of data sets using PFL. A, graphs show log2 SILAC ratio plots of total proteins identified from co-IP using GFP-Pol2C as bait before normalization or threshold analysis (i) and after application of a 40% Sepharose PFL threshold filter with the plot now showing only putative contaminants (light green), i.e., proteins with PFL values over 40% (ii) and total data set replotted after normalization to set the median SILAC log2 ratio value of predicted contaminants to 0 (iii). Predicted contaminants are shaded in light green, and other proteins are shown in dark green. The effect of this normalization procedure on the Gaussian ratio distribution curves for the three separate M/L, H/L, and H/M values recorded in the triple SILAC analysis is shown in parallel on the right for i–iii in the form of SILAC ratio distribution histograms (as in Fig. 2B). B, repeat of the normalization procedure shown above using data from a separate triple SILAC co-IP experiment using antibodies to an endogenous protein (Pol2A) rather than a GFP-tagged bait.
Comparative Analysis of Normalized Data Sets
Next we used the data analysis work flow described above to analyze normalized GFP-Pol2C pulldown and endogenous Pol2A co-IP triple SILAC experiments. A customized Sepharose PFL combined with a frequency threshold of 40% was used (i) to highlight putative nonspecific contaminants and (ii) to normalize the data sets. Fig. 8 shows the GFP-Pol2C and endogenous Pol2A data sets plotted as log2(H/M) against log2(M/L) ratios after normalization (Fig. 8, A and B, respectively). The predicted contaminant-enriched group, i.e. proteins that show a frequency of detection above 40% in the Sepharose PFL, are shaded in light green, and all other proteins are shown in dark green.
Fig. 8.
Analysis of protein interaction dynamics using normalized data sets. A and B, graphs are log2 SILAC M/L versus H/M ratios comparing normalized data sets from triple SILAC experiments analyzing proteins specifically interacting with either GFP-Pol2C (A) or endogenous Pol2A (B). Each point represents the normalized median SILAC ratio value for all quantified peptides assigned to that protein. Bait proteins are shown in red. Core subunits of RNA polymerase II are shown in blue. A threshold PFL value of 40% was used, and all proteins with a 40% or greater frequency value are shown in light green. The dotted red line shows an alternative x axis defined by the behavior of the bait protein. Proteins within red ovals are RNA polymerase II subunits whose specific interaction with the bait shows a decrease of 2-fold or more. C, identification of specific protein interaction partners with low M/L SILAC ratios using PFL frequencies. The graph shows each protein group identified in the Pol2C data set plotted on the x axis and the normalized median SILAC value for that protein group plotted on the y axis (similar to Fig. 2A). It has been color-coded to highlight all protein groups with a PFL value below 40% in dark green, whereas protein groups showing a frequency value above 40% are shown in light green. Proteins belonging to the R2TP/prefoldin-like complex (11) are highlighted in purple. D, same graph as A with the proteins of the R2TP/prefoldin-like complex highlighted in purple.
GFP-Pol2C interaction partners were analyzed in U2OS cells either with or without α-amanitin treatment (Fig. 8A), whereas endogenous Pol2A interaction partners were analyzed in U2OS cells either with or without combined α-amanitin and leptomycin B treatment. RNA polymerase II subunits are marked in blue, and the bait protein is highlighted in red (Fig. 8, A and B). In the Pol2C co-IP experiment, 11 of the 12 known RNA polymerase II subunits (Pol2A–Pol2L) were detected with high sequence coverage, and a large number of peptides were identified and quantified (see Table I for GFP-Pol2C data set). All 12 RNA polymerase II subunits were identified in the Pol2A co-IP experiment.
Table I. Comparison of peptide data quality for RNA polymerase II subunits.
All known RNA polymerase II subunits (Pol2A–Pol2L) except Pol2F were identified and quantified in the SILAC co-IP using GFP-Pol2C as bait. They all show high sequence coverage and a large number of peptides identified and quantified, underlining the quality of the data. The bait protein, GFP-Pol2C, is bold. log2(H/M) ratios of all subunits are normalized in the table so that log2(H/M) of Pol2C is 0. Subunits are listed above the bait protein when their interaction with the bait was increased upon α-amanitin treatment (log2(H/M) versus 2C > 0), whereas subunits are listed below when their interaction with the bait protein was decreased upon α-amanitin treatment (log2(H/M) versus 2C < 0). Interactions are considered significantly affected when log2(H/M) versus 2C > 1 or log2(H/M) versus 2C < −1 (equivalent to a change in value of 2-fold or greater).
| Gene names | Accession numbers | No. of unique peptides | Sequence coverage | No. of peptides quantified | SILAC M/L ratio | log2(H/M) vs. 2C | (H/M) S.D. |
|---|---|---|---|---|---|---|---|
| % | % | ||||||
| POLR2K | IPI00023975.1 | 3 | 53.4 | 3 | 56.2 | 0.51 | 3.4 |
| POLR2L | IPI00003311.1 | 4 | 74.6 | 9 | 13.9 | 0.45 | 8.5 |
| POLR2J | IPI00003310.2, IPI00873238.1, IPI00291359.3, IPI00884938.1, IPI00878433.1, IPI00553186.2, IPI00744926.1, IPI00472231.7, IPI00556199.1, IPI00016841.3, IPI00748167.1, IPI00879159.1, IPI00880135.1 | 4 | 57.5 | 21 | 21.2 | 0.35 | 15.5 |
| POLR2B | IPI00027808.1, IPI00873948.2, IPI00894355.1, IPI00894141.1, IPI00418797.4, IPI00026445.3, IPI00184886.3, IPI00894524.1, IPI00894248.1 | 81 | 62.1 | 272 | 21.5 | 0.35 | 16.6 |
| POLR2C | IPI00018288.1 | 17 | 85.8 | 73 | 12.4 | 0 | 24.1 |
| POLR2A | IPI00031627.3, IPI00385524.1, IPI00419565.3, IPI00383337.1, IPI00784155.1 | 116 | 61.4 | 215 | 22.2 | −0.97 | 36.1 |
| POLR2D | IPI00007283.1 | 6 | 62 | 5 | 5.7 | −1.14 | 26.4 |
| POLR2H | IPI00003309.4, IPI00791019.1, IPI00790361.1, IPI00791273.1 | 8 | 58 | 12 | 29.9 | −1.15 | 31.5 |
| POLR2G | IPI00218895.6 | 11 | 74.4 | 14 | 11.2 | −1.32 | 35.7 |
| POLR2I | IPI00006113.1 | 7 | 78.4 | 9 | 20.1 | −1.33 | 42.1 |
| POLR2E | IPI00291093.3 | 10 | 58.1 | 23 | 27.6 | −1.34 | 34.8 |
If the pulldown efficiency is the same between the two conditions tested (±α-amanitin), the log2(H/M) ratio should be 0 for the bait protein. In practice, this is often not the case due for example to variations in expression levels, accessibility, and/or fractionation efficiency induced by the treatment. Hence, we used the bait protein as a reference point to draw a second x axis such that proteins falling above the new x axis line indicate increased interaction with the bait and proteins falling below indicate decreased interaction as a result of the treatment. Here, interactions were considered as significantly affected when a 2-fold or greater change was observed upon treatment (Table I). The GFP-Pol2C data set shows partial disassembly of the RNA polymerase II complex after α-amanitin treatment (Fig. 8A) because GFP-Pol2C interaction with many RNA polymerase II subunits, including 2A, 2D, 2E, 2G, 2H, and 2I, is significantly decreased after α-amanitin treatment (Fig. 8A, proteins within the red oval). However, some subunits remain associated, and new protein interaction partners were also identified, suggesting that intermediate subcomplexes are formed upon α-amanitin treatment. The same approach was applied to analyze the Pol2A data set, showing that Pol2A interaction with all RNA polymerase II subunits, except Pol2H, is decreased after treatment with both α-amanitin and leptomycin B (Fig. 8B, proteins within the red oval). A more detailed analysis and discussion of these data characterizing the formation of subcomplexes during RNA polymerase II assembly is presented elsewhere.2
Importantly, although high SILAC M/L ratios unambiguously identify specific interaction partners, the application of the PFL to the data set can help identify additional specific interaction partners otherwise missed because their lower SILAC ratios overlap with nonspecific contaminants. This overlap is particularly visible for proteins with a SILAC M/L ratio <3 (Fig. 8C, dark and light green columns). By highlighting all predicted contaminants (frequency of detection >40%), the PFL approach helps to focus on the remaining putative specific interaction partners. For example, many proteins of the R2TP/prefoldin-like complex, i.e. UXT, RUVBL1/2, PFDN2/6, and PDRG1, were not identified in the Pol2C data set with high SILAC M/L ratios but show a frequency value below 40% (see Table II and Fig. 8D, purple data points). Interestingly, the R2TP/prefoldin-like complex has been connected to the RNA polymerase II complex (11, 35). This shows that these proteins are indeed bona fide interaction partners of Pol2C that would have been overlooked in the analysis without the PFL. In summary, the PFL approach combined with triple SILAC experiments has been shown to provide an effective and flexible work flow for the detection and analysis of specific interactions within multiprotein complexes.
Table II. Embedding of putative specific interaction partners within contaminants at low SILAC M/L ratios.
A selection of protein groups from the Pol2C data set with low SILAC M/L ratios (<5) are listed with their PFL frequencies and ranked by M/L ratio from highest to lowest. Protein groups with a PFL value below the threshold (40%) and belonging to the R2TP/prefoldin-like complex are bold.
| Gene names | Accession numbers | Normalized SILAC M/L ratio | PFL frequency value |
|---|---|---|---|
| KRT19 | IPI00479145.2 | 5.03 | 100 |
| UXT | IPI00170862.1, IPI00002646.1, IPI00553080.1 | 3.85 | 16 |
| ACTN4 | IPI00013808.1, IPI00908458.1, IPI00845465.1, IPI00908776.1, IPI00793285.1, IPI00903019.1, IPI00018829.2, IPI00217047.4, IPI00217048.1, IPI00217044.1 | 3.43 | 52 |
| RUVBL2 | IPI00009104.7, IPI00909925.1 | 2.70 | 26 |
| TUBB8 | IPI00292496.1 | 2.01 | 77 |
| PDRG1 | IPI00027887.4 | 1.81 | 16 |
| HIST2H2AB | IPI00216730.3, IPI00829588.1 | 1.77 | 81 |
| PFDN2 | IPI00006052.3 | 1.7 | 26 |
| VIM | IPI00418471.6, IPI00552689.1, IPI00465084.6, IPI00793184.1, IPI00013164.4, IPI00910602.1, IPI00021751.5, IPI00217507.5, IPI00869219.1, IPI00853115.1, IPI00908745.1, IPI00237671.9, IPI00868727.1, IPI00166205.2, IPI00477227.3, IPI00001453.2, IPI00853283.1, IPI00744385.2, IPI00909238.1 | 1.62 | 97 |
| RUVBL1 | IPI00021187.4, IPI00788942.1, IPI00902501.1, IPI00796459.1 | 1.61 | 29 |
| FLNC | IPI00178352.5, IPI00413958.4, IPI00455021.3 | 1.44 | 77 |
| PFDN6 | IPI00005657.1 | 1.28 | 13 |
DISCUSSION
In this study, we have introduced the use of data analysis technology adapted from the field of BI to improve the reliability of discriminating specific from nonspecific protein interaction partners. Although this approach is broadly applicable to a wide range of protein interaction analyses, we focus here on describing an enhanced methodology for the analysis of triple SILAC immunoaffinity purification experiments. This identifies genuine protein interaction partners more efficiently and also aids the characterization of changes in protein complexes that can arise either as a result of varied biological conditions or in response to specific perturbations. To date, there are still relatively few studies that have explored the dynamics of protein-protein interactions using quantitative proteomics-based approaches (17, 36). A major aim of the methodology described in this study is to facilitate such analyses. In contrast with other common approaches, our work flow discourages the premature removal of putative contaminant proteins either experimentally or in silico. Instead, we adopt a comprehensive and inclusive approach that takes advantage of the high sensitivity of protein detection now possible using MS-based identification of proteins from model organisms. We use interactive analysis that integrates several objective criteria to annotate, rather than discard, all proteins in every data set. This is of particular importance for the detection and characterization of low affinity and/or low abundance specific protein interaction partners that would otherwise remain undetected among the large excess of background contaminants and nonspecific interactors.
An important issue in all MS-based protein identification studies is the reliability of protein identification and quantification. Although analyses of biological responses are mostly concerned with comparing the differential behavior of individual proteins, the MS analysis and SILAC procedures directly measure peptides. It must be remembered that the quality of data can differ considerably between separate proteins in the data set, which can vary in the number of peptides identified and quantified, the total sequence coverage, and the accuracy and similarity in the SILAC ratios measured for separate peptides assigned to the same protein. Consideration of these parameters can assist with drawing reliable conclusions, and they can be incorporated also into the visualizations of the experiments to provide further depth to the analysis of the MS data.
A key feature of the approach we describe is the generation of a PFL that provides a dynamic list of all proteins identified in co-IP experiments and annotation of their frequency of detection. As opposed to the static bead proteome, the PFL benefits from continuously being updated with every new experiment that is performed. Thus, addition of new data sets will improve both its reliability and its coverage. The PFL described here contains 10,623 IPI numbers; this corresponds to ∼12% of the IPI human proteome. However, this can expand in the future to cover the entire human proteome as more data sets from additional co-IP experiments are added, incorporating different conditions and other cell types. In contrast with the previous notion of characterizing a set of putative contaminants to eliminate them from the data set, the PFL approach does not stigmatize any protein as a contaminant. This more accurately reflects the fact that a given protein can interact specifically with certain baits and nonspecifically with others. Instead, the PFL provides an objective annotation for all proteins that predicts their probability of being a contaminant under a defined set of experimental conditions. Applying this annotation to co-IP data sets facilitates discrimination between proteins with high versus low probabilities of being either specific or nonspecific interaction partners. This is further enhanced by the use of powerful visualization tools, including the use of color coding to focus attention on selected sets of proteins identified for further analysis. Furthermore, it provides the ability to flexibly adjust threshold values as determined by the user to create optimal settings for each individual experiment.
Another advantage of the PFL approach is that it can be filtered for the parameters from the data set under analysis, e.g. cell extract, type of affinity matrix, etc., to create a customized PFL that more accurately predicts contaminants relevant to each new experiment. The spectrum of parameters available for customization of the PFL includes all of the dimensions and metadata recorded in the data repository. PepTracker is designed to incorporate a laboratory management tool to facilitate the detailed and consistent recording of metadata from each experiment that can be used directly to generate customized PFLs. Although the spectrum of dimensions and experimental conditions incorporated in PepTracker is currently focused on human cells, this can in the future be expanded to include a wider range of data, such as other model organisms, and new dimensions, such as detailed genotypes of the cells or organisms being analyzed. In addition, the PFL is applicable also to other types of MS analyses not involving SILAC data. For example, it can enhance the analysis of label-free experiments by adding additional objective criteria to identify putative nonspecific contaminants.
The generation of the PFL involved adapting advanced techniques from the BI field that deal well with the efficient analysis of large data sets. The core concept of BI revolves around understanding and modeling data in an appropriate format that makes analysis easier and more intuitive for end users. BI technology is designed for rapid interactive response and works particularly well for train-of-thought analysis whereby response times from queries are rapid enough (1–2 s) to allow a user to follow a sequence of ideas where each answer can prompt another question. The advantages of rapid response times on productivity have been well understood for many years (34). To our knowledge, this is the first direct application of such BI technology in cell biology or proteomics research. BI techniques facilitate the analysis of complex data and are essentially discipline-agnostic. They have recently been successfully applied, for example, to the analysis of historical science data, which has enhanced our understanding of how Darwin developed the theory of evolution by natural selection (37). We suggest that wider application of these techniques will be of great utility not only for proteomics research but also for other research areas involving the collection and mining of very large data sets as is now common in biomedical science.
Our work flow highlights the need for automation that can deal with the integrated analysis of many large data sets that are inherently multidimensional. We have described how the PFL approach can be applied to objectively normalize data and facilitate comparisons of information from separate experiments. The PepTracker environment is capable of storing many consistently annotated data sets and thus presents the opportunity to integrate these data sets, along with associated metadata, to perform what we term a “superexperiment.” The PFL represents an example of a superexperiment that incorporates data from a large number of separate immunoaffinity purification experiments. By using this approach to encompass other types of quantitative proteomics experiments, we aim to expand the superexperiment concept. For example, other types of SILAC and MS analyses provide information about the dynamics of distinct protein properties, such as subcellular localization, turnover, and post-translational modifications (38). Future work will therefore develop the use of BI technology within the PepTracker environment to normalize and mine these combined data sets. It is also envisaged that a web-based interface can be developed to provide the wider community with access to the PFL and related tools.
Acknowledgments
We thank Drs. Douglas Lamont and Kenneth Beattie of the Fingerprints Proteomics Facility at the University of Dundee for technical assistance with the MS analysis. We thank members of the Lamond laboratory for advice and suggestions.
* This work was supported in part by Wellcome Trust Program Grant 073980/Z/03/Z (to A. L.) with additional support from European Union (EU) FP7 Grant Proteomics Specification in Time and Space (PROSPECTS), EU Network of Excellence Grant European Alternative Splicing Network (EURASNET), and an interdisciplinary Radical Solutions for Researching the Proteome (RASOR) initiative, which is supported by the Biotechnology and Biological Sciences Research Council (BBSRC), Engineering and Physical Sciences Research Council, Scottish Higher Education Funding Council, and Medical Research Council (MRC).
2 S. Boulon, B. Pradet-Balade, C. Verheggen, D. Molle, M. Georgieva, K. Azzag, Y. Ahmad, H. Neel, A. I. Lamond, and E. Bertrand, submitted manuscript.
1 The abbreviations used are:
- SILAC
- stable isotope labeling with amino acids in cell culture
- PFL
- protein frequency library
- Pol2C
- RNA polymerase II subunit C
- Pol2A
- RNA polymerase II subunit A
- BI
- business intelligence
- OLAP
- on-line analytical processing
- IP
- immunoprecipitation
- IPI
- International Protein Index
- GFP
- green fluorescent protein
- L
- light
- M
- medium
- H
- heavy
- dH2O
- distilled H2O
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- LTQ
- linear trap quadrupole.
REFERENCES
- 1.Charbonnier S., Gallego O., Gavin A. C. (2008) The social network of a cell: recent advances in interactome mapping. Biotechnol. Annu. Rev 14, 1–28 [DOI] [PubMed] [Google Scholar]
- 2.Cox J., Mann M. (2007) Is proteomics the new genomics? Cell 130, 395–398 [DOI] [PubMed] [Google Scholar]
- 3.Collins M. O., Choudhary J. S. (2008) Mapping multiprotein complexes by affinity purification and mass spectrometry. Curr. Opin. Biotechnol 19, 324–330 [DOI] [PubMed] [Google Scholar]
- 4.Gingras A. C., Gstaiger M., Raught B., Aebersold R. (2007) Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol 8, 645–654 [DOI] [PubMed] [Google Scholar]
- 5.Oeljeklaus S., Meyer H. E., Warscheid B. (2009) New dimensions in the study of protein complexes using quantitative mass spectrometry. FEBS Lett 583, 1674–1683 [DOI] [PubMed] [Google Scholar]
- 6.Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., Superti-Furga G. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 7.Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 8.Babu M., Krogan N. J., Awrey D. E., Emili A., Greenblatt J. F. (2009) Systematic characterization of the protein interaction network and protein complexes in Saccharomyces cerevisiae using tandem affinity purification and mass spectrometry. Methods Mol. Biol 548, 187–207 [DOI] [PubMed] [Google Scholar]
- 9.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 10.Blow N. (2009) Systems biology: untangling the protein web. Nature 460, 415–418 [DOI] [PubMed] [Google Scholar]
- 11.Cloutier P., Al-Khoury R., Lavallée-Adam M., Faubert D., Jiang H., Poitras C., Bouchard A., Forget D., Blanchette M., Coulombe B. (2009) High-resolution mapping of the protein interaction network for the human transcription machinery and affinity purification of RNA polymerase II-associated complexes. Methods 48, 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewing R. M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M. D., O'Connor L., Li M., Taylor R., Dharsee M., Ho Y., Heilbut A., Moore L., Zhang S., Ornatsky O., Bukhman Y. V., Ethier M., Sheng Y., Vasilescu J., Abu-Farha M., Lambert J. P., Duewel H. S., Stewart I. I., Kuehl B., Hogue K., Colwill K., Gladwish K., Muskat B., Kinach R., Adams S. L., Moran M. F., Morin G. B., Topaloglou T., Figeys D. (2007) Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol 3, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 14.Ong S. E., Mann M. (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc 1, 2650–2660 [DOI] [PubMed] [Google Scholar]
- 15.Ranish J. A., Brand M., Aebersold R. (2007) Using stable isotope tagging and mass spectrometry to characterize protein complexes and to detect changes in their composition. Methods Mol. Biol 359, 17–35 [DOI] [PubMed] [Google Scholar]
- 16.Vermeulen M., Hubner N. C., Mann M. (2008) High confidence determination of specific protein-protein interactions using quantitative mass spectrometry. Curr. Opin. Biotechnol 19, 331–337 [DOI] [PubMed] [Google Scholar]
- 17.Blagoev B., Kratchmarova I., Ong S. E., Nielsen M., Foster L. J., Mann M. (2003) A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol 21, 315–318 [DOI] [PubMed] [Google Scholar]
- 18.Selbach M., Mann M. (2006) Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK). Nat. Methods 3, 981–983 [DOI] [PubMed] [Google Scholar]
- 19.Trinkle-Mulcahy L., Andersen J., Lam Y. W., Moorhead G., Mann M., Lamond A. I. (2006) Repo-Man recruits PP1 gamma to chromatin and is essential for cell viability. J. Cell Biol 172, 679–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., Morrice N. A., Swift S., Rothbauer U., Leonhardt H., Lamond A. I. (2008) Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol 183, 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brand M., Ranish J. A., Kummer N. T., Hamilton J., Igarashi K., Francastel C., Chi T. H., Crabtree G. R., Aebersold R., Groudine M. (2004) Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat. Struct. Mol. Biol 11, 73–80 [DOI] [PubMed] [Google Scholar]
- 22.Tackett A. J., DeGrasse J. A., Sekedat M. D., Oeffinger M., Rout M. P., Chait B. T. (2005) I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J. Proteome Res 4, 1752–1756 [DOI] [PubMed] [Google Scholar]
- 23.Lam Y. W., Lamond A. I., Mann M., Andersen J. S. (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol 17, 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cristea I. M., Williams R., Chait B. T., Rout M. P. (2005) Fluorescent proteins as proteomic probes. Mol. Cell. Proteomics 4, 1933–1941 [DOI] [PubMed] [Google Scholar]
- 25.Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M. C., Leonhardt H. (2008) A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics 7, 282–289 [DOI] [PubMed] [Google Scholar]
- 26.Harsha H. C., Molina H., Pandey A. (2008) Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nat. Protoc 3, 505–516 [DOI] [PubMed] [Google Scholar]
- 27.Ong S. E., Mann M. (2007) Stable isotope labeling by amino acids in cell culture for quantitative proteomics. Methods Mol. Biol 359, 37–52 [DOI] [PubMed] [Google Scholar]
- 28.Mousson F., Kolkman A., Pijnappel W. W., Timmers H. T., Heck A. J. (2008) Quantitative proteomics reveals regulation of dynamic components within TATA-binding protein (TBP) transcription complexes. Mol. Cell. Proteomics 7, 845–852 [DOI] [PubMed] [Google Scholar]
- 29.Wang X., Huang L. (2008) Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol. Cell. Proteomics 7, 46–57 [DOI] [PubMed] [Google Scholar]
- 30.Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 31.Wiœniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 32.Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 33.Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 34.Lambert G. N. (1984) A comparative study of system response time on program developer productivity. IBM Syst. J 23, 36–43 [Google Scholar]
- 35.Gstaiger M., Luke B., Hess D., Oakeley E. J., Wirbelauer C., Blondel M., Vigneron M., Peter M., Krek W. (2003) Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science 302, 1208–1212 [DOI] [PubMed] [Google Scholar]
- 36.Foster L. J., Rudich A., Talior I., Patel N., Huang X., Furtado L. M., Bilan P. J., Mann M., Klip A. (2006) Insulin-dependent interactions of proteins with GLUT4 revealed through stable isotope labeling by amino acids in cell culture (SILAC). J. Proteome Res 5, 64–75 [DOI] [PubMed] [Google Scholar]
- 37.Kohn D., Murrell G., Parker J., Whitehorn M. (2005) What Henslow taught Darwin. Nature 436, 643–645 [DOI] [PubMed] [Google Scholar]
- 38.Boisvert F. M., Lam Y. W., Lamont D., Lamond A. I. (December21, 2010) A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol. Cell. Proteomics 9(3):457.70. [DOI] [PMC free article] [PubMed] [Google Scholar]