Abstract
Intracellular bacteria cause serious infectious diseases such as tuberculosis, shigellosis, and listeriosis. The Drosophila peptidoglycan recognition protein (PGRP)-LE functions as an important host pattern recognition receptor against intracellular bacteria such as Listeria monocytogenes. One PGRP-LE-mediated intracellular response against L. monocytogenes infection is the induction of autophagy, a conserved intracellular degradation system. Here, to further elucidate PGRP-LE-mediated intracellular innate immune responses, we performed a strategic microarray analysis and identified the Listericin gene, whose expression is induced in response to L. monocytogenes infection in a PGRP-LE-dependent manner. RNA interference and overexpression experiments demonstrated that Listericin gene induction is cooperatively regulated by PGRP-LE and the JAK-STAT (Janus kinase-signal transducers and activators of transcription) pathway. An in vitro cell culture assay showed that Listericin is secreted as processed forms and suppresses the growth of L. monocytogenes and Gram-negative bacteria. A colony formation unit assay clearly demonstrated that induction of the Listericin gene suppresses not only the growth of L. monocytogenes but also the growth of Gram-negative bacteria in vivo. Based on these findings, we propose that the Listericin gene encodes a novel antibacterial peptide-like protein whose induction is cooperatively regulated by PGRP-LE and the JAK-STAT pathway.
Keywords: Antimicrobial Peptides, Immunology/Innate Immunity, Organisms/Bacteria, Organisms/Drosophila, Receptors, Signal Transduction/JAK-STAT
Introduction
Intracellular pathogens cause infectious diseases such as tuberculosis, acquired immune deficiency syndrome, and malaria, which have high worldwide morbidity and mortality rates. These intracellular pathogens survive in the host cells by manipulating basic host processes or immune systems to establish their own intracellular niche (1).
Listeria monocytogenes, one of the best studied facultative intracellular Gram-positive bacteria, causes the serious food-borne disease listeriosis, which affects mainly immunocompromised individuals, pregnant women, and neonates (2, 3). Extensive in vitro cell culture studies have defined the life cycle and virulence factors that allow these pathogens to thrive in host cells (3–5). Upon entry into either phagocytotic or non-phagocytotic cells, L. monocytogenes secrete a cholesterol-dependent pore-forming cytotoxin, listeriolysin O, that disrupts the phagosome membrane and allows the bacteria to escape from vacuoles and proliferate in the cytosol (6–8). Cytosolic L. monocytogenes express an actin-nucleating protein, ActA, that facilitates host actin polymerization to form a scaffold that allows the bacteria to move into the cytosol and spread to neighboring cells (9). Although several microbiologists have identified the key pathogenic factors in this multistep process of L. monocytogenes infection (3, 10), the underlying mechanisms in terms of host defense systems remain unclear.
Drosophila is an excellent model system to decipher the precise molecular mechanisms of host innate immune responses to microbial infections due to the availability of powerful genetic techniques combined with molecular and biochemical approaches and RNA interference (RNAi) tools that can be used in these organisms (11, 12). In addition to the practical experimental advantages, high conservation of pathogen recognition, signaling pathways, and effector mechanisms between Drosophila and mammals (13, 14) also contributes to the biologic significance of the innate immune mechanisms of Drosophila.
Upon microbial infection, Drosophila recognize pathogens with germ line-encoded pattern recognition receptors that are highly conserved from insects to animals (12, 13, 15). A representative pattern recognition receptor is the peptidoglycan recognition protein (PGRP)2 family, which specifically distinguishes bacteria-derived peptidoglycans (PGN) and drives the activation of innate immune signaling pathways such as the Toll and immune deficiency (imd) pathways (12, 16, 17). The Toll pathway is mainly activated by fungal and lysine-type PGN-containing Gram-positive bacterial infection and activates the nuclear factor κB (NF-κB) transcription factors Dorsal and Dif (Dorsal-related immunity factor), whereas the imd pathway is predominantly activated by diaminopimelic acid (DAP)-type PGN-containing bacteria (mainly Gram-negative bacteria) and activates the NF-κB homolog Relish (11, 12, 18). Subsequently, these activated NF-κB factors drive numerous effector genes, including the expression of seven distinct types of antimicrobial peptides (AMP; e.g. Attacin, Cecropin, Defensin, Diptericin, Drosocin, Drosomycin, and Metchnikowin), which are effective against Gram-negative and Gram-positive bacteria and fungi (19–22). Recent studies have provided strong evidence that the JAK-STAT pathway, originally reported to be responsible for classical developmental processes (23–25), is also involved in other aspects of the innate immune response, such as defense against viral infection (26), tissue damage recovery, hemocyte proliferation and differentiation (27), and gut immunity (28).
Recent in vitro genome-wide RNAi screening (29, 30) and in vivo genetic screening (31, 32) identified many novel host innate factors involved in the defense against L. monocytogenes infection. Nevertheless, how L. monocytogenes are recognized by pattern recognition receptors and how they are eliminated in the host cell cytosol remains unknown.
In addition to the extracellular and intracellular functions of PGRP-LE to induce AMP after recognizing DAP-type PGN (18, 33), Yano et al. (34) recently demonstrated a novel role of PGRP-LE as an intracellular receptor against L. monocytogenes with a DAP-type PGN. Survival experiments indicate that PGRP-LE mutant flies die rapidly after L. monocytogenes infection. Consistently, the data from an in vitro cell culture also support findings from in vivo studies that intracellular growth of L. monocytogenes is much higher in Drosophila S2 cells without PGRP-LE expression than in S2 cells with PGRP-LE expression (34). Moreover, PGRP-LE has a crucial role inducing autophagy, which is a highly conserved cellular process involved in lysosomal degradation of cytoplasmic components. This infection-induced autophagy occurs independently of the Toll and imd pathways and directly promotes host survival, providing other avenues of intracellular innate immunity (34).
The present study aimed to obtain more details about the PGRP-LE-mediated host innate immune responses against L. monocytogenes infection. Based on strategic microarray analyses of different L. monocytogenes infection combinations, either in the absence or presence of PGRP-LE, and comparison of those data with those of different Gram-positive and -negative bacteria injected into adult flies, we identified CG9080 (referred to here as the Listericin gene) as a previously uncharacterized gene that is induced in response to L. monocytogenes infection in a PGRP-LE-dependent manner. Functional RNAi and overexpression studies demonstrated that Listericin gene induction depends on both PGRP-LE and the JAK-STAT pathway. Further biochemical and in vivo overexpression studies revealed that Listericin is secreted and is capable of suppressing the growth of L. monocytogenes as well as Gram-negative bacteria in vitro and in vivo. Therefore, the Listericin gene encodes a novel AMP-like protein whose induction is cooperatively regulated by PGRP-LE and the JAK-STAT pathway.
EXPERIMENTAL PROCEDURES
L. monocytogenes Infection of Drosophila S2 Cells
7.5 × 105 Drosophila S2 cells or metallothionein promoter-PGRP-LE-expressing S2 cells (Inducible-LE (Ind-LE)) (34) per well were seeded in 24-well plates. After 30 min of 50-μm water-soluble cholesterol treatment, a suspension of L. monocytogenes (approximately ∼40 bacteria/cell unless otherwise described) or heat-killed Escherichia coli was added, and immune stimulation continued for 1.5 h at 28 °C. Cells were then washed with phosphate-buffered saline, reseeded onto new plates, and incubated for 8 h in Drosophila Schneider's medium containing 100 μm CuSO4 and 10 μg/ml gentamicin (Nakalai Tesque, Kyoto, Japan). Infection conditions for the preparation of microarray samples using S2 cells and S2* cells stably expressing yellow fluorescent protein (YFP)-tagged PGRP-LE induced by an actin-promoter (YFP-LE cells) (18) in 6-well plates were described previously (34).
DNA Microarray Analysis
Total RNA from S2 cells and Drosophila adult flies homogenized in TRIzol (Invitrogen) was isolated using an RNeasy kit (Qiagen, Valencia, CA). The RNA quality was checked using the Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Total RNA (1 μg) was amplified and labeled as complementary RNA (cRNA) using an IVT labeling kit (Affymetrix, Santa Clara, CA). Affymetrix Drosophila Genome 2.0 arrays were hybridized with 30 μg of labeled cRNA, washed, stained, and scanned. All microarray procedures were performed according to the Affymetrix protocol. Two independent experiments were performed for each duplicated condition of S2 cells and various bacteria-injected flies (supplemental Fig. 1). These samples allowed for four comparisons, infected versus control samples (supplemental Fig. 1). Changes in expression are shown as the signal log ratio. The most widely used alternative transformation of the ratio is the logarithm base 2, which has the advantage of producing a continuous spectrum of values and treating up- and down-regulated genes similarly (35). It is important to remember that logarithms treat numbers and their reciprocals symmetrically: log2(1) = 0, log2(2) = 1 log2(1/2) = −1, log2(4) = 2, log2(1/4) = −2, and so on. Raw data of all processed microarray analyses are available at the Drosophila Microarray Database website at Tohoku University.
Overexpression and RNAi in Drosophila S2 Cells
Cells (1.5 × 106/ml) were transfected in 24-well plates using a calcium phosphate precipitation kit (Invitrogen) with 1 μg of either a Upd-GFP (36), PGRP-LC (37), or TollΔLRR (38) expression vector for overexpression and 1 μg of either dsGFP, dsKey, dsDif, dsdorsal, or dsStat92E for RNAi analysis. After 18 h of transfection, cells were washed with phosphate-buffered saline, incubated for another 24 h, and used for L. monocytogenes infection. To make double-stranded RNAs, single-stranded RNAs were synthesized with the T7 RNA polymerase (Novagen, Madison, WI). Annealed double-stranded RNAs were precipitated in ethanol and dissolved in injection buffer (0.1 mm sodium phosphate, pH 6.8, 5 mm KCl) (39).
Stable Cell Lines
To establish stable S2 cell lines expressing green fluorescent protein (GFP), which were used as a negative control, and with or without C-terminal V5-HIS-tagged Listericin, cDNA fragments were amplified using the following primers: forward (5′-CCC GGG GAA TTC GAT AAT TCC CGC CAT GAG TAA AGG AGA AGA AC-3′) and reverse (5′-CCC GGG CTC GAG TTA TTT GTA TAG TTC ATC CAT G-3′) for GFP; forward (5′-CCC GGG GAA TTC GAT AAT TCC CGC CAT GAA ACA GTA CCT GGT GC-3′) and reverse (5′-CCC GGG CTC GAG TTT ACG TCC CCA ACT GGA ACT G-3′) for Listericin-V5-HIS, and forward (5′-CCC GGG GAA TTC GAT AAT TCC CGC CAT GAA ACA GTA CCT GGT GC-3′) and reverse (5′-CCC GGG CTC GAG TTA TTT ACG TCC CCA ACT GGA ACT G-3′) for Listericin without tags and subcloned into EcoRI-XhoI sites of the pAC5.1-V5-HIS vector (Invitrogen). These vectors were transfected together with pCoHygro for GFP and Listericin-V5-HIS and pCoBlast for mock vector (pAC5.1 vector only), and Listericin without tags and stable transfected cells were selected with Hygromycin B or Blasticidin S according to the manufacturer's protocol (Invitrogen).
Semiquantitative Reverse Transcription (RT)-PCR and Real-time PCR
Total RNA was isolated from each genotype of ∼20 flies or S2 cells with TRIzol reagent (Invitrogen). Total RNA (1 μg) was used for cDNA synthesis with ReverTraAce reverse transcriptase (Toyobo, Osaka) and oligo(dT)15 primer (Promega). Using the first-strand cDNA (0.5 μl), real-time PCR was performed using a LightCycler (Roche Diagnostics). Primers used for semiquantitative RT-PCR and real-time PCR are described in supplemental Table 1.
Fly Strains and Transgenic Flies
Flies were grown on standard medium at 25 °C. All other strains used have been published as follows: UAS-GFP and Collagen (Cg)-Gal4 (40). A PCR fragment of the Listericin gene without tags was amplified with the same primers to make the stable cell lines described above and subcloned into the EcoRI-XhoI sites of the pUAST vector. Transgenic UAS-Listericin flies were obtained by standard embryo microinjection of the pUAST-Listericin vector in the w118 strain.
Immunoblot Analysis
After 3 to 4 days of culture in conditioned medium, Listericin-V5-HIS expressing or non-expressing cells were lysed in buffer containing 50 mm Tris-HCl, 2% (w/v) sodium dodecyl sulfate, 10% (v/v) glycerol, and 100 mm β-mercaptoethanol. Samples were separated by 15% SDS-PAGE, transferred to a Hybond-P polyvinylidene fluoride membrane (GE Healthcare), and reacted with anti-V5 epitope antibody (Invitrogen). The same amount of proteins used for immunoblot analysis was visualized by Coomassie Brilliant Blue (CBB) staining.
Survival Experiments and Bacterial Strains
Flies 3–5 days after eclosion were infected with a ∼69-nl injection of L. monocytogenes suspension per fly. The optical density at 600 nm for the wild-type (10403S) and Δhly L. monocytogenes (DP-L2161) (8) suspension was 0.00001 (∼10 bacteria/fly) or 0.001 (∼1000 bacteria/fly). All survival experiments were performed using 30 flies of each genotype at 28 °C. Surviving flies were counted daily by transferring them to fresh vials. The following pathogens were used for infection: E. coli (K-12), Erwinia carotovora carotovora 15 (Ecc15), Staphylococcus aureus (ATCC10801, wood46), and Staphylococcus saprophyticus (GTC0205).
Affinity Purification of Secreted Listericin-V5-HIS
A total of 400 ml of 5–6-day cultured conditioned medium from Listericin-V5-HIS-expressing cells in serum-free SFX insect medium (HyClone) was first dialyzed in 10 mm sodium phosphate buffer, pH 6.4, overnight, and then the pH was shifted to 8.0 for 6 h. The dialyzed medium containing 300 mm NaCl, 20 mm imidazole, and an appropriate concentration of protease inhibitor cocktails (Roche Diagnostics) was subjected to affinity chromatography on a nickel nitrilotriacetic acid-agarose bead column (Qiagen) at 4 °C, washed, and eluted with 0.25 m imidazole. Eluted fractions were precipitated by 10% trichloroacetic acid, washed in cold acetone, and dissolved in SDS sample buffer. The samples were applied to SDS-PAGE and blotted onto a polyvinylidene fluoride membrane (Millipore). The N-terminal amino acid sequence of the CBB-stained band detected in immunoblot analysis using anti-V5 epitope antibody was analyzed by Edman degradation protein sequencing (HiPep Laboratories).
Colony Forming Unit (CFU) Assay
For in vitro cell culture assay, conditioned medium collected from cells expressing S2-GFP (used as control) or S2-Listericin cultured for 3–4 days was mixed with the appropriate number of L. monocytogenes, E. coli, or S. saprophyticus cells diluted serially after 0, 1, or 2 h at 29 °C for L. monocytogenes and at 37 °C for E. coli and S. saprophyticus and plated onto the appropriate plates. For the in vivo assays, 24 h after a ∼70-nl injection of each bacterial strain, the flies were collected and sterilized with 70% ethanol. A total of 10 flies of each genotype were homogenized in 500 μl of the appropriate bacterial medium, serially diluted, and plated onto the appropriate plates (brain heart infusion medium for L. monocytogenes; Luria Bertani medium for E. coli and Ecc15; nutrient broth medium for S. aureus and S. saprophyticus). The next day, colony numbers were counted to calculate the CFU per ml or per fly. The optical density at 600 nm for each injected bacterial suspension was as follows: L. monocytogenes (0.0001), S. aureus (0.0001), E. coli (1.0), and Ecc15 (1.0).
RESULTS
Identification of CG9080 as Listericin
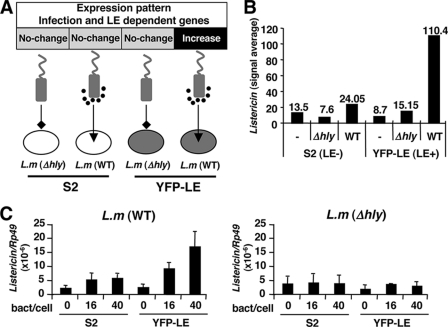
To identify uncharacterized PGRP-LE-dependent host defense genes against L. monocytogenes infection, we performed a strategic microarray analysis for four different combinations of infections (Fig. 1A). We used normal S2 cells and YFP-LE cells (YFP-tagged PGRP-LE-expressing cells) as representative PGRP-LE absent or present host cells, respectively, and infected them with a cytoplasm non-escapable Δhly mutant lacking listeriolysin O or escapable wild-type L. monocytogenes strains. Microarray analysis identified 68 candidate genes that were specifically induced with an at least a 5-fold up-regulation upon wild-type L. monocytogenes infection of YFP-LE cells (supplemental Fig. 2; see the Drosophila Microarray Database web site). To further identify innate factors common between in vitro and in vivo infection, we performed other rounds of microarray analysis of adult flies injected with bacteria in vivo. Both wild-type and Δhly L. monocytogenes as well as other Gram-positive (Enterococcus faecalis, Micrococcus luteus, S. aureus, and Bacillus subtilis) and Gram-negative (E. coli) bacteria were injected into adult flies, and whole gene expression profiles were analyzed by microarray analysis (see the Drosophila Microarray Database web site). Analysis of all microarray data from adult flies and comparison with the 68 candidate genes from the in vitro S2 assay led to the identification of a previously uncharacterized gene, CG9080 (named the Listericin), that was predominantly induced upon wild-type L. monocytogenes infection in only YFP-LE cells (Fig. 1B). To further confirm the microarray results, we verified Listericin gene expression at different multiplicities of infection (bacteria:host = 16:1, 40:1) with real-time RT-PCR analysis. The results clearly indicated that the Listericin gene is predominantly induced by infection by wild-type, but not Δhly, L. monocytogenes of YFP-LE, but not S2, cells (Fig. 1C). These results suggest that the Listericin gene is induced in response to cytoplasmic infection by L. monocytogenes in a PGRP-LE-dependent manner.
FIGURE 1.
Identification of the Listericin gene by strategic microarray. A, schematic presentation of four microarray samples is shown. S2 and YFP-LE cells were infected by either Δhly mutant lacking listeriolysin O or wild-type (WT) L. monocytogenes (L.m.). B, shown is a signal average of Listericin gene expression from microarray analysis. C, real-time PCR analysis of Listericin gene expression in S2 cells and YFP-LE cells with different multiplicities of infection of L. monocytogenes (WT or Δhly) is shown. Ribosomal protein Rp49 was used as an internal control. Bars indicate S.D.
Cooperative Regulation of Listericin Induction by PGRP-LE and the JAK-STAT Pathway
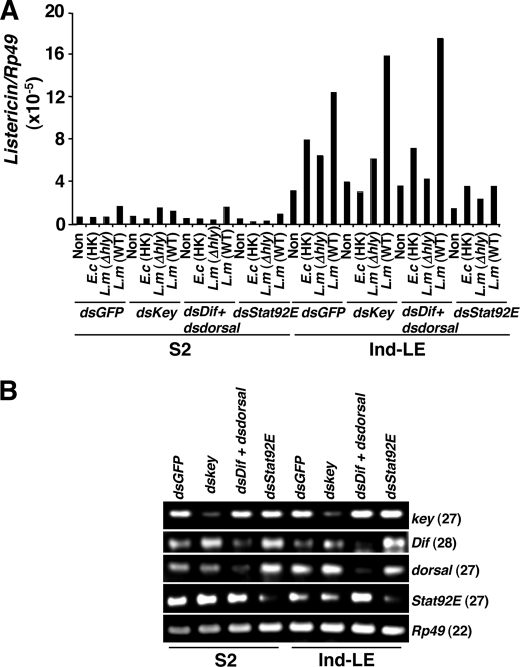
PGRP-LE acts as an intracellular pattern recognition receptor against DAP-type PGN and L. monocytogenes to induce AMPs such as Diptericin and Attacin and autophagy in an imd pathway-dependent and -independent manner, respectively (18, 34). We next examined which signaling pathway controls Listericin gene induction upon L. monocytogenes infection. We targeted three major innate immune signaling pathways, imd, Toll, and JAK-STAT, by RNAi targeting of the critical components of each pathway; inhibitor of NF-κB γ homologue key (41) for the imd pathway, NF-κB transcription factors Dif and Dorsal (42) for the Toll pathway, and transcription factor Stat92E (43) for the JAK-STAT pathway (Fig. 2). In this assay we used Ind-LE-derived S2 cells (34) instead of YFP-LE-expressing S2* cells (17) to eliminate the possibility of different expression levels in different cell lines. We again observed PGRP-LE-dependent Listericin gene induction in Ind-LE cells in response to L. monocytogenes infection with even stronger expression compared with YFP-LE cells. Stat92E RNAi significantly suppressed the PGRP-LE-dependent Listericin gene induction against immune stimulation by heat-killed E. coli and L. monocytogenes infections (Fig. 2A). Key RNAi suppressed heat-killed E. coli-dependent Listericin gene induction but not L. monocytogenes-dependent Listericin gene induction, suggesting a key independent regulation of Listericin gene induction specific to L. monocytogenes infection. We also confirmed gene-specific RNAi effects in both S2 and Ind-LE cells (Fig. 2B).
FIGURE 2.
Control of PGRP-LE-dependent Listericin gene induction in response to immune stimulation by the JAK-STAT pathway. A, dsGFP (used as a negative double-stranded RNA control), dskey (for the imd pathway), dsDif and dsdorsal (for the Toll pathway), and dsStat92E (for the JAK-STAT pathway) were transfected into either S2 cells or Ind-LE cells (metallothionein promoter-dependent inducible-PGRP-LE), and the cells were stimulated by heat-killed E. coli (E.c (HK)), Δhly, and wild type (WT) L. monocytogenes (L.m). Listericin gene expression was then measured at 8 h after immune stimulation. Representative data of two independent experiments are shown. B, semiquantitative RT-PCR analysis to check RNAi effects on each gene is shown. Numbers in parentheses indicate PCR cycle number. Ribosomal protein Rp49 was used as an internal control.
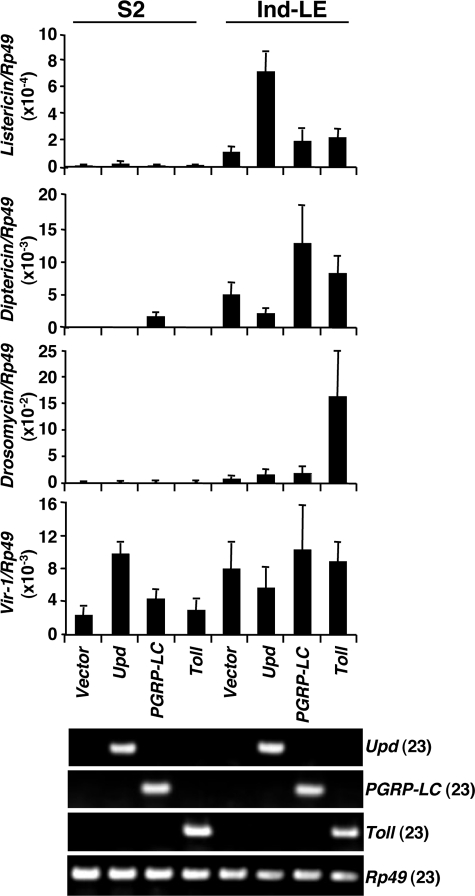
We next confirmed the results of RNAi experiments using the converse experiments in which each pathway was constitutively activated by the expression of unpaired (Upd), a ligand for the domeless receptor in the JAK-STAT pathway (36), PGRP-LC, an upstream receptor of the imd pathway (37), and TollΔLRR, a Toll receptor lacking the extracellular leucine-rich repeat (LRR) domain (38) in cell culture (Fig. 3). Expression of Upd induced Listericin gene expression in Ind-LE cells but not in S2 cells, indicating that PGRP-LE and the JAK-STAT pathway have synergistic effects on Listericin gene induction. The expression of Upd, however, induced the expression of Vir-1, a target of the JAK-STAT pathway (26), in S2 cells. A similar synergistic effect of PGRP-LE and the Toll pathway was observed on Drosomycin induction. Diptericin expression was induced by both PGRP-LE and PGRP-LC, consistent with previous reports (11, 12, 33). Toll, PGRP-LC, and Upd expression was verified by semiquantitative RT-PCR. Taken together, these results indicate that Listericin gene induction is cooperatively regulated by PGRP-LE and the JAK-STAT pathway, at least in cell culture.
FIGURE 3.
Synergistic effect of activation of JAK-STAT pathway and expression of PGRP-LE on the expression of the Listericin gene. The ligand of the JAK-STAT pathway (Upd), the receptor for the imd pathway (PGRP-LC), and activated Toll receptor of the Toll pathway (TollΔLRR) were each expressed in S2 or Ind-LE cells, and Listericin, Diptericin, Drosomycin, and Vir-1 expression was measured by real-time PCR. The lower panel shows the forced expression of each gene analyzed by RT-PCR. Numbers in parentheses indicate PCR cycle number. Rp49 was used as the internal control. The mean of three independent experiments is shown. Bars indicate S.D.
Secreted Listericin Suppresses L. monocytogenes Growth in Vitro
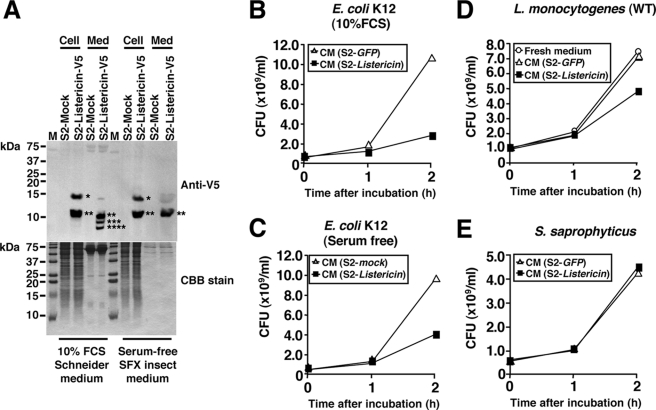
The Listericin gene encodes a protein of 121 amino acids with a signal peptide at the N terminus, a glycine-rich region, and an amidation site at the C terminus (supplemental Fig. 3), similar to the properties of AMPs such as Diptericins, Attacins, Sarcotoxin II, etc. (44). To determine whether Listericin is secreted, cell lysate and the conditioned medium of S2 cells expressing the V5 and histidine epitope-tagged Listericin at the C terminus (Listericin-V5-HIS) was analyzed by immunoblotting using the anti-V5 antibody. We detected two major signals at ∼15 and ∼10 kDa in the cell lysate and three signals at ∼10, ∼8, and ∼6 kDa in the conditioned medium (Fig. 4A, left). A 15-kDa signal corresponding to the full-length Listericin-V5-HIS product (151 amino acids) was only detected in the cell lysate, whereas 3 bands smaller than 15 kDa were detected in the conditioned medium (see asterisks in Fig. 4A). In contrast to the total amount of proteins in the conditioned medium visualized by CBB staining, the intensity of anti-V5 signals was stronger than that in the cell lysate, suggesting that Listericin was actively secreted into the medium after proteolytic cleavage. To determine the actual cleavage site of secreted Listericin, we purified Listericin-V5-HIS from serum-free conditioned medium using nickel nitrilotriacetic acid affinity chromatography. This serum-free medium is lower in protein content than serum-supplemented medium, which simplifies the purification process and increases the yield of the end product. Immunoblot analysis showed that in contrast to the three major bands detected in 10% fetal calf serum Schneiders medium (Fig. 4A, left), only a single ∼10-kDa band was detected in the serum-free conditioned medium (Fig. 4A, right). We then purified the Listericin-V5-HIS from serum-free conditioned medium and determined its N-terminal amino acid sequence. The CBB-stained band had the N-terminal sequence as HFGGGFG. Thus, we concluded that the cleavage site of secreted Listericin was at the 49th amino acid (AAR↓HFG) (supplemental Fig. 3).
FIGURE 4.
Secretion of Listericin into the cell culture medium and suppression of L. monocytogenes and E. coli growth. A, cell lysate (Cell) and conditioned medium (Med) of 10% fetal bovine serum containing Schneider or serum-free SFX insect medium from either mock (pAC5.1 vector only) or Listericin-V5-HIS transfected cells were analyzed by immunoblotting using anti-V5 antibody. Asterisks indicate the positions of detected signals. The same amount of proteins used for the immunoblot was also visualized by CBB staining. B–E, conditioned medium from the Listericin gene-overexpressed S2 cell line was mixed with the appropriate numbers of E. coli K12 in 10% fetal bovine serum (FBS) medium (B), E. coli K12 in serum-free medium (C), L. monocytogenes (D), and S. saprophyticus (E), and the bacterial growth was analyzed by CFU assay. Stable GFP-expressing S2 cells were used as a negative control. The graph shows representative results from three independent experiments. WT, wild type.
To further characterize the biochemical properties of Listericin, conditioned medium of Listericin gene-expressing cells was reacted with the appropriate numbers of bacteria including L. monocytogenes, and bacterial growth was monitored (Fig. 4, B–E). Compared with medium from GFP-expressing S2 cells, Listericin-containing medium suppressed the growth of L. monocytogenes and E. coli, but not S. saprophyticus.
Listericin Overexpression Confers Resistance against L. monocytogenes Infection
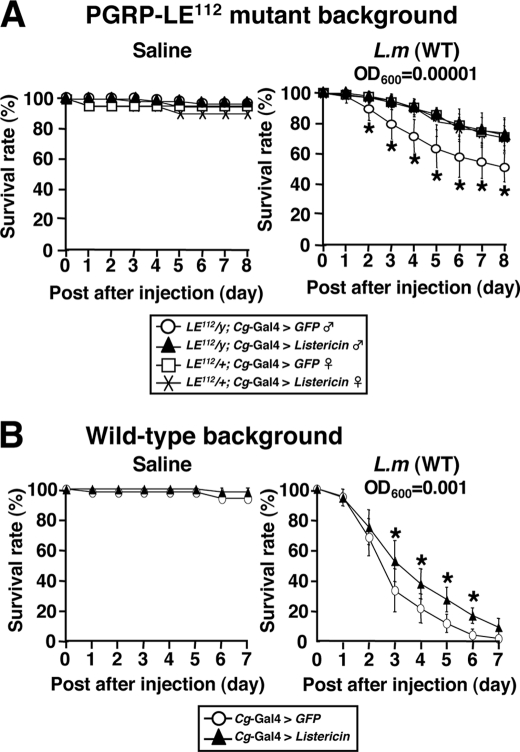
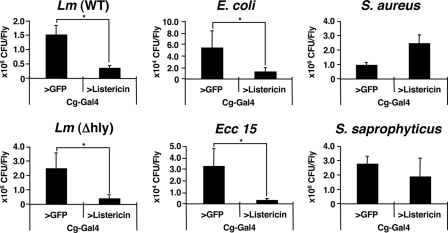
Because Listericin-RNAi flies (45), driven by a heat-shock-Gal4 driver, did not show any susceptible phenotype against L. monocytogenes infection (data not shown), we characterized Listericin function in an in vivo gain-of-function study. Using a Cg-Gal4 driver, the Listericin gene was overexpressed in immune organs (fat body and hemocytes), and susceptibility against L. monocytogenes infection was tested. Survival experiments showed that Listericin gene overexpression restored the survival phenotype against L. monocytogenes infection in both the PGRP-LE112 mutant (Fig. 5A, ∼10 bacteria/fly) and wild-type background (Fig. 5B, ∼1000 bacteria/fly). To gain further insight into the functional properties of Listericin in vivo, the hemolymph bacterial loads of the flies after various types of bacterial infection were examined. CFU assays clearly demonstrated that, compared with control GFP, Listericin gene overexpression significantly suppressed both wild-type and Δhly L. monocytogenes bacterial loads in the hemolymph (Fig. 6). We further extended this assay to other Gram-positive (S. aureus and S. saprophyticus) and Gram-negative (E. coli and Ecc 15) bacteria, showing that overexpression of the Listericin gene in flies uniquely suppresses the bacterial loads of Gram-negative, but not Gram-positive bacteria, in the hemolymph (Fig. 6).
FIGURE 5.
Effect of overexpression of the Listericin gene on host survival against L. monocytogenes infection. Survival rates of Cg-Gal4-driven GFP as a control and Listericin gene-overexpressed flies were tested after injecting saline (used as a bacteria-free control) or L. monocytogenes (L.m (WT)) at 29 °C in a PGRP-LE112 mutant (A) and wild-type (B) background. The 69-nl injection of A600 = 0.00001 and A600 = 0.001 L. monocytogenes suspension approximately corresponds to ∼10 and ∼1000 bacteria/fly, respectively. *, p < 0.05 (Wilcoxon-Mann-Whitney test).
FIGURE 6.
Effect of overexpression of the Listericin gene on growth of infected bacteria in vivo. At 24 h after injection of L. monocytogenes (Lm (WT)), L. monocytogenes (Δhly), E. coli, Ecc 15, S. aureus, and S. saprophyticus into Collagen (Cg)-Gal4-driven GFP as a control, Listericin gene-overexpressed flies were sterilized by ethanol on the surface, homogenized, and diluted serially in the appropriate medium. Bacterial growth in each fraction was quantified by determining CFU by plate assay. Data represent the mean of at least three independent experiments with more than 30 flies of each genotype examined. Bars indicate S.D. *, p < 0.05.
DISCUSSION
In the present study, microarray analyses identified the Listericin gene whose expression is induced in response to L. monocytogenes infection in a PGRP-LE-dependent manner. Expression studies using cell cultures demonstrated that the Listericin gene is cooperatively induced by PGRP-LE and the JAK-STAT pathway. Consistent with PGRP-LE-dependent induction of the Listericin gene, further functional studies revealed that Listericin suppresses the growth of Gram-negative bacteria and L. monocytogenes, which are recognized by PGRP-LE.
A Blast search revealed that Listericin is restricted to Drosophila species, based on the sequence similarity analyses with other species (supplemental Fig. 3). This finding might be due to the extreme diversity of AMP primary structures among different species (46). As a single mutation of the relatively short AMP sequence can drastically alter the biologic activity (46–48), Listericin may have uniquely evolved in Drosophila species to adapt to their particular microbial environments. Despite the low sequence similarity, Listericin possesses features representative of AMPs, such as overrepresentation of glycine residues, which is observed in other AMPs of Diptera (Diptericins, Attacins, or Sarcotoxin II), Coleoptera (Coleoptericin, Holotricin II and III, and Tenecin III), and Hemiptera (Hemiptericin) (44) and C-terminal amidation (47, 48) as well as secretion as a proteolytically processed form. Although sequence alignments of 12 Drosophila Listericins predicted at least four potential conserved proteolytic cleavage sites at Arg-46, Arg-49, Arg-69, and Arg-70, our N-terminal sequence analysis together with the immunoblotting data clearly identified its cleavage site at Arg-49 (supplemental Fig. 3). This, by analogy to many other AMPs, could be a protecting prodomain that has to be cleaved to activate the Listericin. In addition to this structural feature, Listericin also has biologic activity potentially acting as an AMP. Consistent with previous findings that one of the targets of glycine-rich AMPs is bacterial cell wall synthesis, Listericin suppresses the growth of DAP-type PGN-containing bacteria, Gram-negative bacteria, and L. monocytogenes but does not suppress the growth of Lys-type PGN-containing Gram-positive bacteria.
The JAK-STAT pathway is involved in immune reactions including hematopoiesis (49) in addition to its critical roles in development, such as segmentation, sex determination, larval imaginal disc development, and oogenesis (23). For example, upon bacterial infection the JAK-STAT pathway transcription factor STAT92E (41) is translocated in mosquitoes (50) and Drosophila, leading to the expression of Tep and Tot family genes (51). Supporting the notion that the Listericin gene is regulated by the JAK-STAT pathway, the Listericin promoter contains a putative STAT binding site (data not shown). Further analysis is required, however, to elucidate the molecular mechanisms of cooperative regulation of the Listericin gene by PGRP-LE and the JAK-STAT pathway.
Although constitutive Listericin gene expression promoted host survival (Fig. 5) and suppressed the growth of Gram-negative bacteria and L. monocytogenes both in vitro (Fig. 4) and in vivo (Fig. 6), Listericin-RNAi flies (45), which had a ∼70% reduction in Listericin gene mRNA were not susceptible to L. monocytogenes infection compared with GFP-expressing control flies (data not shown). Although this might be due simply to the insufficient RNAi effects, it is more likely that the expression of other AMPs compensates for host survival in these conditions. Supporting this possibility, it was previously reported that single constitutive Drosophila AMP expression restores host resistance against infection even in a Toll- and imd-deficient background (52).
We previously reported that PGRP-LE-dependent induction of autophagy is crucial to eliminate intracellular L. monocytogenes in Drosophila (34). Although many mammalian in vitro studies have demonstrated that autophagic control functions as an important innate immune reaction for the elimination of intracellular pathogens such as group A Streptococcus (53), Mycobacterium tuberculosis (54, 55), Legionella pneumophila (56), Coxiella burnetti (57), and Porphyromonas gingivalis (58), some pathogens such as L. monocytogenes and Shigella, which are more motile within the host cytoplasm, can evade the autophagic control in mammals (59–61). Further analysis of the induction mechanisms of the Listericin gene and autophagy in response to L. monocytogenes infection in Drosophila might provide clues to aid in the prevention of intracellular bacterial infections in humans.
Supplementary Material
Acknowledgments
We thank D. A. Portnoy (University of California, Berkeley, CA), D. E. Higgins (Harvard Medical School), and M. Mitsuyama (Kyoto University) for supplying the Listeria strains, J. M. Reichhart and J. L. Imler (CNRS, Institut de Biologie Moleculaire et Cellulaire, Strasbourg, France) for supplying pAC-PGRP-LC and pPAC-TollΔLRR, M. Boutros and N. Pelte (German Cancer Research Center) for supplying pAC-Upd-GFP, and the Bloomington Stock Center, Drosophila Genetic Resource Center at the Kyoto Institute of Technology, and Genetic Strain Research Center of the National Institute of Genetics for the fly stocks.
This work was supported, in whole or in part, by National Institutes of Health Grant AI07495. This work was also supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Japan Society for the Promotion of Sciences, the Program for the Promotion of Basic Research Activities for Innovative Biosciences, Strategic International Cooperative program from Japan Science and Technology Agency, the Uehara Foundation, the Naito Foundation, and a Global COE Research Grant (Tohoku University Ecosystem Adaptability).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Table 1.
- PGRP
- peptidoglycan recognition protein
- AMP
- antimicrobial peptide
- CBB
- Coomassie Brilliant Blue
- CFU
- colony forming unit
- DAP
- diaminopimelic acid
- Dif
- Dorsal-related immunity factor
- GFP
- green fluorescent protein
- imd
- immune deficiency
- Ind-LE
- inducible LE
- JAK-STAT
- Janus kinase-signal transducers and activators of transcription
- NF-κB
- nuclear factor κ B
- PGN
- peptidoglycan
- Upd
- unpaired
- YFP
- yellow fluorescent protein
- LRR
- leucine-rich repeat
- RT
- reverse transcription.
REFERENCES
- 1.Alonso A., García-del Portillo F. (2004) Int. Microbiol. 7, 181–191 [PubMed] [Google Scholar]
- 2.Vázquez-Boland J. A., Kuhn M., Berche P., Chakraborty T., Domínguez-Bernal G., Goebel W., González-Zorn B., Wehland J., Kreft J. (2001) Clin. Microbiol. Rev. 14, 584–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamon M., Bierne H., Cossart P. (2006) Nat. Rev. Microbiol. 4, 423–434 [DOI] [PubMed] [Google Scholar]
- 4.Cossart P., Lecuit M. (1998) EMBO J. 17, 3797–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Portnoy D. A., Jacks P. S., Hinrichs D. J. (1988) J. Exp. Med. 167, 1459–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gedde M. M., Higgins D. E., Tilney L. G., Portnoy D. A. (2000) Infect. Immun. 68, 999–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Riordan M., Yi C. H., Gonzales R., Lee K. D., Portnoy D. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13861–13866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng L. W., Portnoy D. A. (2003) Cell. Microbiol. 5, 875–885 [DOI] [PubMed] [Google Scholar]
- 9.Cossart P. (2000) Cell. Microbiol. 2, 195–205 [DOI] [PubMed] [Google Scholar]
- 10.Pizarro-Cerdá J., Cossart P. (2006) J. Pathol. 208, 215–223 [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann J. A. (2003) Nature 426, 33–38 [DOI] [PubMed] [Google Scholar]
- 12.Ferrandon D., Imler J. L., Hetru C., Hoffmann J. A. (2007) Nat. Rev. Immunol. 7, 862–874 [DOI] [PubMed] [Google Scholar]
- 13.Janeway C. A. (1989) Cold Spring Harbor Symp. Quant. Biol. 54, 1–13 [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann J. A., Reichhart J. M. (2002) Nat. Immunol. 3, 121–126 [DOI] [PubMed] [Google Scholar]
- 15.Akira S., Uematsu S., Takeuchi O. (2006) Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 16.Takehana A., Katsuyama T., Yano T., Oshima Y., Takada H., Aigaki T., Kurata S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13705–13710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goto A., Kurata S. (2006) Invertebr. Surv. J. 3, 103–110 [Google Scholar]
- 18.Kaneko T., Yano T., Aggarwal K., Lim J. H., Ueda K., Oshima Y., Peach C., Erturk-Hasdemir D., Goldman W. E., Oh B. H., Kurata S., Silverman N. (2006) Nat. Immunol. 7, 715–723 [DOI] [PubMed] [Google Scholar]
- 19.Tzou P., De Gregorio E., Lemaitre B. (2002) Curr. Opin. Microbiol. 5, 102–110 [DOI] [PubMed] [Google Scholar]
- 20.Engström Y. (1999) Dev. Comp. Immunol. 23, 345–358 [DOI] [PubMed] [Google Scholar]
- 21.Lehrer R. I., Ganz T. (1999) Curr. Opin. Immunol. 11, 23–27 [DOI] [PubMed] [Google Scholar]
- 22.Silverman N., Maniatis T. (2001) Genes Dev. 15, 2321–2342 [DOI] [PubMed] [Google Scholar]
- 23.Zeidler M. P., Bach E. A., Perrimon N. (2000) Oncogene 19, 2598–2606 [DOI] [PubMed] [Google Scholar]
- 24.Hou S. X., Zheng Z., Chen X., Perrimon N. (2002) Dev. Cell 3, 765–778 [DOI] [PubMed] [Google Scholar]
- 25.Bach E. A., Perrimon N. (2003) in Signal Transducers and Activators of Transcription (STATs); Activation and Biology (Sehgal P. B., Levy D. E., Hirano T. eds) pp. 87–104, Kluwer Academic Publishers Group, Dordrecht, Netherlands [Google Scholar]
- 26.Dostert C., Jouanguy E., Irving P., Troxler L., Galiana-Arnoux D., Hetru C., Hoffmann J. A., Imler J. L. (2005) Nat. Immunol. 6, 946–953 [DOI] [PubMed] [Google Scholar]
- 27.Agaisse H., Perrimon N. (2004) Immunol. Rev. 198, 72–82 [DOI] [PubMed] [Google Scholar]
- 28.Buchon N., Broderick N. A., Poidevin M., Pradervand S., Lemaitre B. (2009) Cell Host Microbe 5, 200–211 [DOI] [PubMed] [Google Scholar]
- 29.Cheng L. W., Viala J. P., Stuurman N., Wiedemann U., Vale R. D., Portnoy D. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13646–13651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agaisse H., Burrack L. S., Philips J. A., Rubin E. J., Perrimon N., Higgins D. E. (2005) Science 309, 1248–1251 [DOI] [PubMed] [Google Scholar]
- 31.Ayres J. S., Freitag N., Schneider D. S. (2008) Genetics 178, 1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordon M. D., Ayres J. S., Schneider D. S., Nusse R. (2008) PLoS Pathog. 4, e1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takehana A., Yano T., Mita S., Kotani A., Oshima Y., Kurata S. (2004) EMBO J. 23, 4690–4700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yano T., Mita S., Ohmori H., Oshima Y., Fujimoto Y., Ueda R., Takada H., Goldman W. E., Fukase K., Silverman N., Yoshimori T., Kurata S. (2008) Nat. Immunol. 9, 908–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quackenbush J. (2002) Nat. Genet. 32, 496–501 [DOI] [PubMed] [Google Scholar]
- 36.Harrison D. A., McCoon P. E., Binari R., Gilman M., Perrimon N. (1998) Genes Dev. 12, 3252–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goto A., Matsushita K., Gesellchen V., El Chamy L., Kuttenkeuler D., Takeuchi O., Hoffmann J. A., Akira S., Boutros M., Reichhart J. M. (2008) Nat. Immunol. 9, 97–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tauszig-Delamasure S., Bilak H., Capovilla M., Hoffmann J. A., Imler J. L. (2002) Nat. Immunol. 3, 91–97 [DOI] [PubMed] [Google Scholar]
- 39.Goto A., Blandin S., Royet J., Reichhart J. M., Levashina E. A. (2003) Nucleic Acids Res. 31, 6619–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asha H., Nagy I., Kovacs G., Stetson D., Ando I., Dearolf C. R. (2003) Genetics 163, 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rutschmann S., Jung A. C., Zhou R., Silverman N., Hoffmann J. A., Ferrandon D. (2000) Nat. Immunol. 1, 342–347 [DOI] [PubMed] [Google Scholar]
- 42.Rutschmann S., Jung A. C., Hetru C., Reichhart J. M., Hoffmann J. A., Ferrandon D. (2000) Immunity 12, 569–580 [DOI] [PubMed] [Google Scholar]
- 43.Yan R., Small S., Desplan C., Dearolf C. R., Darnell J. E., Jr. (1996) Cell 84, 421–430 [DOI] [PubMed] [Google Scholar]
- 44.Imler J. L., Bulet P. (2005) Chem. Immunol. Allergy 86, 1–21 [DOI] [PubMed] [Google Scholar]
- 45.Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., Couto A., Marra V., Keleman K., Dickson B. J. (2007) Nature 448, 151–156 [DOI] [PubMed] [Google Scholar]
- 46.Zasloff M. (2002) Nature 415, 389–395 [DOI] [PubMed] [Google Scholar]
- 47.Simmaco M., Mignogna G., Barra D. (1998) Biopolymers 47, 435–450 [DOI] [PubMed] [Google Scholar]
- 48.Boman H. G. (2000) Immunol. Rev. 173, 5–16 [DOI] [PubMed] [Google Scholar]
- 49.Bina S., Zeidler M. (2008) A JAK-STAT Pathway in Disease (Stephanou A. ed) pp. 24–42, Landes Bioscience, University College London, London UK [Google Scholar]
- 50.Barillas-Mury C., Han Y. S., Seeley D., Kafatos F. C. (1999) EMBO J. 18, 959–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agaisse H., Petersen U. M., Boutros M., Mathey-Prevot B., Perrimon N. (2003) Dev. Cell 5, 441–450 [DOI] [PubMed] [Google Scholar]
- 52.Tzou P., Reichhart J. M., Lemaitre B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2152–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakagawa I., Amano A., Mizushima N., Yamamoto A., Yamaguchi H., Kamimoto T., Nara A., Funao J., Nakata M., Tsuda K., Hamada S., Yoshimori T. (2004) Science 306, 1037–1040 [DOI] [PubMed] [Google Scholar]
- 54.Gutierrez M. G., Master S. S., Singh S. B., Taylor G. A., Colombo M. I., Deretic V. (2004) Cell 119, 753–766 [DOI] [PubMed] [Google Scholar]
- 55.Deretic V., Singh S., Master S., Harris J., Roberts E., Kyei G., Davis A., de Haro S., Naylor J., Lee H. H., Vergne I. (2006) Cell. Microbiol. 8, 719–727 [DOI] [PubMed] [Google Scholar]
- 56.Otto G. P., Wu M. Y., Clarke M., Lu H., Anderson O. R., Hilbi H., Shuman H. A., Kessin R. H. (2004) Mol. Microbiol. 51, 63–72 [DOI] [PubMed] [Google Scholar]
- 57.Romano P. S., Gutierrez M. G., Berón W., Rabinovitch M., Colombo M. I. (2007) Cell. Microbiol. 9, 891–909 [DOI] [PubMed] [Google Scholar]
- 58.Dorn B. R., Dunn W. A., Jr., Progulske-Fox A. (2001) Infect. Immun. 69, 5698–5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ogawa M., Yoshimori T., Suzuki T., Sagara H., Mizushima N., Sasakawa C. (2005) Science 307, 727–731 [DOI] [PubMed] [Google Scholar]
- 60.Birmingham C. L., Canadien V., Gouin E., Troy E. B., Yoshimori T., Cossart P., Higgins D. E., Brumell J. H. (2007) Autophagy 3, 442–451 [DOI] [PubMed] [Google Scholar]
- 61.Birmingham C. L., Canadien V., Kaniuk N. A., Steinberg B. E., Higgins D. E., Brumell J. H. (2008) Nature 451, 350–354 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.